Significance
Mass cytometry enables the measurement of nearly 40 different proteins at the single-cell level, providing an unprecedented level of multidimensional information. Because of the complexity of these datasets across diverse populations of cells, new computational tools are needed to glean useful biological insights. Here we describe ACCENSE (Automatic Classification of Cellular Expression by Nonlinear Stochastic Embedding), a tool that computes a two-dimensional nonlinear distillation of the raw data, and automatically stratifies cells into phenotypic subpopulations based on their distribution of markers. Applying this tool to murine CD8+ T-cell data recovers known naive and memory subpopulations, and reveals additional diversity within these. In particular, we identify a novel subpopulation with a distinct multivariate phenotype, but which is not distinguishable on a biaxial plot of conventional markers.
Keywords: immunophenotyping, machine learning, class discovery, CyTOF, FACS
Abstract
Mass cytometry enables an unprecedented number of parameters to be measured in individual cells at a high throughput, but the large dimensionality of the resulting data severely limits approaches relying on manual “gating.” Clustering cells based on phenotypic similarity comes at a loss of single-cell resolution and often the number of subpopulations is unknown a priori. Here we describe ACCENSE, a tool that combines nonlinear dimensionality reduction with density-based partitioning, and displays multivariate cellular phenotypes on a 2D plot. We apply ACCENSE to 35-parameter mass cytometry data from CD8+ T cells derived from specific pathogen-free and germ-free mice, and stratify cells into phenotypic subpopulations. Our results show significant heterogeneity within the known CD8+ T-cell subpopulations, and of particular note is that we find a large novel subpopulation in both specific pathogen-free and germ-free mice that has not been described previously. This subpopulation possesses a phenotypic signature that is distinct from conventional naive and memory subpopulations when analyzed by ACCENSE, but is not distinguishable on a biaxial plot of standard markers. We are able to automatically identify cellular subpopulations based on all proteins analyzed, thus aiding the full utilization of powerful new single-cell technologies such as mass cytometry.
The immune system comprises many cell types that perform highly diverse functions and interact in complex ways during an immune response. The functional capabilities of individual cells are inextricably linked with their phenotypes, as defined by the expression levels of different proteins. These phenotypes are dynamic and alterations often occur, for example during the differentiation of lymphocytes from naive to memory cells upon encountering their specific antigens (1). Understanding which immune cell phenotypes exist is thus important for understanding the functional properties of the immune system as a whole. Flow cytometry, where cells are stained with fluorescently labeled antibodies and their protein targets quantified by light emission signals at single-cell resolution, has been the gold-standard technology for many years (2). Using this technique, hundreds of different immune cell populations have been defined based on differential protein expression. For example, T lymphocytes have been subdivided into helper T cells and killer T cells based on the expression of the coreceptors CD4 and CD8, respectively. In mice, these T-cell populations have also been further subdivided into antigen-naive cells (CD44−CD62L+) and multiple subpopulations of antigen-exposed cells [e.g., central memory (TCM, CD44+CD62L+), effector memory (TEM, CD44+CD62L−), and short-lived effector cells (TSLEC, CD44+KLRG1+CD122+)]. Corresponding populations also exist in humans, although the defining markers differ. In both species, these T-cell subpopulations also exhibit functional differences in their proliferative potential, killing capacity, and cytokine production (3).
Flow cytometry is currently constrained to 12–16 parameters per cell due to the limited light spectra and overlapping emission signals. In contrast, mass cytometry allows up to 42 parameters to be quantified on individual cells using metal-chelated probes without any significant signal overlap, thus resolving cellular phenotypes at an unprecedented level of detail (4). Using this technology, Newell et al. recently showed a continuous distribution of human CD8+ T-cell phenotypes and a previously unexpected level of functional diversity among these cells (5).
The high-dimensional data (Fig. 1A) generated by mass cytometry are challenging to interpret in biologically meaningful ways. Conventional flow cytometry analysis involves manual analysis through a laborious and highly subjective process known as “gating” (Fig. 1B) (6). As the number of biaxial plots to analyze increases combinatorially with the number of markers analyzed, this process becomes intractable beyond 10–12 parameters. Important advances have been made toward developing better analytic tools for multivariate cytometry data (7). Many of these tools cluster cells with similar protein expression, like the recently developed spanning-tree progression analysis of density normalized events (SPADE) algorithm, which has been applied to mass cytometry data (8, 9). SPADE uses multivariate information to define cellular clusters and displays the underlying phenotypic hierarchy in a tree-like structure. The main drawbacks of clustering approaches are the loss of single-cell resolution and the requirement for prespecification of the number of target clusters desired, introducing bias regarding a quantity that is rarely known.
Fig. 1.
