Abstract
Successful infection by enteric bacterial pathogens depends on the ability of the bacteria to colonise the gut, replicate in host tissues and disseminate to other hosts. Pathogens such as Salmonella, Shigella and enteropathogenic and enterohaemorrhagic E. coli (EPEC and EHEC), utilise a type III secretion system (T3SS) to deliver virulence effector proteins into host cells during infection that promote colonisation and interfere with antimicrobial host responses 1-3. Here we report that the T3SS effector NleB1 from EPEC binds to host cell death domain containing proteins and thereby inhibits death receptor signalling. Protein interaction studies identified FADD, TRADD and RIPK1 as binding partners of NleB1. NleB1 expressed ectopically or injected by the bacterial T3SS prevented Fas ligand or TNF-induced formation of the canonical death inducing signalling complex (DISC) and proteolytic activation of caspase-8, an essential step in death receptor induced apoptosis. This inhibition depended on the N-GlcNAc transferase activity of NleB1, which specifically modified Arg117 in the death domain of FADD. The importance of the death receptor apoptotic pathway to host defence was demonstrated using mice deficient in the FAS signalling pathway, which showed delayed clearance of the EPEC-like mouse pathogen Citrobacter rodentium and reversion to virulence of an nleB mutant. The activity of NleB suggests that EPEC and other attaching and effacing (A/E) pathogens antagonise death receptor induced apoptosis of infected cells, thereby blocking a major antimicrobial host response.
Keywords: EPEC, apoptosis, caspase-8, death domain
Members of the death receptor family such as FAS and TNFR1 contain an intracellular death domain (DD) and require caspase-8 for the activation of effector caspases and consequent cell death by the extrinsic apoptotic pathway 4, 5. Within seconds of engagement by Fas ligand (FasL), pre-associated FAS trimers undergo multimerisation and recruit the DD containing adaptor protein FADD via heterotypic DD interactions 5. FADD then recruits pro-caspase-8 forming the death-inducing signalling complex (DISC), whereupon caspase-8 undergoes conformational change, auto-proteolytic processing and release from the DISC to allow cleavage of cytosolic substrates, including pro-caspase-3 and Bid 5. Similarly, upon stimulation by TNF, TNFR1 recruits the adaptor protein TRADD and the receptor kinase, RIPK1, leading to NF-κB activation. Alternatively, cell death may be induced via TNFR1 internalisation and formation of a secondary complex that contains TRADD/RIPK1/FADD and caspase-8 6, 7.
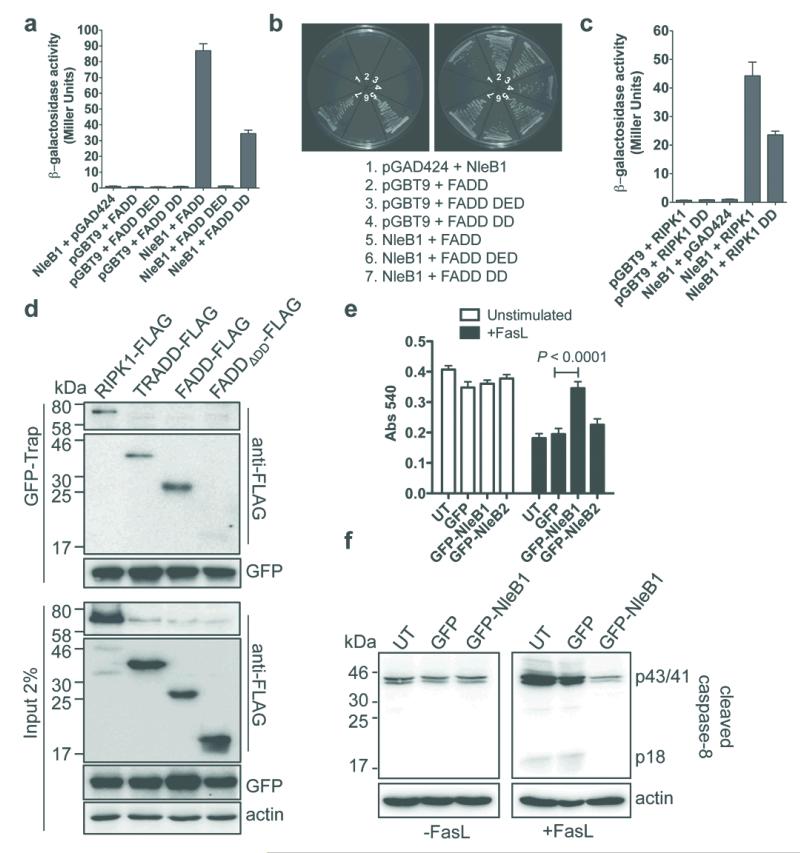
Previously, we reported that the EPEC T3SS effector NleB1 inhibited IκB degradation in cells upon stimulation with TNF but not IL-1β, suggesting that NleB1 interfered with death receptor signalling 8. To identify host cell binding partners of NleB1, we screened a yeast-2-hybrid cDNA library derived from HeLa cells using NleB1 as bait. FADD and RIPK1, were recovered multiple times. Transformation into a different yeast strain demonstrated that the interaction of NleB1 with FADD and RIPK1 occurred through their respective DD (Fig. 1a-c). Studies in transfected HEK293T cells showed that FADD-FLAG, TRADD-FLAG and RIPK1-FLAG co-purified with a GFP-NleB1 fusion protein as well as 2HA-tagged NleB1 translocated during EPEC E2348/69 infection (Fig. 1d and Extended Data Fig. 1a, b). The DD of FADD was required for binding to NleB1 from different A/E pathogens (Fig. 1d and Extended Data Fig. 1b). As TRADD and RIPK1 are critical components of the TNFR1 signalling complex, we investigated the influence of NleB1 on inhibition of the NF-κB dependent cytokine, IL-8, during EPEC infection. Unlike NleE, a known inhibitor of NF-κB activation 8, 9, neither NleB1 nor its close homologue NleB2 appeared to inhibit IL-8 production during EPEC infection (Extended Data Fig. 1c). Therefore we focussed our attention on FADD-dependent apoptosis signalling. After treatment with FasL, HeLa cells transfected with pGFP-NleB1 showed significantly higher levels of MTT reduction (mitochondrial activity) compared to pEGFP transfected cells (Fig. 1e), suggesting that NleB1 blocked cell death. Immunoblot analysis with an antibody that recognises only cleaved caspase-8, revealed that FasL-induced caspase-8 cleavage was substantially reduced in HeLa cells transfected with pGFP-NleB1 (Fig. 1f). By immunofluorescence microscopy (IF), fewer cells expressing GFP-NleB1 contained cleaved caspase-8 after treatment with FasL (Extended Data Fig. 1d, e). In contrast, a comparable number of cells contained cleaved caspase-3 in untransfected, GFP and GFP-NleB1 expressing cells treated with tunicamycin (Extended Data Fig. 1e). This indicated that NleB1 had no effect on activation of the intrinsic apoptotic pathway.
