Abstract
Prior genome-wide association studies (GWAS) of major depressive disorder (MDD) have met with limited success. We sought to increase statistical power to detect disease loci by conducting a GWAS mega-analysis for MDD. In the MDD discovery phase, we analyzed more than 1.2 million autosomal and X chromosome single-nucleotide polymorphisms (SNPs) in 18 759 independent and unrelated subjects of recent European ancestry (9240 MDD cases and 9519 controls). In the MDD replication phase, we evaluated 554 SNPs in independent samples (6783 MDD cases and 50 695 controls). We also conducted a cross-disorder meta-analysis using 819 autosomal SNPs with P< 0.0001 for either MDD or the Psychiatric GWAS Consortium bipolar disorder (BIP) mega-analysis (9238 MDD cases/8039 controls and 6998 BIP cases/7775 controls). No SNPs achieved genome-wide significance in the MDD discovery phase, the MDD replication phase or in pre-planned secondary analyses (by sex, recurrent MDD, recurrent early-onset MDD, age of onset, pre-pubertal onset MDD or typical-like MDD from a latent class analyses of the MDD criteria). In the MDD-bipolar cross-disorder analysis, 15 SNPs exceeded genome-wide significance (P<5×10−8), and all were in a 248 kb interval of high LD on 3p21.1 (chr3:52 425 083–53 822 102, minimum P= 5.9×10−9 at rs2535629). Although this is the largest genome-wide analysis of MDD yet conducted, its high prevalence means that the sample is still underpowered to detect genetic effects typical for complex traits. Therefore, we were unable to identify robust and replicable findings. We discuss what this means for genetic research for MDD. The 3p21.1 MDD-BIP finding should be interpreted with caution as the most significant SNP did not replicate in MDD samples, and genotyping in independent samples will be needed to resolve its status.
Keywords: genetics, genome-wide association study, major depressive disorder, mega-analysis, meta-analysis
Introduction
Major depressive disorder (MDD) is a genetically complex trait. The lifetime prevalence of MDD is ~15%.1,2 As a recurrent course is most common,3 MDD is accompanied by considerable morbidity4–6 excess mortality5,7 and substantial costs.8 The World Health Organization projects MDD to be the second leading cause of disability by 2020.9
The heritability of MDD is 31–42%,10 although certain subsets of MDD may be more heritable (for example, recurrent, early-onset MDD or clinically ascertained MDD).11,12 The modest heritability of MDD could reasonably be expected to complicate attempts to identify genetic loci that confer risk or protection. However, heritability is not necessarily a key determinant for the identification of strong and replicable genetic associations.13 For example, there have been notable successes in genome-wide searches14 for susceptibility loci for breast cancer (heritability ~25%), lung cancer (26%), Type 2 diabetes mellitus (26%), Parkinson’s disease (34%), multiple sclerosis (41%), systemic lupus erythematosus (44%) and age-related macular degeneration (46%).15–20
The most important determinant of success in identifying associations for complex traits is the underlying genetic architecture (that is, the number of loci and their frequencies, effect sizes, modes of action and interactions with other genetic loci and environmental factors). Heritability alone reveals little about genetic architecture. In the absence of a detailed understanding of genetic architecture, sample size and phenotypic homogeneity are the critical determinants of discovering robust and replicable genetic associations. Eight genome-wide association studies (GWAS) for MDD have been published,21–28 with one locus of possible genome-wide significance. 26 When these studies were planned, there were few data to guide sample size requirements. Several had historically notable sample sizes and far more comprehensive genomic coverage than any prior study. However, it has become clear that the effects of common genetic variants for most complex human diseases are considerably smaller than many had anticipated.14 This implies that sample sizes necessary for identification of common genetic main effects were far larger than could be attained by single-research groups or existing consortia.
Meta-analysis has thus become essential in human complex trait genetics. There are now many examples where meta-analyses combining dozens of primary data sets have illuminated the genetic architecture of complex traits such as height,29 body mass,30 Crohn’s disease31 and Type 2 diabetes mellitus.32 Following this proven model, we created the Psychiatric GWAS Consortium (PGC)33,34 to conduct field-wide combined analyses for MDD as well as ADHD,35 bipolar disorder (BIP),36 schizophrenia37 and autism. Our goal was to evaluate the evidence for common genetic variation in the etiology of MDD using the largest and most comprehensively genotyped sample hitherto collected.
Materials and methods
Overview
In the discovery phase, we conducted mega-analysis for MDD using nine primary samples. All groups uploaded individual genotype and phenotype data to a central computer cluster, and the PGC Statistical Analysis Group conducted uniform quality control, imputation and association analyses. Mega-analysis and meta-analysis yield essentially identical results in theory38 and in practice.37 However, mega-analysis of individual phenotype and genotype data was used to allow more consistent quality control and analysis, disentangle the issue of control subjects used by multiple studies, allow conditional analyses and to enable efficient secondary analyses. In the replication phase, we evaluated the top loci in seven independent MDD samples and in the PGC BIP megaanalysis36 given the phenotypic and genetic overlap between MDD and BIP.39,40 Finally, we conducted exploratory analyses of MDD sub-phenotypes in an attempt to index clinical heterogeneity. Most of the primary genotype data and the results have been deposited in the NIMH Human Genetics Initiative Repository (Supplementary Methods).
Samples
Full sample details are given in the Supplementary Methods. For the discovery phase, we included all identified primary MDD samples21–25,27,28,41 that conducted genome-wide genotyping (> 200K single-nucleotide polymorphisms (SNPs)) on individual subjects of European ancestry. Cases were required to have diagnoses of DSM-IV lifetime MDD established using structured diagnostic instruments from direct interviews by trained interviewers (two studies required recurrent MDD and one recurrent, early-onset MDD) or clinician-administered DSM-IV checklists. Most studies ascertained cases from clinical sources, and most controls were randomly selected from the population and screened for lifetime history of MDD. The sample sizes reported here differ from the primary reports due to different quality control procedures and apportioning of overlapping controls. We determined the relatedness of all pairs of individuals using genotypes of SNPs present on all platforms, and excluded one of each duplicate or closely related pair. The discovery mega-analysis consists of 18 759 independent and unrelated subjects of recent European ancestry (9240 MDD cases and 9519 controls).
There were two sets of analyses conducted on additional samples. For MDD replication, we used meta-analysis to combine the autosomal discovery results (554 SNPs with P< 0.001) with summary association results from independent samples42–48 (6783 MDD cases and 50 695 controls). The discovery SNP results were grouped into regions defined by linkage disequilibrium using an iterative process after ranking all SNPs by association P-value: for SNPs with r2 > 0.2 in a 1Mb window (based on HapMap3 CEU+TSI), the most strongly associated SNP was retained. In addition, given the close genetic and phenotypic relationships between MDD and BIP, we combined the MDD discovery sample and the PGC BIP mega-analysis36 to evaluate 819 autosomal SNPs with P < 0.0001 in either of the separate analyses. (See Sklar et al.36 for complete description). In effect, we tested for associations with a more broadly defined mood disorder phenotype. After resolving overlapping control samples, there were 32 050 independent subjects (9238 MDD cases/8039 controls and 6998 BIP cases/7775 controls).
SNP genotyping
SNP genotyping is described in the Supplementary Methods and summarized in Supplementary Table S2. Briefly, all samples were genotyped with SNP arrays intending to provide genome-wide coverage of common variation. Imputation was performed within each study in batches of 300 individuals. Batches were randomly assigned to keep the same case–control ratios as in the primary studies. We used Beagle 3.0.4 [ref. 49] with the CEU+TSI HapMap3 data as reference (410 phased haplotypes)50 to impute 1 235 109 autosomal SNP allele dosages. We had previously evaluated this approach by masking and then imputing genotyped loci and found a high correlation between the genotyped and imputed allele dosages (Pearson r > 0.999).37
Quality control
Genotyping coordinates are given in NCBI Build 36/UCSC hg18. For the discovery phase, quality control was conducted separately for each resolved sample. SNPs were removed for missingness ≥0.02, case–control difference in SNP missingness ≥0.02, SNP frequency difference from HapMap3 [ref. 50] ≥0.15, or exact Hardy–Weinberg equilibrium test in controls <1×10−6. Subjects were removed for excessive missingness (≥0.02), identical or closely related to any subject in any sample (π̂> 0.2 based on common autosomal SNPs) and if there was evidence for diverging ancestry. Ancestry was estimated using multidimensional scaling applied to 8549 SNPs directly genotyped in all samples and in approximate linkage equilibrium.
Statistical analysis
We used logistic regression to test the association of MDD diagnosis with imputed SNP dosages under an additive model. This test has correct type 1 error with imputed data.51 Covariates included study indicators and five principal components reflecting ancestry. For the MDD replication samples, the top SNP in each region was tested for association, and fixed-effect meta-analysis was used for the replication samples, and for the combination of PGC discovery and replication data.
Chromosome X
Female sex is an established risk factor for MDD, and analysis of chromosome X is particularly salient (although not included in many GWAS). Imputation using HapMap3 reference genotypes (as in the primary analysis) was not possible due to persisting difficulties with the phased chromosome X data, but we were able to impute using 1000 Genomes Project data.52 Chromosome X imputation was conducted for subjects passing QC for the autosomal analysis and with SNP call rates > 0.95 for chrX SNPs. SNPs with missingness ≥0.05 or HWE P<10−6 (females) were excluded. Phasing was conducted using MACH53 in female subjects. Imputation was performed separately for males and females using MINIMAC with haplotypes from 381 European samples from the 1000 Genomes Project as reference (1.45 million chrX SNPs, but many were monomorphic in our sample). Chromosome X SNPs in HapMap2 and HapMap3 with r2≥0.3 were carried forward for further analysis (122 602 SNPs). Association was tested under an additive logistic regression model implemented in PLINK (meta-analysis of male and female association results) using the same covariates as for the autosomal analysis.
