Abstract
Prefoldin (PFD) is a jellyfish-shaped molecular chaperone that has been proposed to play a general role in de novo protein folding in archaea and is known to assist the biogenesis of actins, tubulins, and potentially other proteins in eukaryotes. Using point mutants, chimeras, and intradomain swap variants, we show that the six coiledcoil tentacles of archaeal PFD act in concert to bind and stabilize nonnative proteins near the opening of the cavity they form. Importantly, the interaction between chaperone and substrate depends on the mostly buried interhelical hydrophobic residues of the coiled coils. We also show by electron microscopy that the tentacles can undergo an en bloc movement to accommodate an unfolded substrate. Our data reveal how archael PFD uses its unique architecture and intrinsic coiled-coil properties to interact with nonnative polypeptides.
Coiled coils consist of two or more parallel or antiparallel amphipathic α-helices that twist around one another to form supercoils (1). The primary sequences of the helices display a heptad repeat (abcdefg), where apolar residues are found preferentially in the first (a) and fourth (d) positions. Although the knobs-into-holes packing of the hydrophobic residues is the predominant stabilizing force for a coiled coil, inter- and intrahelical ionic interactions can act to further stabilize or destabilize its supersecondary structure (1). Coiled coils are found in several molecular chaperones, a diverse family of proteins whose collective cellular role is to ensure the quality control (e.g., folding, assembly, and transport) of nonnative proteins (2, 3). Archaeal prefoldin (PFD) is a chaperone that contains six canonical antiparallel coiled coils whose N- and C-terminal helices project outward from a double β-barrel oligomerization domain; the overall shape of the hexameric protein complex, assembled from two PFDα and four PFDβ subunits (α2β4), resembles a jellyfish with six tentacles (4). In solution, its tentacles are likely to be fully solvated and independently mobile (4). A lower-resolution electron microscope image of recombinant human PFD, which consists of six different proteins (two α class and four β class subunits), reveals that it possesses the same overall structure (5).
Like other chaperones, archaeal PFD can selectively interact with and stabilize nonnative (unfolded) polypeptides that expose hydrophobic surfaces in vitro, helping to prevent their aggregation (4, 6, 7). Preliminary studies have shown that deletion of the distal coiled-coil regions in either the α or β subunit abrogates chaperone activity in vitro, implying that PFD grasps its substrates in a multivalent manner (4). Similarly, the eukaryotic PFD–actin complex recently visualized by EM shows that non-native actin, one of its substrates, makes multiple contacts with the distal regions of the tentacles (5).
In the crowded cellular environment, eukaryotic PFD is likely to transiently stabilize ribosome-bound nascent polypeptides (8) before shuttling them to a chaperonin (an ATP-dependent cylindrical chaperone) for completion of folding (9). Functional cooperation between PFD and eukaryotic cytosolic Chaperonin Containing TCP-1 (CCT) likely arises through a transient ternary complex with substrate that accelerates folding and prevents aggregation (5, 9–11). The range of substrates bound by eukaryotic PFD overlaps at least in part with CCT, because it includes actins and tubulins (9–13).
Although the in vivo substrates of archaeal PFD are not known, its ability to stabilize a wide array of unfolded proteins in vitro (e.g., rhodanese, actin, lysozyme, firefly luciferase, and GFP) suggests that it performs a general role in recognizing and assisting the biogenesis (folding) of nonnative proteins in the cell (6, 7, 14). Archaeal PFD seems to function as an ATP-independent holdase for nonnative proteins before passing on the substrate to an ATP-dependent chaperonin, much like its eukaryotic counterpart (3, 4, 6, 7, 9). How PFD binds and stabilizes PFD nonnative proteins at the molecular level therefore represents a fundamental question that needs to be explored.
In this study, we characterize the function of archaeal PFD complexes using a panel of α and β subunit variants assembled in different combinations and use EM to visualize the interaction between the chaperone and a nonnative protein. Our data show that archaeal PFD utilizes, in a concerted manner, partially buried hydrophobic residues in the tips of flexible coiled coils to interact with and prevent the aggregation of its nonnative substrate.
Materials and Methods
Preparation of Constructs. PCR-based mutagenesis was used to create point mutant, intradomain swapped, and chimeric prefoldin constructs of both the α and β subunits. For mutations near the N and C termini, one pair of mutagenic primers was used to amplify the product. For mutations further from the termini, two or three sets of nested mutagenic primers were used in successive PCR reactions. PCR products were subcloned into pRSET6a at NdeI and BamHI sites, and the constructs were verified by DNA sequencing (see Fig. 5, which is published as supporting information on the PNAS web site, for the amino acid sequences of all constructs).
Protein Expression and Purification. Wild-type Pyrococcus horikoshii or wild-type and mutant Methanothermobacter thermoautotrophicus PFD subunits were produced in Escherichia coli strain BL21(DE3)pLysS, purified, and assembled into α2β4 complexes as described (4). Purified protein complexes (stored frozen in 25% glycerol) were dialyzed against buffer A (20 mM sodium phosphate, pH 8.0/100 mM NaCl) at 4°C and concentrated with Centriprep YM-10 or Ultra-15 centrifugal filter units (Millipore) before analysis. Protein concentrations were determined by quantitative amino acid analyses (Alberta Peptide Institute, Edmonton, AB) and Bradford protein assays (Bio-Rad). The observed molecular weights of recombinant PFD subunits, determined by matrix-assisted laser desorption ionization time-of-flight mass spectrometry, were as predicted; in the case MtβCePFD6, the initiating methionine was absent. Recombinant polyhistidine-tagged GFP (F99S/M153T/V163A) was expressed and purified as described (15).