Figure 1. NleB1 binds death domain proteins and inhibits caspase-8 activation.
a, Yeast two-hybrid analysis of protein-protein interactions in S. cerevisiae PJ69-4A. Results are mean ± SEM β-galactosidase activity from at least three independent experiments performed in triplicate. b, Growth of S. cerevisiae PJ69-4A on medium to select for protein-protein interactions (left panel) or plasmid maintenance (right panel). Representative images from at least three independent experiments c, Yeast two-hybrid analysis of protein interactions in S. cerevisiae PJ69-4A. Results are mean ± SEM β-galactosidase activity from at least three independent experiments performed in triplicate. d, GFP-Trap® of GFP-NleB1 and detection of FADD-FLAG, TRADD-FLAG and RIPK1-FLAG in HEK293T cells. Actin; loading control. Representative immunoblot from at least three independent experiments e, MTT reduction in HeLa cells expressing GFP, GFP-NleB1 or GFP-NleB2. UT, untransfected. Results are the mean ± SEM of absorbance at 540 nm from three independent experiments performed in triplicate. *P < 0.0001, unpaired, two-tailed t-test. f, Cleaved caspase-8 in HeLa cells expressing GFP or GFP-NleB1. p43/41 and p18 are products of processed caspase-8. UT, untransfected. Representative immunoblot from at least three independent experiments.
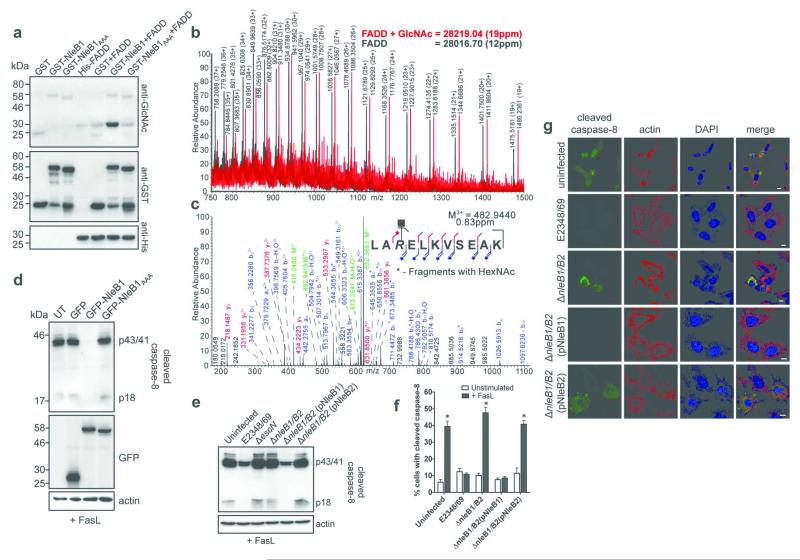
NleB from the murine A/E pathogen C. rodentium (CR) was recently described as a GlcNAc transferase and a member of the glycogenin family of enzymes 10. Given the ability of NleB1 to bind FADD and inhibit proteolytic activation of caspase-8, we examined whether FADD was post-translationally modified by NleB1. Following incubation with GST-NleB1 and UDP-GlcNAc, we observed GlcNAc modification of His-FADD (Fig. 2a). This modification was not present upon incubation with an NleB1 catalytic site mutant (NleB1AAA) 10. Similar modification of FADD-FLAG occurred upon ectopic expression of GFP-NleB1 in HeLa cells (Extended Data Fig. 1f). Intact protein LC-MS of His-FADD incubated with GST-NleB1 and UDP-GlcNAc revealed a mass shift matching a single GlcNAc modification on FADD (Fig. 2b). Peptide sequencing of multiple spectra from in-gel digests unambiguously identified Arg117 as the site of N-GlcNAcylation (Fig. 2c, Extended Data Fig. 2-3, Table S1). This was confirmed by substitution of Arg117 in FADD with alanine, whereas alanine substitution at Ser122 had no impact on NleB-mediated N-GlcNAcylation (Extended Data Fig. 4). Arg117 is located at the interface of the FAS-FADD DD interaction and is critical for assembly of the FAS-FADD oligomeric complex and formation of the DISC 11, 12. Accordingly, GFP-NleB1 but not catalytically inactive GFP-NleB1AAA inhibited caspase-8 activation (Fig. 2d).
Figure 2. Enzymatic activity of NleB1.
a, In vitro assay for NleB1 GlcNAc modification of FADD using recombinant proteins and 1 mM UDP-GlcNAc. Representative immunoblot from at least three independent experiments b, Intact protein mass spectrometry of FADD incubated with GST-NleB1 and UDP-GlcNAc. c, High resolution CID spectrum of the peptide corresponding to FADD115-125. *Diagnostic fragment ions that carry the GlcNAc modification. d, Cleaved caspase-8 in FasL-treated HeLa cells expressing GFP, GFP-NleB1 or GFP-NleB1AAA. UT, untransfected. Actin; loading control. Representative immunoblot from at least three independent experiments e, Cleaved caspase-8 in HeLa cells infected with derivatives of EPEC and treated with FasL. Representative immunoblot from at least three independent experiments f, Quantification of cleaved caspase-8 by immunofluorescence microscopy of HeLa cells infected with derivatives of EPEC and treated with FasL. Results are mean ± SEM percentage cells with cleaved caspase-8 from two independent experiments counting ~200 cells in triplicate. *P < 0.0001 compared to uninfected, unstimulated control, one-way ANOVA g, Immunofluorescence staining for detection of cleaved caspase-8 induced by FasL in HeLa cells infected with derivatives of EPEC. Scale bar 10 μm. Representative images from at least three independent experiments
During infection, NleB1 delivered by the EPEC T3SS inhibited FasL-induced caspase-8 activation in HeLa cells. This inhibition was lost in cells infected with an escN mutant, that has a non-functional T3SS, or an ΔnleB1/B2 double mutant (Fig. 2e-g). Complementation of the ΔnleB1/B2 mutant with nleB1 but not nleB2 restored the ability of EPEC to inhibit caspase-8 activation (Fig. 2e-g), demonstrating that the two NleB proteins have distinct functions. This mirrored our protein interaction studies, which showed that NleB2 did not interact with FADD or TRADD and bound only weakly to RIPK1 (Extended Data Fig. 5a-d).