Secondary analyses
MDD is suspected to have important phenotypic heterogeneity, and association analyses might yield clearer findings if clinical features are incorporated into genetic analyses. Thus, we conducted predefined secondary analyses intended to index plausible sources of phenotypic heterogeneity in MDD cases. (a) Sex. As the lifetime prevalence of MDD is approximately two times greater in females,54,55 we conducted association analyses separately in males and females to evaluate sex-specific genetic risk variants. (b) Recurrence and age of onset. As recurrence and age of onset may index heterogeneity in MDD,10,56 we analyzed early-onset MDD (≤30 years), recurrent MDD (≥2 episodes), pre-pubertal onset MDD (≤12 years, see Weissman et al.57) and age of onset of MDD as a quantitative trait. (c) Symptoms. As MDD is phenotypically heterogeneous, we obtained MDD symptom data from 88% of all MDD cases (the nine DSM-IV ‘A’ criteria disaggregated to code increase and decrease in appetite, weight, sleep and energy level). Latent class cluster models were fit to binary responses for these MDD ‘A’ criteria, and identified three latent classes in MDD cases characterized by weight loss/insomnia, weight gain/insomnia and hypersomnia (see Supplementary Methods for more details). The predominant latent class was consistent with ‘typical’ MDD58,59 and we analyzed cases indexed by this class.
Results
In the discovery stage, we conducted a GWAS megaanalysis for MDD in 18 759 independent and unrelated subjects of recent European ancestry (9240 MDD cases and 9519 controls, Table 1). There were considerable similarities across samples: all subjects were of European ancestry, all cases were assessed with validated methods and met DSM-IV criteria for lifetime MDD, and most controls were ascertained from community samples and screened to remove individuals with lifetime MDD (Supplementary Methods and Supplementary Figures S6–S9).
Table 1.
Cases and controls used in discovery and replication phases
| Phase | Sample | Subjects | MDD case | Control |
|---|---|---|---|---|
| Discovery | GAIN | 3461 | 1696 | 1765 |
| GenRED | 2283 | 1030 | 1253 | |
| GSK | 1751 | 887 | 864 | |
| MDD2000-QIMR_610 | 1184 | 433 | 751 | |
| MDD2000-QIMR_317 | 1977 | 1017 | 960 | |
| MPIP | 913 | 376 | 537 | |
| RADIANT + Bonn/Mannheim | 2225 | 935 | 1290 | |
| RADIANT | 3213 | 1625 | 1588 | |
| STAR*D | 1752 | 1241 | 511 | |
| MDD replication | deCODE | 34 229 | 1067 | 33 162 |
| GenPod/NEWMEDS | 5939 | 477 | 5462 | |
| Harvard i2b2 | 902 | 460 | 442 | |
| PsyCoLaus | 2794 | 1303 | 1491 | |
| SHIP-LEGEND | 1806 | 313 | 1493 | |
| TwinGene | 9562 | 1861 | 7701 | |
| GenRED2/DepGenesNetworks | 2246 | 1302 | 944 | |
| MDD-BIP cross-disorder | PGC MDD | 17 277 | 9238 | 8039 |
| PGC BIP | 14 773 | 6998 | 7775 | |
| Totals | Discovery | 18 759 | 9240 | 9519 |
| MDD replication | 57 478 | 6783 | 50 695 | |
| MDD-BIP cross-disorder | 32 050 | 16 236 | 15 814 |
Abbreviations: BIP, bipolar disorder; MDD, major depressive disorder; PGC, Psychiatric GWAS Consortium.
Sample acronyms are defined in the Supplementary Methods. Sample sizes differ from the primary publications due to varying quality control procedures and re-allocation of controls that were used in multiple studies.
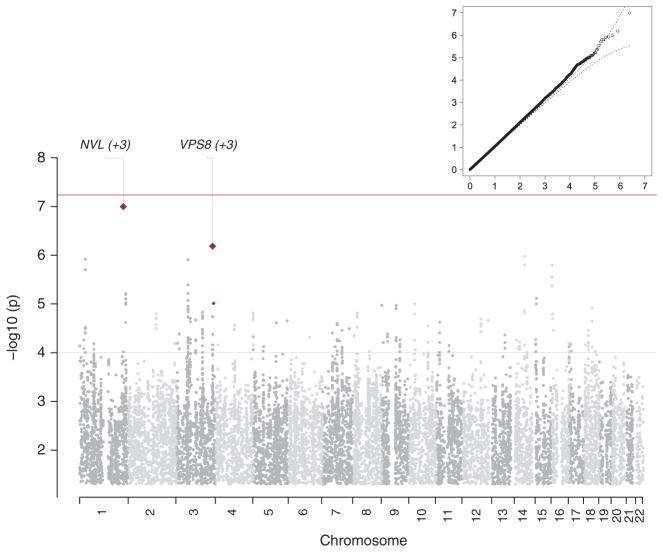
An overview of the results is in Figure 1. The quantile–quantile plot shows conformity of the observed results to those expected by chance. The overall λ [ref. 60] (the ratio of the observed median χ2 to that expected by chance) was 1.056 and λ1000 was 1.006 (that, λ rescaled to a sample size of 1000 cases and 1000 controls).61 The Manhattan plot depicts the association results in genomic context, and no region exceeded genome-wide significance (P<5×10−8).62 We conducted imputation with Hap-Map2 [ref. 63] and 1000 Genomes Project data52 in addition to HapMap3 and obtained similar genomewide association results.
Figure 1.
Overview of results from the discovery genome-wide association study mega-analysis for major depressive disorder. The inset shows the quantile–quantile plot (observed by expected P-values on the −log10scale) showing conformity of the observed results to expectations under the null. The main part of the figure shows the Manhattan plot (−log10 of the P-value by genomic location) of the association results in genomic context. No region exceeded genome-wide significance in the discovery sample.
The minimum P-values for the main analysis were at rs11579964 (chr1: 222 605 563 bp, P = 1.0×10−7) and rs7647854 (chr3:186 359 477 bp, P = 6.5×10−7; Supplementary Tables S16 and S17). Bioinformatic analyses of 201 SNPs with P < 0.0001 and the 1655 SNPs in moderate linkage disequilibrium (LD, r2 > 0.5) showed no overlap with literature findings in the NHGRI GWAS catalog,14 with transcripts differentially expressed in post-mortem brain samples of individuals with MDD,64 or with SNPs that were genome-wide significant or notable in the PGC association analyses of ADHD, BIP, or schizophrenia. We noted that a few of these 201 SNPs were ±20 kb of genes previously studied in MDD (ADCY9 and PDLIM5),65 or notable in prior hypotheses of the etiology of psychiatric disorders (GRM7, HTR7 and RELN).
In the analyses of chrX, no SNP achieved genome-wide significance in analysis of all samples or in separate analyses of females and males. The most significant SNP across all analyses was rs12837650 in the female-only analysis (P = 5.6×10−6).
In the MDD replication phase, 554 SNPs with P < 0.001 from the discovery mega-analysis were evaluated in independent samples totaling 6783 MDD cases and 50 695 controls (Table 1). For these SNPs, the replication samples did not produce logistic regression β coefficients in the same directions as the discovery analysis more frequently than expected by chance (sign test, P = 0.05). No SNP exceeded genome-wide significance for a joint analysis of the discovery and replication samples (Supplementary Table S18). The minimum P-value was for rs1969253 (P = 4.8×10−6, chr3:185 359 206), located in an intron of the disheveled 3 gene (DVL3). Given the probable etiological heterogeneity of MDD, we also conducted replication analyses of subtypes of MDD. For analyses restricted to female cases and controls, the direction of effects tended to be consistent between the discovery and replication samples (sign test, P = 0.006) although no SNP neared genome-wide significance (minimum P = 4.8×10−6 at rs1969253, chr3: 185 359 206). For male cases and controls, the sign test was not significant (P = 0.17), and no SNP was genome-wide significant (minimum P = 3.8×10−7 at rs2498828, chr14:91 491 028). For recurrent MDD, there was greater evidence of consistency of effects between the discovery and replication samples (sign test, P = 0.006), and the minimum P-value was 1.0×10−6 at rs2668193 (chr3:185 419 374).
In the MDD-BIP cross-disorder analyses, we evaluated support for a broader mood disorders phenotype. Due to the need to resolve overlapping subjects, the sample sizes and P-values differ from the numbers given above. There were 32 050 independent subjects (9238 MDD cases/8039 controls and 6998 BIP cases/7775 controls), 160 SNPs with P < 0.0001 in the MDD discovery phase and 659 SNPs in the BIP discovery phase (no SNP had P < 0.0001 for both MDD and BIP). First, in aggregate, SNPs selected from the BIP discovery phase showed evidence of replication in MDD (65 of 100 independent SNPs had logistic regression β-coefficients in the same direction in both BIP and MDD, sign test, P = 0.0018). However, the reverse comparison was near chance level (46 of 76 independent SNPs selected from MDD analyses had consistent effects in BIP, sign test P = 0.042). Second, in the combined analysis of these 819 SNPs, 15 exceeded genome-wide significance (P<5×10−8) and all were in a 248 kb interval of high LD on 3p21.1 (chr3:52 425 083–53 822 102, minimum P=5.9×10−9 at rs2535629; Supplementary Table S19, Supplementary Figure S20). The 116 SNPs in this region were all selected from the BIP sample (P < 0.0001), and none from the MDD sample. The region of strongest signal contained 84 SNPs from rs2878628 to rs2535629 (chr3:52 559 755–52 808 259). This region contains multiple genes: PBRM1 (chromatin remodeling and renal cell cancer), GNL3 (stem cell maintenance and tumorgenesis), GLT8D1, SPCS1, NEK4, the ITIH1-ITIH3-ITIH4 gene cluster (possibly involved in cancer), four micro-RNA and three small nucleolar RNA genes. This region had genome-wide significant findings in three prior GWAS: rs1042779 (chr3:52 796 051) for BIP,66 rs736408 (chr3:52 810 394) for a combined BIP-schizophrenia phenotype36 and rs2251219 (chr3:52 559 827) for a combined MDD-BIP phenotype67 (although a reanalysis suggested most of the signal arose from the BIP group).68 The PGC analyses include nearly all subjects in the prior reports, and thus cannot be considered independent evidence. As discussed below, we advise caution in interpreting this result.