Characterization of PFD Variants. All PFD complexes were characterized by analytical size-exclusion chromatography (SEC) and circular dichroism (CD). For SEC, samples were run on a Superdex S200HR PC 3.2/30 column (Amersham Pharmacia) equilibrated in buffer A. Far-UV CD spectra were recorded on a Jasco (Easton, MD) 710 spectropolarimeter by using 10 accumulations from 260 to 190 nm at room temperature. Protein samples were diluted to 0.4 mg/ml in buffer A. Thermal denaturation experiments were performed essentially as described (16). Stability of PFD variants was examined by monitoring the CD ellipticity at 222 nm as a function of temperature with a heating rate of 1.3°C/min in buffer A and a path length of 20 mm. Melting temperatures (Tm) reported in Table 1, which is published as supporting information on the PNAS web site, correspond to the dissociation of the hexamer containing the indicated PFD variant in a complex with the other wild-type subunit (16).
Prevention of Protein Aggregation Assays. In vitro chaperone activity of PFD variants was determined essentially as described (6). Briefly, hen egg-white lysozyme (Sigma) was dissolved in denaturing buffer (6 M guanidine·HCl/100 mM NaCl/20 mM sodium phosphate, pH 8.0/50 mM DTT) and then diluted 100× to a final concentration of 2 μM into buffer A alone or containing various concentrations of either wild-type or mutant PFD complexes. Aggregation of substrate was monitored spectrophotometrically at 360 nm for 10 min at 25°C. Raw absorbance data were normalized, and relative aggregation was defined as the fraction of the final absorbance value observed in the buffer A alone control. Conalbumin aggregation assays were performed as above, except the protein was diluted to a final concentration of 0.75 μM. Each sample was run at least in duplicate for a given experiment, and each experiment was repeated at least twice on separate occasions. The data presented are representative trials.
Formation and Analysis of PFD–GFP Complexes. Native GFP in 20 mM Tris·Cl/100 mM NaCl/1 mM DTT, pH 8.0 (buffer B) was acid-denatured by adding HCl to a final concentration of 12.5 mM. For SEC analysis, 116 μM denatured GFP was diluted into buffer A or buffer A containing 11 μM prefoldin to a final concentration of 11 μM and mixed rapidly. The mixture was incubated on ice for 1 hour, centrifuged for 5 min at 16,100 × g, and 50 μl was analyzed by SEC as described above. Peptide backbone absorbance was monitored at 222 nm and GFP excitation at 396 nm. Fractions analyzed by SDS/PAGE were PFD Peak A, 1.25- to 1.5-ml elution volume, and GFP Peak B, 1.60- to 1.85-ml elution volume. Before EM, PFD was dialyzed against buffer B and mixed with acid-denatured GFP as above without column purification.
EM. Aliquots of substrate-free PFD or PFD–GFP complexes were applied to carbon grids and negatively stained with 2% uranyl acetate. Images were taken under low-dose conditions at a ×60,000 nominal magnification in a JEOL 1200EX-II electron microscope operated at 100 kV and recorded on Kodak SO-163 film. For image processing, 1,265 substrate-free and 1,926 GFP-bound PFD particles displaying U-shaped side views were selected from independent samples. U-shaped views were selected among other views (i.e., W-shaped views) essentially as described (5). The presence or absence of GFP in the sample did not affect the distribution of the two PFD views. Particles were centered and aligned by using a free-pattern algorithm (17) and subsequently subjected to a neural network classification procedure (18). This procedure served to discriminate, when analyzing the putative PFD/GFP complexes, between particles containing a stain-excluding region between the tentacles (i.e., GFP-bound particles; 20% of the population) and those possessing a stain-penetrating region (i.e., the substrate-free particles; 80% of the population). The homogeneous populations from the two independent samples were subsequently processed and averaged.
Miscellaneous. All molecular graphics were generated by using pymol (DeLano Scientific, San Carlos, CA). Molar amounts refer to hexamers for PFD complexes (84 kDa) and monomers for lysozyme (14 kDa), conalbumin (75 kDa), and GFP (27 kDa). Multiple sequence alignments were performed by using clustalx (ftp://ftp-igbmc.u-strasbg.fr/pub/clustalx) software followed by manual editing. Similarity scores were assigned by using a Gonnet PAM250 similarity matrix.