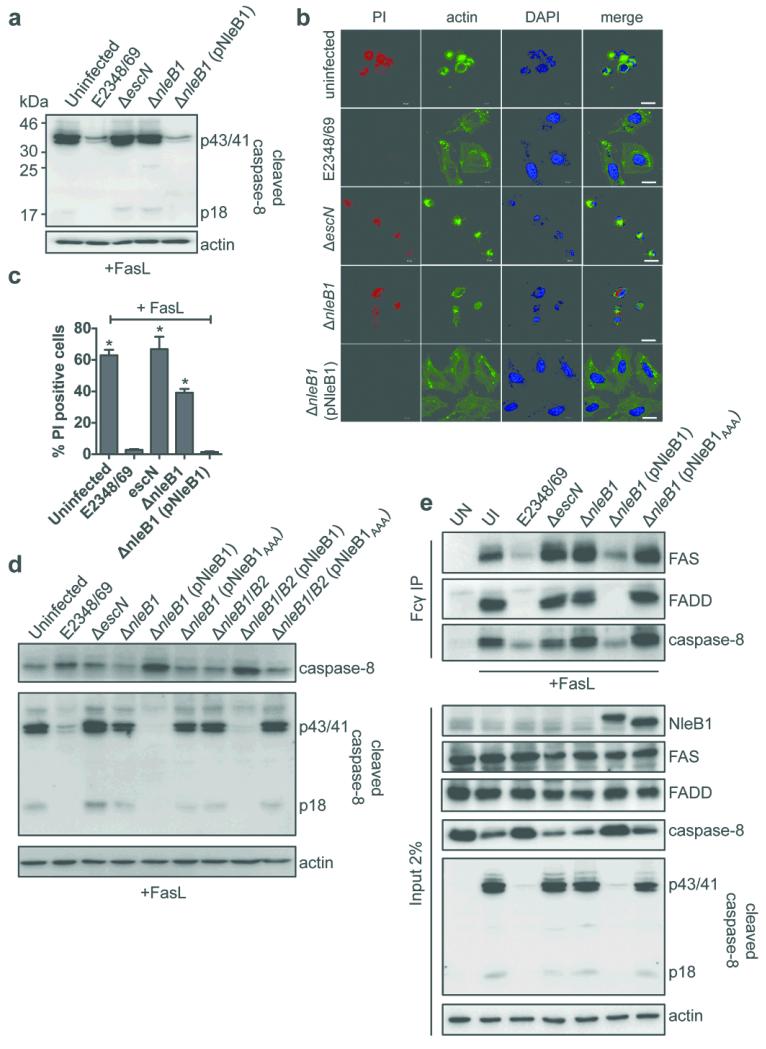
Arg117 in FADD is essential for FAS-FADD and TRADD-FADD DD interactions 11, 12. Consistent with the role of NleB1 in inhibition of FADD-dependent caspase-8 activation, an nleB1 mutant was unable to inhibit FasL- or TNF-induced activation of caspase-8 while the complemented strain was as effective as WT EPEC (Fig. 3a and Extended Data Fig. 5e, f). EPEC expressing NleB1 also inhibited FasL-induced apoptosis as measured by propidium iodide (PI) staining (Fig. 3b, c). These phenotypes were not due to differences in adherence (Extended Data Fig. 5g).
Figure 3. Inhibition of FasL-induced DISC formation and cell death by EPEC.
a, Cleaved caspase-8 in HeLa cells infected with derivatives of EPEC. Actin; loading control. Representative immunoblot from at least three independent experiments b, Cell death visualised by PI staining in HeLa cells infected with derivatives of EPEC and treated with FasL. Scale bar 20 μm. Representative images from at least three independent experiments c, Quantification of PI staining and microscopic analysis in HeLa cells infected with derivatives of EPEC and treated with FasL. Results are mean ± SEM percentage cells with PI staining from two independent experiments counting ~200 cells in triplicate. *P < 0.0001 compared to E2348/69 infected cells, one-way ANOVA. d, Cleaved and full-length caspase-8 in HeLa cells infected with derivatives of EPEC and treated with FasL. Actin; loading control. Representative immunoblot from at least three independent experiments e, DISC components induced by FasL coupled to Fcδ and precipitated with protein G beads. UN, untreated and uninfected; UI, uninfected. Actin; loading control. Representative immunoblot from at least three independent experiments.
NleB1 GlcNAc transferase activity was required for inhibition of FasL-induced caspase-8 processing during infection (Fig. 3d). To examine the impact of NleB1 on FAS DISC assembly, which precedes caspase-8 activation, we immunoprecipitated the FAS receptor complex from infected HeLa cells treated with FasL (Fig. 3e). Prior infection of cells with EPEC substantially inhibited FasL-induced DISC formation and this required NleB1 GlcNAc transferase activity (Fig. 3e).
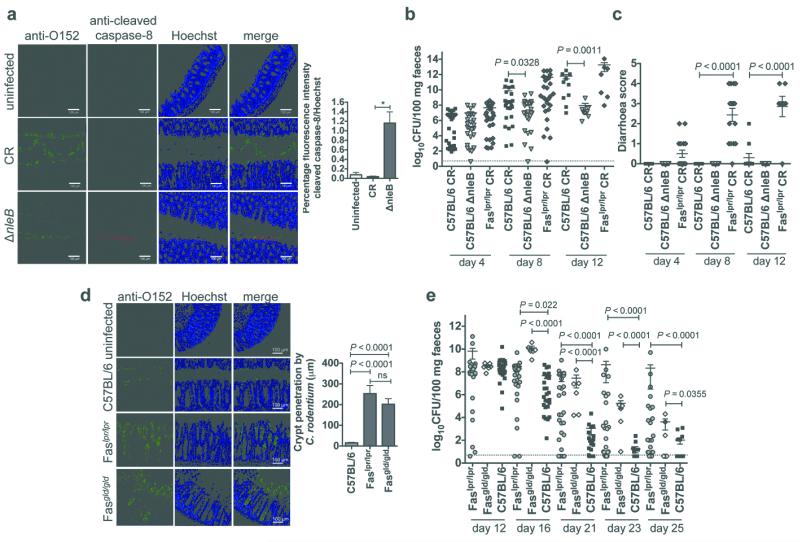
We next examined the effect of NleB on inhibition of caspase-8 processing in vivo by infecting WT C57BL/6 mice with CR or an nleB mutant of CR and staining tissue sections for cleaved caspase-8. While few cells were positive for cleaved caspase-8 during CR infection, significant numbers of cells were positive for cleaved caspase-8 during infection with the nleB mutant (Fig. 4a). Many of these caspase-8 positive cells were present as sloughed cells in the gut lumen (Extended Data Fig. 6a).