We conducted a set of pre-planned secondary analyses using the discovery samples. These analyses presume that observable clinical features allow the ability to index etiological genetic heterogeneity. The clinical features we chose—sex, age of onset, recurrence and typicality—had a rationale from genetic epidemiological studies, and were comparably assessed in most of the discovery samples (Supplementary Methods). The results are summarized in Table 2, and detail on regions with P<1×10−5 provided in Supplementary Table S21. Parallel analyses of chrX SNPs for these secondary phenotypes also failed to identify convincing associations. Given the level of resolution afforded by our sample size and genotyping, none of these clinical features successfully indexed the clinical heterogeneity of MDD (all λ1000 values were small and no P-value approached genome-wide significance). However, we note that the total samples available for these analyses were small for a GWAS of a complex and modestly heritable trait. Moreover, as described above, SNPs identified in analyses by sex and for recurrent MDD did not yield genome-wide significance in replication in external samples.
Table 2.
Summary of secondary analyses
| Secondary analysis | Cases | Controls | λ1000 | Best finding |
|---|---|---|---|---|
| Primary analyses as reference | ||||
| Discovery phase | 9240 | 9519 | 1.008 | rs11579964, chr1:222,605,563, P = 1.0 × 10−7 |
| Combined discovery plus replication | 16 023 | 60 214 | NA | rs1969253, chr3:185,359,206, P = 3.4 × 10−6 |
| (a) By sex | ||||
| Females | 6118 | 5366 | 1.005 | rs1969253, chr3:185,359,206, P = 1.0 × 10−7 |
| Males | 3122 | 4153 | 0.999 | rs7296288, chr12:47,766,235, P = 2.3 × 10−7 |
| (b) Onset and recurrence | ||||
| Recurrent | 6743 | 9519 | 1.006 | rs4478239, chr4:188,428,300, P = 4.7 × 10−7 |
| Recurrent early onset (≤ 30 years) | 4710 | 9519 | 1.007 | rs1276324, chr18:19,172,417, P = 6.7 × 10−7 |
| Childhood onset (≤ 12 years) | 774 | 6077 | 1.015 | rs4358615, chr6:27,106,546, P = 2.3 × 10−6 |
| Age of onset as a continuous trait | 8920 | — | 0.998 | rs16948388, chr17:45,242,175, P = 1.0 × 10−6 |
| (c) Sub-type analysis | ||||
| Latent class 1 (weight loss and insomnia) | 3814 | 9519 | 1.007 | rs9830950, chr3:61,097,358, P = 1.0 × 10−7 |
λ1000 is the genomic inflation factor scaled to a constant sample size of 1000 cases and 1000 controls. Age of onset analyzed using a square root transformation.
Finally, under the assumptions that MDD is highly polygenic and that power is not optimal,69,70 we conducted risk profile analyses using the MDD discovery phase samples. We split these samples into two sets and used 80% to develop a risk profile to predict case–control status in the remaining 20% of the samples (Supplementary Methods). These analyses showed a modest (R2 = 0.6%) but highly significant (P<10−6) predictive capacity.
Discussion
This is the largest and most comprehensive genetic study of MDD. There were 18 759 subjects in the MDD discovery phase, 57 478 subjects in the MDD replication phase and 32 050 subjects in cross-disorder analyses of MDD and BIP. Analyses included the primary phenotype of MDD, three sets of autosomal imputation data (HapMap3, HapMap2 and 1000 Genomes), analysis of chrX, and multiple sub-phenotypes selected based on prior epidemiological and genetic epidemiological studies (Table 2).
The primary finding of this paper is that no locus reached genome-wide significance in the combined discovery and replication analysis of MDD. Our results are consistent with null results from other MDD meta-analyses using subsets of the present sample.22,23,25,28 The risk profile analyses are consistent with the presence of genetic effects, which our analysis was underpowered to detect. Although not significant, several analyses (that is, MDD, femalesonly and recurrent MDD) pointed at a region on chr3:185.3Mb near the gene (DVL3) encoding the Wnt-signaling phosphoprotein disheveled 3. DVL3 transcripts are decreased in the nucleus accumbens of individuals with MDD71 and are overexpressed in the leukocytes of individuals reporting social isolation,72 and the DVL3 protein product is upregulated in rats after treatment with antipsychotics.73,74 The chr3:185.3 Mb region also contains several serotonin receptors (HTR3D, HTR3C and HTR3E). However, none of these analyses were strongly compelling.
We advise caution in interpreting the evidence for association of SNPs on 3p21.1 with a broad mood disorder phenotype based on the combined PGC MDD and BIP discovery samples (minimum P = 5.9×10−9 at rs2535629, chr3:52808259). Evidence to date suggests that this locus is associated with BIP66 and schizophrenia,36 and an even broader association was suggested by a PGC meta-analysis of MDD, BIP, schizophrenia, ADHD and autism. This separate PGC analysis included nearly all of the samples reported here, and the top finding was again for rs2535629 (P = 2.5×10−12).75 The BIP sample made the strongest contribution to the combined analysis (OR = 1.15) followed by schizophrenia (OR = 1.10), MDD (OR = 1.10), ADHD (OR = 1.05) and autism (OR = 1.05). Although a five-disorder model was statistically the most likely and significant heterogeneity of ORs across disorders was not detected, the MDD replication data reported here raise some questions whether MDD also has an association in this region. We obtained MDD replication data for two SNPs on 3p21.1 (Supplementary Table S18), and observed no additional support for association for rs2535629 (discovery P = 0.0001, replication P = 0.56, combined P = 0.002) or rs3773729 (discovery P = 0.00022, replication P = 0.022 with different direction of association, combined P = 0.0095). Similarly, replication samples for the PGC BIP study36 provided little additional evidence for two SNPs in this region (rs736408 and rs3774609). In contrast, stronger evidence for association was observed in the PGC SCZ study after adding data from replication samples (rs2239547, chr3:52 830 269; discovery P = 2.2×10−6, replication P = 0.003, combined P=6×10−8).37 The PGC analyses reported here include most samples used in previous reports of genome-wide significant association in this region for BIP,66 BIP-SCZ36 and MDD-BIP,67 underscoring the need for analysis of independent samples.
Thus, this locus has produced genome-wide significant evidence for association to BIP,66 with evidence for broader set of associated phenotypes (especially SCZ).36,75 The inconsistency of results in large MDD and BIP replication samples suggests that the current finding should be viewed with caution. If specific genetic variants can be identified that underlie the BIP association in this region, it will be possible to evaluate their degree of association with other phenotypes including MDD. A continuing challenge in this field is the differentiation between true pleiotropy (genetic risk factors associated with distinct phenotypes) versus diagnostic misclassification (phenotypic overlap in cases with different genetic risk factors, leading to diagnostic ‘error’). There is a robust and evolving literature in psychiatric genetic epidemiology regarding the degree of independence versus co-segregation of current diagnostic categories, as well as the occurrence and familial risks of cases with mixed syndromes and changes in clinical syndromes over time. It is likely that analyses of large-scale genomic data will provide new perspectives on these issues.
On the whole, these results for MDD are in sharp contrast to the now substantial experience with GWAS for other complex human traits. GWAS has been a widely applied (> 860 studies) and remarkably successful technology in the identification of > 2200 strong associations for a wide range of biomedical diseases and traits.14 The vast majority of GWAS with sample sizes > 18 000 found at least one genome-wide significant finding (178/189 studies, 94.2%),14 and yet we found no such associations for MDD. What implications do these null results have for research into the genetics of MDD? Why might the results have turned out this way? We frame our discussion around a series of implications and hypotheses for future research.
Caveat: genome coverage
The genotyping chips used by the primary studies had good coverage of common variation across the genome. It is possible that genetic variation important in the etiology of MDD was missed if LD was insufficient with genotyped variants. In particular, we had suboptimal or poor coverage of uncommon variation (MAF 0.005–0.05), and we have not yet analyzed copy number variation (PGC analyses of copy number variants are underway). In addition, the discovery studies used eight genotyping platforms, and it is possible that causal common variation was missed because not all platforms had good coverage in the same regions. However, these caveats should be interpreted in the context of the many successful GWAS meta-analyses that faced similar limitations.
Implication: exclusions
For the phenotype of MDD, we can exclude combinations of MAF and effect size with 90% power. The exclusionary regions are genotypic relative risks (GRRs) ≥1.16 for MAF 0.30–0.50, ≥1.18 for MAF 0.20–0.25, ≥1.21 for MAF 0.15, ≥1.25 for MAF 0.10 and ≥1.36 for MAF 0.05. The technologies we used for genotyping probably captured the more common variation well, but were progressively less comprehensive at lower MAF. These exclusion GRRs equate to a variance in liability of ~0.5%. Since this study was conceived, we have gained considerable knowledge about the likely effect sizes of variants contributing to common complex disease. Therefore, these exclusion architectures are not unexpected.
Implication: future sample sizes
Association studies in psychiatry have traditionally had small sample sizes (< 1000 total subjects). For even a modest amount of genotyping in a candidate gene (10 SNPs), 90% power to detect a genotypic relative risk of 1.16 at MAF 0.30 requires 3600 cases and 3600 controls. It is possible to speculate that larger genetic effects exist at smaller MAF (0.005–0.05). Investigators, reviewers and editors need to be cognizant of these requirements, as smaller samples may be difficult to interpret due to inadequate power.
Hypothesis: suboptimal phenotype
MDD is defined descriptively without reference to any underlying biology, biomarker or pathophysiology.76,77 Genetic epidemiological studies have suggested that subtypes of MDD might be more familial or have higher heritability (for example, recurrent MDD,10 recurrent early-onset MDD11 and clinically ascertained MDD12). It is possible that well-powered genetic studies of these less common and arguably more heritable forms of MDD would have greater success. However, a sizable fraction of our cases were from hospital sources and our analyses of recurrent MDD and recurrent early-onset MDD were unrevealing, although these observations are qualified by the smaller sample sizes. The selection of a phenotype for genetic studies presents a dilemma for MDD researchers: larger samples which are more representative of the population can be achieved for broadly defined MDD, whereas restricted phenotypes may be more familial but are more difficult to recruit in large numbers from the population. Some other forms of MDD can only be defined using methods that are difficult to operationalize in large samples (for example, extensive clinical interviews, biological assays like repeated hormone measures or brain imaging).