Results and Discussion
Properties of PFD Coiled Coils. In an attempt to understand the attributes of the coiled coils that confer the ability of PFD to interact with and stabilize nonnative proteins, we evaluated the amino acid sequence conservation of the coiled coils in relation to the 3D structure of the chaperone. We constructed multiple sequence alignments of 11 PFD α and β subunits from different genera and assigned a score for the degree of conservation at each residue (Fig. 6, which is published as supporting information on the PNAS web site). We noted, based on the crystal structure, that the N-terminal helices of the archaeal α and β subunit coiled coils face into the rectangular cavity, ostensibly forming the binding surface for nonnative proteins; in contrast, C-terminal helices localize mainly to the outside surface of the cavity (Fig. 7, which is published as supporting information on the PNAS web site). It was thus surprising to find that the primary sequences of the N-terminal helices were not more conserved on the whole than those of the C-terminal helices if they indeed contained the substrate-binding site (Fig. 6). More importantly, when we mapped the amino acid conservation onto the structure of PFD, there were few highly conserved surface-exposed residues that were preferentially found inside the cavity (Fig. 7). A large proportion of the conserved residues are within the partially buried hydrophobic core (a/d residues) of the coiled coils. Together, these observations led us to hypothesize that the basis of action of PFD likely depends on the unique spatial arrangement, and intrinsic properties of the coiled coils rather than on the presence of conserved patches of substrate binding surface-exposed residues.
Cavity Surface Formed by the Coiled Coils. To test for the potential presence of an essential solvent-exposed binding site on the cavity surface of PFD, we designed mutants in which the C-terminal helix comprises the cavity surface and the N-terminal helix faces the external solvent. To this end, the amino acid sequences of the distal N- and C-terminal coiled-coil helices were switched relative to the wild-type subunits (Figs. 1A and 5). We chose a crossover point near the middle of the coiled coils, because removal of protein sequences beyond this point (i.e., the distal region) negates substrate binding by PFD (4). Therefore, the proximal-to-distal sequence of side chains in both of the switched N- and C-terminal helices is identical to that of the wild-type subunits. Although the backbone polarity within the swapped helical regions is reversed relative to the wild-type sequence, the overall structural properties of the switched coiled coils appear to be essentially unchanged (see below).
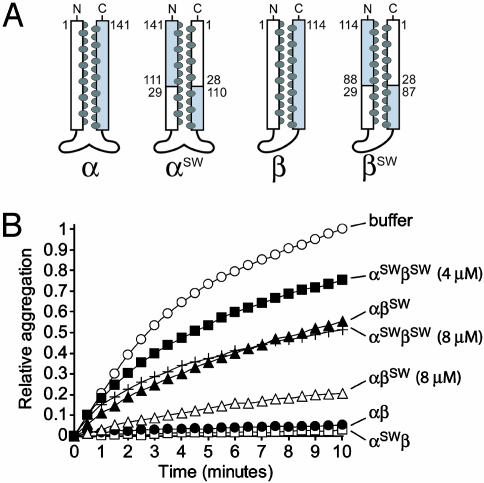
Fig. 1.
Chaperone activity of intradomain swap (switch) mutant complexes. (A) Schematic representations of α and β switch mutants. For the wild-type subunits, the N- and C-terminal helices are colored white and gray, respectively; the interhelical a/d residues are represented as dark ovals and correspond to those shown in the PFD subunit alignments (Fig. 6). For the switch mutants, the numbering scheme and colors used correspond to the wild-type sequences and show where the crossover points occur. (B) Effect of PFD switch mutants on the aggregation of denatured lysozyme. Relative aggregation of 2 μM lysozyme (monitored at 360 nm) during 10 min in buffer alone or in the presence of wild-type or prefoldin variants (as shown on the right of each curve). PFD complexes were at 2 μM unless otherwise indicated.
The recombinant α and β switch (SW) mutant subunits (αSW and βSW) were assembled with each other or with wild-type subunits (α or β). To assess the structural integrity of these complexes, we performed analytical SEC, far-UV CD, and measured thermal stability by CD, as described (4, 16). SEC indicated that the mutant and wild-type complexes had an identical Stoke's radius, implying the same overall shape and coiled-coil length (data not shown). The far-UV CD spectra revealed that the secondary structure content of the mutant complexes was indistinguishable from wild type, confirming there were no gross structural defects (data not shown). We also monitored thermal stability by CD at 222 nm and found that the complexes possess melting temperatures (Tm) comparable to their wild-type counterpart, which denatures at 61°C (Table 1). Therefore, the switch mutations alter the nature of the cavity surface without affecting the amino acid composition or significantly altering the structure or stability of the PFD hexamer.
Each PFD complex containing PFDα and -β variants was tested for chaperone activity in a standard prevention-of-aggregation assay by using lysozyme as a model substrate (4, 6). Importantly, the aggregation of lysozyme is not affected by the presence of irrelevant proteins, including aldolase and native lysozyme, even at elevated concentrations (data not shown); moreover, inactive PFD variants have no effect on the aggregation of denatured proteins (4). Last, the assay is performed at a temperature (25°C) well below the melting points of the αSW- and βSW-containing complexes (59°C and 53°C, respectively; Table 1).
A PFD complex assembled from α switch and wild-type β subunits (αSWβ) prevented the aggregation of denatured lysozyme as efficiently as wild-type PFD (Fig. 1B). The β switch mutant, when combined with wild-type α subunit (αβSW), displayed reduced yet significant activity at the same 1:1 molar ratio over denatured lysozyme and almost full activity at a 4:1 ratio over substrate (Fig. 1B). Finally, a complex containing both α and β switch mutants (αSWβSW), in which all six coiled coils are switched, still had detectable chaperone activity at a 2:1 molar ratio over substrate and significant activity at a 4:1 ratio. This concentration-dependent prevention of aggregation activity shows that the switch mutations do not abolish all PFD activity, although their activities are significantly reduced (Fig. 1B).