Figure 4. Infection of mice deficient for FAS signalling with CR.
a, Immunofluorescence staining and quantification of cleaved caspase-8 in colonic sections from C57BL/6 mice infected with CR or a CR nleB mutant. Representative images from at least three separate sections of colon at least 100 μm apart (transverse or longitudinal), per animal from five individual mice per group. Results are mean ± SEM percentage fluorescence intensity of cleaved caspase-8 staining relative to Hoechst staining of five fields with CR infection from three mice per group. *P = 0.0001, unpaired two tailed t-test. b, Bacterial load in the faeces during acute infection with CR. Each data point represents log10CFU/100 mg faeces per individual animal on days 4, 8 and 12 after infection. Mean ± SEM are indicated, dotted line represents detection limit. P values from Mann Whitney U. c, Diarrhoea score at day 4, 8 and 12 post-infection. Scoring system is described in the Supplementary Methods. Mean ± SEM are indicated. P values from one-way ANOVA. d, Immunofluorescence staining and quantification of CR penetration into intestinal crypts in colonic sections from C57BL/6, Faslpr/lpr and Fasgld/gld mice. Representative images from at least two separate sections of colon at least 100 μm apart (transverse or longitudinal), per animal from five individual mice per group. Results are the mean ± SEM maximum distance of CR staining from the epithelial surface (in μm) of five independent sections with at least three measurements per section. P = 0.0001, unpaired two tailed t-test. e, Bacterial load in the faeces during the resolving phase of CR infection. Each data point represents log10CFU/100 mg faeces per individual animal on days 12, 16, 21, 23 and 25 post-infection. Mean ± SEM are indicated, dotted line represents detection limit. P values from Mann Whitney U.
The observation that NleB1 inhibited FasL-induced caspase-8 cleavage suggested that FAS-mediated apoptosis contributes to host defence against A/E pathogens. We tested this by infecting FAS-deficient Faslpr/lpr mutant mice with CR13. As shown previously, nleB was essential for efficient colonisation of WT C57BL/6 mice 14 (Fig. 4b). Although there was no difference in faecal shedding of CR between C57BL/6 and Faslpr/lpr mice during the acute phase of infection (first 12 days) (Fig. 4b), Faslpr/lpr mice developed severe, watery diarrhoea (Fig. 4c). In addition, CR penetrated the crypts in Faslpr/lpr mice, unlike in C57BL/6 mice where the bacteria were predominantly located at the tips of the villi (Fig. 4d). To study the resolving phase of infection (post day 12), C57BL/6 and Faslpr/lpr mice were infected with a lower dose of CR to avoid the rapid onset of severe diarrhoea and monitored for up to 33 days. Faslpr/lpr mice were impaired in their ability to clear CR compared to C57BL/6 mice (Fig. 4e and Extended Data Fig. 6b).
The development of severe diarrhoea in CR-infected Faslpr/lpr mice was unexpected and sections of colon from mice infected with CR for 10 days were examined to identify differences in pathology between Faslpr/lpr mice and C57BL/6 mice. Histological analysis of colons from Faslpr/lpr mice revealed greater erosion of the epithelium (tissue damage) as well as increased colon weight and crypt height compared to C57BL/6 mice (measures of CR-induced intestinal hyperplasia 14) (Extended Data Fig. 7a-d). In addition, Faslpr/lpr mice showed more pronounced neutrophil infiltration, which penetrated across the muscularis mucosa (Extended Data Fig. 7e). Similar phenotypes were observed in FasL-deficient Fasgld/gld mice and Bid-deficient mice (Fig. 4d, e and Extended Data Fig. 8). Bid is not required for FasL-induced apoptosis in lymphoid cells, but is required for FasL-induced apoptosis in several other cell types 15, 16. This suggests that FAS signalling in epithelial cells rather than lymphoid cells limits CR-induced colitis.
We reasoned that if NleB interfered with FAS-mediated apoptosis signalling during infection, then CR nleB mutant bacteria would no longer be attenuated in mice that carry defects in this apoptosis pathway. Indeed, Faslpr/lpr mice infected with the nleB mutant showed comparable levels of bacterial colonisation and severity of diarrhoea compared to their littermates infected with CR (Extended Data Fig. 6b and 9a, b). The same was not true for espI or espF mutants of CR, which retained their known colonisation defects in Faslpr/lpr mice (Extended Data Fig. 9c, d) 17, 18.
During infection with A/E pathogens, the predominant site of NleB activity is in enterocytes where the translocation of bacterial effector proteins occurs 1. NleB-mediated inhibition of FasL/FAS-induced apoptosis may prolong host cell attachment and faecal shedding of the pathogen. Indeed NleB is associated with the transmission of CR amongst littermates 19 and is epidemiologically associated with outbreaks of HUS caused by EHEC 20, 21. Conversely, the reduced colonisation of C57BL/6 mice by CR nleB mutants and increased numbers of activated caspase-8 positive cells may reflect partial protection by the FAS apoptotic pathway through the elimination of infected cells 14. This is consistent with the prolonged shedding of CR in FAS-deficient animals compared to C57BL/6 mice. The increased intestinal pathology observed in CR-infected FAS-, FasL- and Bid-deficient mice has also revealed a function for the FAS apoptotic pathway in limiting the extent of colitis during gastrointestinal infection. This aggravation of disease may be mirrored in humans, as nucleotide polymorphisms in the FASLG gene, encoding FasL, have been associated with the development of inflammatory bowel disease 22. In summary, the discovery of a bacterial T3SS effector that specifically modifies DD containing proteins through a novel post translational modification and thereby inhibits death receptor mediated apoptosis has revealed a new virulence mechanism in bacterial infection.
METHODS
Bacterial strains, plasmids, yeast strains and growth conditions
The bacterial strains, yeast strains and plasmids used in this study are listed in Table S2. All PCR primers are listed in Table S3. Bacteria were grown at 37°C in Luria-Bertani (LB) medium or Dulbecco’s modified Eagle’s medium (DMEM) (GIBCO, NY) where indicated and supplemented with ampicillin (100 μg/mL), kanamycin (100 μg/mL), nalidixic acid (50 μg/mL) or chloramphenicol (25 μg/mL) when necessary. Yeast strains were grown at 30°C in YPD (yeast extract/peptone/dextrose) medium or yeast nitrogen minimal medium supplemented with 2% glucose and amino acids including histidine (20 μg/mL), methionine (20 μg/mL), tryptophan (20 μg/mL), adenine (20 μg/mL), uracil (20 μg/mL) and leucine (30 μg/mL) when necessary. For infection of HeLa cells, overnight cultures of EPEC grown in LB medium were subcultured 1:50 into DMEM and grown statically for 3-4 h at 37°C with 5% CO2.The optical density (A600) of the bacterial cultures was measured to standardize the inoculum before infection. Cultures were induced with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) for 30 min prior to infection.