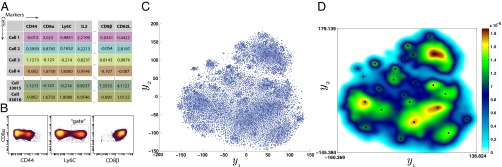
ACCENSE applied to high-dimensional, high-throughput mass cytometry data. (A) Illustration of a sample mass cytometry dataset. Rows correspond to different cells whereas columns correspond to the different markers (cell-surface antigens and intracellular proteins) whose expressions are measured using metal-chelated antibodies. Entries correspond to transformed values (arcsinh) of mass–charge ratios that indicate expression levels of each marker. (B) Biaxial plots showing the expression of CD8α (y axis) against the expression of CD44 (Left), Ly6C (Middle), and CD8β (Right), respectively, and an illustration of the manual gating approach to identify cells expressing Ly6C (Middle). This group of cells is conventionally described as Ly6C+, whereas the remaining cells are tagged as Ly6C−. A similar approach is applied to classify cells based on other markers of interest. (C) The 2D t-SNE map of CD8+ T cells derived from SPF B6 mice. Each point represents a cell from the training set (M = 18,304) derived by down-sampling the original dataset. (D) A composite map depicting the local probability density of cells as embedded in panel C, computed using a kernel-based transform (Eq. 2 with γ = 7). Local maxima in this 2D density map represent centers of phenotypic subpopulations and were identified using a standard peak-detection algorithm (14).
As an alternative, dimensionality reduction approaches aim at finding low-dimensional representations of high-dimensional data to allow easier visualization and interpretation, while retaining single-cell resolution. The spatial organization of datapoints in the low-dimensional space can be used to group cells into subpopulations with similar protein expression. Newell et al. applied principal component analysis (PCA) to 25-parameter mass cytometry data of human CD8+ T cells, and used the top three principal components (3D-PCA) to separate subpopulations (5). 3D-PCA represents the data in terms of three summary variables, each a linear combination of the original dimensions, defined so as to maximally capture the underlying variance in the data. That PCA finds the most optimal representation within the set of possible linear projections of the data is, however, also an important limitation––a linear projection may be too restrictive to yield accurate representations (10). To address this limitation, Amir et al. recently applied a nonlinear dimensionality reduction approach to visualize mass cytometry data (11). By using t-distributed stochastic neighbor embedding (or t-SNE) (12), multivariate cellular data could be represented on a 2D plot, similar to conventional biaxial flow plots. However, in contrast with these plots, wherein distance between cells reflects expression differences between only the two markers, distances on the t-SNE plot account for differences across all of the markers. Amir et al. demonstrated that t-SNE could effectively capture phenotypic relationships between cells, such as normal and leukemic bone marrow cells (11).
Here we combine t-SNE with density-based partitioning into a single tool, ACCENSE (Automatic Classification of Cellular Expression by Nonlinear Stochastic Embedding), and use it to identify murine CD8+ T-cell subpopulations (SI Appendix, Tables S1 and S2) from high-dimensional mass cytometry data (SI Appendix 1) without having to predefine the number of expected populations. The work of Newell et al. (5) pertaining to human CD8+ T cells inspired us to ask to what extent a similar scenario was applicable in laboratory mice, which have been extensively used to advance our understanding of basic immunology over the years. Our analysis not only recovers well-known naive and memory CD8+ T-cell populations, but also identifies phenotypically distinct subpopulations within and outside of these. We believe that ACCENSE will be important for exploratory analysis by automatically extracting and quantifying cell populations, based not on only a few, but on the combined expression of the many different proteins measured by mass cytometry.
Results
Computational Methods.
Here, we provide a high-level overview of the embedding (using t-SNE) and clustering steps in ACCENSE (see also SI Appendix 2). Let x(i) represent the normalized N-dimensional protein expression vector encoding the phenotype of cell i (= 1, 2,…,M cells). We seek corresponding 2D vectors {y(i)} such that T cells with similar phenotype are embedded close to each other in the map, whereas phenotypically dissimilar cells are embedded far apart. t-SNE employs pairwise probabilities {pi,j} between cells i and j such that pi,j is large if x(i) and x(j) are similar, and small otherwise. Let qi,j represent the corresponding quantity in the 2D map, encoding similarity between the embeddings y(i) and y(j). The embeddings {y(i)} that maximally conserve the information between the high-dimensional and low-dimensional representations are found by minimizing the Kullback–Leibler divergence (13) between {pi,j} and {qi,j},
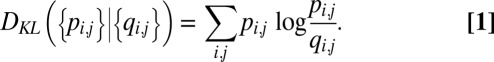 |
Thus, we compute a 2D distillation that faithfully preserves neighborhood relationships present in the high-dimensional data. Furthermore, {y(i)} can encode nonlinear relationships because they are not constrained to be linear combinations of {x(i)}, as in standard PCA. The optimal embeddings are estimated by a numerical gradient descent procedure (SI Appendix 3), which, owing to the nonconvex objective function in Eq. 1, only guarantees a local minimum. Due to the 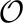 (M2) computational and memory complexity of t-SNE, we down-sampled the original dataset in a density-dependent manner (SI Appendix 1.5) to extract a smaller-size “training set,” which we explicitly embedded using the t-SNE algorithm.
(M2) computational and memory complexity of t-SNE, we down-sampled the original dataset in a density-dependent manner (SI Appendix 1.5) to extract a smaller-size “training set,” which we explicitly embedded using the t-SNE algorithm.
