Angiotensin II stimulation of HDAC2 production, phosphorylation by CK2, and resulting modulation of target genes, which promote cardiac hypertrophy, are opposed by estrogen/ERβ. Angiotensin II also represses class II HDAC4 and 5 production and stimulates their phosphorylation, which expels them from the nucleus, and estrogen prevents this.
Abstract
The development and progression of cardiac hypertrophy often leads to heart failure and death, and important modulators of hypertrophy include the histone deacetylase proteins (HDACs). Estrogen inhibits cardiac hypertrophy and progression in animal models and humans. We therefore investigated the influence of 17-β-estradiol on the production, localization, and functions of prohypertrophic (class I) and antihypertrophic (class II) HDACs in cultured neonatal rat cardiomyocytes. 17-β-Estradiol or estrogen receptor β agonists dipropylnitrile and β-LGND2 comparably suppressed angiotensin II–induced HDAC2 (class I) production, HDAC-activating phosphorylation, and the resulting prohypertrophic mRNA expression. In contrast, estrogenic compounds derepressed the opposite effects of angiotensin II on the same parameters for HDAC4 and 5 (class II), resulting in retention of these deacetylases in the nucleus to inhibit hypertrophic gene expression. Key aspects were confirmed in vivo from the hearts of wild-type but not estrogen receptor β (ERβ) gene–deleted mice administered angiotensin II and estrogenic compounds. Our results identify a novel dual regulation of cardiomyocyte HDACs, shown here for the antihypertrophic sex steroid acting at ERβ. This mechanism potentially supports using ERβ agonists as HDAC modulators to treat cardiac disease.
INTRODUCTION
Cardiac hypertrophy arises from a variety of stimuli that cause vascular or cardiac stress forces, resulting in cardiac remodeling to preserve function (Dunn and Pfeffer, 1999; Wagenaar et al., 2002; Berridge, 2006; Heineke and Molkentin, 2006). Because the cardiomyocyte is postmitotic, the response of the cells is to enlarge. However, if the underlying stress persists, the heart often undergoes fibrosis, myocyte thinning due to apoptosis, and progression to heart dilation. The result is an inability of the myocardium to pump sufficient blood to oxygenate tissues, leading to progressive organ dysfunction and often death.
Estrogen is a sex steroid that binds to both estrogen receptor α (ERα) and ERβ in cardiomyocytes (Pedram et al., 2008), cardiac fibroblasts (Pedram et al., 2010), epicardial vascular endothelial cells (Favre et al., 2010), and large-artery endothelial and smooth muscle cells (Zhu et al., 2002; Kumar et al., 2007). Several groups showed that estrogen (17-β-estadiol [E2]) prevents cardiac hypertrophy, fibrosis, and progression to heart failure, mainly from binding to ERβ in vitro and in vivo (van Eickels et al., 2002; Skavdahl et al., 2005; Babiker et al., 2006). Preventive effects of E2/ERβ result from activating the MCIP1 gene to block calcineurin activation in myocytes (Pedram et al., 2005, 2008), stimulating atrial and brain natriuretic peptide production (Pedram et al., 2008), lowering elevated blood pressure (Zhu et al., 2002; Jazbutyte et al., 2007), and preventing fibroblast-to-myofibroblast transition and collagen formation to inhibit cardiac fibrosis (Pedram et al., 2010).
In older individuals, myocardial hypertrophy frequently develops in postmenopausal women and exceeds that of age-matched men (Agabiti-Rosei and Muiesan, 2002). However, this can be reversed by sex steroid replacement (Miya et al., 2002). Sex steroid use after menopause results in a significant decrease in left ventricular mass (Lim et al., 1999), especially in hypertensive women (Light et al., 2001; van Eickels et al., 2002). However, despite the foregoing studies, the mechanisms by which estrogen inhibits cardiac hypertrophy are not fully understood.
Whereas class II histone deacetylases (HDACs) are important for inhibiting cardiac hypertrophy, class I HDACs promote this disorder (Zhang et al., 2002; Backs et al., 2006; Haberland et al., 2009). These proteins act primarily in the nucleus to deacetylate both histone and nonhistone proteins and modify the actions of transcription factors, such as myocyte-enhancing factors. The potential dual regulation of both classes of HDACs by antihypertrophic endogenous substances has not been described. We speculated that the antihypertrophic properties of E2 and ERβ result in part from the modulation of HDAC(s) production and action. Using angiotensin II (AngII) and endothelin-1 (ET-1) as stimulants for cardiomyocyte and in vivo cardiac hypertrophy, we investigated whether E2/ERβ prevents the effects of these peptides to modulate HDAC(s) production, cellular localization, and function. Such results would identify novel estrogen actions and could offer additional insight to facilitate the design of HDAC modulators to treat cardiac diseases.
RESULTS
Angiotensin II and E2/ERβ regulate class II HDAC production
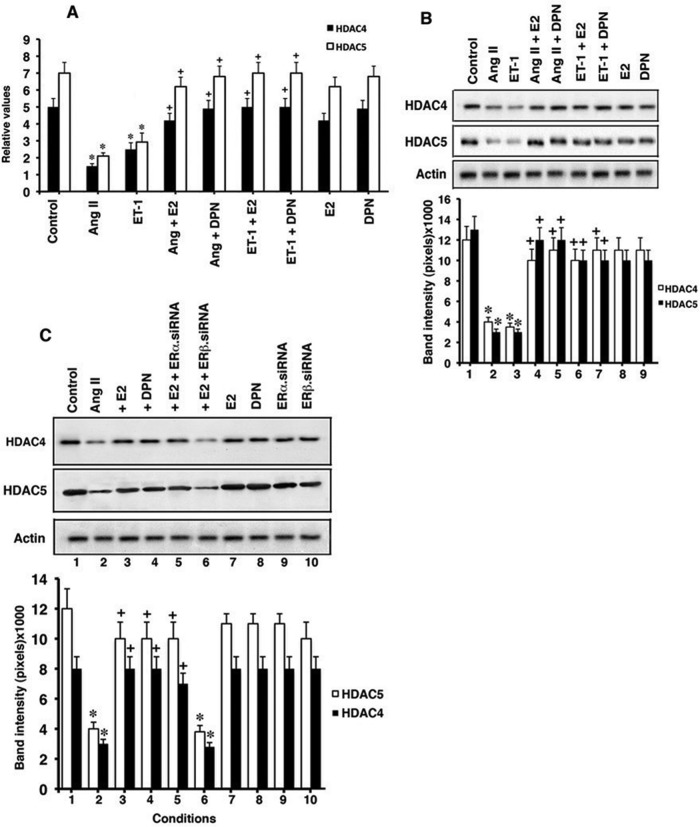
We first determined whether the hypertrophic peptides AngII and ET-1, and E2 or an ERβ-selective agonist, dipropylnitrile (DPN), modulate HDAC4 and 5 expression. In cultured neonatal rat cardiomyocytes, AngII or ET-1 (100 nM) strongly inhibited mRNA expression of both HDACs, determined by quantitative PCR (qPCR). However, relevant concentrations of E2 and DPN comparably and significantly prevented these actions of AngII and ET-1 but had little effect in the absence of AngII (Figure 1A). Protein levels of HDAC4 and 5 were also suppressed by AngII or ET-1, and the estrogenic compounds nearly completely prevented inhibition of HDAC protein production caused by the hypertrophic peptides (Figure 1B). These results also support ERβ as mediating estrogen actions. To further implicate ERβ, we knocked down ERα or ERβ with small interfering RNA (siRNA; validation of the protein knockdowns from the siRNAs is shown in Supplemental Figure S1, A and B). Only the knockdown of ERβ prevented the reversal by E2 of AngII inhibition of HDAC proteins (Figure 1C, lanes 2, 3, 5, and 6). These results indicate that in our model, E2/ERβ serve as derepressors of the antihypertrophic class II HDACs rather than as stimulators of their production. This occurs in the setting of prohypertrophic peptides that are implicated in human cardiovascular diseases.
FIGURE 1:
Modulation of class II HDAC expression. (A) Cardiomyocytes were incubated with AngII (100 nM), ET-1 (10 nM), or each + E2 (10 nM) or equimolar DPN (ERβ ligand) for 24 h. mRNA expression for HDAC4 and 5 was determined by qPCR. Data (mean ± SEM) are from three combined experiments. *p < 0.05 for control vs. AngII or ET-1 by ANOVA + Schefe's test, +p < 0.05 for AngII or ET-1 vs. same + E2 or DPN. (B) Protein expression from same experiments. (C) ERβ mediates estrogenic compound inhibition of HDAC expression. siRNAs to ERα or ERβ were expressed in cardiomyocytes for 24 h before the described experiments. *p < 0.05 for control vs. condition, +p < 0.05 for AngII vs. condition.
HDAC phosphorylation and subcellular localization are regulated by AngII and E2/ERβ
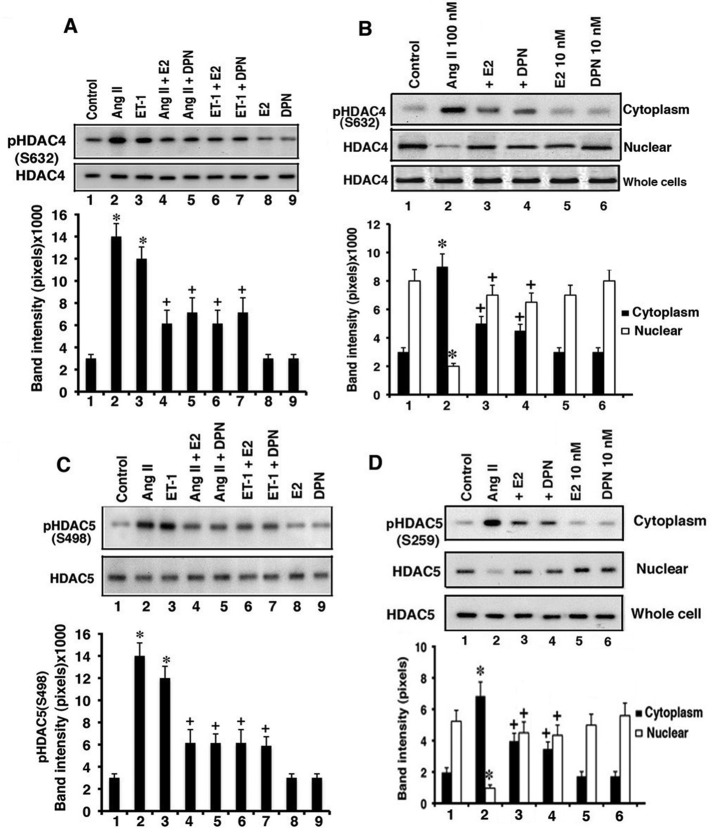
It is well established that AngII causes the phosphorylation of class II antihypertrophic HDACs at specific serine residues. As a result, these deacetylases are exported from the nucleus to cytoplasm, where they no longer serve antihypertrophic functions (Zhang et al., 2002; Backs et al., 2006; Haberland et al., 2009). For HDAC4, hypertrophic agent signaling through calcium and calcium/calmodulin-dependent protein kinase II (CaMKII) causes Ser-632 phosphorylation, resulting in nuclear exclusion of the deacetylase (Backs et al., 2006). Here we found that AngII and ET-1 each stimulated HDAC4 S632 phosphorylation in whole-cell lysates, but this was significantly prevented by E2 or DPN (Figure 2A). HDAC4 total protein levels were unchanged, as phosphorylation was determined at 15 min of cell incubation.
FIGURE 2:
HDAC phosphorylation and cell localization. (A) AngII (100 nM) and ET-1 (10 nM) both stimulate HDAC4 Ser-632 phosphorylation, inhibited by E2 or DPN. Cardiomyocytes were incubated with agents for 30 min. The bar graph is from three experiments. *p < 0.05 for control vs. AngII or ET-1, +p < 0.05 for AngII or ET-1 vs. same + E2 (10 nM) or DPN (10 nM). (B) Cell fractionation of Ser-632 phosphorylated HDAC4. Cardiomyocytes were incubated as described and then lysed and separated into nuclear and cytoplasmic fractions. The study was repeated twice more. (C) Ser-498 phospho-HDAC5 is stimulated by AngII and ET-1 and inhibited by E2 or DPN. *p < 0.05 for control vs. AngII or ET-1, +p < 0.05 for AngII or ET-1 vs. same + E2 or DPN. (D) Phospho-S259 HDAC5 in cell fractions at 30 min. Total HDAC5 protein is also shown, and the study was repeated. Statistical analysis as in B.