Construction of NleB, TRADD, RIPK1 and FADD expression vectors
The plasmids and primers used in this study are listed in Table S2 and S3, respectively. The nleB1 gene from EPEC E2348/69 (GenBank Accession CAS10779), nleB1 from EHEC O157:H7 EDL933 (GenBank Accession NP_289553.1) and nleB from C. rodentium (GenBank Accession NC_013716.1) were amplified from genomic DNA by PCR using the primer pairs NleB1F/NleB1R, NleB1EHF/NleB1EHR or NleBCRF/NleBCRR respectively and ligated into EcoRI/BamHI digested pEGFP-C2 to generate N-terminal GFP fusions to NleB (GFP-NleB1, GFP-NleB1EHEC, GFP-NleBCR). The nleB2 gene was amplified from EPEC E2348/69 genomic DNA (GenBank Accession CAS08589) using primer pairs NleB2F/NleB2R and ligated into EcoRI/BamHI digested pEGFP-C2. To generate the complementing vectors, pNleB1 and pNleB2, nleB1 and nleB2 were amplified from EPEC E2348/69 genomic DNA by PCR using the primer pairs NleB1F/NleB1R and NleB2F/NleB2R and ligated into EcoRI/BamHI digested pTrc99A. To create pNleB1-2HA and pNleB2-2HA, nleB1 or nleB2 was amplified from EPEC E2348/69 genomic DNA by PCR using the primer pairs NleB1F/NleB1R-2HA and NleB2F/NleB2R-2HA, respectively, and ligated into EcoRI/BamHI digested pTrc99A. To generate GST tagged NleB, nleB1 was amplified from EPEC E2348/69 genomic DNA by PCR using the primer pairs NleB1F/NleB1PGEXR and ligated into EcoRI/SalI digested pGEX-4T-1.
TRADD-FLAG was kindly provided by Dr. Jürg Tschopp (University of Lausanne, Lausanne, Switzerland). FADD-FLAG and RIPK1-FLAG were provided by Dr. Ashley Mansell. To generate FADDΔDD-FLAG, the coding region for amino acids 1-96 of FADD was amplified by PCR from FADD-FLAG using the primer pairs FADDF/FADDΔDDR and ligated into EcoRI/BamHI digested p3XFLAG-Myc-CMV to generate an N-terminal 3xFLAG fusion to FADDaa1-96 (FADDΔDD-FLAG). To generate His-FADD, FADD was amplified from FADD-FLAG using the primer pairs FADDHISF/FADDHISR and ligated into BamHI/SalI digested pET28a.
To generate pGBT-NleB1, nleB1 was amplified from EPEC E2348/69 or EHEC O157:H7 EDL933 genomic DNA by PCR using primer pairs NleB1EPGBTF/NleB1R or NleB1EHGBTF/NleB1EHR (pGBT-NleB1 and pGBT-NleB1EH). The PCR products were digested with SmaI/EcoRI and EcoRI/BamHI, respectively and ligated into pGBT9. The plasmids pGAD-FADDDED and pGAD-FADDDD were generated from the template pGAD-FADD recovered from the cDNA library screen using primer pairs FADDGADF/FADDDEDR and FADDDDF/FADDGADR, respectively. The FADDDED and FADDDD PCR products were digested with EcoRI/HindIII and EcoRI/SalI, respectively, and ligated into pGAD424. The pGAD-RIPK1DD plasmid was generated using primers RIPK1DDF/RIPK1GADR using pGAD-RIPK1 recovered from the cDNA library screen as template DNA, and cloned via the EcoRI and BamHI restriction sites.
Construction of non-polar deletion mutants
C. rodentium strain ICC169 ΔnleB (ICC911) was constructed using the lambda Red-based mutagenesis system23. The nleB gene was replaced by a kanamycin resistance cassette following integration of a linear PCR product generated using the primers pair crNleB-pKD4-Fw/crNleB-pKD4-Rv, which contain 50 nucleotide homology extensions corresponding to regions in 5′ and 3′ of the nleB open reading frame and priming sequences for the kanamycin cassette of plasmid pKD4 (underlined)23. The PCR product was DpnI-digested to remove residual template, gel purified and electroporated into ICC169 carrying the temperature-sensitive lambda Red helper plasmid pKD46 following induction of Lambda Red recombinase with 10 mM L-arabinose at 30°C. Deletion mutant of C. rodentium nleB was confirmed by PCR and DNA sequencing and the growth rates of the mutant and parental strains were assessed in rich and minimal media. The C. rodentium strains ICC169 ΔespI (ICC179) and ICC169 ΔespF (ICC177) has been described previously 17. EspI binds to Sec24 and inhibits COPII-dependent vesicle trafficking while EspF is a multifunctional effector that has been associated with destabilisation of mitochondria, induction of apoptosis, activation of sorting nexin 9 and disruption of tight junctions1. These mutants were chosen as controls as they exhibit varying degrees of attenuation in the mouse model of CR infection and have functions unrelated to NleB.
The EPEC E2348/69 nleB1 mutant was created with the one step PCR lambda Red method using primers NleB1 E69FRTF155 and NleB1 E69FRTR156. pKD3 was used as template DNA to amplify the chloramphenicol cassette. The cassette was electroporated into WT EPEC E2348/69 and positive clones were selected on LB agar with 5 μg/mL chloramphenicol. The mutations were checked by PCR using primer pairs from outside and inside the mutation. The EPEC E2348/69 nleB2 mutant was created using primers NleB2 E69FRTF115 and NleB2 E69FRTR116. pKD4 was used as template DNA to amplify the kanamycin cassette. The cassette was electroporated into WT EPEC E2348/69 and positive clones selected for on LB agar with 50 μg/mL kanamycin. The mutations were checked by PCR with a combination of primers from outside and inside the mutation. The EPEC E2348/69 nleB1/nleB2 double mutant was generated using the one step PCR method by amplifying the nleB2 mutation from the nleB2 mutant with primers NleB2212 and NleB2213. The PCR product was transferred into the nleB1 mutant strain by electroporation. Mutants were first selected on LB agar plates with 20 μg/mL of kanamycin and then grown with chloramphenicol (25 μg/mL) and kanamycin (50 μg/mL) to select for nleB1 and nleB2 allelic replacement.
Site-directed mutagenesis
The His-FADDR117A and His-FADDS122A mutants, and the NleB1 mutants, pNleB1AAA, GFP-NleB1AAA and pGEX-NleB1AAA were generated using the Stratagene Quickchange II Site-Directed Mutagenesis Kit. FADDR117A and FADDS122A were generated by PCR using the primer pairs FADD(R117A)F/FADD(R117A)R and FADD(S122A)F/FADD(S122A)R, respectively with His-FADD as template DNA. For the NleB mutants, pNleB1, GFP-NleB1 or pGEX-NleB1 were used as template DNA and amplified by PCR using the primer pair pNleB1(AAA)F/pNleB1(AAA)R. Plasmids were digested with DpnI at 37°C overnight before subsequent transformation into the appropriate E. coli strain.