Because of the α2β4 stoichiometry of the PFD hexamer, the αSWβ, αβSW, and αSWβSW variants represent complexes in which increasing amounts of the cavity surface (i.e., two, four, or six coiled-coil tentacles, respectively) are affected, and this coincides with a gradual decrease in activity (Fig. 1B). The additional loss in activity of αSWβSW relative to αβSW appears to reveal a defect in the αSW variant and a contribution from both subunit types toward chaperone activity. It is therefore possible that some of the residues that normally face into the cavity do in fact contribute to substrate binding. These residues do not appear to be essential for interaction with a nonnative protein to occur, although their absence may reduce the substrate-binding affinity of the chaperone. These data represent a previously undescribed and interesting finding that is consistent with our observation that the interior N-terminal helix of PFD is not detectably more conserved than the outward-facing C-terminal helix (Fig. 6).
Intrinsic Properties of the Coiled-Coil Motif. Given both the relative paucity of conserved solvent-exposed residues in the putative substrate-binding site and the fact that the switch mutations retain partial chaperone activity, we hypothesized that PFD function may depend to a large degree on intrinsic properties of the coiled-coil motif. We therefore predicted that heterologous coiled-coil sequences should at least partially support the chaperone activity of the complex.
To test our hypothesis, we engineered PFD chimeras in which coiled-coil regions of the myosin II heavy chain, Rad50 zinc hook domain, and Caenorhabditis elegans PFD6 (one of the four eukaryotic β class PFD subunits) were fused to shortened coiled coils of the PFDβ subunit (βMyosin, βRad50, and βCePFD6), and a similar region of Rad50 was fused to the PFDα subunit (αRad50) (Fig. 2A). The heterologous sequences have very low sequence identity (9–11%) and similarity (35–36%) to the corresponding archaeal sequences and were designed to form a coiled coil with the same number and register of heptad repeats as that of wild-type PFD. As with the switch mutant complexes, the chimeric complexes were found to be indistinguishable from wild-type PFD hexamers by SEC and far-UV CD, eliminating the possibility of major structural perturbations. Thermal denaturation of the chimeras revealed similar stabilities to wild-type PFD, with the exception of the αβCePFD6 chimera, which melted at a lower temperature (Tm = 43°C). This behavior is consistent with the low optimal growth temperature of C. elegans (15–20°C) but is still significantly above the assay temperature used (Table 1).
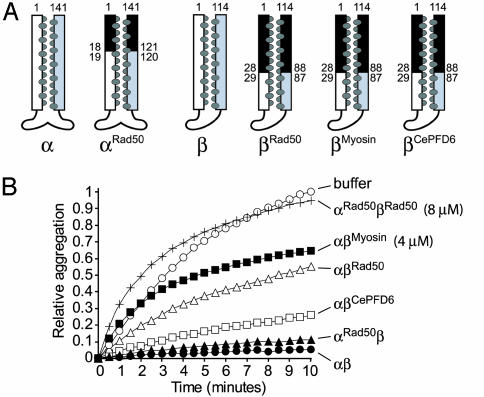
Fig. 2.
Chaperone activity of chimeric complexes. (A) Schematic representations of chimeric PFD subunits. The exogenous coiled-coil region is colored black, and the numbers adjacent to the helical regions refer to the amino acid position at which the fusion has taken place. (B) Effect of chimeric PFD mutants on the aggregation of 2 μM denatured lysozyme. PFD complexes (as shown on the right of each curve) were at 2 μM unless otherwise indicated.
When tested for chaperone activity, the αRad50β chimeric complex displayed near-wild-type activity (Fig. 2B). PFD complexes of the wild-type α subunit with chimeric β subunits (αβCePFD6, αβMyosin, and αβRad50) were also able to bind and stabilize denatured lysozyme and denatured conalbumin (Fig. 2B and data not shown, respectively), although these variants had a range of intermediate chaperone activities relative to wild-type. As with the PFD switch mutants, that a PFD complex in which only the α subunit is chimeric has more activity than β subunit chimeras (i.e., αRad50β vs. αβRad50; Fig. 2B) is consistent with the presence of four potential binding sites for the β subunits and only two for the α subunits. Therefore, even though both subunit types may be similarly compromised, the defect is expected to be more apparent in the β chimeras. Importantly, the activities of the chimeras are greater than that observed when the corresponding subunit is truncated, even though the truncations remove less of the coiled coil than is replaced in the chimeras (4). This partial rescue of activity shows that the chimeric coiled-coil regions, which are devoid of archaeal PFD sequence, can contribute to the chaperone activity of the PFD complex.
Remarkably, the αRad50 and βRad50 chimeric subunits, which are partially active in a wild-type background, had no measurable chaperone activity when assembled together (Fig. 2B). Thus, either wild-type subunit confers a significant level of activity in the context of a mutation in the other subunit, whereas the same mutations in all six subunits seem to eliminate function. This cooperation between α and β subunits is an important concept for PFD function and reflects its ability to bind substrates multivalently (4, 19). Altogether, these results are consistent with the notion that prefoldin coiled coils have evolved specific features particular to their chaperone function, which do not occur to the same extent in any heterologous coiled-coil sequence. In this respect, it is notable that αβCePFD6, which contains a eukaryotic β class subunit, was more efficient at stabilizing denatured lysozyme than the other chimeras (αβMyosin or αβRad50) (Fig. 2B), despite the lack of conserved sequence (Fig. 5) or amino acid composition (data not shown). This suggests that certain unique properties of the coiled coils are likely shared by eukaryotic and archaeal PFD subunits. Indeed, archaeal PFDβ can partially complement the cytoskeletal phenotype caused by the lack of the Saccharomyces cerevisiae β class (gim1 or gim4) genes (6).