We also determined the phosphorylation of HDAC4 in the intact cell. Cardiomyocytes were incubated with AngII ± E2 or DPN and microscopically visualized by immunofluorescence. Using a phospho-S632 HDAC4 specific antibody, we found that AngII caused increased Ser-632 phosphorylation and localized the modified HDAC4 to the cytoplasmic/perinuclear compartment (Supplemental Figure S2A). In contrast, E2 and DPN each inhibited AngII-induced HDAC4 phosphorylation.
To further understand the effect of phosphorylation for HDAC4 cell localization, we carried out similar short-exposure experiments and determined phospho-HDAC4 levels in cytoplasmic and nuclear cell fractions by immunoblot (Figure 2B). Compared to control, in which most HDAC4 was not phosphorylated and was found in the nucleus, AngII stimulated the trafficking of phosphorylated HDAC4 to cytoplasm. Because nuclear (nonphosphorylated) HDAC4 protein was markedly reduced by AngII exposure, we propose that most HDAC4 undergoes this posttranslational modification in response to the hypertrophic peptide. Validation of subcellular fraction purity is shown in Supplemental Figure S3A. In contrast, E2 and DPN markedly reduced the amount of phosphorylated HDAC4 (Figure 2B) and derepressed protein production (Figure 1B), resulting in relocalization to and enhanced expression of this enzyme in the nucleus.
We also determined AngII and ERβ interactions affecting another class II deacetylase, HDAC5. Ser-259/498 phosphorylations within this deacetylase protein occur from protein kinase D (PKD) activation and result in HDAC nuclear-cytoplasmic trafficking (Backs and Olson, 2006). As seen in Figure 2C, AngII and ET-1 each caused enhanced phosphorylation of HDAC5 at Ser-498. Phosphorylation was markedly reduced from concomitant exposure of the cardiomyocytes to either E2 or DPN. From subcellular fractions, AngII also caused Ser-259 phosphorylation of HDAC5 and the relocation of the modified deacetylase from nucleus to cytoplasm compared with control cells (Figure 2D). However, coexposure of the cells to E2 or DPN significantly reversed these effects of AngII, resulting in more HDAC5 localization in the nuclear compartment. To further support these findings, we visualized whole cells by microscopy. As seen in Supplemental Figure S2B, AngII induced the phosphorylation of HDAC5, and the phospho-HDAC primarily resided in the perinuclear (cytoplasmic) area, but phosphorylation was prevented by E2/ERβ. Therefore, both class II HDACs respond to hypertrophic factors with the expected phosphorylations and relocalization to cytoplasm. As a novel mechanism, E2/ERβ prevents both the specific phosphorylations and resulting nuclear exclusion of both class II HDACs, consistent with the antihypertrophic functions of estrogen (Pedram et al., 2005, 2008).
Signaling mechanisms that regulate HDAC phosphorylation
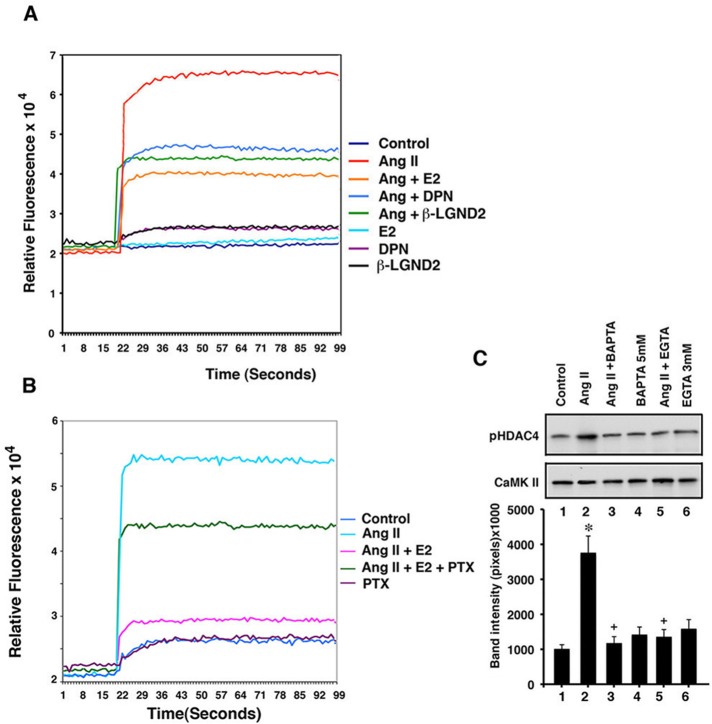
HDAC4 is phosphorylated predominantly through CaMKII in response to hypertrophic factors (Backs et al., 2006), and so we determined whether E2/ERβ inhibited this kinase. Calcium is an early and important signal for CaMKII activation, and we measured calcium flux by modified Fura-2 fluorescence in the cardiomyocytes. AngII stimulated a rapid and sustained increase in calcium over the length of the experiment, significantly and comparably reduced by E2, DPN, or a second ERβ-specific agonist, β-LGND2 (Yepuru et al., 2010; Figure 3A). We postulated that E2/ERβ might cause inhibition of calcium flux through a Gαi-dependent mechanism. Estrogen stimulates the Gαi subunit (Kumar et al., 2007), and Gαi inhibits calcium flux (Zhao et al., 2010; Wang et al., 2011). We found that pertussis toxin, which inhibits estrogen activation of Gαi (Kumar et al., 2007), strongly reversed E2 inhibition of AngII-induced calcium flux (Figure 3B).
FIGURE 3:
Signaling to HDAC4 phosphorylation. (A) Calcium flux in cardiomyocytes. Cells were loaded with Fluor-4 and incubated with AngII ± E2, DPN, or β-LGND2, and calcium changes over 100 s were determined by spectrophotometer. The study was repeated a second time. (B) Pertussis toxin (100 ng/ml) inhibits AngII-induced calcium flux. The study was repeated a second time. (C) HDAC4 S632 phosphorylation by CaMKII is calcium dependent. CaMKII was immunoprecipitated from cardiomyocytes for use in an in vitro kinase assay. The bar graph is from three experiments. *p < 0.05 for control vs. AngII, +p <0.05 for AngII vs. same + BAPTA or EGTA. IgG immunoprecipitation showed no bands.
To implicate calcium in the regulation of CAMKII, we performed an in vitro kinase assay in which CaMKII was immunoprecipitated from cardiomyocytes under various conditions and incubated with HDAC4 substrate protein. Phosphorylation of HDAC4 at S632 (a CaMKII phosphorylation site) was then determined by specific immunoblotting. AngII caused enhanced calcium-dependent, CaMKII-induced S632 phosphorylation of HDAC4, since the calcium chelators 1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid (BAPTA) and ethylene glycol tetraacetic acid (EGTA) suppressed this action (Figure 3C). Thus E2/ERβ inhibition of calcium, probably through Gαi signaling, accounts for the sex steroid receptor inhibiting CaMKII-induced HDAC4 phosphorylation and resulting nuclear exclusion (Figures 2A and 3).
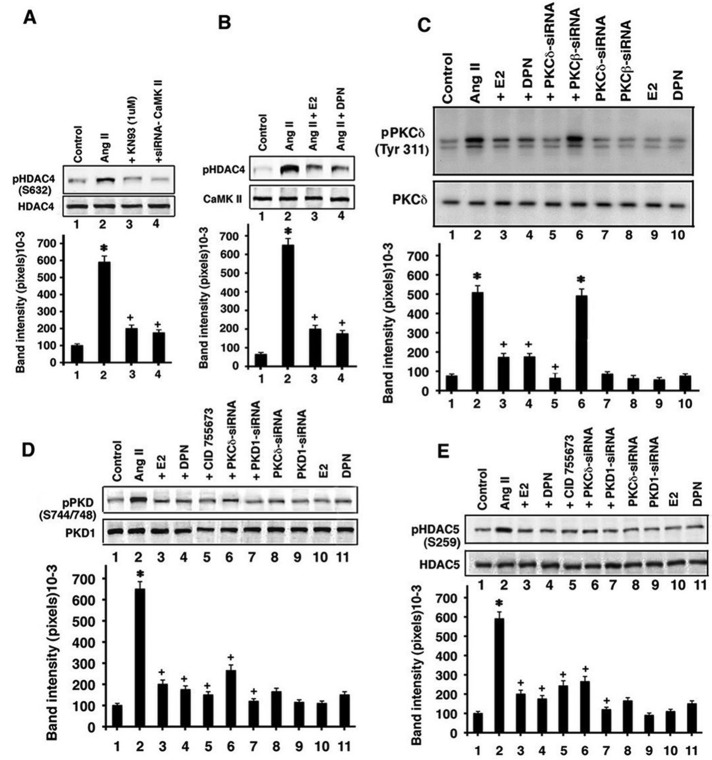
Providing support for this idea, we performed additional HDAC4 S632 phosphorylation experiments. We first showed that AngII stimulation of HDAC4 S632 phosphorylation in the cardiomyocytes at 60 min was significantly prevented by coincubation with a soluble inhibitor of CaMKII, KN93 (Figure 4A). We also knocked down this kinase with specific siRNA (siRNA validation in Supplemental Figure S1C) and found a significant loss of HDAC4 S362 phosphorylation. These results strongly implicate CaMKII for AngII action in this respect. We then incubated the cardiomyocytes with AngII ± E2 or DPN, immunoprecipitated the CaMKII from each condition, and added equal protein amounts of the kinase to HDAC4 substrate protein for in vitro kinase activity determination. E2 or DPN comparably caused a significant inhibition of CaMKII-dependent, AngII-induced HDAC4 S362 phosphorylation (Figure 4B)
FIGURE 4:
Estrogen modulation of signaling. (A) CaMKII mediates AngII-induced S632HDAC4 phosphorylation. *p < 0.05 for control vs. AngII, +p < 0.05 for AngII vs. AngII + KN93 or siRNA to CaMKII. The data are from three experiments. (B) E2 and DPN inhibit CaMKII phosphorylation of HDAC4. (C) AngII stimulates and E2 and DPN inhibit PKCδ activation. The bar graph is from three experiments. *p < 0.05 for control vs. AngII or AngII + PKCβ siRNA, +p < 0.05 for AngII vs. AngII + E2 or DPN, ++p < 0.05 for AngII vs. AngII + siRNA PKCδ. (D) PKD-activating phosphorylations are stimulated by AngII through a PKCδ-dependent mechanism and are opposed by E2 and DPN. *p < 0.05 for control vs. AngII, +p < 0.05 for AngII vs. AngII + E2 or DPN, CID755763 (50 μM), PKC, or PKD siRNAs. (E) HDAC5 S259 phosphorylation is stimulated by AngII through a PKCδ and PKD–related mechanism and opposed by E2 and DPN. *p < 0.05 for control vs. AngII, +p < 0.05 for AngII vs. AngII + E2 or DPN, CID755763, PKCδ, or PKD siRNAs.
We also determined HDAC5 phosphorylation as regulated by prohypertrophic peptides and antihypertrophic steroids. Phosphorylation of HDAC5 at S259 occurs by hypertrophic factor signaling through protein kinase Cδ (PKCδ) and PKD. It has been reported that S498 phosphorylation results from CaMKII activity, but this has not been consistently found (Zhang et al., 2002; Backs et al., 2006; Backs and Olson, 2006; Kong et al., 2006). We therefore investigated the former pathway.