Yeast two-hybrid screening and β-galactosidase assay
NleB1 from EPEC strain E2348/69 (GenBank Accession CAS10779) and NleB1 from EHEC O157:H7 strain EDL933 (GenBank Accession NP_289553.1) were used as bait. The BD Matchmaker pre-transformed HeLa cDNA library (Clontech, CA, USA) was screened according to the manufacturer’s protocols (Clontech PT3183-1 manual) to identify HeLa proteins interacting with NleB. The yeast strain AH109 (MATα) was transformed with pGBT-NleB using the LiAc method and mated with Y187 (MATα) carrying the cDNA library in pGADT7 Rec plasmid. The mating mixtures were plated onto quadruple drop-out plates (Trp−, Leu−, Ade−, His−) to select for diploids expressing reporter genes. The pGADT7-Rec-cDNA plasmids were selectively rescued from those diploids with positive protein interactions into E. coli KC8. The pGADT7-Rec-cDNA plasmids were then sequenced using primer Rec744 to identify the cDNA inserts.
β-galactosidase assays were performed according to the manufacturer’s protocols (Clontech PT3024-1 manual). Briefly, pGADT7-Rec plasmid alone or with pGBT-NleB (or pGBT9, pGAD-FADD, pGAD-FADDDED, pGAD-FADDDD when necessary) were transformed into Saccharomyces cerevisiae strain PJ69-4A using the LiAc method. Transformants were selected on Trp− Leu− plates and grown to an optical density (A600) of 0.6 before lysis and assay for the level of β-galactosidase activity using ONPG as a substrate. Data are from at least three biological replicates performed in triplicate.
Bacterial adherence assay
Standard lines of HeLa and HEK293T cells are maintained in our laboratory and regularly tested for mycoplasma contamination. To assess the level of bacterial replication during infection and treatment with FasL, bacterial cultures grown in DMEM for were standardised by measurement of OD600 and plated on selective media for assessment by viable count. HeLa cell monolayers were infected in duplicate with the same cultures for 3 h before being incubated for 1.5 h in media supplemented with or without 20 ng/mL Fcγ-FasL. Following treatment, cells were washed three times and attached bacteria were resuspended in DMEM by pipetting and subsequently spread on selective media for assessment of bacterial viable count. Data are from at least three biological replicates performed in triplicate.
Immunoprecipitation and detection of HA-, GFP-, FLAG-tagged proteins
All immunoprecipitation experiments were performed as at least three biological replicates. For immunoprecipitation by GFP-Trap®-M (Chromotek, Germany), HEK293T cells were grown in 10 cm tissue culture dishes (Greiner Bio One) and co-transfected with GFP-NleB1 or GFP-NleB2 in combination with RIPK1-FLAG, TRADD-FLAG, FADD-FLAG or FADDΔDD-FLAG. Following transfection (after 18-24 h), lysis and immunoprecipitation were carried out according to instructions for immunoprecipitation of GFP-fusion proteins provided by the GFP-Trap®-M supplier.
For immunoprecipitation of 2 × hemagglutinin (HA) tagged proteins, HeLa cells cultured in T-75cm2 tissue culture flasks (Corning, USA) were first transfected with FADD-FLAG, RIPK1-FLAG or TRADD-FLAG expression constructs for 18 h followed by infection with various EPEC derivatives for 90-120 min. Cells were then washed three times with cold PBS and lysed in 1 mL cold lysis buffer (1% Triton X-100, 50 mM Tris-HCl pH 7.4, 1 mM EDTA, 150 mM NaCl and Complete Protease Inhibitor (Roche)) and incubated on ice for 10 min before collection. Cell debris was pelleted and equal volumes of supernatant collected. 100 μL of the supernatant was kept as input and 60 μL of monoclonal Anti-HA-agarose (Clone HA-7, Sigma-Aldrich) was added to the remainder of the supernatant which was incubated on a rotating wheel at 4°C overnight. The agarose was washed three times with lysis buffer and resuspended in 50 μL of 2× SDS-sample buffer.
For immunoprecipitation of Fas receptor complexes (DISC) by protein G, HeLa cells were grown in 10 cm tissue culture dishes and infected with various EPEC derivatives for 2 h. Cells were then treated with 50 μg/mL gentamycin and 1 μg of Fcγ-FasL for 1 h. An uninfected control was left untreated. Cells were lysed in 400 μL of 2 × DISC lysis buffer (20 mM Tris-HCl pH 7.4, 150 mM NaCl, 2 mM EDTA, 1% TritonX-100, 10% glycerol, and Complete Protease Inhibitor (Roche)) and incubated on ice for 10 min prior to centrifugation to remove cell debris. An aliquot (80 μL) of the supernatant was kept as input and 40 μL of Protein G Dynabeads (Invitrogen) was added to the remainder of the sample which was placed on a rotating wheel at 4°C overnight. Beads were washed three times with lysis buffer and finally resuspended in 75 μL of DISC lysis buffer with 25 μL of 5× SDS-sample buffer.
All samples were boiled for 5 min, subjected to SDS-PAGE and transferred to nitrocellulose membranes. Membranes were probed with the following primary antibodies where necessary: mouse monoclonal anti-HA.11 (16B12) (Covance, USA), mouse monoclonal anti-FLAG M2-HRP (Sigma-Aldrich), mouse monoclonal anti-FADD (BD Transduction Laboratories), rabbit monoclonal anti-Fas (C18C12) (Cell Signalling), rabbit polyclonal anti-caspase-8 (D35G2) (Cell Signalling), mouse monoclonal anti-GFP (7.1 and 13.1) (Roche) or mouse monoclonal anti-β-actin (AC-15) diluted 1:1000 in TBS with 5% BSA and 0.1% Tween. Detection and visualization were performed as previously mentioned.
Preparation of GST and His tagged proteins, demonstration of GlcNAcylation of recombinant FADD by NleB1 and detection by immunoblotting
Overnight cultures of BL21 (pGEX-4T-1), BL21 (pGEX-NleB1), BL21 (pGEX-NleB1AAA) and BL21 (pET-FADD), BL21 (pET-FADDR117A), BL21 (pET-FADDS122A) grown in LB were diluted 1:100 in 200 mL of LB supplemented with either kanamycin (pET) or ampicillin (pGEX) (100 mg/mL) with shaking to an optical density of (A600) 0.6 at 37°C. Cells were incubated with 1mM IPTG and grown for a further 2 h then pelleted by centrifugation. Proteins were purified by either nickel or glutathione affinity chromatography in accordance with the manufacturer’s instructions (Novagen, EMD4Biosciences, USA). Protein concentrations were determined using a BCA kit (Thermo Scientific).