The loss of chaperone activity in the chimeric complexes could in principle be accounted for by the absence of specific solvent-exposed residues that normally contribute to substrate binding in wild-type PFD. Alternatively, or in addition, the chimeras could be deficient in subtle properties that affect the accessibility of their interhelical hydrophobic residues. The second possibility led us to hypothesize that the common a/d hydrophobic residues may, in large part, form the intrinsic property of coiled coils involved in substrate recognition and binding by PFD.
Hydrophobic Interface of the Coiled Coils. Archaeal PFD, like some well-characterized molecular chaperones that bind nonnative proteins promiscuously (for example, Hsp70 and the bacterial chaperonin GroEL; refs. 2, 20) likely recognizes its substrates mainly because they expose normally buried hydrophobic surfaces (6). Unlike other chaperones, however, the surface displayed within the cavity of PFD, where substrate binding is expected to occur, is almost entirely devoid of distinctly solvent-exposed hydrophobic residues (4). The apolar interhelical interface of the coiled coils may therefore be directly responsible for interactions with substrates.
To test this hypothesis, we engineered serine-substituted PFD variants at one to four pairs of the predominantly hydrophobic a/d heptad repeat residues in the distal ends of the α and β subunit coiled coils. Serine was chosen because of its relatively small size, polar character, and tolerance for inclusion into α-helices (21, 22). The intended effect of the sequential substitutions is to gradually remove the potential hydrophobic-binding site while retaining intact helices and the overall rod-like structure of the tentacle. PFD α subunits mutated in up to three a/d pairs (αHM1, αHM2, and αHM3), and β subunits mutated in up to four a/d pairs (βHM1, βHM2, βHM3, and βHM4) were constructed (Fig. 3A; HM, hydrophobic mutant). The overall structures and stabilities of these mutants were essentially identical to wild-type PFD, as judged by SEC, far-UV CD, and thermal denaturation studies (data not shown and Table 1).
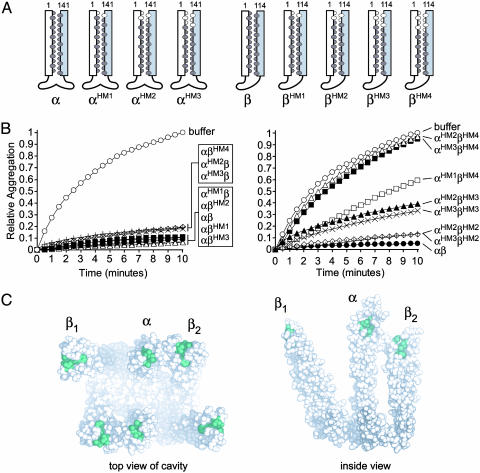
Fig. 3.
Hydrophobic a/d coiled-coil residues are required for chaperone activity. (A) Schematic representations of the a/d residue point mutants. Hydrophobic residues between the coiled coils are dark ovals; residues mutated to serine are colored white. (B) Effect of hydrophobic a/d point mutants on the aggregation of 2 μM denatured lysozyme. Each PFD complex, shown on the right, was tested at 2 μM. (C) Mutated hydrophobic residues shown on the PFD crystal structure. PFD is shown looking into the cavity (Left), and from inside, viewing the cavity surface (Right). Residues that were mutated are colored green. Not shown is one pair of hydrophobic residues (L3 and A113) in theβ subunit termini and one amino acid (D3) in the N terminus of the α subunit, which were not resolved in the crystal structure but may also form part of the coiled coil (4, 6).
PFD complexes containing a wild-type subunit and any hydrophobic point mutant were found to have near-wild-type activities at a 1:1 ratio of chaperone hexamer to denatured lysozyme (Fig. 3B Left). To test for functional cooperation between the α and β subunits, we assayed the activity of a complex containing three substituted pairs in the α subunit and four substitutions in the β subunit (αHM3βHM4). Remarkably, this PFD variant was unable to prevent the aggregation of denatured lysozyme (Fig. 3B Right) or conalbumin (Fig. 4D). We verified the structural integrity of this inactive complex as described above and also examined it by EM (data not shown). The latter analysis showed that the ultrastructural features of the mutant were indistinguishable from wild-type archaeal (e.g., see Fig. 4B) and eukaryotic PFD (5).
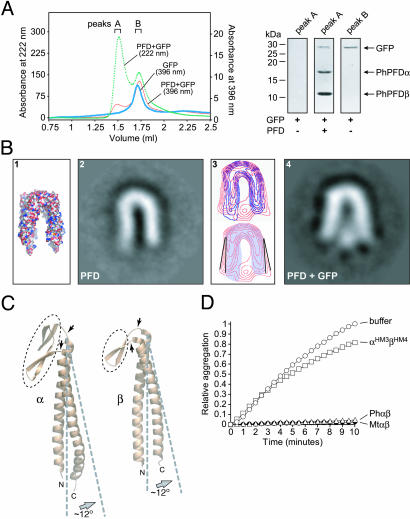
Fig. 4.