Hypothesis: MDD is particularly heterogeneous
An early criticism of GWAS meta-analysis was that combining samples from multiple sites to increase sample size would introduce crippling heterogeneity. This concern was not borne out by experience. Indeed, the number of significant associations has increased as more individual studies have been combined using meta-analysis for other heterogeneous diseases such as Type 2 diabetes mellitus,32 inflammatory bowel disease78 and multiple cancers79,80 along with anthropometric traits like height29 and body mass.30 It is possible that MDD might be exceptional, and have greater clinical and etiological heterogeneity, as well as non-genetic phenocopies. The different endorsement rates of the MDD criteria between cohorts may support this conjecture (Supplementary Table S12). Higher heterogeneity implies reduced statistical power as the genetic effect size distribution will be diluted. Higher heterogeneity— that is, many different ‘types’ of MDD—would suggest that identifying more optimal MDD-related phenotypes may be a practical step forward if adequate sample sizes could be achieved.
Hypothesis: MDD has a divergent genetic architecture
The unquestionable success of GWAS in identifying strong and replicable associations for so many human diseases is intriguing given that the additive logistic regression model generally used is rudimentary. The dependent variable is disease status (1 = yes, 0 = no), the continuous independent variable is a SNP genotype (coded as the number of copies of the minor allele or as the imputed allelic dosage, 0–2), plus covariates like principal components to adjust for ancestry. It is possible that MDD is distinctive, and that the additive logistic model is not an adequate approximation of the genetic architecture of MDD (see Kohli et al.26). There are numerous alternative genetic architectures, although many are at least partly detectable using an additive model.
There has been considerable speculation that gene–environment interactions are particularly salient for MDD. It is possible that MDD can only be understood if genetic and environmental risk factors are modeled simultaneously. The most prominent example for MDD is the moderation of environmental stress by genetic variation in a functional polymorphism near the serotonin transporter (5-HTTLPR).81 As in the initial report in 2003, some evidence has supported this GxE interaction82,83 other analyses have not84,85 and the original finding (from a longitudinal study in Dunedin, New Zealand) did not replicate in an independent longitudinal study in Christchurch, New Zealand.86 A practical issue is again the tradeoff between relatively inexpensive, cross-sectional assessments of MDD case and control status and the detailed longitudinal data required to accurately characterize environmental stressors.
Hypothesis: insufficient power
Although this is one of the largest GWAS analyses ever conducted in psychiatry (second only to the PGC schizophrenia study),37 the sample size may still have been too small. The very small but highly significant variance explained in the polygenic risk score analyses (P<10−6 testing one hypothesis) is consistent with a hypothesis of insufficient power in this study.
The overlapping hypotheses listed above imply that an association study for MDD has less power than for studies of many other complex genetic disorders. However, even if the hypotheses listed above were not the contributing factors, we may still conclude that insufficient power underpins the dearth of results from this mega-analysis by considering the epidemiology of MDD. MDD is highly prevalent in the population, implying that cases are less extreme in the population compared with the controls and therefore larger sample sizes are required. For example, we have calculated that sample sizes 2.4 times larger are needed for GWAS of MDD (prevalence 0.15) compared with schizophrenia (prevalence 0.007).25,87 Furthermore, if we assume as a first approximation that the number and frequency distribution of risk alleles is the same for MDD and schizophrenia, then samples sizes five times larger are needed to account for the lower heritability of MDD (0.37)10 compared with schizophrenia (0.81),88 implying lower effect sizes at each locus (see Wray et al.25 and Yang et al.87 for details). Obtaining a total sample size on the order of 100 000 MDD cases plus controls would require a significant investment for ascertainment, phenotyping, DNA collection and genotyping, but could be accomplished using national registers or via electronic medical records of large health care organizations. Such sample sizes have been achieved in studies of quantitative traits and yielded large numbers of genome-wide significant results.29,30
Conclusion
This report contributes important new data about the nature of MDD.33 Unlike a large number of other GWAS that provide precious etiological clues, our analyses are more informative about what MDD is not. The path to progress is likely to be more difficult for MDD, but there are a number of rational next steps. We have offered some ideas about how progress might be achieved. The PGC is conducting GWAS metaanalyses across ADHD, autism, BIP, MDD and schizophrenia, and these very large analyses could identify genetic variants that predispose or protect to psychiatric disorders in general, and thus provide key initial findings that could be used to disentangle the etiology of MDD. Analysis of copy number variation has provided important leads for autism and schizophrenia, and might prove informative for MDD.
Supplementary Material
Acknowledgments
We thank the thousands of people with MDD who donated time and effort to make this research possible. The PGC was funded by NIMH Grants MH085520 (lead PI PFS) and MH080403. We thank our colleagues in the PGC Bipolar Disorder Working Group who allowed pre-publication access to their GWAS mega-analysis results for the MDD-BIP crossdisorder analyses. The Bonn/Mannheim (BoMa) GWAS was supported by the German Federal Ministry of Education and Research, within the context of the National Genome Research Network 2 (NGFN-2), the National Genome Research Network plus (NGFNplus) and the Integrated Genome Research Network (IG) MooDS (Grant 01GS08144 to S Cichon and MM Nöethen, and Grant 01GS08147 to M Rietschel). The work at deCODE was funded by European Union Grants LSHM-CT-2006-037761 (Project SGENE), PIAP-GA-2008-218251 (Project PsychGene) and HEALTH-F2-2009-223423 (Project PsychCNVs). GenPod was funded by the Medical Research Council (UK) and supported by the Mental Health Research Network. Genotyping of the GenPod sample was funded by the Innovative Medicines Initiative Joint Undertaking under Grant Agreement number 115008 (NEWMEDS). The GenRED GWAS project was supported by NIMH R01 Grants MH061686 (DF Levinson), MH059542 (WH Coryell), MH075131 (WB Lawson), MH059552 (JB Potash), MH059541 (WA Scheftner) and MH060912 (MM Weissman). We acknowledge the contributions of Dr George S Zubenko and Dr Wendy N Zubenko, Department of Psychiatry, University of Pittsburgh School of Medicine, to the GenRED I project. The NIMH Cell Repository at Rutgers University and the NIMH Center for Collaborative Genetic Studies on Mental Disorders made essential contributions to this project. Genotyping was carried out by the Broad Institute Center for Genotyping and Analysis with support from Grant U54 RR020278 (which partially subsidized the genotyping of the GenRED cases). Collection and quality control analyses of the control data set were supported by grants from NIMH and the National Alliance for Research on Schizophrenia and Depression. We are grateful to Knowledge Networks (Menlo Park, CA, USA) for assistance in collecting the control data set. We express our profound appreciation to the families who participated in this project, and to the many clinicians who facilitated the referral of participants to the study. The Depression Genes and Networks ARRA grant was funded by RC2MH089916. Funding for the Harvard i2b2 sample was provided by a subcontract to RH Perlis and JW Smoller as part of the i2b2 Center (Informatics for Integrating Biology and the Bedside), an NIH-funded National Center for Biomedical Computing based at Partners HealthCare System (U54LM008748, PI: IS Kohane), and by an NIMH Grant to RH Perlis (MH086026). Max Planck Institute of Psychiatry MARS study was supported by the BMBF Program Molecular Diagnostics: Validation of Biomarkers for Diagnosis and Outcome in Major Depression (01ES0811). Genotyping was supported by the Bavarian Ministry of Commerce, and the Federal Ministry of Education and Research (BMBF) in the framework of the National Genome Research Network (NGFN2 and NGFN-Plus, FKZ 01GS0481 and 01GS08145). The Netherlands Study of Depression and Anxiety (NESDA) and the Netherlands Twin Register (NTR) contributed to GAIN-MDD and to MDD2000. Funding was from: the Netherlands Organization for Scientific Research (MagW/ZonMW Grants 904-61-090, 985-10-002, 904-61-193, 480-04-004, 400-05-717, 912-100-20; Spinozapremie 56-464-14192; Geestkracht program Grant 10-000-1002); the Center for Medical Systems Biology (NWO Genomics), Biobanking and Biomolecular Resources Research Infrastructure, VU University’s Institutes for Health and Care Research and Neuroscience Campus Amsterdam, NBIC/BioAssist/RK (2008.024); the European Science Foundation (EU/QLRT-2001-01254); the European Community’s Seventh Framework Program (FP7/2007-2013); ENGAGE (HEALTH-F4-2007-201413); and the European Science Council (ERC, 230374). Genotyping was funded in part by the Genetic Association Information Network (GAIN) of the Foundation for the US National Institutes of Health, and analysis was supported by grants from GAIN and the NIMH (MH081802). CM Middeldorp was supported by the Netherlands Organization for Scientific Research (NOW-VENI grant 916-76-125). The PsyCoLaus study was supported by grants from the Swiss National Science Foundation (#3200B0–105993, #3200B0-118′308, 33CSC0-122661) and from GlaxoSmithKline (Psychiatry Center of Excellence for Drug Discovery and Genetics Division, Drug Discovery - Verona, R&D). We express our gratitude to the Lausanne inhabitants who volunteered to participate in the PsyCoLaus study. We also thank V Mooser, G Weaber and P Vollenweider who initiated the CoLaus project. Funding for the QIMR samples was provided by the Australian National Health and Medical Research Council (241944, 339462, 389927, 389875, 389891, 389892, 389938, 442915, 442981, 496675, 496739, 552485, 552498, 613602, 613608, 613674, 619667), the Australian Research Council (FT0991360, FT0991022), the FP-5 GenomEUtwin Project (QLG2-CT-2002-01254) and the US National Institutes of Health (AA07535, AA10248, AA13320, AA13321, AA13326, AA14041, MH66206, DA12854, DA019951), and the Center for Inherited Disease Research (Baltimore, MD, USA). We thank the twins and their families registered at the Australian Twin Registry for their participation in the many studies that have contributed to this research. RADIANT was funded by: a joint grant from the UK Medical Research Council and GlaxoSmithKline (G0701420); the National Institute for Health Research (NIHR) Specialist Biomedical Research Centre for Mental Health at the South London and Maudsley NHS Foundation Trust and the Institute of Psychiatry, King’s College London; and the UK Medical Research Council (G0000647). The GENDEP study was funded by a European Commission Framework 6 grant, EC Contract Ref.: LSHB-CT-2003-503428. SHIP is part of the Community Medicine Research net of the University of Greifswald, Germany, which is funded by the Federal Ministry of Education and Research (Grants no. 01ZZ9603, 01ZZ0103 and 01ZZ0403), the Ministry of Cultural Affairs and the Social Ministry of the Federal State of Mecklenburg-West Pomerania. Genome-wide data have been supported by the Federal Ministry of Education and Research (Grant no. 03ZIK012) and a joint grant from Siemens Healthcare, Erlangen, Germany and the Federal State of Mecklenburg-West Pomerania. The University of Greifswald is a member of the ‘Center of Knowledge Interchange’ program of the Siemens AG. SHIPLEGEND is funded by the German Research Foundation (DFG: GR 1912/5-1). Genotyping of STAR*D was supported by an NIMH Grant to SP Hamilton (MH072802). STAR*D was funded by the National Institute of Mental Health (contract N01MH90003) to the University of Texas Southwestern Medical Center at Dallas (AJ Rush, principal investigator). The TwinGene study was supported by the Swedish Ministry for Higher Education, the Swedish Research Council (M-2005-1112), GenomEUtwin (EU/QLRT-2001-01254; QLG2-CT-2002-01254), the Swedish Foundation for Strategic Research and the US National Institutes of Health (U01 DK066134). This study makes use of data generated by the Wellcome Trust Case–Control Consortium. A full list of the investigators who contributed to the generation of the data is available from http://www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under awards 076113 and 085475.
