Abstract
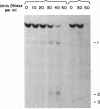
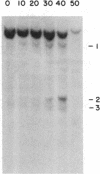
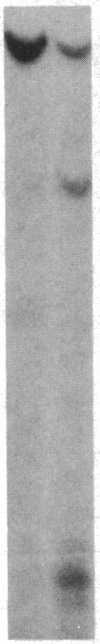
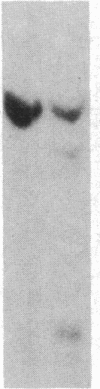
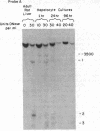
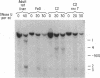
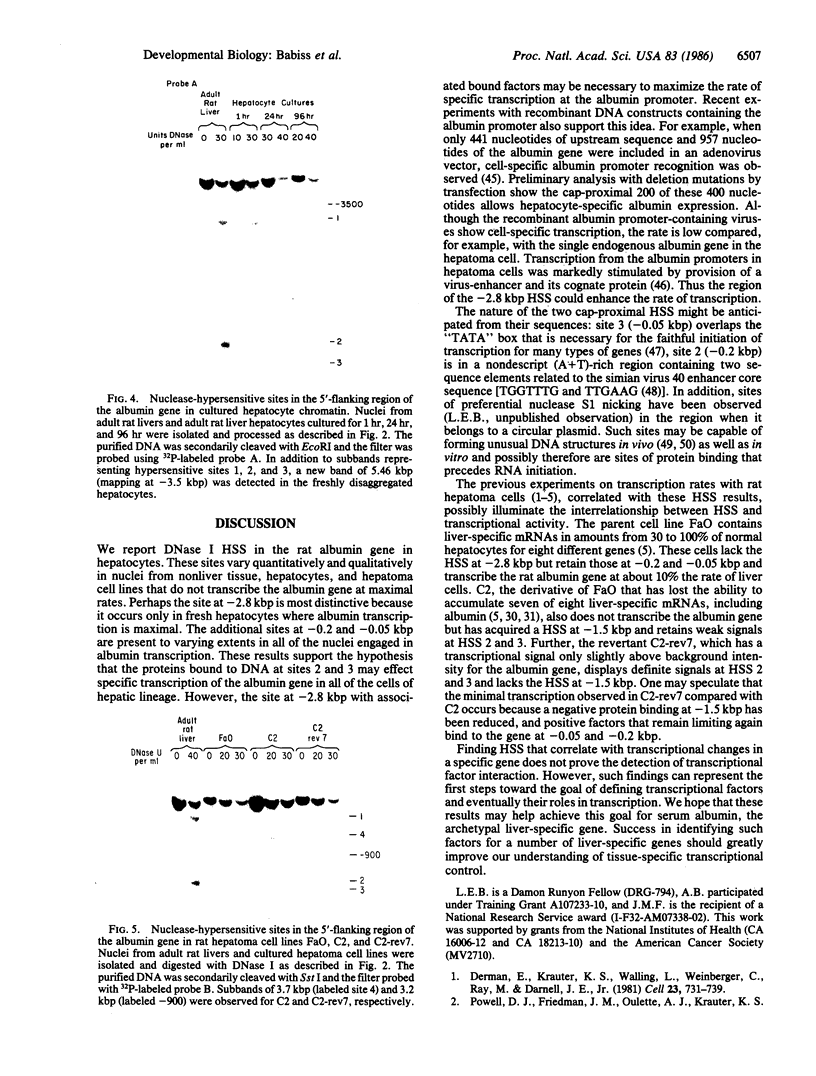
As tested by DNase I digestion, the chromatin structure in several regions 5' to the rat serum albumin gene varies in tissues and cell lines that differ in transcription rate of this gene. Three DNase I-hypersensitive regions were found in hepatocyte nuclei but not in kidney cell nuclei. The sites were approximately 2.8 kbp (site 1), 0.2 kbp (site 2), and 0.05 kbp (site 3) upstream from the cap site of the gene. In rat fetal liver tissue and rat hepatoma cell lines (FaO, C2, and C2-rev7), as well as in cultured primary hepatocytes where the rate of albumin gene transcription is lower than in adult liver, hypersensitive site (HSS) 1 was absent while sites 2 and 3 were present. In addition, the C2 cell line, which does not express albumin mRNA, contains a different HSS at position -1.5 kbp. Factors (proteins) bound to sites 2 and 3 may allow cell-specific transcription, but the additional factor interaction at site 1 could be required for a maximal rate of albumin gene transcription.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Burch J. B., Weintraub H. Temporal order of chromatin structural changes associated with activation of the major chicken vitellogenin gene. Cell. 1983 May;33(1):65–76. doi: 10.1016/0092-8674(83)90335-5. [DOI] [PubMed] [Google Scholar]
- Cartwright I. L., Abmayr S. M., Fleischmann G., Lowenhaupt K., Elgin S. C., Keene M. A., Howard G. C. Chromatin structure and gene activity: the role of nonhistone chromosomal proteins. CRC Crit Rev Biochem. 1982;13(1):1–86. doi: 10.3109/10409238209108709. [DOI] [PubMed] [Google Scholar]
- Clayton D. F., Darnell J. E., Jr Changes in liver-specific compared to common gene transcription during primary culture of mouse hepatocytes. Mol Cell Biol. 1983 Sep;3(9):1552–1561. doi: 10.1128/mcb.3.9.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton D. F., Harrelson A. L., Darnell J. E., Jr Dependence of liver-specific transcription on tissue organization. Mol Cell Biol. 1985 Oct;5(10):2623–2632. doi: 10.1128/mcb.5.10.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costlow N., Lis J. T. High-resolution mapping of DNase I-hypersensitive sites of Drosophila heat shock genes in Drosophila melanogaster and Saccharomyces cerevisiae. Mol Cell Biol. 1984 Sep;4(9):1853–1863. doi: 10.1128/mcb.4.9.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derman E., Krauter K., Walling L., Weinberger C., Ray M., Darnell J. E., Jr Transcriptional control in the production of liver-specific mRNAs. Cell. 1981 Mar;23(3):731–739. doi: 10.1016/0092-8674(81)90436-0. [DOI] [PubMed] [Google Scholar]
- Deschatrette J., Moore E. E., Dubois M., Weiss M. C. Dedifferentiated variants of a rat hepatoma:reversion analysis. Cell. 1980 Apr;19(4):1043–1051. doi: 10.1016/0092-8674(80)90095-1. [DOI] [PubMed] [Google Scholar]
- Deschatrette J., Weiss M. C. Characterization of differentiated and dedifferentiated clones from a rat hepatoma. Biochimie. 1974;56(11-12):1603–1611. doi: 10.1016/s0300-9084(75)80286-0. [DOI] [PubMed] [Google Scholar]