Next, we used a kernel-based estimate of the 2D probability density Kγ(y) (SI Appendix 4, Fig. S1) of cells in the embedding,
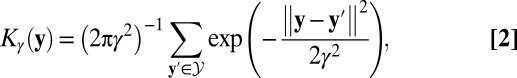 |
where the sum is over the locations of all cell locations in the embedding. Local maxima in Kγ(y) correspond to phenotypic subpopulations (Fig. 1 C and D) and were identified using a 2D peak-finding algorithm (14). Upon comparing the results produced by different choices of the kernel-bandwidth γ, we found a value that provided an accurate coarse-grained representation of the local and global features present in the phenotypic space (see SI Appendix 4 and Fig. S2). Although heuristic, this approach allows us to approximately identify clusters of CD8+ T cells in a data-driven manner without having to prespecify their number. We also note that directly applying a 35-dimensional kernel to the original space of protein expression data to find cellular subpopulations without first performing dimensionality reduction is fraught with challenges, and is not practical (SI Appendix 2.2).
Analyzing CD8+ T-Cell Populations in Specific Pathogen-Free Mice Using t-SNE.
CD8+ T cells derived from the blood of six specific-pathogen free (SPF) B6 mice (SI Appendix, Table S1) were assessed for expression of 35 markers, which included cell-surface and intracellular proteins (SI Appendix, Table S3). Each mouse was sampled twice, one of which (S) was stimulated for 5 h with phorbol-12-myristate-13-acetate (PMA) and Ionomycin (SI Appendix 1) while the other sample (U) was analyzed without any treatment. The complete dataset consisted of 36,309 cells, which we down-sampled in a density-dependent manner to obtain a training set of 18,304 cells (see SI Appendix 1.5). Fig. 1C shows the 2D embedding depicting the phenotypic space occupied by SPF mice T cells. The remaining cells were embedded onto this map based on their similarity to the training set (SI Appendix 5), which did not alter the global density profile of the original map (SI Appendix, Fig. S3). Different down-sampled datasets produced qualitatively similar maps.
The contiguous organization of mouse CD8+ T cells in Fig. 1B is consistent with human CD8+ T-cell data (5). The distribution of phenotypes exhibits a high degree of stereotypy, as is expected in these isogenic mice with similar environmental exposure (SI Appendix, Fig. S4). Because our samples were derived from mice of different ages, we also found age-related patterns (vide infra).
Despite the continuous organization of phenotypes, however, the nonuniform distribution of cells in Fig. 1C suggests that not all phenotypes are equally frequent among CD8+ T cells. Density-based partitioning of the t-SNE map identified 24 distinct subpopulations (Fig. 1D; Fig. S2) labeled S1−S24 (Fig. 2A). In contrast, projecting the same data along the top two principal components revealed only three distinct subpopulations, with >80% of the cells within one population (SI Appendix 6). Moreover, this representation captured only 21% of the underlying variance, and the spectrum of the covariance matrix indicated that the top 19 principal components altogether captured only 75% of the overall variance in the data (SI Appendix, Fig. S5). These observations underscore the limitations of linear dimensionality reduction and the need for an approach that can account for nonlinearities abundant in cytometry data.
Fig. 2.
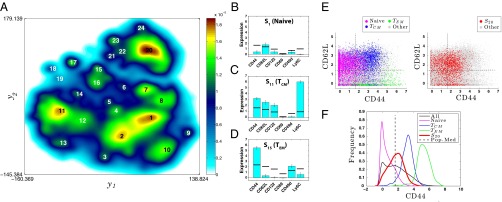
ACCENSE identifies CD8+ T-cell subpopulations in SPF B6 mice. (A) Subpopulations S1−S24 identified as local maxima in the density map are marked. (B–D) Median expression values of CD44, CD62L, CD122, CD69, CD49d, and Ly6C within the most populous subpopulations that are part of the putative naive, TCM, and TEM compartments within SPF CD8+ T cells, identified using ACCENSE. The complete phenotypic signatures of these subpopulations are described in SI Appendix, Figs. S6, S16, and S20, respectively. Bar heights indicate the median expression for each marker within the subpopulation, estimated from cells sampled close to the locations of the subpopulation peaks identified in Fig. 1D (1,500 cells were sampled for the large naive and TCM subpopulations S1 and S11, whereas 200 cells were sampled for the smaller TEM subpopulation S15). For each marker k, the width of the error bar equals 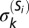 , the median-absolute deviation (MAD) of the distribution of marker expression within the subpopulation (SI Appendix 7). The blue horizontal bar for each marker corresponds to its median expression
, the median-absolute deviation (MAD) of the distribution of marker expression within the subpopulation (SI Appendix 7). The blue horizontal bar for each marker corresponds to its median expression 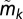 across all the cells in the dataset. (E) CD44 vs. CD62L expression of cells in the dataset. (Left) Individual naive (S1, S2), TCM (S11, S12), and TEM (S15, S17) subpopulations are shown in magenta, blue, and green dots, respectively. Gray dots represent T cells from the remaining subpopulations. Dashed lines represent median protein expression. The location of the colored cells belonging to these subpopulations are consistent with their conventionally associated phenotype ––e.g., naive, CD44−CD62L+; TCM, CD44+CD62L+; and TEM, CD44+CD62L−. (Right) Cells from subpopulation S20, shown as red circles, are not (as a group) clearly distinguishable as “+” or “−” for either marker. Gray circles represent T cells from all other subpopulations. (F) Distribution of CD44 expression within different subpopulations. “All” represents the CD44 expression distribution across all cells in the data (M = 36,309). For the naive (magenta), TCM (blue), and TEM (green) subpopulations, the corresponding distributions largely fall to one side of the population median (dashed black line), and therefore can be unequivocally classified as either a “+” or a “−” phenotype. In contrast, the CD44 distribution of S20 (red) peaks close to the population median and classifies as an “int” phenotype according to our convention.