Appendix
Collaborators
| Study | Title | First name | Last name | Affiliation | Country | Role on primary study |
|---|---|---|---|---|---|---|
| mpip, mdd2000, statistical genetics | Dr | Stephan | Ripke | Harvard University/Broad Institute | USA | Principal analyst |
| QIMR, mdd2000, statistical genetics | Dr | Naomi R | Wray | Queensland Institute of Medical Research/University of Queensland | Australia | Co-PI, analyst |
| radiant, statistical genetics | Prof | Cathryn M | Lewis | Institute of Psychiatry, King’s College London | UK | Co-investigator |
| stard | Assoc Prof | Steven P | Hamilton | University of California, San Francisco | USA | PI |
| genred, GenRED2, DepGenesNetworks | Prof | Myrna M | Weissman | Columbia University | USA | PI |
| radiant | Dr | Gerome | Breen | Institute of Psychiatry, King’s College London | UK | Co-investigator |
| statistical genetics | Dr | Enda M | Byrne | Queensland Institute of Medical Research | Australia | Genetic analyses |
| mdd2000 | Prof | Douglas HR | Blackwood | University of Edinburgh | UK | Co-PI |
| gain, mdd2000 | Prof | Dorret I | Boomsma | VU University, Amsterdam | The Netherlands | Co-PI GAIN, PI NTR |
| boma | Prof | Sven | Cichon | University of Bonn | Germany | PI |
| QIMR, mdd2000 | Prof | Andrew C | Heath | Washington University, St Louis | USA | QIMR sample and funding |
| mpip, gsk | Prof | Florian | Holsboer | Max Planck Institute of Psychiatry | Germany | PI |
| mpip | Dr | Susanne | Lucae | Max Planck Institute of Psychiatry | Germany | PI |
| QIMR, mdd2000 | Prof | Pamela AF | Madden | Washington University, St Louis | USA | QIMR sample and funding |
| QIMR, mdd2000 | Prof | Nicholas G | Martin | Queensland Institute of Medical Research | Australia | QIMR sample and funding |
| radiant | Prof | Peter | McGuffin | Institute of Psychiatry, King’s College London | UK | PI |
| gsk, radiant | Dr | Pierandrea | Muglia | GlaxoSmithKline | Italy | Co-PI, co-investigator |
| boma | Prof | Markus M | Noethen | University of Bonn | Germany | PI |
| gain, mdd2000 | Prof | Brenda P | Penninx | VU University Medical Center, Amsterdam | The Netherlands | Co-PI GAIN, PI NESDA |
| QIMR, mdd2000 | Dr | Michele L | Pergadia | Washington University, St Louis | USA | Co-investigator |
| genred, GenRED2, DepGenesNetworks | Prof | James B | Potash | University of Iowa | USA | PI |
| boma | Prof | Marcella | Rietschel | Central Inst Mental Health, University of Heidelberg | Germany | PI |
| gain, statistical genetics | Prof | Danyu | Lin | University of North Carolina | USA | Statistical geneticist |
| mpip | Prof | Bertram | Müller-Myhsok | Max Planck Institute of Psychiatry | Germany | Collaborator |
| genred, GenRED2, DepGenesNetworks | Dr | Jianxin | Shi | National Cancer Institute | USA | Statistical geneticist |
| deCODE | Dr | Stacy | Steinberg | deCODE Genetics | Iceland | Genetic analyses |
| SHIP-LEGEND | Prof | Hans J | Grabe | University of Greifswald | Germany | PI |
| TwinGene | Prof | Paul | Lichtenstein | Karolinska Institutet | Sweden | PI |
| TwinGene | Dr | Patrik | Magnusson | Karolinska Institutet | Sweden | PI |
| Harvard i2b2 | Assoc Prof | Roy H | Perlis | Massachusetts General Hospital | USA | PI |
| PsyCoLaus | Prof | Martin | Preisig | University of Lausanne | Switzerland | PI |
| Harvard i2b2 | Assoc Prof | Jordan W | Smoller | Massachusetts General Hospital | USA | Co-PI |
| deCODE | Dr | Kari | Stefansson | deCODE Genetics | Iceland | PI |
| genpod | Dr | Rudolf | Uher | Institute of Psychiatry, King’s College London | UK | PI |
| PsyCoLaus | Dr | Zoltan | Kutalik | University of Lausanne | Switzerland | Genetic analyses |
| genpod | Dr | Katherine E | Tansey | Institute of Psychiatry, King’s College London | UK | Co-investigator |
| SHIP-LEGEND | Dr | Alexander | Teumer | University of Greifswald | Germany | Genetic analyses |
| TwinGene | Alexander | Viktorin | Karolinska Institutet | Sweden | Genetic analyses | |
| radiant | Dr | Michael R | Barnes | GlaxoSmithKline | UK | Co-investigator |
| mpip | Dr | Thomas | Bettecken | Max Planck Institute of Psychiatry | Germany | Collaborator |
| mpip | Dr | Elisabeth B | Binder | Max Planck Institute of Psychiatry | Germany | Collaborator |
| boma | René | Breuer | Central Inst Mental Health, University of Heidelberg | Germany | Collaborator | |
| Harvard i2b2 | Victor M | Castro | Partners HealthCare System | USA | Bioinformatician | |
| Harvard i2b2 | Dr | Susanne E | Churchill | Partners HealthCare System | USA | Co-investigator |
| genred, GenRED2 | Prof | William H | Coryell | University of Iowa | USA | PI |
| radiant | Prof | Nick | Craddock | Cardiff University | UK | Co-investigator |
| radiant | Prof | Ian W | Craig | Institute of Psychiatry, King’s College London | UK | Co-investigator |
| mpip | Dr | Darina | Czamara | Max Planck Institute of Psychiatry | Germany | Collaborator |
| gain, mdd2000 | Prof | Eco J | De Geus | VU University, Amsterdam | The Netherlands | Co-investigator GAIN, co-PI NTR |
| boma | Franziska | Degenhardt | University of Bonn | Germany | Collaborator | |
| radiant | Prof | Anne E | Farmer | Institute of Psychiatry, King’s College London | UK | Co-investigator |
| Harvard i2b2 | Prof | Maurizio | Fava | Massachusetts General Hospital | USA | Co-investigator |
| boma | Josef | Frank | Central Inst Mental Health, University of Heidelberg | Germany | Collaborator | |
| Harvard i2b2 | Vivian S | Gainer | Partners HealthCare System | USA | Bioinformatician | |
| Harvard i2b2 | Patience J | Gallagher | Massachusetts General Hospital | USA | Research Coordinator | |
| QIMR, mdd2000 | Dr | Scott D | Gordon | Queensland Institute of Medical Research | Australia | Genetic analyses |
| Harvard i2b2 | Sergey | Goryachev | Partners HealthCare System | USA | Bioinformatician | |
| boma | Magdalena | Gross | University of Bonn | Germany | Collaborator | |
| genpod | Dr | Michel | Guipponi | University of Geneva | Switzerland | Co-investigator |
| QIMR, mdd2000 | Anjali K | Henders | Queensland Institute of Medical Research | Australia | QIMR project manager | |
| boma | Stefan | Herms | University of Bonn | Germany | Collaborator | |
| QIMR, mdd2000 | Prof | Ian B | Hickie | University of Sydney, Sydney | Australia | QIMR funding |
| boma | Susanne | Hoefels | University of Bonn | Germany | Collaborator | |
| gain | Prof | Witte | Hoogendijk | Erasmus Medical Center | The Netherlands | Co-investigator |
| gain, mdd2000 | Dr | Jouke Jan | Hottenga | VU University, Amsterdam | The Netherlands | Genetic analyses |
| Harvard i2b2 | Assoc Prof | Dan V | Iosifescu | Massachusetts General Hospital | USA | Co-investigator |
| mpip | Dr | Marcus | Ising | Max Planck Institute of Psychiatry | Germany | Collaborator |
| radiant | Dr | Ian | Jones | Cardiff University | UK | Co-investigator |
| radiant | Dr | Lisa | Jones | University of Birmingham | UK | Co-investigator |
| gain | Assoc Prof | Tzeng | Jung-Ying | North Carolina State University | USA | Statistical geneticist |
| genred, GenRED2 | Prof | James A | Knowles | University of Southern California | USA | Co-PI |
| Harvard i2b2 | Prof | Isaac S | Kohane | Brigham and Women’s Hospital | USA | Co-investigator |
| mpip | Dr | Martin A | Kohli | Max Planck Institute of Psychiatry | Germany | Collaborator |
| radiant | Dr | Ania | Korszun | Queen Mary University of London | UK | Co-investigator |