- Dorsch-Häsler K., Fisher P. B., Weinstein I. B., Ginsberg H. S. Patterns of viral DNA integration in cells transformed by wild type or DNA-binding protein mutants of adenovirus type 5 and effect of chemical carcinogens on integration. J Virol. 1980 May;34(2):305–314. doi: 10.1128/jvi.34.2.305-314.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EAGLE H. Amino acid metabolism in mammalian cell cultures. Science. 1959 Aug 21;130(3373):432–437. doi: 10.1126/science.130.3373.432. [DOI] [PubMed] [Google Scholar]
- Emerson B. M., Felsenfeld G. Specific factor conferring nuclease hypersensitivity at the 5' end of the chicken adult beta-globin gene. Proc Natl Acad Sci U S A. 1984 Jan;81(1):95–99. doi: 10.1073/pnas.81.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson B. M., Lewis C. D., Felsenfeld G. Interaction of specific nuclear factors with the nuclease-hypersensitive region of the chicken adult beta-globin gene: nature of the binding domain. Cell. 1985 May;41(1):21–30. doi: 10.1016/0092-8674(85)90057-1. [DOI] [PubMed] [Google Scholar]
- Fritton H. P., Igo-Kemenes T., Nowock J., Strech-Jurk U., Theisen M., Sippel A. E. Alternative sets of DNase I-hypersensitive sites characterize the various functional states of the chicken lysozyme gene. Nature. 1984 Sep 13;311(5982):163–165. doi: 10.1038/311163a0. [DOI] [PubMed] [Google Scholar]
- Fritton H. P., Sippel A. E., Igo-Kemenes T. Nuclease-hypersensitive sites in the chromatin domain of the chicken lysozyme gene. Nucleic Acids Res. 1983 Jun 11;11(11):3467–3485. doi: 10.1093/nar/11.11.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greengard O. Enzymic differentiation in mammalian liver injection of fetal rats with hormones causes the premature formation of liver enzymes. Science. 1969 Feb 28;163(3870):891–895. doi: 10.1126/science.163.3870.891. [DOI] [PubMed] [Google Scholar]
- Greengard O., Federman M., Knox W. E. Cytomorphometry of developing rat liver and its application to enzymic differentiation. J Cell Biol. 1972 Feb;52(2):261–272. doi: 10.1083/jcb.52.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAM R. G. CLONAL GROWTH OF MAMMALIAN CELLS IN A CHEMICALLY DEFINED, SYNTHETIC MEDIUM. Proc Natl Acad Sci U S A. 1965 Feb;53:288–293. doi: 10.1073/pnas.53.2.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson D. M., Clayton D. F., Darnell J. E., Jr, Reid L. M. Posttranscriptional modulation of gene expression in cultured rat hepatocytes. Mol Cell Biol. 1984 Sep;4(9):1929–1934. doi: 10.1128/mcb.4.9.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye J. S., Pratt-Kaye S., Bellard M., Dretzen G., Bellard F., Chambon P. Steroid hormone dependence of four DNase I-hypersensitive regions located within the 7000-bp 5'-flanking segment of the ovalbumin gene. EMBO J. 1986 Feb;5(2):277–285. doi: 10.1002/j.1460-2075.1986.tb04210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet I., Lieman-Hurwitz J., Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986 Feb 28;44(4):535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- Kunnath L., Locker J. DNaseI sensitivity of the rat albumin and alpha-fetoprotein genes. Nucleic Acids Res. 1985 Jan 11;13(1):115–129. doi: 10.1093/nar/13.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen A., Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982 Jun;29(2):609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Wood W. I., Dolan M., Engel J. D., Felsenfeld G. A 200 base pair region at the 5' end of the chicken adult beta-globin gene is accessible to nuclease digestion. Cell. 1981 Nov;27(1 Pt 2):45–55. doi: 10.1016/0092-8674(81)90359-7. [DOI] [PubMed] [Google Scholar]