across all the cells in the dataset. (E) CD44 vs. CD62L expression of cells in the dataset. (Left) Individual naive (S1, S2), TCM (S11, S12), and TEM (S15, S17) subpopulations are shown in magenta, blue, and green dots, respectively. Gray dots represent T cells from the remaining subpopulations. Dashed lines represent median protein expression. The location of the colored cells belonging to these subpopulations are consistent with their conventionally associated phenotype ––e.g., naive, CD44−CD62L+; TCM, CD44+CD62L+; and TEM, CD44+CD62L−. (Right) Cells from subpopulation S20, shown as red circles, are not (as a group) clearly distinguishable as “+” or “−” for either marker. Gray circles represent T cells from all other subpopulations. (F) Distribution of CD44 expression within different subpopulations. “All” represents the CD44 expression distribution across all cells in the data (M = 36,309). For the naive (magenta), TCM (blue), and TEM (green) subpopulations, the corresponding distributions largely fall to one side of the population median (dashed black line), and therefore can be unequivocally classified as either a “+” or a “−” phenotype. In contrast, the CD44 distribution of S20 (red) peaks close to the population median and classifies as an “int” phenotype according to our convention.
Phenotypic Coarse-Graining.
We sought to extract the marker expression patterns of each of the CD8+ T-cell subpopulations depicted in Fig. 2A. For each subpopulation we compared the median expression for a marker within that subpopulation to the median expression of the same marker across all cells in the original dataset (see SI Appendix 7). Naively, one might be tempted to label a subpopulation as “+” for a particular marker if its median intrasubpopulation expression is higher than its median expression across all of the cells, and “−” if it is lower. However, such a rigid classification of phenotypes can be misleading for subpopulations identified here based on multivariate protein expression. This is because expression values of a particular marker k within a subpopulation Si follow a distribution––therefore, labeling the subpopulation strictly according to the subpopulation median 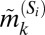 will not accurately capture the true phenotype if
will not accurately capture the true phenotype if 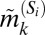 is close to the population median
is close to the population median 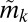 , and if the underlying intrasubpopulation distribution of protein expression is wide (e.g., see the discussion on S20 below). To alleviate this, we classified subpopulation phenotypes such that the width of the marker distributions
, and if the underlying intrasubpopulation distribution of protein expression is wide (e.g., see the discussion on S20 below). To alleviate this, we classified subpopulation phenotypes such that the width of the marker distributions 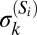 is incorporated in the simplest manner––namely, subpopulation Si is “+” for marker k if
is incorporated in the simplest manner––namely, subpopulation Si is “+” for marker k if 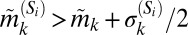 and “−” for marker k if
and “−” for marker k if 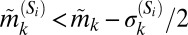 , else it is “int” (for intermediate). Using three ordinal categories in this manner, which incorporate the first two moments of the marker distribution, enables us to achieve a higher degree of precision in cell classification while avoiding the complexity of the entire distribution. The resulting coarse-grained “phenotypic signatures” of S1−S24 are shown in SI Appendix, Figs. S6–S29 and summarized in SI Appendix, Table S4. To test whether our procedure yielded subpopulations with distinct phenotypes, we analyzed each pair of subpopulations to compute the number of markers between them that were significantly different (see SI Appendix, Table S5).
, else it is “int” (for intermediate). Using three ordinal categories in this manner, which incorporate the first two moments of the marker distribution, enables us to achieve a higher degree of precision in cell classification while avoiding the complexity of the entire distribution. The resulting coarse-grained “phenotypic signatures” of S1−S24 are shown in SI Appendix, Figs. S6–S29 and summarized in SI Appendix, Table S4. To test whether our procedure yielded subpopulations with distinct phenotypes, we analyzed each pair of subpopulations to compute the number of markers between them that were significantly different (see SI Appendix, Table S5).
Analysis of Identified CD8+ T-Cell Subpopulations in Mice.
Phenotypic diversity among naive T cells.