| TwinGene | Dr | Mikael | Landen | Karolinska Institutet | Sweden | Co-investigator |
| genred, GenRED2 | Prof | William B | Lawson | Howard University | USA | PI |
| genpod | Prof | Glyn | Lewis | University of Bristol | UK | PI |
| mdd2000 | Dr | Donald | MacIntyre | University of Edinburgh | UK | Co-investigator |
| boma | Prof | Wolfgang | Maier | University of Bonn | Germany | Collaborator |
| boma | Dr | Manuel | Mattheisen | University of Bonn | Germany | Collaborator |
| stard | Prof | Patrick J | McGrath | Columbia University | USA | Co-PI |
| mdd2000 | Dr | Andrew | McIntosh | University of Edinburgh | UK | Co-investigator |
| mdd2000 | Dr | Alan | McLean | University of Edinburgh | UK | Co-investigator |
| gain, mdd2000 | Dr | Christel M | Middeldorp | VU University, Amsterdam | The Netherlands | Phenotype collection |
| radiant | Dr | Lefkos | Middleton | Imperial College | UK | Co-investigator |
| QIMR, mdd2000 | Prof | Grant M | Montgomery | Queensland Institute of Medical Research | Australia | QIMR sample and funding |
| Harvard i2b2 | Asst Prof | Shawn N | Murphy | Massachusetts General Hospital | USA | Co-investigator |
| SHIP-LEGEND | Prof | Matthias | Nauck | University of Greifswald | Germany | Biobanking |
| gain, mdd2000 | Prof | Willem A | Nolen | Groningen University Medical Center | The Netherlands | Site PI NESDA |
| QIMR, mdd2000 | Dr | Dale R | Nyholt | Queensland Institute of Medical Research | Australia | Genetic analyses |
| genpod | Prof | Michael | O’Donovan | Cardiff University | UK | Co-investigator |
| deCODE | Dr | Högni | Oskarsson | Therapeia, University Hospital | Iceland | Phenotype collection |
| TwinGene | Dr | Nancy | Pedersen | Karolinska Institutet | Sweden | Co-investigator |
| genred, GenRED2 | Prof | William A | Scheftner | Rush University Medical Center | USA | PI |
| SHIP-LEGEND | Andrea | Schulz | University of Greifswald | Germany | Phenotype collection | |
| boma | Prof | Thomas G | Schulze | University of Goettingen | USA | Collaborator |
| stard | Asst Prof | Stanley I | Shyn | University of Washington | USA | Collaborator |
| deCODE | Dr | Engilbert | Sigurdsson | Landspitali University Hospital | Iceland | Phenotype collection |
| stard | Assoc Prof | Susan L | Slager | Mayo Clinic | USA | Co-PI |
| gain, mdd2000 | Prof | Johannes H | Smit | VU University Medical Center, Amsterdam | The Netherlands | Co-investigator GAIN, phenotype collection NESDA |
| deCODE | Dr | Hreinn | Stefansson | deCODE Genetics | Iceland | Co-investigator |
| boma | Dr | Michael | Steffens | University of Bonn | Germany | Collaborator |
| deCODE | Dr | Thorgeir | Thorgeirsson | deCODE Genetics | Iceland | Co-investigator |
| gsk | Dr | Federica | Tozzi | GlaxoSmithKline | Italy | Co-investigator |
| boma | Dr | Jens | Treutlein | Central Inst Mental Health, University of Heidelberg | Germany | Collaborator |
| mpip | Dr | Manfred | Uhr | Max Planck Institute of Psychiatry | Germany | Collaborator |
| mdd2000 | Prof | Edwin JCG | van den Oord | Virginia Commonwealth University | USA | Funding |
| gain, mdd2000 | Gerard | Van Grootheest | VU University Medical Center, Amsterdam | The Netherlands | Phenotype collection | |
| SHIP-LEGEND | Prof | Henry | Völzke | University of Greifswald | Germany | PI |
| Harvard i2b2 | Dr | Jeffrey B | Weilburg | Massachusetts General Hospital | USA | Co-investigator |
| gain, mdd2000 | Dr | Gonneke | Willemsen | VU University, Amsterdam | The Netherlands | Phenotype collection |
| gain, mdd2000 | Prof | Frans G | Zitman | Leiden University Medical Center, Leiden | The Netherlands | Site PI NESDA |
| statistical genetics | Dr | Benjamin | Neale | Harvard University/Broad Institute | USA | Collaborator |
| statistical genetics | Assoc Prof | Mark | Daly | Harvard University/Broad Institute | USA | Statistical geneticist |
| genred, GenRED2, DepGenesNetworks | Prof | Douglas F | Levinson | Stanford University | USA | PI |
| gain, mdd2000 | Prof | Patrick F | Sullivan | University of North Carolina | USA | PI |
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
Author contributions
All collaborators reviewed and approved the final version of the manuscript. Overall coordination: Patrick F Sullivan. Statistical analysis: Mark J Daly (lead), Stephan Ripke, Cathryn M Lewis, Dan-Yu Lin, Naomi R Wray, Benjamin Neale, Douglas F Levinson, Gerome Breen, Enda M Byrne. Phenotype analysis: Naomi R Wray (lead), Douglas F Levinson, Marcella Rietschel, Witte Hoogendijk, Stephan Ripke. Writing committee: Patrick F Sullivan (lead), Steven P Hamilton, Douglas F Levinson, Cathryn M Lewis, Stephan Ripke, Myrna M Weissman, Naomi R Wray. MDD discovery sample data contributed by: Bonn/Mannheim study—René Breuer, Sven Cichon, Franziska Degenhardt, Josef Frank, Magdalena Gross, Stefan Herms, Susanne Hoefels, Wolfgang Maier, Manuel Mattheisen, Markus M Nöethen, Marcella Rietschel, Thomas G Schulze, Michael Steffens, Jens Treutlein; GAIN MDD study—Dorret I Boomsma, Eco J De Geus, Witte Hoogendijk, Jouke Jan Hottenga, Tzeng Jung-Ying, Dan-Yu Lin, Christel MMiddeldorp, Willem A Nolen, Brenda P Penninx, Johannes H Smit, Patrick F Sullivan, Gerard van Grootheest, Gonneke Willemsen, Frans G Zitman; GenRED study—William H Coryell, James A Knowles, William B Lawson, Douglas F Levinson, James B Potash, William A Scheftner, Jianxin Shi, Myrna M Weissman; GlaxoSmithKline study—Florian Holsboer, Pierandrea Muglia, Federica Tozzi; MDD2000 study—Douglas HR Blackwood, Dorret I Boomsma, Eco J De Geus, Jouke Jan Hottenga, Donald J MacIntyre, Andrew McIntosh, Alan McLean, Christel M Middeldorp, Willem A Nolen, Brenda P Penninx, Stephan Ripke, Johannes H Smit, Patrick F Sullivan, Gerard van Grootheest, Gonneke Willemsen, Frans G Zitman, Edwin JCG van den Oord; Max Planck Institute of Psychiatry Study—Florian Holsboer, Susanne Lucae, Elisabeth Binder, Bertram Müller-Myhsok, Stephan Ripke, Darina Czamara, Martin A Kohli, Marcus Ising, Manfred Uhr, Thomas Bettecken; RADIANT study— Michael R Barnes, Gerome Breen, IanWCraig, Anne E Farmer, Cathryn M Lewis, Peter McGuffin, Pierandrea Muglia; Queensland Institute for Medical Research study—Enda Byrne, Scott D Gordon, Andrew C Heath, Anjali K Henders, Ian B Hickie, Pamela AF Madden, Nicholas G Martin, Grant M Montgomery, Dale R Nyholt, Michele L Pergadia, Naomi R Wray; and STAR*D study—Steven P Hamilton, Patrick J McGrath, Stanley I Shyn, Susan L Slager.
MDD replication sample data contributed by: deCODE Genetics—Högni Oskarsson, Engilbert Sigurdsson, Hreinn Stefansson, Kari Stefansson, Stacy Steinberg, Thorgeir Thorgeirsson; Depression Genes Networks study—Douglas F Levinson, James B Potash, Jianxin Shi, Myrna M Weissman; GENPOD—Michel Guipponi, Glyn Lewis, Michael O’Donovan, Katherine E Tansey, Rudolf Uher; GenRED2 study—William H Coryell, James A Knowles, William B Lawson, Douglas F Levinson, James B Potash, William A Scheftner, Jianxin Shi, Myrna M Weissman; Harvard i2b2 study—Victor M Castro, Susanne E Churchill, Maurizio Fava, Vivian S Gainer, Patience J Gallagher, Sergey Goryachev, Dan V Iosifescu, Isaac S Kohane, Shawn N Murphy, Roy H Perlis, Jordan W Smoller, Jeffrey B Weilburg; PsyCoLaus study—Zoltan Kutalik, Martin Preisig; SHIP-LEGEND study—Hans J Grabe, Matthias Nauck, Andrea Schulz, Alexander Teumer, Henry Völzke; and TwinGene study—Mikael Landen, Paul Lichtenstein, Patrik Magnusson, Nancy Pedersen, Alexander Viktorin.