In cultured cardiomyocytes, AngII caused significantly enhanced activation of PKCδ, reflected as Tyr-311 phosphorylation, which was prevented by E2 or DPN (Figure 4C). AngII also caused activating phosphorylations of PKD (S744/748), which were comparably and significantly inhibited by estrogenic compounds (Figure 4D). Furthermore, PKCδ siRNA prevented PKD phosphorylations as induced by AngII, linking the two kinases (validation of specific PKC siRNA(s) in Supplemental Figure S1, D and E, where PKCβ serves as a negative control, and Figure 4C). However, calcium chelation with BAPTA did not affect the ability of AngII to stimulate PKCδ or PKD activity (Supplemental Figure S3B). As shown in Figure 4E, S259 HDAC5 phosphorylation was stimulated by AngII and inhibited by E2, DPN, or a PKD inhibitor (CID 755673; validated in Figure 4D). PKCδ or PKD1 knockdown with siRNA also significantly impaired AngII-induced HDAC5 phosphorylation (PKD protein inhibition by siRNA is shown in Supplemental Figure S1F). Our results indicate novel functions of E2/ERβ to inhibit the linked pathways from Gαi, calcium, and CaMKII to HDAC4 phosphorylation and from PKC to PKD, which is important for HDAC5 phosphorylation, with phosphorylations of both deacetylases resulting from hypertrophic peptide actions. These results indicate a new mechanism for endogenous steroids to prevent exclusion from the nucleus of class II HDAC(s), underlying the antihypertrophic functions of estrogen.
Functional roles of class II HDACs
An important transition during the development of cardiac hypertrophy is switching of the myosin heavy chain (MHC) isoform from α-MHC in the normal heart to β-MHC in the hypertrophied organ (Lompre et al., 1979). This change embodies reorganization of the ventricle and sarcomere. We previously showed that AngII stimulates this gene/protein isoform transition in the heart and that estrogen significantly prevents this hypertrophic factor action (Pedram et al., 2005, 2008). Estrogen signaling from the membrane receptor pool has been shown by others and us to modulate the epigenetic regulation of transcription, most often working in conjunction with the nuclear ER pool (Pedram et al., 2012). We therefore determined whether AngII induces acetylation of histone 3 at the βMHC promoter, an effect that likely results from the exclusion of class II HDACs in the nucleus (Figure 2, B and D), thereby contributing to the development of cardiac hypertrophy.
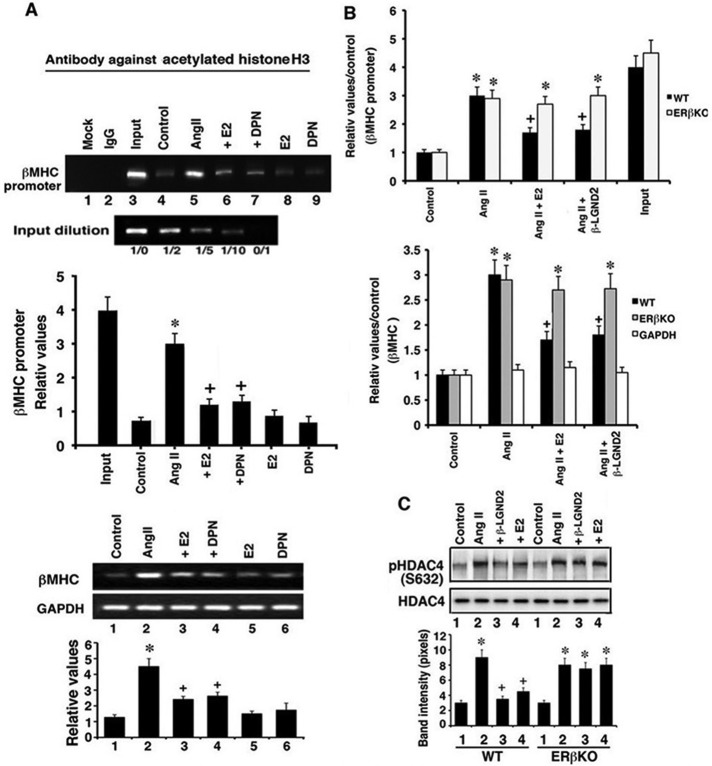
Using cultured cardiomyocytes and chromatin immunoprecipitation (ChIP), we found that AngII caused significant acetylation of histone 3 (H3) at the β-MHC promoter, an epigenetic mark that strongly correlates with induction of genes (Zhang et al., 2002; Suganuma et al., 2010; Figure 5A, top). H3 acetylation induced by AngII also correlated with increased βMHC mRNA expression stimulated by the hypertrophic peptide (Figure 5A, bottom). E2 and DPN significantly blocked all of these effects of AngII. These results are consistent with the ability of E2/ERβ to prevent the nuclear exclusion of HDAC4 and 5 shown here.
FIGURE 5:
Functional roles of HDACs. (A) Top, ChIP for histone 3 acetylation at the βMHC promoter in cardiomyocytes cultured for 24 h. The procedure is described in Materials and Methods. Bar graph of band densitometry is from three experiments. *p < 0.05 for control vs. AngII, +p < 0.05 for AngII vs. AngII + E2 or DPN. Bottom, βMHC mRNA expression normalized to GAPDH expression by qPCR. *p < 0.05 for control vs. AngII, +p <0.05 for AngII vs. AngII + E2 or DPN. (B) Top, ChIP of H3 acetylation at the βMHC promoter from WT or mouse ventricular DNA. *p < 0.05 for control vs. AngII or AngII plus condition in ERβKO mice, +p < 0.05 for AngII vs. AngII + E2 or DPN. Data as reflected in the bar graph are from the pooled samples of four to six mice per condition. Bottom, βMHC mRNA expression by qPCR in WT and ERβKO mouse ventricles. *p < 0.05 for control vs. AngII or AngII plus condition in ERβKO mice, +p < 0.05 for AngII vs. AngII + E2 or DPN. GAPDH data are from WT mice. (C) HDAC4 S632 phosphorylation in mouse ventricles. Data are from the pooled samples of four to six mice per condition. *p < 0.05 for control vs. AngII or AngII plus condition in ERβKO mice, +p < 0.05 for AngII vs. AngII + E2 or DPN.
To determine in vivo relevance to our studies, we used material from investigations of cardiac hypertrophy that we recently reported (Pedram et al., 2013). Wild-type (WT) and ERβKO ovariectomized female mice were infused by osmotic minipump with AngII over 4 wk in the absence or presence of coinfused β-LGND2 or an E2 pellet inserted under the skin. The mice were then examined for multiple cardiac parameters. E2 or β-LGND2 substantially prevented AngII-induced hypertension, cardiac hypertrophy, and cardiac fibrosis (Pedram et al., 2013). Here, using DNA from the left ventricles of WT mice, we determined that AngII stimulated H3 acetylation at the β-MHC promoter by ChIP assay, which was significantly inhibited upon coinfusion of the mice with β-LGND2 or exposed to E2 (Figure 5B, top). However, estrogenic compound effects were active in the WT but not the ERβKO mouse, consistent with mediation of estrogen's antihypertrophic effects through ERβ. These results correlate with βMHC gene expression in the ventricles of WT (but not ERβKO) mice exposed to the same conditions (Figure 5B, bottom). Thus the epigenetic and genetic data are consistent and correlate with the regulation of class II HDAC function/phosphorylation and cell localization.
To further support the latter conclusions in vivo, we determined the phosphorylation of HDAC4 at S632 in ventricles from the various mice. As mentioned, phosphorylation at this residue leads to nuclear exclusion of HDAC4. In WT mice, AngII infusion resulted in an increased phosphorylation of HDAC4 S632, which was significantly prevented by E2 or β-LGND2 administration (Figure 5C). However, the estrogenic compounds opposed only phosphorylation stimulated by AngII in WT mice.
Modulation of HDAC2 by AngII and E2/ERβ
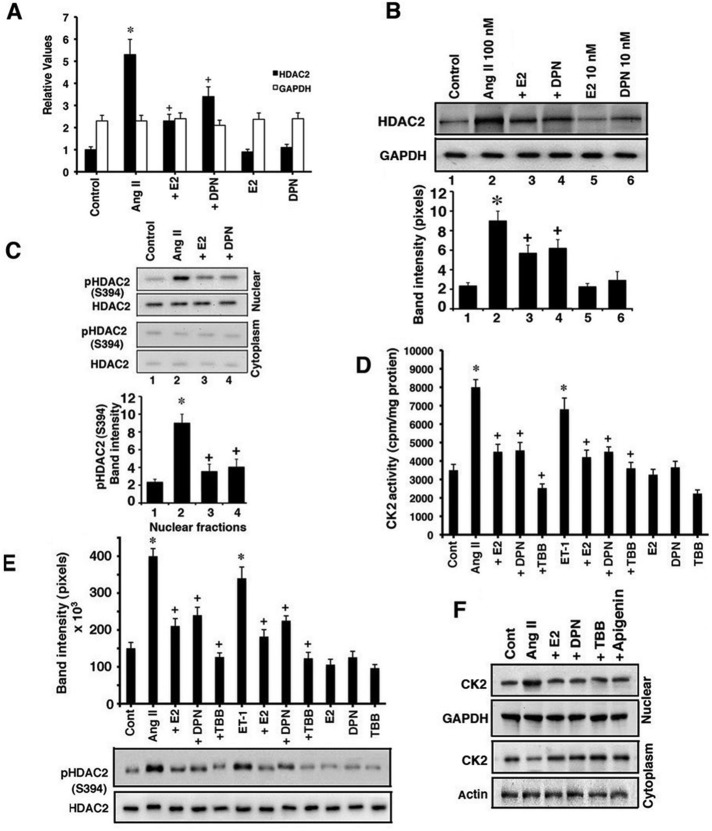
Class I HDACS such as HDAC2 have been strongly implicated as prohypertrophic enzymes, promoting the transcription of genes such as Nppa and Myh7 (Trivedi et al., 2007) that are associated with the cardiac phenotype (Backs and Olson, 2006; Trivedi et al., 2007). We determined whether AngII and E2/ERβ regulate this HDAC. In cultured cardiomyocytes, AngII significantly stimulated HDAC2 mRNA expression (Figure 6A) and protein (Figure 6B) at 24 h, each significantly inhibited by E2 or DPN. We also investigated the phosphorylation and cell localization of the deacetylase, since HDAC2 is an effective hypertrophy-promoting protein when it is phosphorylated in the nucleus of the cardiomyocyte (Kee et al., 2006; Trivedi et al., 2007).
FIGURE 6:
AngII and E2/ERβ regulate HDAC2. (A) HDAC2 mRNA expression is determined by qPCR from cardiomyocytes incubated under various conditions for 24 h. GAPDH is determined as a control. Data are from three experiments. *p < 0.05 for control vs. AngII, +p < 0.05 for AngII vs. AngII + E2 or DPN. (B) HDAC2 protein abundance. *p < 0.05 for control vs. AngII or AngII plus condition in ERβKO mice, +p < 0.05 for AngII vs. AngII + E2 or DPN. (C) HDAC2 S394 phosphorylation in nuclear and cytosolic cell fractions. Total HDAC2 is also shown, and the bar graph is from three experiments. *p < 0.05 for control vs. AngII, +p < 0.05 for AngII vs. AngII + E2 or DPN. (D) CK2 activity in cardiomyocytes. *p < 0.05 for control vs. AngII or ET-1, +p < 0.05 for AngII or ET-1 vs. same + E2, DPN, or TBB. (E) HDAC2 S394 phosphorylation is stimulated by hypertrophic peptides in a CK2-dependent manner and inhibited by E2/ERβ. HDAC2 total protein is shown as loading control. *p < 0.05 for control vs. AngII or ET-1, +p < 0.05 for AngII or ET-1 vs. same+ E2, DPN, or TBB. (F) AngII stimulates CK2 abundance in the nucleus of cardiomyocytes, which is inhibited by E2, DPN, and CK2 inhibitors. Subcellular fractionation of CK2 protein is shown. This study was repeated.