CD8+ T cells are conventionally divided into naive, central memory (TCM), and effector memory subpopulations (TEM) based on their expression of homing receptors and their propensity to traffic into secondary lymphoid organs (15). Naive T cells are characterized by their low and high expression of the cell-surface glycoprotein CD44 and the homing receptor CD62L, respectively (i.e., CD44−CD62L+). Studies have also associated a low expression of CD122 (subunit of a cytokine receptor), CD69 (an activation marker), CD49d (integrin subunit), and Ly6C (accessory glycoprotein) with naive cells (3). Traditionally, it was believed that naive cells differentiate to acquire a memory phenotype only upon antigen encounter, but this may also occur in the context of homeostatic proliferation (16), or T-cell receptor (TCR) cross-reactivity (17). Subpopulation 1 or S1 on the t-SNE map (Fig. 2 A and B) fulfills the traditional criteria for naive marker expression (SI Appendix, Fig. S6). Within the six mice, this subpopulation comprised 12–17% of the cells in the unstimulated samples and 6–8% of the stimulated samples (SI Appendix, Fig. S30A).
Adjacent to S1 in the t-SNE map is S2 (Fig. 2A), which occurred at a frequency of 12–17% in the unstimulated samples and 5–11% in the stimulated samples (SI Appendix, Fig. S30B). S2 also had the “naive” phenotype CD44−CD62L+; nonetheless, it exhibited significant differences compared with S1 across many markers. S2 was CD45RB+CD45RC+, both isoforms of the phosphatase CD45, and CD8α+CD8β+ (SI Appendix, Fig. S7), whereas S1 was either “−” or “int” for these markers. Additionally, the smaller subpopulations S3, S4, S5 also exhibit the naive phenotype CD44−CD62L+, but had significant differences across other markers compared with S1 (SI Appendix, Figs. S8–S10). Two additional subpopulations S7 and S8, located proximal to the naive subpopulations 1–5 (Fig. 2A), had the intriguing phenotype CD44−CD62L− (SI Appendix, Figs. S12 and S13). Together these results show a previously unappreciated heterogeneity within the naive CD8+ T-cell compartment.
Memory CD8+ T-cell subpopulations.
S11 exhibited the phenotype CD44+CD62L+, which is characteristic of CD8+ TCM cells in mice. Additionally this subpopulation was CD122+CD69−CD49d−Ly6C+ (Fig. 2C), which is also consistent with TCM function (3). S11 was present at a much higher frequency in the two 22-mo-old mice (24% and 17%) compared with the two 5-mo-old mice (7% and 9%) within the unstimulated samples (see SI Appendix, Fig. S31E), consistent with the increase of the memory pool with age in humans (16, 18). In the two 7.5-mo-old mice S11 accounted for 22% and 14% of all cells. A neighboring subpopulation S12 had a similar phenotype across all markers but had considerably lower median expression of CD44 and CD122, and higher CD49d compared with S11 (see SI Appendix, Figs. S16 and S17). S12 is also proximal to the “naive-like” subpopulation S2. The organization of the naive subpopulations S1−S2 and the TCM subpopulations S11−S12 in close proximity to each other is suggestive of a phenotypic continuum of known subpopulations, in line with the human data (5).
Effector memory T-cells (TEM) circulate in the periphery and execute immediate effector functions, typically expressing CD44 but lacking the lymphoid homing receptor CD62L. S15 and S17 were CD44+CD62L−, consistent with the TEM phenotype (see Fig. 2D; SI Appendix, Figs. S20 and S22). However, 21 markers were significantly different between S15 and S17 (SI Appendix, Table S5). Interestingly, these subpopulations were embedded further away from the naive subpopulations than the TCM subpopulations, suggesting that the TCM and naive phenotypes are more similar. Newell et al. also reported a continuous phenotypic progression from naive to TCM and finally to TEM in humans (5).
A large CD8+ T-cell population with CD44int phenotype.
The t-SNE map (Fig. 2A) also showed a populous CD44int group S20 which was distinct from the naive, TCM, and TEM subpopulations described above. S20 was present at a frequency of 11.5–18.5% in all unstimulated samples, with a discernible decrease in older animals (SI Appendix, Fig. S33B). Compared with the naive subpopulation (S1) these T cells had, most notably, a significantly reduced expression of CD8β and (to a lesser extent) CD8α, and the CD45RB and -RC isoforms, while having a significantly up-regulated expression of TCRβ, CD3ε, and CD5, as well as other differences (see SI Appendix, Figs. S6 and S25). The organization of subpopulations depicted in Fig. 2A clearly shows that S20 is phenotypically distinct from the canonical naive, TCM, and TEM subpopulations. Interestingly, when we focused only on the expression of markers conventionally used to classify cells into naive and memory phenotypes, the distinctiveness of S20 was less obvious. In particular, we observed that the median expression within S20 of CD44, the typical naive–memory distinguishing marker, falls in an intermediate range (Fig. 2E, Right and Fig. 2F) in contrast with the conventional naive, TCM, and TEM subpopulations (Fig. 2E, Left). In a standard biaxial plot involving CD44 and CD62L, S20 can be conflated with the conventional subpopulations despite being phenotypically distinct. This example illustrates the value of incorporating information across multiple markers while defining phenotypic subpopulations of cells.
Stimulation-associated phenotypes.