Conflict of interest
Elisabeth Binder received grant support from Pharmaneuroboost. Hans J Grabe reports receiving funding from: German Research Foundation; Federal Ministry of Education and Research Germany; speakers honoraria from Bristol-Myers Squibb, Eli Lilly, Novartis, Eisai, Wyeth, Pfizer, Boehringer Ingelheim, Servier and travel funds from Janssen-Cilag, Eli Lilly, Novartis, AstraZeneca and SALUS-Institute for Trend-Research and Therapy Evaluation in Mental Health. Florian Holsboer is a shareholder of Affectis Pharmaceuticals and co-founder of HolsboerMaschmeyer-NeuroChemie. James A Knowles is on the Scientific Advisory Committee for Next-Generation Sequencing of Life Technologies and is a technical advisor to SoftGenetics. Pierandrea Muglia was a full-time employee of GSK when the work was performed. Bertram Müller-Myhsok consulted for Affectis Pharmaceuticals. Matthias Nauck reports funding from: the Federal Ministry of Education and Research Germany, Bio-Rad Laboratories, Siemens AG, Zeitschrift für Laboratoriumsmedizin, Bruker Daltronics, Abbott, Jurilab Kuopio, Roche Diagnostics, Dade Behring, DPC Biermann and Becton Dickinson. Rudolf Uher has received funding from a number of pharmaceutical companies as part of the European Union Innovative Medicine Initiative funded NEWMEDS project. Federica Tozzi was a full-time employee of GSK when the work was performed. Henry Völzke reports funding from: Sanofi-Aventis, Biotronik, the Humboldt Foundation, the Federal Ministry of Education and Research (Germany) and the German Research Foundation. No other author reports a conflict of interest.
References
- 1.Hasin DS, Goodwin RD, Stinson FS, Grant BF. Epidemiology of major depressive disorder: results from the National epidemiologic survey on alcoholism and related conditions. Arch Gen Psychiatry. 2005;62:1097–1106. doi: 10.1001/archpsyc.62.10.1097. [DOI] [PubMed] [Google Scholar]
- 2.Kessler RC, Berglund P, Demler O, Jin R, Koretz D, Merikangas KR, et al. The epidemiology of major depressive disorder: results from the National Comorbidity Survey Replication (NCS-R) JAMA. 2003;289:3095–3105. doi: 10.1001/jama.289.23.3095. [DOI] [PubMed] [Google Scholar]
- 3.Mueller TI, Leon AC, Keller MB, Solomon DA, Endicott J, Coryell W, et al. Recurrence after recovery from major depressive disorder during 15 years of observational follow-up. Am J Psychiatry. 1999;156:1000–1006. doi: 10.1176/ajp.156.7.1000. [DOI] [PubMed] [Google Scholar]
- 4.Judd LL. The clinical course of unipolar major depressive disorders. Arch Gen Psychiatry. 1997;54:989–991. doi: 10.1001/archpsyc.1997.01830230015002. [DOI] [PubMed] [Google Scholar]
- 5.Lopez AD, Mathers CD, Ezzati M, Jamison DT, Murray CJ. Global and regional burden of disease and risk factors, 2001: systematic analysis of population health data. Lancet. 2006;367:1747–1757. doi: 10.1016/S0140-6736(06)68770-9. [DOI] [PubMed] [Google Scholar]
- 6.Wittchen HU, Jacobi F, Rehm J, Gustavsson A, Svensson M, Jonsson B, et al. The size and burden of mental disorders and other disorders of the brain in Europe 2010. Eur Neuropsychopharmacol. 2011;21:655–679. doi: 10.1016/j.euroneuro.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 7.Angst F, Stassen HH, Clayton PJ, Angst J. Mortality of patients with mood disorders: follow-up over 34–38 years. J Affective Disord. 2002;68:167–181. doi: 10.1016/s0165-0327(01)00377-9. [DOI] [PubMed] [Google Scholar]
- 8.Gustavsson A, Svensson M, Jacobi F, Allgulander C, Alonso J, Beghi E, et al. Cost of disorders of the brain in Europe 2010. Eur Neuropsychopharmacol. 2011;21:718–779. doi: 10.1016/j.euroneuro.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 9.Murray CJL, Lopez AD. Evidence-based health policy: lessons from the Global Burden of Disease Study. Science. 1996;274:740–743. doi: 10.1126/science.274.5288.740. [DOI] [PubMed] [Google Scholar]
- 10.Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry. 2000;157:1552–1562. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- 11.Levinson DF. The genetics of depression: a review. Biol Psychiatry. 2006;60:84–92. doi: 10.1016/j.biopsych.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 12.McGuffin P, Katz R, Watkins S, Rutherford J. A hospital-based twin register of the heritability of DSM-IV unipolar depression. Arch Gen Psychiatry. 1996;53:129–136. doi: 10.1001/archpsyc.1996.01830020047006. [DOI] [PubMed] [Google Scholar]
- 13.Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, Thomas G, et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 14.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, et al. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 16.Wirdefeldt K, Gatz M, Reynolds CA, Prescott CA, Pedersen NL. Heritability of Parkinson disease in Swedish twins: a longitudinal study. Neurobiol Aging. 2011;32:1923, e1–8. doi: 10.1016/j.neurobiolaging.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hawkes CH, Macgregor AJ. Twin studies and the heritability of MS: a conclusion. Mult Scler. 2009;15:661–667. doi: 10.1177/1352458509104592. [DOI] [PubMed] [Google Scholar]
- 18.Poulsen P, Kyvik KO, Vaag A, Beck-Nielsen H. Heritability of type II (non-insulin-dependent) diabetes mellitus and abnormal glucose tolerance–a population-based twin study. Diabetologia. 1999;42:139–145. doi: 10.1007/s001250051131. [DOI] [PubMed] [Google Scholar]
- 19.Seddon JM, Cote J, Page WF, Aggen SH, Neale MC. The US twin study of age-related macular degeneration: relative roles of genetic and environmental influences. Arch Ophthalmol. 2005;123:321–327. doi: 10.1001/archopht.123.3.321. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Yang S, Chen JJ, Zhou SM, He SM, Liang YH, et al. Systemic lupus erythematosus: a genetic epidemiology study of 695 patients from China. Arch Dermatol Res. 2007;298:485–491. doi: 10.1007/s00403-006-0719-4. [DOI] [PubMed] [Google Scholar]
- 21.Rietschel M, Mattheisen M, Frank J, Treutlein J, Degenhardt F, Breuer R, et al. Genome-wide association, replication, and neuroimaging study implicates HOMER1 in the etiology of major depression. Biol Psychiatry. 2010;68:578–585. doi: 10.1016/j.biopsych.2010.05.038. [DOI] [PubMed] [Google Scholar]
- 22.Sullivan P, de Geus E, Willemsen G, James MR, Smit JH, Zandbelt T, et al. Genomewide association for major depressive disorder: a possible role for the presynaptic protein piccolo. Mol Psychiatry. 2009;14:359–375. doi: 10.1038/mp.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi J, Potash JB, Knowles JA, Weissman MM, Coryell W, Scheftner WA, et al. Genome-wide association study of recurrent early-onset major depressive disorder. Mol Psychiatry. 2011;16:193–201. doi: 10.1038/mp.2009.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muglia P, Tozzi F, Galwey NW, Francks C, Upmanyu R, Kong XQ, et al. Genome-wide association study of recurrent major depressive disorder in two European case-control cohorts. Mol Psychiatry. 2010;15:589–601. doi: 10.1038/mp.2008.131. [DOI] [PubMed] [Google Scholar]
- 25.Wray N, Pergadia M, Blackwood D, Penninx B, Gordon S, Nyholt D, et al. Genome-wide association study of major depressive disorder: new results, meta-analysis, and lessons learned. Mol Psychiatry. 2011;17:36–48. doi: 10.1038/mp.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohli MA, Lucae S, Saemann PG, Schmidt MV, Demirkan A, Hek K, et al. The neuronal transporter gene SLC6A15 confers risk to major depression. Neuron. 2011;70:252–265. doi: 10.1016/j.neuron.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis CM, Ng MY, Butler AW, Cohen-Woods S, Uher R, Pirlo K, et al. Genome-wide association study of recurrent major depression in the UK population. Am J Psychiatry. 2010;167:949–957. doi: 10.1176/appi.ajp.2010.09091380. [DOI] [PubMed] [Google Scholar]
- 28.Shyn SI, Shi J, Kraft JB, Potash JB, Knowles JA, Weissman MM, et al. Novel loci for major depression identified by genome-wide association study of Sequenced Treatment Alternatives to Relieve Depression and meta-analysis of three studies. Mol Psychiatry. 2011;16:202–215. doi: 10.1038/mp.2009.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249 796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Psychiatric GWAS Consortium. A framework for interpreting genomewide association studies of psychiatric disorders. Mol Psychiatry. 2009;14:10–17. doi: 10.1038/mp.2008.126. [DOI] [PubMed] [Google Scholar]
- 34.Psychiatric GWAS Consortium. Genome-wide association studies: history, rationale, and prospects for psychiatric disorders. Am J Psychiatry. 2009;166:540–546. doi: 10.1176/appi.ajp.2008.08091354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neale BM, Medland SE, Ripke S, Asherson P, Franke B, Lesch KP, et al. Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry. 2010;49:884–897. doi: 10.1016/j.jaac.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sklar P, Ripke S, Scott LJ, Andreassen OA, Cichon S, Craddock N, et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schizophrenia Psychiatric Genome-Wide Association Study Consortium. Genome-wide association study of schizophrenia identifies five novel loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin DY, Zeng D. Meta-analysis of genome-wide association studies: no efficiency gain in using individual participant data. Genet Epidemiol. 2010;34:60–66. doi: 10.1002/gepi.20435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsuang MT, Faraone SV. The Genetics of Mood Disorders. The Johns Hopkins University Press; Baltimore: 1990. [Google Scholar]