As shown in the cell images of Supplemental Figure S2C, AngII stimulated the activating phosphorylation of HDAC2 at Ser-394 (S394) that is seen in the nucleus. In addition, E2 and DPN significantly diminished the phosphorylation of nuclear HDAC2. To confirm the cell images, we carried out immunoblotting of HDAC2 S394 phosphorylation in cytosolic and nuclear fractions. AngII and ET-1 significantly stimulated the nuclear HDAC phosphorylation that E2 and DPN inhibited (Figure 6C). Of interest, under all conditions, total HDAC2 remained in the nucleus. Thus E2/ERβ does not cause intracellular translocation of the prohypertrophic HDAC2 protein, in contrast to the steroid effects on class II HDACs.
Creatine kinase (CK2) phosphorylates HDAC2 at S394, resulting in enhanced activity of the enzyme (Eom et al., 2011). CK2 has been reported to be constitutively active in some cells, but we clearly find that in cardiomyocytes, AngII and ET-1 each causes an increase in cytoplasmic CK2 activity, determined as precipitated kinase from cytosolic fraction activity against a substrate protein in vitro (Figure 6D). Furthermore, a kinase family member, CK1, has also been considered to be constitutively active, but recently was found to be regulated by Wnt signaling (Cruciat et al., 2013). The activation of CK2 by AngII was unaffected by calcium chelation with BAPTA (Supplemental Figure S3C). E2 and DPN significantly inhibited AngII and ET-1 activation of the kinase, which was also blocked by the soluble CK2 inhibitor tetrabromobenzotriole (TBB; Kramerov et al., 2011; Figure 6D). These findings correlate with the ability of TBB, E2, and DPN to also block AngII- and ET-1–induced phosphorylation of HDAC2 at S394 at 1-h cell exposure (Figure 6E), implicating CK2 as a hypertrophic factor modulating HDAC2 activity (Kramerov et al., 2011).
We then determined the subcellular phosphorylation and localization of CK2 protein in the cardiomyocytes. AngII stimulated the expression of CK2 protein in the nucleus of the whole cell, which was substantially reduced by E2, DPN, or the CK inhibitors TBB and apigenin (also validating the function of the latter; Supplemental Figure S2D). To determine whether AngII promoted the trafficking of the CK2 protein to the nucleus, we fractionated the cardiomyocytes after 2-h exposure to peptide and steroids. AngII stimulated the trafficking of this protein kinase from cytoplasm to the nucleus compared with the control condition (Figure 6F). This is consistent with the enhanced phosphorylation of nuclear HDAC2 by CK2 upon exposure of the cells to AngII (Figure 6E). The increased trafficking of this kinase to the nucleus was inhibited by E2, DPN, and, of interest, the CK2 inhibitors TBB and apigenin. The latter observation suggests the novel concept that activation of CK2 by AngII, as we show in cytoplasm (Figure 6D), may promote nuclear localization of the kinase and subsequent phosphorylation of HDAC2. It is also possible that inhibition of CK2 activity by estrogenic compounds, TBB, or apigenin promotes nuclear exclusion of the kinase (Figure 6F). Thus it is CK2 and not HDAC2 that traffics in response to AngII but is inhibited by E2/ERβ. There is little knowledge on 1) mechanistically how any substance activates CK2, 2) the structural requirements and protein–protein interactions for CK2 to traffic in or out of the nucleus, and 3) the upstream signaling by membrane E2/ERβ that affects all this, thus illustrating the need for future extensive investigation.
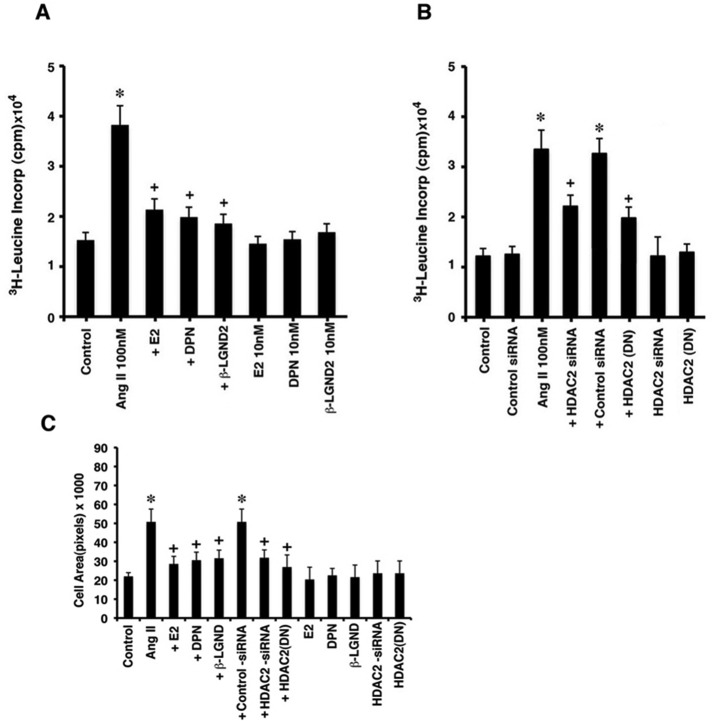
To investigate the functional roles of HDAC2, we incubated the cardiomyocytes with AngII ± estrogenic compounds and determined enhanced protein synthesis, a hallmark of myocyte hypertrophy (Pedram et al., 2005). AngII significantly stimulated leucine incorporation, which was inhibited by E2, DPN, or β-LGND2 (Figure 7A). To implicate HDAC2, the same study was done in cardiomyocytes transfected with control or HDAC2 siRNA or a dominant-negative (DN) HDAC2 plasmid (pcDNA3-HDAC2 S394A) 24 h before AngII exposure. HDAC2 siRNA resulted in a substantial decrease in the endogenous protein (Supplemental Figure S1G) and significantly prevented AngII stimulation of protein synthesis in the cells (Figure 7B). Expressing the DN mutant HDAC2 in cardiomyocytes (Eom et al., 2011; validated in Supplemental Figure S4) also strongly inhibited AngII-stimulated leucine incorporation (Figure 7B). Cell area, another parameter of hypertrophy, was also determined. AngII significantly increased cardiomyocyte area, which was significantly prevented by E2, DPN, or β-LGND2 (Figure 7C). Furthermore, this effect of AngII was also reversed by siRNA to HDAC2 or expression of the dominant-negative deacetylase. These results indicate that HDAC2 plays a significant role in the ability of AngII to stimulate in vitro cardiomyocyte hypertrophy, which is opposed by estrogenic compounds acting through ERβ.
FIGURE 7:
Cardiomyocyte hypertrophy is modulated by HDAC2. (A) Protein synthesis is stimulated by AngII and inhibited by E2 and ERβ agonists in vitro. The [3H]leucine incorporation in cultured cardiomyocytes was done as described in Materials and Methods. The bar graph is from three experiments.*p < 0.05 for control vs. AngII, +p < 0.05 for AngII vs. AngII + E2, DPN, or β-LGND2. (B) HDAC2 protein knockdown or dominant-negative (DN) HDAC2 expression prevents AngII-stimulated protein synthesis. *p < 0.05 for control vs. AngII, +p < 0.05 for AngII vs. AngII + siRNA to HDAC2. (C) Cardiomyocyte surface area was measured as described in Materials and Methods. The bar graph is from three experiments combined. *p < 0.05 for control vs. AngII, +p < 0.05 for AngII vs. AngII + estrogenic compound or siRNA to HDAC2 or DN HDAC2.
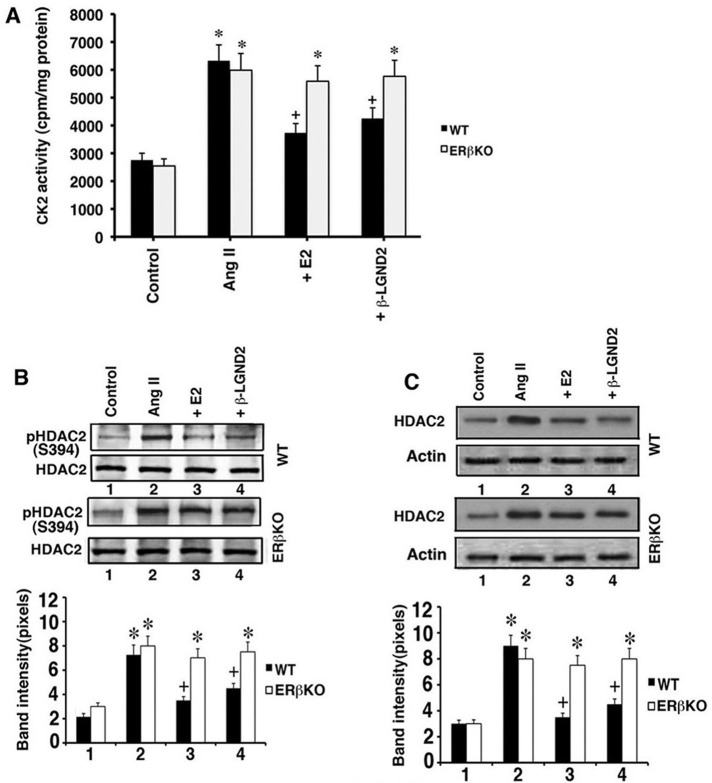
To validate some of these key findings in vivo, we determined CK2 and HDAC2 activity in WT and ERβKO mouse ventricles. As shown in Figure 8A, AngII stimulated and E2 and β-LGND2 prevented CK2 activation, the latter only in WT mice. For HDAC2 S394 phosphorylation, AngII stimulated this phosphorylation in all mice, but E2 and β-LGND2 prevented this only in WT mouse ventricles (Figure 8B). HDAC2 protein abundance was also stimulated by AngII in all mice but singularly inhibited by E2 and β-LGND2 in WT mouse hearts (Figure 8C).
FIGURE 8:
In vivo regulation of CK2 and HDAC2. (A) CK2 activity was determined by immunoprecipitating the kinase from the ventricular proteins for in vitro assay using substrate protein. Bar graph is from four to six mouse samples per condition in WT and ERβKO mice. *p < 0.05 for control vs. AngII or AngII + condition in ERβKO mice, +p < 0.05 for AngII vs. AngII + E2 or β-LGND2 in WT mice. (B) HDAC2 S394 phosphorylation is stimulated by AngII and is inhibited by E2 and β-LGND2, the latter only in WT mice. Total HDAC2 protein is normalization control. *p < 0.05 for control vs. AngII or AngII + condition in ERβKO mice, +p < 0.05 for AngII vs. AngII + E2 or β-LGND2 in WT mice. (C) HDAC2 protein abundance is stimulated by AngII but inhibited by E2 or β-LGND2 in WT mice. Data analysis is done as in B.