Substrate binding occurs near the ends of flexible coiled coils. (A) Coelution of PFD and GFP on a superdex 200 size-exclusion column (Left). The green dashed line is PFD (PhPFD) plus denatured GFP monitored at 222 nm, the thick blue line is GFP alone monitored at its excitation maximum (396 nm), and the red line is PFD plus denatured GFP monitored at 396 nm. Coelution was also demonstrated by SDS/PAGE analysis of peak A (1.25–1.50 ml) with and without PhPFD and of peak B without PhPFD (1.65–1.80 ml) (Right). (B) Interaction of unfolded GFP with PFD. (1) Molecular surface of PFD crystal structure; negatively stained and averaged EM images of substrate-free prefoldin (2), and GFP-bound PFD (4). (3 Upper) Merged contour maps (blue, PFD alone; red, PFD + GFP). (Lower) The approximate tilt angle change (≈12° opening) of the substrate-bound PFD subunits (red) relative to that of PFD alone (the contour map is shaded blue). (C) The putative hinge domains connecting PFD coiled coils and theβ-barrel domain are shown with arrows. Gray dashed lines indicate the (approximate) 12° opening motion, and the β-barrel oligomerization domain regions are circled with narrow black dashed lines. (D) Effect of wild-type (Ph and Mt) and MtαHM3βHM4 PFD complexes on the aggregation of denatured conalbumin (75 kDa). Aggregation assays were performed as for denatured lysozyme except conalbumin was 0.75 μM and PFD or its variants (Right) were added at a 5:1 ratio over substrate (3.75 μM).
Compared to the inactive αHM3βHM4 complex, the presence of two additional pairs of a/d residues in the α subunit (αHM1βHM4) partially restored the activity of the complex (Fig. 3B Right). As might be expected, complexes assembled from less severely mutated variants, lacking 3 a/d pairs in PFDβ, and two or three pairs in PFDα (αHM2βHM3 and αHM3βHM3) showed intermediate activities compared to wild-type PFD (Fig. 3B Right). PFD complexes with the same PFDα backgrounds as above but lacking only two a/d pairs in PFDβ (αHM2βHM2 and αHM3βHM2) showed nearly full activity (Fig. 3B Right).
Altogether, these results demonstrate three important properties of the coiled coils in archaeal PFD. First, the partially buried hydrophobic interface between the amphipathic helices is required for effective interaction and stabilization of a nonnative substrate by the chaperone. We also observed this effect in the partially active βRad50 chimera; replacing the first four pairs of a/d residues with serine (βRad50HM4) impaired its ability to function in the complex (data not shown). It is notable that the residues comprising the binding site are only partially exposed and are at the apex of the coiled-coil tentacles (Fig. 3C), where increased flexibility (indicated by higher B factors in the tip regions; see Protein Data Bank ID 1FXK) may facilitate the exposure of interhelical apolar residues. Indeed, such increased exposure may result from the partial unwinding of the coiled coils, as suggested by Siegert et al. (4). Second, the binding site appears to be diffuse because there is progressive loss of function as more apolar residues are substituted with serine; the binding site could therefore extend somewhat beyond the residues mutated in both the α and β subunits. Last, the coiled coils act in a concerted or multivalent fashion to stabilize a nonnative protein, because alterations in either subunit alone have much less profound effects on chaperone activity than mutations in both subunits.
PFD Functions as a Molecular Clamp. Our mutational analyses of PFD show that hydrophobic residues at the core of each coiled coil comprise the major binding surface. To analyze the manner in which PFD interacts with a substrate, we generated a stable PFD–substrate complex and visualized it by EM.
By mixing acid-denatured GFP (27 kDa) with PFD from P. horikoshii (PhPFD), we could observe a complex by SEC (Fig. 4A). Because PhPFD binds GFP with a somewhat higher affinity than M. thermoautotrophicus PFD, we used PhPFD for subsequent EM studies. EM images were obtained by negative staining of two independent samples, namely substrate-free PhPFD and PhPFD–GFP complexes. For each sample, U-shaped views were selected among other views (i.e., W-shaped views) as described (5) (Fig. 4B 2 and 4). This view permitted the unambiguous identification, processing, and averaging of substrate-free PFD and, after classification of the PFD/GFP complexes according to the absence or presence of stain in the intertentacle area, of the GFP-bound prefoldin (20% of the total population).
The processed EM image of substrate-free PFD (Fig. 4B 2) appears identical in overall structure and geometry to the molecular surface of the PFD crystal structure (Fig. 4B 1). When comparing substrate-free PFD to substrate-bound PFD, a stain-excluding region representing the bound GFP molecule at the distal tips of the coiled coils is immediately apparent (Fig. 4B 4). The location of the GFP confirms the distal coiled-coil regions as the substrate-binding site and explains the sensitivity of the chaperone to mutagenesis in this region.