- 40.Smoller JW, Finn CT. Family, twin, and adoption studies of bipolar disorder. Am J Med Genet C Semin Med Genet. 2003;123C:48–58. doi: 10.1002/ajmg.c.20013. [DOI] [PubMed] [Google Scholar]
- 41.Ising M, Lucae S, Binder EB, Bettecken T, Uhr M, Ripke S, et al. A genomewide association study points to multiple loci that predict antidepressant drug treatment outcome in depression. Arch Gen Psychiatry. 2009;66:966–975. doi: 10.1001/archgenpsychiatry.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis G, Mulligan J, Wiles N, Cowen P, Craddock N, Ikeda M, et al. Polymorphism of the 5-HT transporter and response to antidepressants: randomised controlled trial. Br J Psychiatry. 2011;198:464–471. doi: 10.1192/bjp.bp.110.082727. [DOI] [PubMed] [Google Scholar]
- 43.Firmann M, Mayor V, Vidal PM, Bochud M, Pecoud A, Hayoz D, et al. The CoLaus study: a population-based study to investigate the epidemiology and genetic determinants of cardiovascular risk factors and metabolic syndrome. BMC Cardiovasc Disord. 2008;8:6. doi: 10.1186/1471-2261-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Volzke H, Alte D, Schmidt CO, Radke D, Lorbeer R, Friedrich N, et al. Cohort profile: the study of health in Pomerania. Int J Epidemiol. 2011;40:294–307. doi: 10.1093/ije/dyp394. [DOI] [PubMed] [Google Scholar]
- 45.Preisig M, Waeber G, Vollenweider P, Bovet P, Rothen S, Vandeleur C, et al. The PsyCoLaus study: methodology and characteristics of the sample of a population-based survey on psychiatric disorders and their association with genetic and cardiovascular risk factors. BMC Psychiatry. 2009;9:9. doi: 10.1186/1471-244X-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grabe HJ, Lange M, Wolff B, Volzke H, Lucht M, Freyberger HJ, et al. Mental and physical distress is modulated by a polymorphism in the 5-HT transporter gene interacting with social stressors and chronic disease burden. Mol Psychiatry. 2005;10:220–224. doi: 10.1038/sj.mp.4001555. [DOI] [PubMed] [Google Scholar]
- 47.John U, Greiner B, Hensel E, Ludemann J, Piek M, Sauer S, et al. Study of health in Pomerania (SHIP): a health examination survey in an east German region: objectives and design. Soz Praventivmed. 2001;46:186–194. doi: 10.1007/BF01324255. [DOI] [PubMed] [Google Scholar]
- 48.Lichtenstein P, Sullivan P, Cnattingius S, Gatz M, Johansson S, Carlström C, et al. The Swedish twin registry in the third millennium—an update. Twin Res Hum Genet. 2006;9:875–882. doi: 10.1375/183242706779462444. [DOI] [PubMed] [Google Scholar]
- 49.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu YJ, Lin DY. Analysis of untyped SNPs: maximum likelihood and imputation methods. Genet Epidemiol. 2010;34:803–815. doi: 10.1002/gepi.20527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Gibbs RA, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y, Abecasis GR. Rapid haplotype reconstruction and missing genotype inference. Am J Hum Genet. 2006;S79:2290. [Google Scholar]
- 54.Weissman MM, Bland RC, Canino GJ, Faravelli C, Greenwald S, Hwu HG, et al. Cross-national epidemiology of major depression and bipolar disorder. JAMA. 1996;276:293–299. [PubMed] [Google Scholar]
- 55.Wittchen HU, Jacobi F. Size and burden of mental disorders in Europe–a critical review and appraisal of 27 studies. Eur Neuropsychopharmacol. 2005;15:357–376. doi: 10.1016/j.euroneuro.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 56.Levinson DF, Zubenko GS, Crowe RR, DePaulo RJ, Scheftner WS, Weissman MM, et al. Genetics of recurrent early-onset depression (GenRED): design and preliminary clinical characteristics of a repository sample for genetic linkage studies. Am J Med Genet B Neuropsychiatr Genet. 2003;119:118–130. doi: 10.1002/ajmg.b.20009. [DOI] [PubMed] [Google Scholar]
- 57.Weissman MM, Wolk S, Wickramaratne P, Goldstein RB, Adams P, Greenwald S, et al. Children with prepubertal-onset major depressive disorder and anxiety grown up. Arch Gen Psychiatry. 1999;56:794–801. doi: 10.1001/archpsyc.56.9.794. [DOI] [PubMed] [Google Scholar]
- 58.Sullivan PF, Kessler RC, Kendler KS. Latent class analysis of lifetime depressive symptoms in the National Comorbidity Survey. Am J Psychiatry. 1998;155:1398–1406. doi: 10.1176/ajp.155.10.1398. [DOI] [PubMed] [Google Scholar]
- 59.Sullivan PF, Prescott CA, Kendler KS. The subtypes of major depression: a latent class analysis. J Affective Disord. 2002;68:273–284. doi: 10.1016/s0165-0327(00)00364-5. [DOI] [PubMed] [Google Scholar]
- 60.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 61.de Bakker PI, Ferreira MA, Jia X, Neale BM, Raychaudhuri S, Voight BF. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 63.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, et al. A second generation human haplotype map of over 3. 1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elashoff M, Higgs BW, Yolken RH, Knable MB, Weis S, Webster MJ, et al. Meta-analysis of 12 genomic studies in bipolar disorder. J Mol Neurosci. 2007;31:221–243. doi: 10.1385/jmn:31:03:221. [DOI] [PubMed] [Google Scholar]
- 65.Lopez-Leon S, Janssens AC, Gonzalez-Zuloeta Ladd AM, Del-Favero J, Claes SJ, Oostra BA, et al. Meta-analyses of genetic studies on major depressive disorder. Mol Psychiatry. 2007;13:772–785. doi: 10.1038/sj.mp.4002088. [DOI] [PubMed] [Google Scholar]
- 66.Scott LJ, Muglia P, Kong XQ, Guan W, Flickinger M, Upmanyu R, et al. Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proc Natl Acad Sci USA. 2009;106:7501–7506. doi: 10.1073/pnas.0813386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McMahon FJ, Akula N, Schulze TG, Muglia P, Tozzi F, Detera-Wadleigh SD, et al. Meta-analysis of genome-wide association data identifies a risk locus for major mood disorders on 3p21. 1. Nat Genet. 2010;42:128–131. doi: 10.1038/ng.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Breen G, Lewis CM, Vassos E, Pergadia M, Blackwood D, Boomsma DI, et al. Replication of association of 3p21. 1 with susceptibility to bipolar disorder but not major depression. Nat Genet. 2011;43:3–5. doi: 10.1038/ng0111-3. [DOI] [PubMed] [Google Scholar]
- 69.International Schizophrenia Consortium. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilkinson MB, Dias C, Magida J, Mazei-Robison M, Lobo M, Kennedy P, et al. A novel role of the WNT-dishevelled-GSK3beta signaling cascade in the mouse nucleus accumbens in a social defeat model of depression. J Neurosci. 2011;31:9084–9092. doi: 10.1523/JNEUROSCI.0039-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cole SW, Hawkley LC, Arevalo JM, Sung CY, Rose RM, Cacioppo JT. Social regulation of gene expression in human leukocytes. Genome Biol. 2007;8:R189. doi: 10.1186/gb-2007-8-9-r189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sutton LP, Honardoust D, Mouyal J, Rajakumar N, Rushlow WJ. Activation of the canonical Wnt pathway by the antipsychotics haloperidol and clozapine involves dishevelled-3. J Neurochem. 2007;102:153–169. doi: 10.1111/j.1471-4159.2007.04527.x. [DOI] [PubMed] [Google Scholar]
- 74.Sutton LP, Rushlow WJ. The effects of neuropsychiatric drugs on glycogen synthase kinase-3 signaling. Neuroscience. 2011;199:116–124. doi: 10.1016/j.neuroscience.2011.09.056. [DOI] [PubMed] [Google Scholar]
- 75.Psychiatric GWAS Consortium Cross-Disorder Group. Genomewide analysis of five psychiatric disorders identifies loci with shared effects on psychopathology. (manuscript under review) [Google Scholar]
- 76.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 4. American Psychiatric Association; Washington DC: 1994. [Google Scholar]
- 77.World Health Organization. The ICD-10 Classification of Mental and Behavioural Disorders: Diagnostic Criteria for Research. World Health Organization; Geneva: 1993. [Google Scholar]
- 78.Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Houlston RS, Cheadle J, Dobbins SE, Tenesa A, Jones AM, Howarth K, et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13. 33. Nat Genet. 2010;42:973–977. doi: 10.1038/ng.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, et al. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- 82.Karg K, Burmeister M, Shedden K, Sen S. The serotonin transporter promoter variant (5-HTTLPR), stress, and depression meta-analysis revisited: evidence of genetic moderation. Arch Gen Psychiatry. 2011;68:444–454. doi: 10.1001/archgenpsychiatry.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Uher R, McGuffin P. The moderation by the serotonin transporter gene of environmental adversity in the etiology of depression: 2009 update. Mol Psychiatry. 2010;15:18–22. doi: 10.1038/mp.2009.123. [DOI] [PubMed] [Google Scholar]
- 84.Munafo MR, Durrant C, Lewis G, Flint J. Gene X environment interactions at the serotonin transporter locus. Biol Psychiatry. 2009;65:211–219. doi: 10.1016/j.biopsych.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 85.Risch N, Herrell R, Lehner T, Liang KY, Eaves L, Hoh J, et al. Interaction between the serotonin transporter gene (5-HTTLPR), stressful life events, and risk of depression: a meta-analysis. JAMA. 2009;301:2462–2471. doi: 10.1001/jama.2009.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fergusson DM, Horwood LJ, Miller AL, Kennedy MA. Life stress, 5-HTTLPR and mental disorder: findings from a 30-year longitudinal study. Br J Psychiatry. 2011;198:129–135. doi: 10.1192/bjp.bp.110.085993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang J, Wray NR, Visscher PM. Comparing apples and oranges: equating the power of case-control and quantitative trait association studies. Genet Epidemiol. 2010;34:254–257. doi: 10.1002/gepi.20456. [DOI] [PubMed] [Google Scholar]
- 88.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.