Transcriptional targets for HDAC2
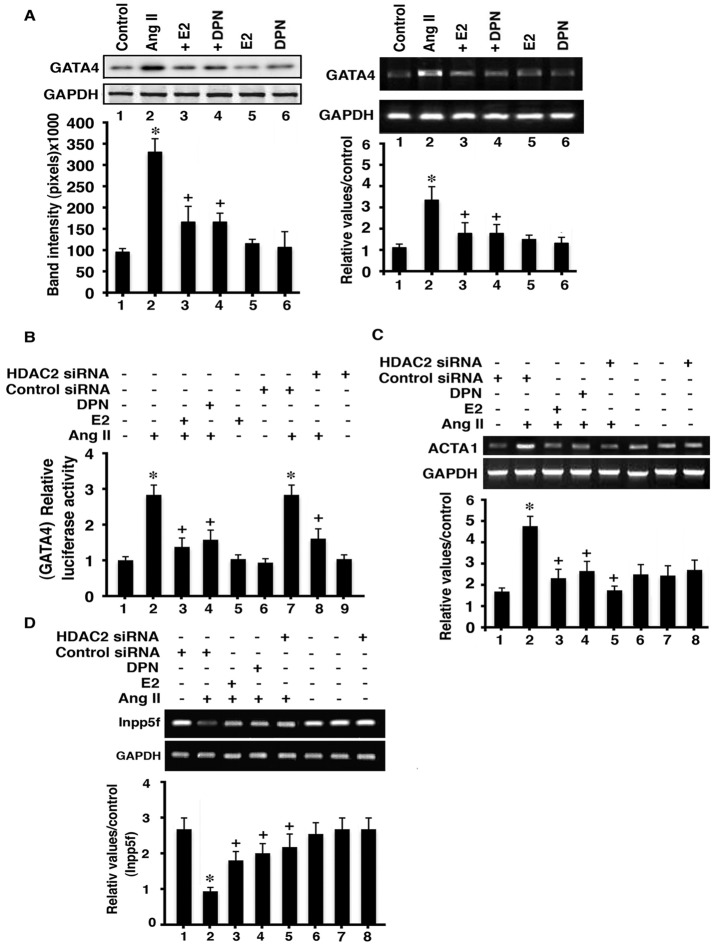
HDAC2 promotes hypertrophy by activating transcription factors and hypertrophic genes and inhibiting antihypertrophic genes. We determined the ability of AngII and E2/ERβ to regulate representative genes in cardiomyocytes. GATA4 is an important transcription factor (TF) and is activated by hypertrophic factor signaling to promote cardiac hypertrophy (Zhou et al., 2012). AngII stimulated the increased mRNA and protein expression of GATA4 in cultured cardiomyocytes, which was prevented by E2 or DPN (Figure 9A). Furthermore, the transcriptional activity of this TF was stimulated by AngII, determined from transfection of a GATA4-luciferase reporter into cardiomyocytes (Figure 9B). GATA4 activity was prevented by HDAC2 siRNA or from coincubation of the myocytes with E2 or DPN. We then determined the response of a hypertrophic gene, ACTA1 (Trivedi et al., 2007), and found that AngII stimulated the expression of this mRNA, dependent upon HDAC2; E2 and DPN also significantly inhibited this action of AngII (Figure 9C). Thus AngII stimulated GATA4 production and activity, the latter dependent upon HDAC2, which was inhibited by E2 or DPN. ACTA1 was shown to be a target for GATA4 (Bisping et al., 2012), and we sequentially linked AngII, HDAC2, and GATA4 to ACTA1 expression.
FIGURE 9:
HDAC and GATA4 interactions. (A) GATA4 protein (left) and mRNA expression (right) in rat cardiomyocytes. The bar graph is from three experiments. *p < 0.05 for control vs. AngII, +p < 0.05 for AngII vs. AngII + E2 or DPN. (B) AngII stimulates GATA4 transcriptional activity in an HDAC2-related manner. The bar graph is from three experiments. *p < 0/05 for control vs. AngII, +p < 0.05 for AngII vs. AngII + E2 or DPN. (C)Target of GATA4 signaling (ACTA1 mRNA) is stimulated by AngII and is inhibited by E2, DPN, or HDAC2 siRNA. (D) Inpp5F mRNA is suppressed by AngII and is reversed by E2, DPN, or HDAC2 siRNA. The bar graph is from three experiments. *p < 0.05 for control vs. AngII, +p < 0.05 for control vs. AngII, +p < 0.05 for AngII vs. AngII + E2, DPN, or siRNA to HDAC2.
As an inhibitor of cardiac hypertrophy, the Inpp5f gene codes for a phosphatase that represses AKT activity in cardiomyocytes (Trivedi et al., 2007). AngII inhibited the expression of the Inpp5f mRNA in cardiomyocytes in an HDAC2-dependent manner, but this was derepressed by coincubation of the cardiomyocytes with E2 or DPN (Figure 9D). AngII signals through AKT to cause an inhibitory phosphorylation of the glycogen synthase kinase 3β (GSK3β) at Ser-9, lifting the repressive phosphorylation by GSK3β of the GATA4 transcription factor and thereby promoting cardiac hypertrophy (Morisco et al., 2001; Antos et al., 2002). Here we implicate HDAC2 in the ability of AngII to repress the Inpp5f mRNA, providing a mechanism by which AngII stimulates AKT to inhibit GSK3β activity and therefore inducing GATA4-mediated hypertrophic gene expression, such as ACTA1 (Figure 9C). We recently showed that ERβ inhibits AngII-induced AKT in cardiomyocytes, in part by derepressing Inpp5F (Pedram et al., 2013). Here we suggest the ability of E2 and DPN to inhibit HDAC2 as a novel mechanism to limit AngII hypertrophic effects that result from GATA4 activation and Inpp5F suppression (Figure 9, B and D).
DISCUSSION
Cardiac hypertrophy results from signaling to the enhanced transcription of key genes that induce structural and muscle sarcomeric reorganization of the ventricle (Heineke and Molkentin, 2006). Important interactions that regulate cardiac hypertrophic gene expression include histone acetyltransferase (HAT) proteins binding to and activating 1) transcription factors such as myocyte enhancing factor (MEF-2) or 2) other coregulator proteins (Backs and Olson, 2006). HATs also acetylate lysines of histone proteins, providing access for transcription factors and coregulators to bind gene promoters. The important transcription factors in cardiac hypertrophy include MEF-2, GATA-4, NFATc3, and CAMTA2, which cooperate to transcribe the structural genes (Wilkins et al., 2002; Akazawa and Komuro, 2003; Song et al., 2006). In the normal heart, the functions of these transcription factors are repressed from binding to HDACs competitively to HATs (Backs and Olson, 2006) or from TF sequestration in cytoplasm (e.g., NFATc3; Wilkins et al., 2002). In response to hypertrophic signals, HATs and HDACs are often posttranslationally modified and sometimes relocalize in the cell, resulting in ventricular enlargement, fibrosis, thinning (cardiomyocyte dropout), and progression to heart failure.
Extensive studies in vitro and in vivo (including knockout [KO] mice) strongly implicate class II HDACs (such as HDAC4, 5, and 9) as preventing cardiac hypertrophy from many (but not all) stimuli (Chang et al., 2004; Backs et al., 2006; Kong et al., 2006; Haberland et al., 2009). Hypertrophic stimuli that regulate HDACs include AngII signaling, which also affects non-HDAC targets, such as the phosphatase calcineurin (Chang et al., 2004; Haberland et al., 2009). In contrast, some class I HDACs (especially HDAC 2) promote cardiac hypertrophy, and these latter targets may be why broad HDAC inhibitors (Kong et al., 2006) or specific class I HDAC inhibitors (Eom et al., 2011) block hypertrophic gene transcription. The balance of prohypertrophic and antihypertrophic HDAC activity may therefore determine the state of the myocardium.
We postulated that endogenous antihypertrophic molecules regulate both classes of HDACs, but little is known about this. Furthermore, the strong cardiac antihypertrophic functions of estrogen (van Eickels et al., 2002; Skavdahl et al., 2005; Babiker et al., 2006; Pedram et al., 2008) have not been previously linked to the modulation of HDAC production, activation, and function. Here we find that AngII and ET-1 signaling to cardiomyocyte hypertrophy in vitro and AngII signaling in vivo reciprocally regulate class I and II HDACs. This regulation is strongly opposed by rapid signaling in response to E2- or ERβ-specific ligands. We showed that ERβ is at the plasma membrane of cardiomyocytes (Pedram et al., 2005), and it is the membrane pool and not the nuclear ER pool that responds to estrogen with rapid signal transduction (Pedram et al., 2009). Thus we propose that membrane ERβ mediates E2 action here.
We report here that the class II HDAC4 and 5 are inhibited by AngII at the gene and protein level, but this is opposed by E2/ERβ. Previous studies suggested that class II HDAC levels do not change in response to stress (Chang et al., 2004; Haberland et al., 2009), but this is likely to depend on the stimulus for HDAC expression. Furthermore, we do not find that estrogenic compounds independently modulate HDAC production: instead, they derepress AngII actions in these regards. We also addressed posttranslational modification of class II HDACs, which is important to their function. Phosphorylation of HDAC4 and 5 by AngII signaling respectively occurs through CaMKII, PKCδ, and PKD, as shown here. AngII induces PKD to associate with HDAC5, causing phosphorylation at the sites we investigated, which is known to cause association with 14-3-3 proteins (Xu et al., 2007). Association of HDAC5 with 14-3-3 proteins is integral to the nuclear export of the deacetylase by CRM1 (Harrison et al., 2004), thereby contributing to derepression of nuclear MEF2 or other transcription factors. HDAC export to the cytoplasm allows nuclear histone acetylases such as p300 to associate with and acetylate TFs (e.g., MEF2, GATA-4) and TF-facilitating proteins such as myocardin, resulting in hypertrophic gene expression (Vega et al., 2004; McKinsey and Olson, 2005). In addition, class II HDACs have been proposed to function in ways that are unrelated to deacetylase activity (Haberland et al., 2009). We report here that E2/ERβ causes deacetylation of histone 3 at the βMHC promoter, in concert with suppression of βMHC expression. Deacetylation likely reflects the ability of E2 and DPN to cause retention of class II HDACs in the nucleus but may also arise in part from suppression of HDAC2 (class I) abundance and activity, which is prohypertrophic.
In this regard, class IIa HDACs in particular form heterodimers with each other and with class I HDAC3. It has been reported that the interaction of HDAC4 with HDAC3 occurs via the N-Cor/SMRT corepressor and is necessary for the deacetylase activity of HDAC4 (Fischle et al., 2002). However, HDAC4 and 5 can prevent MEF2 from associating with p300 in a manner unrelated to their C-terminal domains, which interact with HDAC3, thus serving as inhibitors of p300-mediated acetylation (Zhang et al., 2002). Although the C-terminal domain of class II HDACs has been demonstrated to possess intrinsic deacetylase activity using artificial substrates, in vivo histone or nonhistone substrates for these proteins have not been identified. Our findings involve Gq-mediated stimuli (AngII and ET-1) that modulate HDACs and resulting hypertrophy. It is well established that estrogen and ERβ also suppress pressure-overload models of cardiac dysfunction (Dent et al., 2010; Fliegner et al., 2010; Gardner et al., 2010). However, there are no data in these models regarding HDAC regulation by sex steroids, so we can only speculate that the mechanisms we report here may be applicable to other causes of non–ischemia-related hypertrophy.
Some data suggest that specific inhibitors of PKC and PKD, but not CaMKII, repress the nuclear export of HDAC5, indicating that PKCδ and PKD may be particularly important in this regard (Vega et al., 2004). However, CaMKII causes selective phosphorylation of HDAC4 at Ser-632 and subsequent nuclear export in response to AngII (or ET-1), as we show here. These actions of the hypertrophic peptides are blocked by E2 via ERβ and pertussis toxin–sensitive Gαi, resulting in decreased calcium flux and inhibition of CaMKII. It has also been reported that heterodimerization occurs between HDAC4 and other class II HDACs such as HDAC5 (Kong et al., 2006), and CaMKII phosphorylation appears important in these circumstances. Here we show that E2/ERβ significantly suppress all of these kinases, supporting a novel mechanism of the inhibition of cardiac hypertrophy by this endogenous steroid. More broadly, we are not aware of any antihypertrophic, endogenous molecule that causes a reciprocal/dual regulation of class I and II HDACs. A diagram of these pathways is shown in Supplemental Figure S5.
Regarding HDAC2, we found that the ability of AngII to activate CK2 in cytoplasm is important to the nuclear localization of this kinase. In the nucleus, CK2 phosphorylates HDAC2 at S394 (Eom et al., 2011), resulting in enhanced prohypertrophic actions of this deacetylase (Trivedi et al., 2007). We establish here that inhibition of CK2 activity by a soluble inhibitor of CK2 activity, E2, or DPN also prevents nuclear localization of this kinase, suggesting the linkage of these events sequentially. We also determined downstream mRNA targets of HDAC2 that are oppositely regulated by AngII and E2/ERβ. These include GATA4, ACTA1, and Inpp5f.