- McKeon C., Pastan I., de Crombrugghe B. DNase I sensitivity of the alpha 2(I) collagen gene: correlation with its expression but not with its methylation pattern. Nucleic Acids Res. 1984 Apr 25;12(8):3491–3502. doi: 10.1093/nar/12.8.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahon J. L., Gal A., Erdos T., Sala-Trepat J. M. Differential DNase I sensitivity of the albumin and alpha-fetoprotein genes in chromatin from rat tissues and cell lines. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5031–5035. doi: 10.1073/pnas.81.16.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. A., Ferris R. C., Zhang D. E., Ferenz C. R. The beta-globin domain in immature chicken erythrocytes: enhanced solubility is coincident with histone hyperacetylation. Nucleic Acids Res. 1986 Feb 25;14(4):1667–1682. doi: 10.1093/nar/14.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M. O., Sperling L., Cassio D., Levilliers J., Sala-Trepat J., Weiss M. C. Undermethylation at the 5' end of the albumin gene is necessary but not sufficient for albumin production by rat hepatoma cells in culture. Cell. 1982 Oct;30(3):825–833. doi: 10.1016/0092-8674(82)90287-2. [DOI] [PubMed] [Google Scholar]
- PITOT H. C., PERAINO C., MORSE P. A., Jr, POTTER V. R. HEPATOMAS IN TISSUE CULTURE COMPARED WITH ADAPTING LIVER IN VIVO. Natl Cancer Inst Monogr. 1964 Apr;13:229–245. [PubMed] [Google Scholar]
- Parker C. S., Topol J. A Drosophila RNA polymerase II transcription factor binds to the regulatory site of an hsp 70 gene. Cell. 1984 May;37(1):273–283. doi: 10.1016/0092-8674(84)90323-4. [DOI] [PubMed] [Google Scholar]
- REUBER M. D. A transplantable bile-secreting hepatocellular carcinoma in the rat. J Natl Cancer Inst. 1961 Apr;26:891–899. [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Ruiz-Carrillo A. The histone H5 gene is flanked by S1 hypersensitive structures. Nucleic Acids Res. 1984 Aug 24;12(16):6473–6492. doi: 10.1093/nar/12.16.6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sweet R. W., Chao M. V., Axel R. The structure of the thymidine kinase gene promoter: nuclease hypersensitivity correlates with expression. Cell. 1982 Dec;31(2 Pt 1):347–353. doi: 10.1016/0092-8674(82)90128-3. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. J., Sundin O., Bohn M. A stretch of "late" SV40 viral DNA about 400 bp long which includes the origin of replication is specifically exposed in SV40 minichromosomes. Cell. 1979 Feb;16(2):453–466. doi: 10.1016/0092-8674(79)90021-7. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiher H., König M., Gruss P. Multiple point mutations affecting the simian virus 40 enhancer. Science. 1983 Feb 11;219(4585):626–631. doi: 10.1126/science.6297005. [DOI] [PubMed] [Google Scholar]
- Weintraub H. A dominant role for DNA secondary structure in forming hypersensitive structures in chromatin. Cell. 1983 Apr;32(4):1191–1203. doi: 10.1016/0092-8674(83)90302-1. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weisbrod S. Active chromatin. Nature. 1982 May 27;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]
- Wu C. Activating protein factor binds in vitro to upstream control sequences in heat shock gene chromatin. Nature. 1984 Sep 6;311(5981):81–84. doi: 10.1038/311081a0. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Wu C. Two protein-binding sites in chromatin implicated in the activation of heat-shock genes. Nature. 1984 May 17;309(5965):229–234. doi: 10.1038/309229a0. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Yamamoto K. R. Reversible and persistent changes in chromatin structure accompany activation of a glucocorticoid-dependent enhancer element. Cell. 1984 Aug;38(1):29–38. doi: 10.1016/0092-8674(84)90523-3. [DOI] [PubMed] [Google Scholar]