In addition, the EM images show that the PFD tentacles have flexed outward to accommodate the nonnative GFP (Fig. 4B 3 and 4). An overlay contour map shows that compared to the unbound state, the observed expansion of the cavity corresponds to an outward motion of ≈12° for the tentacles (Fig. 4B 3). This conformational change appears to represent an en bloc movement of the coiled coil. We suggest that a hinge region, consisting of the loops connecting the coiled coils to the β-barrel oligomerization domain, could be responsible for the observed flexibility and outward movement of the α and β supercoils (Fig. 4C). Interestingly, the recent EM image reconstruction of actin within the cavity of eukaryotic PFD showed no apparent movement of the coiled coils to accommodate this larger (45-kDa) nonnative protein (5). In this reconstruction, actin appears to be in a nonglobular conformation that spans the entire opening of the cavity. Together, these observations suggest that the PFD tentacles can move independently to create the cavity shape needed for efficient interaction with substrates of different conformations and/or sizes.
Archaeal PFD is known to interact with proteins as small as 14 kDa (lysozyme) and as large as 62 kDa (firefly luciferase) (4, 6). Fig. 4D shows that both PhPFD and MtPFD are able to completely prevent the aggregation of the 75-kDa protein conalbumin at a 5-fold molar excess over the substrate. By comparison, MtPFD lacking its distal a/d residues is essentially inactive at the same concentration (Fig. 4D). This finding extends the upper size limit of proteins known to interact with PFD. Furthermore, the EM results, which show that PFD tentacles are flexible, may explain how the chaperone can interact with proteins of such diverse sizes.
The wide range of substrate sizes bound by archaeal PFD is consistent with an in vivo role in binding and stabilizing a large repertoire of nascent proteins. Indeed, during synthesis, many polypeptides must be stabilized cotranslationally before spontaneous, or chaperone-assisted, folding (2). In bacteria, the chaperones DnaK (an Hsp70 homolog) and trigger factor cooperate to perform this general stabilizing function (23, 24). It has been suggested that prefoldin could functionally replace these chaperones in archaea, where trigger factor, and often Hsp70, are conspicuously absent (6, 14). If this is the case, it is not surprising that PFD displays a general ability to recognize nonnative proteins, and that those proteins can vary greatly in size and shape, as they would in vivo.
Conclusion
In the present study of the archaeal molecular chaperone, prefoldin, we uncovered a singular ability of its coiled coils to interact with and stabilize nonnative proteins. The substrate-binding mechanism of PFD appears to depend on at least three distinct properties of the coiled coils.
A flexible molecular clamp-like en bloc motion, presumably as a means to grip substrates of varying shapes and/or sizes.
Interhelical hydrophobic residues at the distal tips that are likely to directly contact exposed apolar patches in nonnative substrates.
A concerted action of multiple weak binding sites, where the four outer β subunits appear to contribute more to binding than the two central α subunits.
In contrast to archaeal PFD, it has been suggested that eukaryotic PFD may interact specifically with a limited number of substrates given that it has evolved six divergent and potentially specialized subunits (6, 8–11); however, it is conceivable that some of its six subunits rely on the same properties as PFD to bind exposed hydrophobic patches on its substrates. Indeed, the C. elegans PFD6 coiled coil complemented the function of the chimeric archaeal chaperone more efficiently than other exogenous coiled coils. Moreover, because of their comparable quaternary structures (4, 5), it is possible that eukaryotic PFD could alter its cavity shape to accommodate different substrates, in the same manner as the archaeal chaperone. Thus, eukaryotic PFD may recognize a broader range of substrates than is currently known (12). Evidence for this notion stems from the finding of additional putative substrates of the eukaryotic chaperone, including the tumor suppressor protein VHL (25), the DNA mismatch repair protein MutH4 (26), and the c-Myc transcription factor (27).
Coiled coils are highly abundant in the proteomes of all organisms, accounting for an estimated 2–3% of all protein residues (28). Therefore, in addition to shedding light into the chaperone function of archaeal, and potentially eukaryotic, PFD, our findings are of particular significance because all coiled coils share a nonpolar core. The binding property observed for this region in PFD could extend to other coiled-coil-containing proteins, including molecular chaperones or those that interact with any molecule exposing an apolar surface. Cofactor A, for example, is a tubulin-specific three-stranded coiled-coil chaperone that appears to stabilize quasinative β-tubulin on its mostly hydrophilic surface (29). The contribution of the interhelical apolar residues may play an unrecognized yet important role in substrate binding. The five-stranded coiled-coil protein, COMPcc, is an interesting case of a nonchaperone protein that binds vitamin D within the network of apolar a/d residues (30). Although this latter interaction is highly specific, other coiled coils could conceivably bind a range of hydrophobic molecules using their interhelical hydrophobic residues. In conclusion, our findings provide significant insight into the mechanism of the molecular chaperone function of PFD and extend the known functions of coiled coils to include molecular recognition via their common hydrophobic interface.
Supplementary Material
Acknowledgments
We thank Dr. H. Taguchi (Tokyo Institute of Technology, Tokyo) for the gift of a GFP plasmid. Funds for this research were provided by Canadian Institutes of Health Research (CHR) and Natural Sciences and Engineering Research Council (NSERC) (M.R.L.) and Ministerio di Ciencia y Tecnología (J.M.V.). M.R.L. is the recipient of Michael Smith Foundation for Health Research and CIHR scholar awards, and P.C.S. holds an NSERC scholarship.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: EM, electron microscopy; SEC, size-exclusion chromatography; CD, circular dichroism; PFD, prefoldin.