A recent study found that transverse aortic constriction–induced cardiac hypertrophy or Gαq-dependent hypertrophy involves phospholipase Cε bound to the Golgi/nuclear envelope, which generates diacylglycerol to activate nuclear PKD (Zhang et al., 2013). This pathway was also implicated in ET-1 generating nuclear calcium elevation, which therefore might activate a pool of nuclear CaMKII. However, linkage of this pathway to HDAC phosphorylation was not established in this study. We find that PKCδ activation of PKD is an important pathway for AngII-stimulated HDAC5 phosphorylation, inhibited by E2 and DPN. However, we cannot rule out an effect of extranuclear ERβ on the phospholipase Cε pathway described, especially since the mentioned in vivo studies were carried out in male mice (Zhang et al., 2013). Much additional investigation would be needed to implicate this pathway as part of the AngII-ERβ interaction in female mice or cardiomyocytes. In addition, CaMKII signaling in mitochondria is activated in response to several severe cardiac stress, causing calcium entry into this organelle, which results in cardiomyocyte death (Joiner et al., 2012). We reported that E2 and mitochondrial ERβ inhibit the cellular toxicity of radiation and tamoxifen therapies in breast cancer cells by inhibiting oxidative stress and activation of the intrinsic, mitochondrial apoptotic program (Pedram et al., 2006; Razandi et al., 2013). Mitochondrial ERβ has been identified in cardiomyocytes (Yang et al., 2004). We speculate that in the acutely stressed heart, this ER pool may serve a similar survival function to that in breast cancer, perhaps by inhibiting mitochondrial CaMKII activity, which would be consistent with the general inhibition of this kinase that we report here.
Estrogen-induced cardiac protection was found to occur in HDAC5 and 9–deleted female mice subjected to myocardial infarction (Rooij et al., 2010). Class II HDAC5 and 9 bound to ERα and the MEF-2 transcription factor, preventing ERα expression and steroid receptor–mediated transcription. On deletion of HDAC5 and 9, estrogen and ERα stimulated neoangiogenesis from vascular endothelial growth factor transcription, contributing to enhanced survival of mice after myocardial infarction (Rooij et al., 2010). Postinfarct remodeling of the heart is very different from pathology caused by AngII infusion, and in the model of Rooij et al. (2010), HDAC5 and 9 opposed the protective effects of E2. We previously reported that estrogen provides strong cardiac protection in WT and ERα gene–deleted mice, arising from steroid action at ERβ in the AngII model (Pedram et al., 2005, 2008). Nevertheless, estrogen action through both receptors and several cellular ER pools appears to involve HDACs, depending on the insult, and results in cardiac protection.
Finally, the use of HDAC inhibitors to prevent progression of cardiac disease has been a research focus generating much interest. HDAC inhibitors potently repress the ability of cardiac hypertrophic factors to induce this pathology (Antos et al., 2003; Zhao et al., 2010; McKinsey, 2011). However, repression of cardiac disease likely occurs from a predominant effect on class I HDACs such as HDAC2 (Trivedi et al., 2007) rather than from inhibiting the antihypertrophic class II HDACs, which was seen from using nonspecific inhibitors in the early stages of this research. There also are toxicities in noncardiac systems that result from inhibiting multiple HDACs (McKinsey, 2011). We suggest that administration of an ERβ agonist that has no apparent in vivo toxicity yet provides benefit in multiple organ systems (Yepuru et al., 2010) may be an approach that warrants further investigation. Here we establish that such a compound, β-LGND2, reciprocally regulates in vivo both pro– and anti–cardiac hypertrophic HDACs, which was recently shown by us to prevent AngII-induced hypertension, cardiac hypertrophy, and cardiac fibrosis through multiple mechanisms (Pedram et al., 2013). By avoiding E2 ligation of ERα, which promotes breast and uterine cancer, ERβ-selective agonists may be a new approach to intervening in cardiovascular diseases.
MATERIALS AND METHODS
Cell culture and reagents
Neonatal rat cardiomyocytes were isolated as described (Pedram et al., 2005) and cultured in DMEM/F12 supplemented with 10% fetal bovine serum, 1× insulin-transferrin-selenium (Sigma, St. Louis, MO) antibiotic and antimycotic, and 10 μg/ml fibronectin (to aid adherence). Angiotensin II, endothelin-1, E2, BAPTA, EGTA, and pertussis toxin were all obtained from Sigma, and DPN was from Tocris Pharmaceuticals (Minneapolis, MN). KN93 (CaMKII inhibitor) was from EMD-Millipore (Billerica, MA), and apigenin, TBB, and CID 755673 were from Tocris. HDAC2, 4, and 5 and CaMKII total and phospho-antibodies and MHC isoform antibodies were from Abcam (Cambridge, MA). Phospho and/or total protein antibodies to CK2, PKCδ, and PKCβ were from Santa Cruz Biotechnology (Santa Cruz, CA), and PKD and phospho-PKD antibodies were from Cell Signaling (Danvers, MA). Control, ERα, ERβ, PKC, and PKD siRNAs were from Qiagen (Germantown, MD) or Santa Cruz Biotechnology. All primers were from Invitrogen (Carlsbad, CA). Dominant-negative HDAC2 (pcDNA3-HDAC2 S394A) and WT-HDAC2–encoding plasmids were kind gifts from Chinmay Trivedi, University of Massachusetts Medical School (Worcester, MA).
Measurement of protein synthesis, protein secretion, and cell area
After 24 h in media without serum, the cells were treated with 100 nM AngII in the presence or absence of 10 nM E2 or ERβ agonists. New protein synthesis, a marker of hypertrophy, was determined as described (Pedram et al., 2005). The cells were cultured as described with the addition of 1 μCi/ml [3H]leucine for 48 h. In some conditions, siRNA (control or to HDAC2) or a dominant-negative HDAC2 was expressed for 24 h before AngII exposure. After 48 h, the cells were rinsed in DMEM/F12, and lysed with TRIzol. The protein and RNA were extracted, the protein fraction counted in a scintillation counter, and the mean ± SEM of the counts plotted. Differences between treatments were compared for combined data from three experiments and the results analyzed by analysis of variance (ANOVA) plus Schefe's test (p < 0.05 was significant). Cell surface area was determined under the various conditions after culturing on cover slips and imaging by confocal microscopy. This was done at 40–50% cell confluency. Surface area was quantified by imaging to the complete boundary of 30 individual cells/condition using ImageJ software (National Institutes of Health, Bethesda, MD). From this, a mean ± SEM value was calculated for surface area in each condition from each experiment (Pedram et al., 2005). The planar aspect and deviations of individual cells in the z-axis were determined and adjusted.
Animal studies
All studies were approved by the Animal Care and Research and Development Committees at the Department of Veterans Affairs Medical Center, Long Beach, CA. The 10- to 12-wk-old female C57/BJ6 mice, intact or ovariectomized, were obtained from Harlan/Sprague Dawley, housed in 12-h on/off lighting, and fed rodent chow devoid of soy or most plant products. AngII (1.1 mg/kg per day) in saline or saline alone–filled osmotic minipumps (Alzet, Cupertino, CA; DURECT, Cupertino, CA) or AngII plus 100 μl of β-LGND2 (0.5 mg; Yepuru et al., 2010) was provided by 21-d infusion after subcutaneous insertion under inhaled chlorofluorane anesthesia. In some mice, an E2 pellet (0.5 mg, 21-d release pellets; Innovative Research of America, Sarasota, FL) or placebo pellet was inserted under the skin, but these mice did not receive β-LGND2. This pellet is well documented to produce physiological levels of E2 in the serum of mice (Pedram et al., 2010).
At 21 d, the hearts were removed and weighed, and the ratio of heart to total body weight was determined. RNA, DNA, and protein were extracted and the heart sectioned for further studies. For comparison, ovariectomized female wild-type and ERβ gene–deleted mice were obtained from Ken Korach, National Institute of Environmental Health Sciences (Research Triangle Park, NC) and Harlan Laboratories (Hayward, CA). These mice were subjected to the same conditions as described, and all mice were identically housed and fed the same chow.
Cellular calcium/Fura assay
Calcium activity reflected both extracellular calcium influx through membrane channels and intracellular calcium mobilization from stores and was determined as follows. Calcium was measured in neonatal cardiomyocyte cells by loading with Fluo-4 NW (Molecular Probes, Eugene, OR). The cells were cultured in 96-well microplates (poly-d-lysine [PDL] coated) to subconfluence (40,000–50,000 cells/well), and then cells were synchronized overnight in medium lacking fetal bovine serum and phenol red. The medium was removed to eliminate sources of baseline fluorescence, particularly esterase activity. Fluo-4 NW (100 μl) in loading solution was carefully added to each well, and the cells were incubated at 37°C for 60 min. The microplate was then transferred into a 37°C prewarmed Nowastar spectrofluorometer (BMG Labtech, Offenburg, Germany). The instrument was preset and loaded with test solutions at 2× concentration, with subsequent injection of 50 μl of control or AngII ± estrogenic compounds, followed immediately by 250 cycles of reading over 140 s. In one condition, pertussis toxin (100 mg/ml) was added at the same time as AngII + E2. Each condition was done in triplicate, and the study was repeated a second time. Fluorescence was measured at excitation of 494 nm and emission at 516 nm.
CK2 kinase assays
Synchronized cardiomyocytes were exposed to various treatments (e.g., ANG II ± E2) for 60 min. Cells for each condition (∼1 million) were then washed with DMEM/F12 medium and lysed in buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 50 mM NaF, 5 mM EDTA, 0.5% Triton X-100, 40 mM β-glycerophosphate, 200 mM sodium orthovanadate, 40 mM p-nitrophenol phosphate, 100 mM phenylmethylsulfonyl fluoride (PMSF), and protease inhibitor cocktail [Sigma]). The lysates were centrifuged at 14,000 rpm for 10 min, and the supernatants were exposed to 50 μl of suspended protein A agarose beads in microcentrifuge tubes and rotated for 30 min at 4°C. The cell lysate/protein A bead complex was centrifuged at 6000 rpm for 30 s at 4°C. The supernatant was transferred to a new tube and stored on ice. For the CK2 activity assay, 10 μl of CK2 antibody (dilution 1:50; Santa Cruz Biotechnology) was conjugated to 50 μl of protein A beads for 2 h at room temperature, and the bead complex was washed. Then 1 ml of precleared whole-cell extract from each experimental condition was added to the CK2 antibody/protein A bead complex in lysis buffer (which also serves as the immune-precipitation buffer) and was rotated end over end overnight at 4°C. Beads were washed once with lysis buffer and twice with HEPES buffer (25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], 10 mM Mg acetate). To the lysates/antibody/bead complexes, 40 μl of stock mixture, made up as 20 μl of 3× kinase buffer (25 mM HEPES, pH 7.5, 10 mM Mg acetate, 2 mM dithiothreitol, 40 mM ATP), and 10 μl of H2O was added to 2 μg of CK2 substrate peptide (RRRADDSDDDDD; Signal Chem, Richmond, Canada) in 9 μl of water with 1 μl of [γ-32P]ATP. Each tube was vortexed and incubated at 30°C for 30 min, and the reaction was stopped by adding 200 μg of casein and 100 μl of 4% trichloroacetic acid. Aliquots (50 μl) of supernatants were spotted on P81 phosphocellulose paper (Whatman) and washed with 0.5% phosphoric acid, and the radioactivity of the samples was counted in 5 ml of scintillation liquid. For in vivo determination of CK2 activity, the kinase protein was immunoprecipitated from the ventricular protein lysates of treated WT and ERβKO female mice, and the assay was carried out as described.
PKC, PKD, and CaMKII assays
PKCδ activity was determined at 30 min of incubation as phosphorylation of the protein isoform at the active site, Tyr-311. Cells were exposed to various conditions, and the cells were lysed, separated by SDS–PAGE, transferred to nitrocellulose, and immunoblotted with antibodies to Tyr-311 or antibodies to determine total PKC protein (Santa Cruz Biotechnology). PKD activity at 30 min was determined as Ser-744/748 phosphorylation by immunoblots from cells exposed to the various conditions, using phospho and total antibodies from Cell Signaling. CaMKII activity was determined at 60 min by immunoblot of phosphorylated Ser-632 of HDAC4 from the cardiomyocytes and by an in vitro tube assay, using [γ-32P]ATP, immunoprecipitated CaMKII, and exogenous recombinant human HDAC4 substrate peptide (amino acids 612–1084; Abcam) to reflect HDAC4 Ser-467/632 phosphorylation by CaMKII. All studies were done three times for the combined data.