Because we stimulated the T cells with PMA–Ionomycin, we were able to address phenotypic and functional changes resulting from this broad and unspecific stimulus. The S10, S14, and S24 subpopulations, which were proximal in the t-SNE map to S1, S11, and S20, respectively, were present in the stimulated samples in high proportions, but occurred with negligible frequency in the unstimulated controls across all mice (SI Appendix, Figs. S31D, S32B, and S33F). S10 was also the most populous subpopulation in the stimulated samples occurring at a frequency of 20–50% in all mice. These responsive subpopulations were characterized by significantly up-regulated expression of CD69, IL-2, CD107a (a marker of degranulation upon stimulation), CTLA-4, and loss of CD62L. That S10, S14, and S24 are closer to S1, S11, and S20 than to each other suggests that T cells might still retain their basal phenotypic characteristics across most markers, and only a few (e.g., CD69, IL-2, etc.) might change upon activation through such an unspecific stimulus. The degree of phenotypic change upon activation has not been addressed previously at this level of detail to the best of our knowledge.
Comparison of CD8 T-Cell Subpopulations Between SPF and Germ-Free Mice.
Recent studies have found developmental defects in CD8+ T cells in germ-free (GF) mice, which lack commensal bacterial microbiota (19). We wanted to obtain a more detailed view of the phenotypic profile of T cells in GF mice compared with the SPF mice described above. We collected protein expression data using the same staining panel (SI Appendix, Table S3) and focused on CD8+ T cells (M = 4,086) derived from the blood of three GF mice (SI Appendix, Table S2). As in any automated cytometry analysis technique the presence of “batch effects” due to variations in staining intensity from day to day precluded direct merging of the SPF and GF datasets (SI Appendix 1.4). Instead, we elected to work around batch effects by computing a separate map for the GF data, which revealed eight subpopulations labeled  (see Fig. 3A). Each subpopulation was phenotypically classified as “+,” “−,” or “int” for each protein (see SI Appendix, Table S6). We then compared the presence or absence of specific CD8+ T-cell subpopulations in GF mice with those identified in SPF mice. Importantly, we did not compare a GF subpopulation
(see Fig. 3A). Each subpopulation was phenotypically classified as “+,” “−,” or “int” for each protein (see SI Appendix, Table S6). We then compared the presence or absence of specific CD8+ T-cell subpopulations in GF mice with those identified in SPF mice. Importantly, we did not compare a GF subpopulation  with an SPF subpopulation Sj by the absolute marker expression values (which we expect to be corrupted by batch effects), but rather by their coarse-grained phenotypic signature across each marker (i.e., “+,” “−,” or “int”).
with an SPF subpopulation Sj by the absolute marker expression values (which we expect to be corrupted by batch effects), but rather by their coarse-grained phenotypic signature across each marker (i.e., “+,” “−,” or “int”).
Fig. 3.
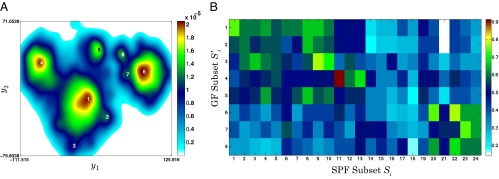
GF subpopulations and their phenotypic similarity with SPF subpopulations. (A) t-SNE map computed from CD8+ T cells derived from the blood of GF mice (M = 4,086) indicating subpopulations 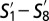 . (B) Phenotypic similarity
. (B) Phenotypic similarity 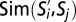 depicted as a heatmap. Rows are subpopulations
depicted as a heatmap. Rows are subpopulations 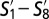 in GF mice and columns are subpopulations S1−S24 in SPF mice. The color of the pixel at location (i, j) represents the fraction of marker phenotypes that are similar across
in GF mice and columns are subpopulations S1−S24 in SPF mice. The color of the pixel at location (i, j) represents the fraction of marker phenotypes that are similar across 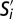 and Sj.
and Sj.
For a GF subpopulation 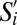 and SPF subpopulation Sj, we computed the phenotypic similarity
and SPF subpopulation Sj, we computed the phenotypic similarity 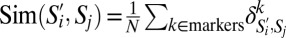 , where n = 35 markers. Here
, where n = 35 markers. Here 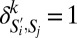 if
if 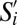 and Sj have the same coarse-grained phenotype for marker k, else
and Sj have the same coarse-grained phenotype for marker k, else 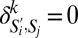 .
. 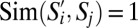 if the subpopulations have the same phenotype across all markers and 0 if they share no common phenotypes.
if the subpopulations have the same phenotype across all markers and 0 if they share no common phenotypes.