References
- 1.Lupas, A. (1996) Trends Biochem. Sci. 21, 375–382. [PubMed] [Google Scholar]
- 2.Hartl, F. U. & Hayer-Hartl, M. (2002) Science 295, 1852–1858. [DOI] [PubMed] [Google Scholar]
- 3.Stirling, P. C., Lundin, V. F. & Leroux, M. R. (2003) EMBO Rep. 4, 565–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegert, R., Leroux, M. R., Scheufler, C., Hartl, F. U. & Moarefi, I. (2000) Cell 103, 621–632. [DOI] [PubMed] [Google Scholar]
- 5.Martin-Benito, J., Boskovic, J., Gomez-Puertas, P., Carrascosa, J. L., Simons, C. T., Lewis, S. A., Bartolini, F., Cowan, N. J. & Valpuesta, J. M. (2002) EMBO J. 21, 6377–6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leroux, M. R., Fändrich, M., Klunker, D., Siegers, K., Lupas, A. N., Brown, J. R., Schiebel, E., Dobson, C. M. & Hartl, F. U. (1999) EMBO J. 18, 6730–6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okochi, M., Yoshida, T., Maruyama, T., Kawarabayasi, Y., Kikuchi, H. & Yohda, M. (2002) Biochem. Biophys. Res. Commun. 291, 769–774. [DOI] [PubMed] [Google Scholar]
- 8.Hansen, W. J., Cowan, N. J. & Welch, W. J. (1999) J. Cell Biol. 145, 265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vainberg, I. E., Lewis, S. A., Rommelaere, H., Ampe, C., Vandekerckhove, J., Klein, H. L. & Cowan, N. J. (1998) Cell 93, 863–873. [DOI] [PubMed] [Google Scholar]
- 10.Geissler, S., Siegers, K. & Schiebel, E. (1998) EMBO J. 17, 952–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siegers, K., Waldmann, T., Leroux, M. R., Grein, K., Shevchenko, A., Schiebel, E. & Hartl, F. U. (1999) EMBO J. 18, 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leroux, M. R. & Hartl, F. U. (2000) Curr. Biol. 10, R260–R264. [DOI] [PubMed] [Google Scholar]
- 13.Rommelaere, H., De Neve, M., Neirynck, K., Peelaers, D., Waterschoot, D., Goethals, M., Fraeyman, N., Vandekerckhove, J. & Ampe, C. (2001) J. Biol. Chem. 276, 41023–41028. [DOI] [PubMed] [Google Scholar]
- 14.Leroux, M. R. (2000) Adv. Appl. Microbiol. 50, 219–277. [DOI] [PubMed] [Google Scholar]
- 15.Sakikawa, C., Taguchi, H., Makino, Y. & Yoshida, M. (1999) J. Biol. Chem. 274, 21251–21256. [DOI] [PubMed] [Google Scholar]
- 16.Fändrich, M., Tito, M. A., Leroux, M. R., Rostom, A. A., Hartl, F. U., Dobson, C. M. & Robinson, C. V. (2000) Proc. Natl. Acad. Sci. USA 97, 14151–14155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Penczek, P., Radermacher, M. & Frank, J. (1992) Ultramicroscopy 40, 33–53. [PubMed] [Google Scholar]
- 18.Marabini, R. & Carazo, J. M. (1994) Biophys. J. 66, 1804–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simons, C. T., Staes, A., Rommelaere, H., Ampe, C., Lewis, S. A. & Cowan, N. J. (2004) J. Biol. Chem. 279, 4196–4203. [DOI] [PubMed] [Google Scholar]
- 20.Bukau, B. & Horwich, A. L. (1998) Cell 92, 351–366. [DOI] [PubMed] [Google Scholar]
- 21.Lawrence, J. R. & Johnson, W. C. (2002) Biophys. Chem. 101–102, 375–385. [DOI] [PubMed] [Google Scholar]
- 22.Myszka, D. G. & Chaiken, I. M. (1994) Biochemistry 33, 2363–2372. [DOI] [PubMed] [Google Scholar]
- 23.Teter, S. A., Houry, W. A., Ang, D., Tradler, T., Rockabrand, D., Fischer, G., Blum, P., Georgopoulos, C. & Hartl, F. U. (1999) Cell 97, 755–765. [DOI] [PubMed] [Google Scholar]
- 24.Deuerling, E., Schulze-Specking, A., Tomoyasu, T., Mogk, A. & Bukau, B. (1999) Nature 400, 693–696. [DOI] [PubMed] [Google Scholar]
- 25.Tsuchiya, H., Iseda, T. & Hino, O. (1996) Cancer Res. 56, 2881–2885. [PubMed] [Google Scholar]
- 26.Her, C., Wu, X., Griswold, M. D. & Zhou, F. (2003) Cancer Res. 15, 865–872. [PubMed] [Google Scholar]
- 27.Mori, K., Maeda, Y., Kitaura, H., Taira, T., Iguchi-Ariga, S. M. & Ariga, H. (1998) J. Biol. Chem. 273, 29794–29800. [DOI] [PubMed] [Google Scholar]
- 28.Wolf, E., Kim, P. S. & Berger, B. (1997) Protein Sci. 6, 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinbacher, S. (1999) Nat. Struct. Biol. 6, 1029–1032. [DOI] [PubMed] [Google Scholar]
- 30.Ozbek, S., Engel, J. & Stetefeld, J. (2002) EMBO J. 21, 5960–5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.