For detection of GlcNAcylation of FADD by NleB1, 2 μg of purified recombinant proteins were incubated either alone or in combination at 37°C for 4 h in the presence of 1 mM UDP-GlcNAc. 5× SDS sample buffer was added to the samples before boiling for 5 min. Samples were subjected to SDS-PAGE and transferred to nitrocellulose membranes which were subsequently probed with the following primary antibodies: mouse monoclonal anti-GlcNAc (CTD110.6) (Cell Signalling), which recognises O-linked and N-linked GlcNAc24, rabbit polyclonal anti-GST (Cell, Signalling), or mouse monoclonal anti-His (AD1.1.10) (AbD Serotech) diluted 1:1000 in TBS with 5% BSA and 0.1% Tween. Detection and visualization were performed as previously mentioned. Detection of GlcNAcylated recombinant FADD was performed as three technical replicates.
For detection of GlcNAcylation during transient transfection with NleB1 and FADD, HEK293T cells were grown in 10 cm tissue culture dishes (Greiner Bio One) and co-transfected with GFP-NleB1 or GFP-NleB1AAA in combination with FADD-FLAG. Cells were washed three times with cold PBS and lysed in 600 μL of cold lysis buffer (1% Triton X-100, 50 mM Tris-HCl pH 7.4, 1 mM EDTA, 150 mM NaCl and Complete Protease Inhibitor (Roche)) and incubated on ice for 10 min before collection. Cell debris was pelleted and equal volumes of supernatant collected. 80 μL of the supernatant was kept as input and 40 μL of monoclonal anti-FLAG M2 magnetic beads (Sigma-Aldrich) was added to the remainder of the supernatant which was incubated on a rotating wheel at 4°C overnight. The beads were washed three times with cold lysis buffer followed by elution of bound proteins with 80 μg of FLAG peptide (Sigma-Aldrich) to which 16 μL of 5× SDS sample buffer was added. Samples were boiled for 5 min, subjected to SDS-PAGE and transferred to nitrocellulose membranes. Membranes were probed with the following primary antibodies where necessary: mouse monoclonal anti-O/N-GlcNAc (CTD110.6) (Cell Signalling), mouse monoclonal anti-FLAG M2-HRP (Sigma-Aldrich), mouse monoclonal anti-GFP (7.1 and 13.1) (Roche) or mouse monoclonal anti-β-actin (AC-15) diluted 1:1000 in TBS with 5% BSA and 0.1% Tween. Detection and visualization were performed as previously mentioned.
Glyco-site specific analysis of FADD
Intact protein analysis was performed by the Proteomics Laboratory at the Walter and Eliza Hall Institute (Fig. 2B and Supp Fig. S7). 10 μg of intact recombinant His-FADD, His-FADDR117A or HisFADDS122A incubated with GST-NleB1 and UDP-GlcNAc, was injected and separated by nano-flow reversed-phase liquid chromatography on a nano LC system (1200 series, Agilent, USA) using a custom packed C4 capillary column (15cm × 0.15mm I.D., packed with Resprosil 300Å C4 5μm resin, Dr Maisch GmbH, Germany) using a linear 45 min gradient from 5 to 100% buffer B at a flow rate of 1.2ul/min (A: 0.1% Formic acid in Milli-Q water B: 0.1% Formic acid, 80% acetonitrile, (Mallinckrodt Baker, New Jersey, USA) 20% Milli-Q water). The nano HPLC was coupled on-line to a Q-Exactive mass spectrometer equipped with a nano-electrospray ion source (ThermoFisher Scientific) set to acquire full scan and all-ion-fragmentation (AIF) sequentially. Mass accuracy of the intact protein was achieved by summing multiple lower resolution (35,000) full scan MS events. The intact mass and error calculation was facilitated by the open source program mMass.
Tryptic peptide analysis was performed by the Proteomics Laboratory at the Walter and Eliza Hall Institute (Fig. 2C, Supp Fig. S5). Reacted and unreacted mixtures of recombinant FADD and NleB1 were purified by SDS-PAGE and bands corresponding to FADD were excised and manual in-gel reduction, alkylation, and tryptic digestion was performed. Extracted peptides were injected and separated by nano flow reversed-phase liquid chromatography on a nano LC system (1200 series, Agilent, USA) using a nanoAcquity C18 150 mm × 0.15 mm I.D. column (Waters, USA) with a linear 45-min gradient from 5 to 100% buffer B set at a flow rate of 1.2 μl/min (A: 0.1% Formic acid in Milli-Q water, B: 0.1% Formic acid, 80% acetonitrile, (Mallinckrodt Baker, New Jersey, USA) 20% Milli-Q water). The nano HPLC was coupled on-line to an LTQ-Orbitrap XL mass spectrometer equipped with a nano-electrospray ion source (ThermoFisher Scientific) for automated MS/MS using multi-stage activation (MSA) setting. The Orbitrap was run in a data-dependent acquisition mode with the Orbitrap resolution set at 30,000 and the top-three multiply charged species selected for fragmentation in the linear ion trap by collision-induced dissociation (CID) (single charged species were ignored). Fragment ions where analysed on the orbitrap with the resolution set at 7, 500 with the ion threshold set to 15,000 counts. The activation time was set to 30 ms, normalized collision energy set to 35. Raw files consisting of full-scan MS and high resolution MS/MS spectra were converted to the MGF data format with Proteome Discoverer 1.4 and searched against UniProt database (2013/02) limiting the search to mouse taxonomy and custom FASTA database containing the protein constructs of FADD and NleB using Mascot v2.4. Mascot parameters for each search included Trypsin/no P enzyme with six missed cleavages, a fixed modification in the form of carbamidomethyl at Cys residues and variable modification of HexNAc at Ser/Thr/Asn/Arg residues, Acetyl at protein N-Terminal, oxidation at Met residues. Spectra were searched with a mass tolerance of 20 ppm in MS mode and 40 mmu in MS/MS mode. The MS/MS fragmentation of modified peptides obtained from the Mascot search was manually sequenced using mMass to assign ion peaks and confirm the site of modification. Detection of GlcNAcylated recombinant FADD by mass spectrometry was performed as two technical replicates.