Fig. 3B, which depicts the matrix 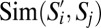 , suggests that the three most populous GF subpopulations––
, suggests that the three most populous GF subpopulations––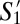 ,
, 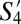 , and
, and 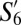 ––have the greatest similarity (>75%) to the three most populous SPF subpopulations––S1 (naive), S11 (TCM), and S20, respectively. Notably, the corresponding “TCM” subpopulations
––have the greatest similarity (>75%) to the three most populous SPF subpopulations––S1 (naive), S11 (TCM), and S20, respectively. Notably, the corresponding “TCM” subpopulations 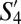 and S11 (both CD44+CD62L+) had >95% phenotypic similarity across all of the 35 markers analyzed.
and S11 (both CD44+CD62L+) had >95% phenotypic similarity across all of the 35 markers analyzed. 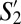 and
and 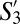 shared >70% phenotypic similarity with S8 and S10, and likely represent phenotypic subdivisions within the naive compartment in GF mice.
shared >70% phenotypic similarity with S8 and S10, and likely represent phenotypic subdivisions within the naive compartment in GF mice. 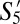 , the fourth most populous GF subpopulation, had a phenotype CD44−CD62L− which was similar to S8 in SPF mice. Our finding of a large TCM subpopulation is consistent with previous reports that T cells with a memory phenotype are found in GF mice (20). Intriguingly however, none of
, the fourth most populous GF subpopulation, had a phenotype CD44−CD62L− which was similar to S8 in SPF mice. Our finding of a large TCM subpopulation is consistent with previous reports that T cells with a memory phenotype are found in GF mice (20). Intriguingly however, none of 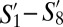 had significant overlap with the TEM subpopulations in SPF mice (S15 or S17), hinting at the possibility that such cells may be reduced or absent in the blood of GF mice.
had significant overlap with the TEM subpopulations in SPF mice (S15 or S17), hinting at the possibility that such cells may be reduced or absent in the blood of GF mice.
Taken together, these results show a great degree of similarity in the phenotypic profile of blood CD8+ T cells between SPF and GF mice. We have shown that the coarse-grained phenotypic signatures can be effectively used to compare cellular expression data collected across different experiments, where expression signals may be corrupted by batch effects. Whereas the number of subpopulations revealed by the t-SNE maps was fewer in GF mice, and an effector memory subpopulation was completely absent, further studies are required to fully establish whether or not these differences are due to the presence or absence of commensal microbiota.
Discussion
Conventional approaches that rely on the manual interpretation of a large number of biaxial plots are unscalable in the context of multiparametric protein expression data across millions of cells. Their reliance on a handful of markers, one or two at a time, can potentially conflate populations that are phenotypically distinct based on multivariate protein expression patterns.
Recently, Amir et al. combined t-SNE with mass cytometry and demonstrated the potential of nonlinear dimensionality reduction in revealing important biological relationships in bone-marrow and leukemia datasets (11). The resulting output is a 2D map, where cells are organized according to their phenotype, taking into account the full protein expression vector in determining their relative positions. Cells with similar protein expression are embedded close to each other in the map and, unlike PCA, this representation effectively captures nonlinear relationships in the high-dimensional data.
Here, we have extended this and proposed a systematic framework for identifying phenotypic subpopulations from high-dimensional mass cytometry data and proposed a straightforward way to coarse-grain their phenotypes automatically, wherein the expression of a specific protein in a particular subpopulation is classified into one of three ordinal categories. Using this tool we termed ACCENSE, we identify phenotypic subpopulations in SPF and GF mice in a data-driven manner without directly prespecifying the number of clusters, and showed that phenotypic coarse-graining allows for comparison of data across batches, even when variations in instrument performance can preclude directly merging datasets for quantitative analysis.
Applying ACCENSE to cells derived from the blood of SPF mice enabled us to recover well-known CD8+ T-cell subpopulations––naive, central, and effector memory, and PMA–Ionomycin stimulation-associated. Phenotypic signatures within markers associated with these subpopulations are consistent with conventional wisdom. The presence of heterogeneous subpopulations and a continuous distribution of cells in the phenotypic space of CD8+ T cells in murine blood are consistent with the findings on human blood CD8+ T cells (5). Our map also elucidated further phenotypic subdivisions within these subsets based on the other markers analyzed. The most populous SPF subpopulations had clear analogs in GF mice, thereby demonstrating a great degree of phenotypic similarity at least among blood-derived CD8+ T cells in these mice. We were, however, unable to detect an effector memory population in the GF samples.
In both SPF and GF mice, we identified a large T-cell subpopulation 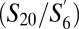 which was distinct from the naive and the memory subpopulations on the t-SNE map and was characterized by intermediate expression of CD44 and CD62L, but distinctly lower expression of the CD8 β-chain and CD45 isoforms. These cells would have escaped notice as a distinct phenotype using standard CD44 vs. CD62L gating, as illustrated in Fig. 2E. This population could either represent T cells wherein the coreceptor CD8 exists predominantly as an α−α homodimer or something completely different. Additional experiments are required to test these possibilities and assess their functional consequences. This result illustrates the potential of taking multivariate protein expression into account while assigning cells to phenotypic subpopulations.
which was distinct from the naive and the memory subpopulations on the t-SNE map and was characterized by intermediate expression of CD44 and CD62L, but distinctly lower expression of the CD8 β-chain and CD45 isoforms. These cells would have escaped notice as a distinct phenotype using standard CD44 vs. CD62L gating, as illustrated in Fig. 2E. This population could either represent T cells wherein the coreceptor CD8 exists predominantly as an α−α homodimer or something completely different. Additional experiments are required to test these possibilities and assess their functional consequences. This result illustrates the potential of taking multivariate protein expression into account while assigning cells to phenotypic subpopulations.