Quantitative real-time PCR
Total RNA was extracted using the Qiagen RNeasy Mini Kit following the manufacturer's protocol. All the samples were treated with DNAse-free (Ambion). RNA purity and concentrations were measured by UV spectrophotometry (A260 and A280). cDNA was synthesized using approximately 500 ng of RNA and Oligo dT primers with the Improm-II reverse transcription system (Promega). Quantitative real-time PCR (qRT-PCR) was used to examine the relative expression of HDAC2, 4, 5, βMHC, Inpp5f, GATA4, and ACTA1, and expression was normalized using the housekeeper glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Primers were designed using Primer3 (http://frodo.wi.mit.edu/) and were blasted to check specificity (primers: HDAC2 forward [F], 5′-ttggcctttctgagctgatt-3′; HDAC2 reverse [R], 5′-agagggtctctgccactgaa-3′; HDAC4 F, 5′-gctctcccagctctccagca-3′; HDAC4 R, 5′-gttgtgagctgctgctgcaccgt-3′;HDAC5 F, 5′-tgagaggcaggcccttcagt-3′; HDAC5 R, 5′-cctccagtgccactcccaac-3′; GATA4 F, 5′-ggctcccagagattcttcct-3′; GATA4 R, 5′-ctctgctacggccagtaagg-3′; β-MHC F, 5′-accaagcagccacgccagta3′; β-MHC R, 5′-tgctttgcctttgcccttgt-3′; Inpp5f F, 5′-aacttgggaaaggcctgg-3′; Inpp5f R, 5′-catggagctgcggatctt-3′; α–smooth muscle actin (ACTA1), 5′-tcgcgaccttactgactacctg-3′, 5′-gcttctctttgatgtcgcgc-3′; GAPDH F, 5′-ccacagtcc atgccatca-3′; GAPDH R, 5′-ggatgaccttgcccacag-3′). Primers were designed to have an annealing temperature of 55°C and to amplify regions of approximately 150 base pairs. PCR amplicon sizes were confirmed by agarose gel electrophoresis before qRT-PCR analysis. For qRT-PCR analysis 500 ng of cDNA was used in a 50-μl reaction consisting of 25 μl of SYBR Green ER qPCR Supermix (Invitrogen), 1 μl of 10 μM forward/reverse primer stocks, and nuclease-free water. Thermocycling was carried out using the iCycler (Bio-Rad) with a melting curve temperature of 60°C. Relative mRNA levels were calculated using the Ct method (Livak and Schmittgen, 2001).
Subcellular fractionation
The subcellular fractionation was performed using the Subcellular Protein Fractionation Kit (78840; Pierce). The extraction buffer for cytoplasmic isolation (500 μl) was added to cell pellets. Cells were incubated at 4°C for 20 min while rotating. Then the homogenates were centrifuged (500 × g) at 4°C for 5 min. The supernatants (cytoplasmic extracts) were transferred to new Eppendorf tubes. The membrane extraction buffer (500 μl) was added to cell pellets, followed by vortexing and incubation at 4°C for 10 min. The homogenates were centrifuged (5600 × g) at 4°C (5 min), and the supernatants (membrane extracts) were transferred to new Eppendorf tubes. The nuclear extraction buffer (250 μl) was added to cell pellets, followed by vortexing and incubation at 4°C for 30 min. The homogenates were centrifuged (7300 × g) at 4°C for 5 min, supernatants (nuclear extracts) were transferred to new Eppendorf tubes, and protein concentrations were estimated by the Bradford assay. Western blotting was performed by loading 20 μg of each cytoplasmic and nuclear extract. Lack of contamination of the cell fractions was accomplished as previously described (Pedram et al., 2006).
Immunofluorescence microscopy
Cardiomyocyte cells (3 × 105) were placed on PDL-coated glass-bottom dishes (MatTek, Ashland, MA). Twenty-four hours later, at a confluency of 70–80%, cells were treated with various agents, washed three times with phosphate-buffered saline (PBS), and fixed with 4% fresh paraformaldehyde in PBS for 10 min at room temperature. After further washing with PBS, the cells were permeabilized with 0.2% Triton Χ-100 in PBS for 5 min and washed with PBS. Before antibody incubation, cells were blocked with serum from the same species as the secondary antibody or 2% bovine serum albumin (BSA) in PBS at room temperature for 30 min. After washing with PBS, cells were incubated overnight at 4°C with primary antibody (dilution 1:50 in 0.5% BSA-PBS). The cells were then incubated for 1 h with fluorescein isothiocyanate–conjugated secondary antibody (dilution, 1:500; Vector Laboratories) at room temperature. Cells were washed three times with PBS, and samples were covered with coverslips and mounting medium and analyzed by immunofluorescence microscopy (Nikon).
Chromatin immunoprecipitation assay
ChIP assays were performed using the Active Motif ChIP-IT kit. For each experimental condition, four confluent 10-cm plates of neonatal cardiomyocytes cells (107cells/dish) were used. After treatments for 24 h, cells were washed with ice-cold PBS and cross-linked for 5 min with 1% formaldehyde, and the reaction was stopped by adding glycine (125 mM). The cells were scraped and washed with cold PBS containing proteinase inhibitor cocktail (Sigma) and 1 mM PMSF. Enzymatic fragmentation of chromatin was performed using the Enzymatic Shearing kit (Active Motif) according to the manufacturer's instructions. For the ChIP assays, 50 μg of digested chromatin was precleared for 1 h with protein A–Sepharose beads. After centrifugation, supernatants were incubated with 10 μg of acetylated histone H3 antibody or rabbit immunoglobulin G (IgG; Cell Signaling) or no antibody (mock condition) and incubated overnight at 4°C under rotation. The immune-precipitated complexes were bound to protein A–Sepharose beads during 3 h at 4°C and washed successively in a lysis buffer (10 mM Tris/HCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, 140 mM NaCl, 0.1% sodium deoxycholate, and 1 mM PMSF, pH 7.4), LiCl buffer (1 mM Tris/HCl, 250 mM LiCl, 0.5% Nonidet P40, 0.5% sodium deoxycholate, and 1 mM EDTA, pH 7.4), and Tris–EDTA buffer (1 mM EDTA and 10 mM Tris/HCl, pH 7.4). Chromatin complexes were eluted by three sequential incubations at room temperature (twice for 15 min and once for 45 min) in 90 μl of elution buffer (100 mM NaHCO3 and 1% [wt/vol] SDS). Proteins were then eliminated by using 200 μg of proteinase K (Promega) in the presence of 10% (wt/vol) SDS by overnight incubation at 37°C. Chromatin DNA was extracted with phenol/chloroform, followed by ethanol precipitation. Samples were dissolved in 80 μl of water. The PCR primers used for the analysis of the rat βMHC promoter (directed against residues between −317 and −255) were 5′-atagcaacagcgaggctctttctg-3′, rat β-MHC, forward; and 5′-ggagctcacctaccagacaga-3′, rat β-MHC, reverse; they were provided by Qiagen. PCR amplification products were analyzed on ethidium bromide–stained 2.5% (wt/vol ) agarose gels.
Supplementary Material
Acknowledgments
The work was supported by the Research Service of the Department of Veteran's Affairs and National Institutes of Health Grant CA-10036 to E.R.L.
Abbreviations used:
- AngII
angiotensin II
- CaMKII
calcium/calmodulin-dependent kinase
- CK2
creatine kinase
- DPN
dipropylnitrile
- E2
17-β-estradiol
- ER
estrogen receptor
- ET-1
endothelin-1
- HDAC
histone deacetylase
- PKC
protein kinase C
- PKD
protein kinase D
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-08-0444) on October 23, 2013.
No conflicts of interest or disclosures were reported by the authors, except R.N. and J.T.D., who are employees of and own stock in GTx.
REFERENCES
- Agabiti-Rosei E, Muiesan ML. Left ventricular hypertrophy and heart failure in women. J Hypertens. 2002;20:S34–S38. [PubMed] [Google Scholar]
- Akazawa H, Komuro I. Roles of cardiac transcription factors in cardiac hypertrophy. Circ Res. 2003;92:1079–1088. doi: 10.1161/01.RES.0000072977.86706.23. [DOI] [PubMed] [Google Scholar]
- Antos CL, McKinsey TA, Dreitz M, Hollingsworth LM, Zhang CL, Schreiber K, Rindt H, Gorczynski RL, Olson EN. Dose-dependent blockade to cardiomyocyte hypertrophy by histone ase inhibitors. J Biol Chem. 2003;278:28930–28937. doi: 10.1074/jbc.M303113200. [DOI] [PubMed] [Google Scholar]
- Antos CL, McKinsey TA, Frey N, Kutsdchke W, McAnally J, Shelton JM, Richardson JA, Hill JA, Olson EN. Activated glycogen synthase-3β suppresses cardiac hypertrophy in vivo. Proc Natl Acad Sci USA. 2002;99:907–912. doi: 10.1073/pnas.231619298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiker FA, Lips D, Meyer R, Delvaux E, Zandberg P, Janssen B, van Eys G, Grohe C, Doevendans PA. Estrogen receptor β protects the murine heart against left ventricular hypertrophy. Arterioscler Thromb Vasc Biol. 2006;26:1524–1530. doi: 10.1161/01.ATV.0000223344.11128.23. [DOI] [PubMed] [Google Scholar]
- Backs J, Olson EN. Control of cardiac growth by histone acetylation/deacetylation. Circ Res. 2006;98:15–24. doi: 10.1161/01.RES.0000197782.21444.8f. [DOI] [PubMed] [Google Scholar]
- Backs J, Song K, Bezprpzvannaya S, Chang S, Olson EN. CAM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. J Clin Invest. 2006;116:1853–1864. doi: 10.1172/JCI27438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge MJ. Remodeling Ca2+ signaling systems and cardiac hypertrophy. Biochem Soc Trans. 2006;34:228–231. doi: 10.1042/BST20060228. [DOI] [PubMed] [Google Scholar]
- Bisping E, Ikeda S, Sedej M, Wakula P, McMullen JR, Tarnavski O, Sedej S, Izumo S, Pu WT, Pieske B. Transcription factor GATA4 is activated but not required for insulin-like growth factor 1 (IGF1)-induced cardiac hypertrophy. J Biol Chem. 2012;287:9827–9834. doi: 10.1074/jbc.M111.338749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, McKinsey TA, Zhang CL, Richardson JA, Hill JA, Olson EN. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol Cell Biol. 2004;24:8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciat C-M, Dolde C, de Groot REA, Ohkawara B, Reinhard C, Korswagen HC, Niehrs C. RNA helicase DDX3 is a regulatory subunit of casein kinase 1 in Wnt-b-catenin signaling. Science. 2013;339:1436–1441. doi: 10.1126/science.1231499. [DOI] [PubMed] [Google Scholar]
- Dent MR, Tappia PS, Dhalla NS. Gender differences in cardiac dysfunction and remodeling due to volume overload. J Card Fail. 2010;16:439–449. doi: 10.1016/j.cardfail.2009.12.017. [DOI] [PubMed] [Google Scholar]
- Dunn FG, Pfeffer MA. Left ventricular hypertrophy in hypertension. N Engl J Med. 1999;340:1279–1280. doi: 10.1056/NEJM199904223401610. [DOI] [PubMed] [Google Scholar]
- Eom GH, et al. Casein kinase-2α1 induces hypertrophic response by phosphorylation of histone deacetylase 2 S394 and its activation in the heart. Circulation. 2011;123:2392–2403. doi: 10.1161/CIRCULATIONAHA.110.003665. [DOI] [PubMed] [Google Scholar]
- Favre J, Gao J, Henry JP, Remy-Jouet I, Fourquaux I, Billon-Gales A, Thulliez C, Arnal JF, Lenfant F, Richard V. Endothelial estrogen receptor alpha plays an essential role in the coronary and myocardial protective effects of estradiol in ischemia/reperfusion. Arterioscler Thromb Vasc Biol. 2010;12:2562–2567. doi: 10.1161/ATVBAHA.110.213637. [DOI] [PubMed] [Google Scholar]
- Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- Fliegner D, et al. Female sex and estrogen receptor-beta attenuate cardiac remodeling and apoptosis in pressure overload. Am J Physiol Regul Integr Comp Physiol. 2010;298:R1597–R1606. doi: 10.1152/ajpregu.00825.2009. [DOI] [PubMed] [Google Scholar]