The studies initiated here provide an important advance in the automatic classification of subpopulations from high-dimensional protein expression data, an important challenge in cytometry. Many algorithms exist for the automated analysis of flow cytometry but generally use only four parameters at most and none of these has yet been applied to mass cytometry (7). The ACCENSE program described here provides a simple way to detect discrete populations in reduced dimensional space. We believe that this will help the full potential of mass cytometry data to be realized.
Materials and Methods
SI Appendix includes detailed descriptions of mouse experiments, mass cytometry, and data-preprocessing (SI Appendix, Sec. 1), mathematical details of t-SNE and density-based subpopulation identification (SI Appendix, Secs. 2–5), PCA on SPF data (SI Appendix, Sec. 6), and phenotypic signatures of SPF and GF subpopulations (SI Appendix, Sec. 7). Implementations of ACCENSE in MATLAB and R are freely available on the website, www.cellaccense.com.
Supplementary Material
Acknowledgments
We are grateful to Dr. G. Berman and Prof. J. Shaevitz for sharing unpublished work, and to Prof. A. Ferguson for a critical reading of the manuscript. We thank Prof. J. Sonnenburg for providing the GF mice. This research was supported by a Poitras predoctoral fellowship (K.S.), the Ragon Institute of MGH, MIT and Harvard (K.S. and A.K.C.), the Wenner-Gren Foundation and the Swedish American Foundation (P.B.), U189 AI 090019 (to M.M.D.), and a National Institutes of Health PO1 AI091580 (to A.K.C.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321405111/-/DCSupplemental.
References
- 1.Kaech SM, Wherry EJ, Ahmed R. Effector and memory T-cell differentiation: Implications for vaccine development. Nat Rev Immunol. 2002;2(4):251–262. doi: 10.1038/nri778. [DOI] [PubMed] [Google Scholar]
- 2.Cantor H, Simpson E, Sato VL, Fathman CG, Herzenberg LA. Characterization of subpopulations of T lymphocytes. I. Separation and functional studies of peripheral T-cells binding different amounts of fluorescent anti-Thy 1.2 (theta) antibody using a fluorescence-activated cell sorter (FACS) Cell Immunol. 1975;15(1):180–196. doi: 10.1016/0008-8749(75)90174-4. [DOI] [PubMed] [Google Scholar]
- 3.Sprent J, Surh CD. Normal T cell homeostasis: The conversion of naive cells into memory-phenotype cells. Nat Immunol. 2011;12(6):478–484. doi: 10.1038/ni.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bendall SC, Nolan GP, Roederer M, Chattopadhyay PK. A deep profiler’s guide to cytometry. Trends Immunol. 2012;33(7):323–332. doi: 10.1016/j.it.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012;36(1):142–152. doi: 10.1016/j.immuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herzenberg LA, Tung J, Moore WA, Herzenberg LA, Parks DR. Interpreting flow cytometry data: A guide for the perplexed. Nat Immunol. 2006;7(7):681–685. doi: 10.1038/ni0706-681. [DOI] [PubMed] [Google Scholar]
- 7.Aghaeepour N, et al. FlowCAP Consortium DREAM Consortium Critical assessment of automated flow cytometry data analysis techniques. Nat Methods. 2013;10(3):228–238. doi: 10.1038/nmeth.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiu P, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29(10):886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bendall SC, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332(6030):687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van der Maaten L, Postma E, Van Den Herik H. Dimensionality reduction: A comparative review. J Mach Learn Res. 2009;10:1–41. [Google Scholar]
- 11.Amir AD, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31(6):545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van der Maaten L, Hinton J. Visualizing data using t-SNE. J Mach Learn Res. 2008;9(85):2579–2605. [Google Scholar]
- 13.Cover TM, Thomas JA. Elements of Information Theory. New York: Wiley; 2012. [Google Scholar]
- 14.Davies ER. Machine Vision. Vol 609. New York: Academic; 1997. [Google Scholar]
- 15.Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401(6754):708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 16.Lee YJ, Jameson SC, Hogquist KA. Alternative memory in the CD8 T cell lineage. Trends Immunol. 2011;32(2):50–56. doi: 10.1016/j.it.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su LF, Kidd BA, Han A, Kotzin JJ, Davis MM. Virus-specific CD4(+) memory-phenotype T cells are abundant in unexposed adults. Immunity. 2013;38(2):373–383. doi: 10.1016/j.immuni.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saule P, et al. Accumulation of memory T cells from childhood to old age: Central and effector memory cells in CD4(+) versus effector memory and terminally differentiated memory cells in CD8(+) compartment. Mech Ageing Dev. 2006;127(3):274–281. doi: 10.1016/j.mad.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 19.Chung H, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149(7):1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Surh CD, Boyman O, Purton JF, Sprent J. Homeostasis of memory T cells. Immunol Rev. 2006;211:154–163. doi: 10.1111/j.0105-2896.2006.00401.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