- Gardner JD, Murray DB, Voloshenyuk TG, Brower GL, Bradley JM, Janicki JS. Estrogen attenuates chronic volume overload induced structural and functional remodeling in male rat hearts. Am J Physiol Heart Circ Physiol. 2010;298:H497–H504. doi: 10.1152/ajpheart.00336.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison BC, Roberts CR, Hood DB, Sweeney M, Gould JM, Bush EW, McKinsey TA. The CRM1 nuclear export receptor controls pathological cardiac gene expression. Mol Cell Biol. 2004;24:10636–10649. doi: 10.1128/MCB.24.24.10636-10649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signaling pathways. Nat Rev Mol Cell Biol. 2006;7:589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- Jazbutyte V, et al. Ligand-dependent activation of ERbeta lowers blood pressure and attenuates cardiac hypertrophy in ovariectomized SHR. Cardiovasc Res. 2007;77:774–781. doi: 10.1093/cvr/cvm081. [DOI] [PubMed] [Google Scholar]
- Joiner MA, et al. CaMKII determines mitochondrial stress responses in heart. Nature. 2012;491:269–273. doi: 10.1038/nature11444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee HJ, et al. Inhibition of histone deacetylation blocks cardiac hypertrophy induced by angiotensin II infusion and aortic banding. Circulation. 2006;113:51–59. doi: 10.1161/CIRCULATIONAHA.105.559724. [DOI] [PubMed] [Google Scholar]
- Kong Y, Tannous P, Lu G, Berenji K, Rothermel BA, Olson EN, Hill JA. Suppression of class I and II histone deacetylases blunts pressure-overload cardiac hypertrophy. Circulation. 2006;113:2579–2588. doi: 10.1161/CIRCULATIONAHA.106.625467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramerov AA, Golub AG, Bdzhola VG, Yarmoluk SM, Ahmed K, Bretner M, Ljubimov AV. Treatment of cultured human astrocytes and vascular endothelial cells with protein kinase CK2 inhibitors induces early changes in cell shape and cytoskeleton. Mol Cell Biochem. 2011;349:125–137. doi: 10.1007/s11010-010-0667-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Wu Q, Chambliss KL, Yuhanna IS, Mumby SM, Mineo C, Tall GG, Shaul PW. Direct interactions with Gαi and Gβγ mediate nongenomic signaling by estrogen receptor β. Mol Endocrinol. 2007;21:1370–1380. doi: 10.1210/me.2006-0360. [DOI] [PubMed] [Google Scholar]
- Light KC, Hinderliter AL, West SG, Grewen KM, Steege JF, Sherwood A, Girdler SS. Hormone replacement improves hemodynamic profile and left ventricular geometry in hypertensive and normotensive postmenopausal women. J Hypertens. 2001;19:269–278. doi: 10.1097/00004872-200102000-00014. [DOI] [PubMed] [Google Scholar]
- Lim WK, Wren B, Jepson N, Roy S, Caplan G. Effect of hormone replacement therapy on left ventricular hypertrophy. Am J Cardiol. 1999;83:1132–1134. doi: 10.1016/s0002-9149(99)00029-6. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time, quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lompre AM, Schwartz K, d'Albis A, Lacombe G, Thiem NV, Swynghedauw B. Myosin isozymes redistribution in chronic heart overloading. Nature. 1979;282:105–107. doi: 10.1038/282105a0. [DOI] [PubMed] [Google Scholar]
- McKinsey TA. Isoform-selective HDAC inhibitors: closing in on translational medicine for the heart. J Mol Cell Cardiol. 2011;51:491–496. doi: 10.1016/j.yjmcc.2010.11.009. [DOI] [PubMed] [Google Scholar]
- McKinsey TA, Olson EN. Toward transcriptional therapies for the failing heart: chemical screens to modulate genes. J Clin Invest. 2005;115:538–546. doi: 10.1172/JCI24144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya Y, Sumino H, Ichikawa S, Nakamura T, Kanda T, Kumakura H, Takayama Y, Mizunuma H, Sakamaki T, Kurabayashi M. Effects of hormone replacement therapy on left ventricular hypertrophy and growth-promoting factors in hypertensive postmenopausal women. Hypertens Res. 2002;25:153–159. doi: 10.1291/hypres.25.153. [DOI] [PubMed] [Google Scholar]
- Morisco C, Seta K, Hardt SE, Lee Y, Vatner SF, Sadoshima J. Glycogen synthase kinase 3β regulates GATA4 in cardiac myocytes. J Biol Chem. 2001;276:28586–28597. doi: 10.1074/jbc.M103166200. [DOI] [PubMed] [Google Scholar]
- Pedram A, Razandi R, Aitkenhead M, Levin ER. Estrogen inhibits cardiomyocyte hypertrophy in-vitro: antagonism of calcineurin-related hypertrophy through induction of MCIP1. J Biol Chem. 2005;280:26339–26348. doi: 10.1074/jbc.M414409200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedram A, Razandi M, Deschenes R, Levin ER. DHHC 7 and 21 are palmitoylacyltranferases for sex steroid receptors. Mol Biol Cell. 2012;23:188–199. doi: 10.1091/mbc.E11-07-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedram A, Razandi M, Kim JK, O'Mahony F, Lee E, Luderer U, Levin ER. Developmental phenotype of a membrane only estrogen receptor α (MOER) mouse. J Biol Chem. 2009;284:3488–3495. doi: 10.1074/jbc.M806249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedram A, Razandi M, Korach KS, Narayanan R, Dalton JT, Levin ER. ERβ selective agonist inhibits angiotensin-induced cardiovascular pathology in female mice. Endocrinology. 2013;154:4352–4364. doi: 10.1210/en.2013-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedram A, Razandi M, Lubahn D, Liu J, Vannan M, Levin ER. Estrogen inhibits cardiac hypertrophy: role of estrogen receptor beta to inhibit calcineurin. Endocrinology. 2008;149:3361–3369. doi: 10.1210/en.2008-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedram A, Razandi M, O'Mahony F, Lubahn D, Levin ER. Estrogen receptor beta prevents cardiac fibrosis. Mol Endocrinol. 2010;24:2152–2165. doi: 10.1210/me.2010-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedram A, Razandi M, Wallace DC, Levin ER. Functional estrogen receptors in the mitochondria of breast cancer cells. Mol Biol Cell. 2006;17:2125–2137. doi: 10.1091/mbc.E05-11-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razandi M, Pedram A, Jordan VC, Fuqua S, Levin ER. Tamoxifen regulates cell fate through mitochondrial estrogen receptor beta in breast cancer. Oncogene. 2013;32:3274–3285. doi: 10.1038/onc.2012.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooij EV, Sutherland FJ, Thijssen VL, Crijns HJ, Dimaio MJ, Shelton J, De Windt LJ, Hill JA, Olson EN. Myocyte enhancer factor 2 and class II histone deacetylases control a gender-specific pathway of cardioprotection mediated by the estrogen receptor. Circ Res. 2010;106:155–165. doi: 10.1161/CIRCRESAHA.109.207084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skavdahl M, Steenbergen C, Clark J, Myers P, Demianenko T, Mao L, Rockman HA, Korach KS, Murphy E. Estrogen receptor-β mediates male-female differences in the development of pressure overload hypertrophy. Am J Physiol Heart Circ Physiol. 2005;288:H469–H476. doi: 10.1152/ajpheart.00723.2004. [DOI] [PubMed] [Google Scholar]
- Song K, Backs J, McAnally J, Qi X, Gerard RD, Richardson JA, Hill JA, Bassel-Duby R, Olson EN. The transcriptional coactivator CAMTA2 stimulates cardiac growth by opposing class II histone deacetylases. Cell. 2006;125:453–466. doi: 10.1016/j.cell.2006.02.048. [DOI] [PubMed] [Google Scholar]
- Suganuma T, Mushegian A, Swanson SK, Abmayr SM, Florens L, Washburn MP, Workman JL. The ATAC acetyltransferase complex coordinates MAP kinases to regulate JNK target genes. Cell. 2010;142:726–736. doi: 10.1016/j.cell.2010.07.045. [DOI] [PubMed] [Google Scholar]
- Trivedi CM, et al. HDAC2 regulates the cardiac hypertrophic response by modulating GSK3β activity. Nat Med. 2007;13:324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- van Eickels M, Grohe C, Cleutjens JP, Janssen BJ, Wellens HJ, Doevendans PA. 17beta-Estradiol attenuates the development of pressure-overload hypertrophy. Circulation. 2002;104:1419–1423. doi: 10.1161/hc3601.095577. [DOI] [PubMed] [Google Scholar]
- Vega RB, Harrison BC, Meadows E, Roberts CR, Papst PJ, Olson EN, McKinsey TA. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol Cell Biol. 2004;24:8374–8385. doi: 10.1128/MCB.24.19.8374-8385.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenaar LJ, Voors AA, Buikema H, van Gilst WH. Angiotensin receptors in the cardiovascular system. Can J Cardiol. 2002;18:1331–1339. [PubMed] [Google Scholar]
- Wang F, Zhang Y, Jiang X, Zhang Y, Zhang L, Gong S, Liu C, Zhou L, Tao J. Neuromedin U inhibits T-type Ca2+ channel currents and decreases membrane excitability in small dorsal root ganaglia neurons in mice. Cell Calcium. 2011;49:12–22. doi: 10.1016/j.ceca.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Wilkins BJ, De Windt LJ, Bueno OF, Braz JC, Glascock BJ, Kimball TF, Molkentin JD. Targeted disruption of NFATc3, but not NFATc4, reveals an intrinsic defect in calcineurin-mediated cardiac hypertrophic growth. Mol Cell Biol. 2002;22:7603–7613. doi: 10.1128/MCB.22.21.7603-7613.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Ha C-H, Wong C, Wang W, Hausser A, Pfizenmaier K, Olson EN, McKinsey TA, Jin Z-G. Angiotensin II stimulates protein kinase D-dependent histone deacetylase 5 phosphorylation and nuclear export leading to vascular smooth muscle cell hypertrophy. Arterioscler Thromb Vasc Biol. 2007;27:2355–2362. doi: 10.1161/ATVBAHA.107.151704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, et al. Mitochondrial localization of estrogen receptor beta. Proc Natl Acad Sci USA. 2004;101:4130–4135. doi: 10.1073/pnas.0306948101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yepuru M, Eswaraka J, Kearby JD, Barrett CM, Raghow S, Veverka KA, Miller DD, Dalton JT, Narayanan R. Estrogen receptor beta selective ligands alleviate high-fat diet and ovariectomy-induced obesity in mice. J Biol Chem. 2010;285:31292–31303. doi: 10.1074/jbc.M110.147850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CL, McKinsey TA, Chang S, Antos CL, Hill JA, Olson EN. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110:479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Malik S, Pang J, Wang H, Park KM, Yule DI, Blaxell BC, Smrcka AV. Phospholipase Cε hydrolyzes perinuclear phosphatidylinositol 4-phosphate to regulate cardiac hypertrophy. Cell. 2013;153:216–227. doi: 10.1016/j.cell.2013.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Fang Q, Starub SG, Lindau M, Sharp GW. Noradrenaline inhibits exocytosis via the G protein bg subunit and refilling of the readily releasable granule pool via the a(i1/2) subunit. J Physiol. 2010;288:3485–3498. doi: 10.1113/jphysiol.2010.190090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, He A, Pu WT. Regulation of GATA4 transcriptional activity in cardiovascular development and disease. Curr Top Dev Biol. 2012;100:143–169. doi: 10.1016/B978-0-12-387786-4.00005-1. [DOI] [PubMed] [Google Scholar]
- Zhu Y, et al. Abnormal vascular function and hypertension in mice deficient in estrogen receptor beta. Science. 2002;295:505–508. doi: 10.1126/science.1065250. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.