The site of modification was verified independently using ETD fragmentation on an Orbitrap Elite mass spectrometer by the Bio21 Mass Spectrometry and Proteomics Facility at the University of Melbourne (Extended Data Fig. 3). The protein complex was digested with Endoproteinase AspN (Roche) at enzyme to protein ratio of 1:5 at 37°C overnight. Reaction contained FADD, NleB1 and UDP-GlcNAc and the incubation was performed as described earlier for anti-GlcNAc immunoblot analysis. LC-MS/MS was carried out on a LTQ Orbitrap Elite (Thermo Scientific) with an EASY nano electrospray interface coupled to an Ultimate 3000 RSLC nanosystem (Dionex). The nanoLC system was equipped with a Acclaim Pepmap nano-trap column (Dionex – C18, 100 Å, 75 μm × 2 cm) and a Thermo EASY-Spray column (Pepmap RSLC C18, 2μm, 100 Å, 75 μm × 25 cm). 4 μl of the peptide mix was loaded onto the enrichment (trap) column at an isocratic flow of 4 μl/min of 3% CH3CN containing 0.1% formic acid for 5 min before the enrichment column is switched in-line with the analytical column. The eluents used for the liquid chromatography were 0.1% (v/v) formic acid (solvent A) and 100% CH3CN/0.1% formic acid (v/v). 0.1% formic acid (v/v). The flow following gradient was used: 3% B to 12% B for 1 min, 12% B to 35% B in 20 min, 35% B to 80% B in 2 min and maintained at 80% B for 2 min and equilibration at 3% B for 7 min before the next sample injection. The LTQ Orbitrap Elite mass spectrometer was operated in the data dependent mode with nano ESI spray voltage of +2.0 kv, capillary temperature of 250°C and S-lens RF value of 60%. A sequential mode whereby spectra were acquired first in positive mode with full scan scanning from m/z 300-1650 in the FT mode at 120,000 resolution followed by Collision induced dissociation (CID), Electron transfer dissociation (ETD) and High energy collision dissociation (HCD). CID in the linear ion trap was carried out in parallel with three most intense peptide ions with charge states ≥2 isolated and fragmented using normalized collision energy of 35 and activation Q of 0.25. The decision tree procedure for ETD activation was also applied whereby peptides with charge state 3 of < 650 m/z, charge state 4 of < 900 m/z and charge state 5 of < 1000 m/z were subjected to 100 ms of ETD activation. HCD at 15,000 resolution using normalized collision energy of 35 and 0.1 ms activation time was carried out on every precursor selected. The mass spectra were searched against the Uniprot database using the Sequest HT (V1.3) search algorithm as part of the Proteome Discoverer 1.4 Workflow (Thermo Scientific). Searching parameters used were: variable modifications (HexNAc of N, S, T and R; 203.079), 2 missed tryptic cleavages, 10 ppm peptide mass tolerance and 0.6 Da fragment ion mass tolerance. The false-discovery rate (derived from corresponding decoy database search) was less than 1%. Peptides of interest were manually inspected and validated.
IL-8 secretion assay
For analysis of IL-8 secretion, HeLa cell monolayers were infected for 3 h before being incubated for 8-12 h in media supplemented with 50 μg/mL gentamycin with or without 20 ng/mL TNF (Calbiochem, EMD4Biosciences, USA). Following this, the HeLa cell supernatant was collected and either used immediately or stored at −20°C for subsequent analysis of IL-8 secretion. IL-8 secretion was measured using the Quantikine Human IL-8 Immunoassay (R&D Systems, MN) according to the manufacturer’s instructions. Samples were measured on a FLUOstar Omega microplate reader (BMG Labtech, Germany). Differences in IL-8 secretion were assessed for significance by one-way analysis of variance (ANOVA) with Dunnett’s Multiple Comparison post-test. Data are from at least three biological replicates performed in triplicate.
Detection of cleaved caspase-8 by immunoblotting
For detection of cleaved caspase-8 during EPEC infection, HeLa cells were grown in 24 well tissue culture plates (Greiner Bio One, Germany) and infected for 3 h with various EPEC derivatives. Cells were then treated with 50 ug/ml gentamicin, and either 20 ng/ml of Fcγ-FasL, 20 ng/ml TNF, or a combination of 20 ng/ml TNF and 10 ug/ml cycloheximide (CHX) for a further 60 min and lysed in 60 ul of 2× SDS-sample buffer. For detection of cleaved caspase-8 by transfection, HeLa cells were grown in 24 well tissue culture plates and either left untransfected or transfected with GFP, GFP-NleB1 or GFP-NleB1AAA for 18-24 h using FuGENE® 6 (Roche, Germany). Cells were then treated with 20 ng/mL of Fcγ-FasL for a further 60 min and collected by lysis in 60 μL of 2×SDS-sample buffer. All samples were boiled for 5 min and subjected to SDS-PAGE followed by transfer to nitrocellulose membranes (PALL, USA). Membranes were incubated at least overnight at 4°C with rabbit polyclonal anti-cleaved caspase-8 (Asp391) (18C8) (Cell Signaling) diluted 1:1000 in TBS with 5% skim milk and 0.1% Tween (Sigma-Aldrich, MO, USA), rabbit polyclonal anti-caspase-8 (D35G2) (Cell Signaling) diluted 1:1000 or mouse monoclonal anti-β-actin (AC-15) (Sigma-Aldrich) diluted 1:5000 in TBS with 5% BSA (Sigma-Aldrich) and 0.1% Tween. Proteins were detected using anti-rabbit or anti-mouse IgG secondary antibodies conjugated to horseradish peroxidase (PerkinElmer, USA) and developed with either ECL Western blotting reagent (Amersham, USA) or Western Lightning Ultra reagent (PerkinElmer). All secondary antibodies were diluted 1:3000 in TBS with 5% BSA and 0.1% Tween. Images were visualized using an MFChemiBis imaging station (DNR, Isreal). At least three biological replicates were performed for all experiments.
Detection of cleaved caspase-8 and -3 in vitro by immunofluorescence microscopy
For visualization of caspase-8 cleavage by immunofluorescence microscopy, HeLa cells were either infected with EPEC E2348/69 derivatives for 3 h or transfected for 16 h followed by an additional 1 h of treatment with 20 ng/mL Fcγ-FasL. One experiment used recombinant rhFasL to treat transfected HeLa cells for comparison with tunicamycin treatment (Extended Data Fig. 1d, e). For this, HeLa cells were transfected for 16 h and treated with 200 ng/mL of rhFasL (R&D Systems, Minneapolis, USA). For visualization of caspase-3 cleavage induced by tunicamycin, HeLa cells were transfected for 16 h, followed by 18 h treatment with 5 μg/mL tunicamycin (Sigma-Aldrich). Cells were then fixed in 3.7% (wt/vol) formaldehyde (Sigma-Aldrich) in PBS for 10 min and permeabilized with 0.2% Triton (Sigma-Aldrich) for 4 min. Following 30 min blocking in PBS with 3% (wt/vol) BSA, samples were exposed to primary antibodies (specific to cleaved caspase-8 or cleaved caspase-3; Cell Signaling Technology) diluted in blocking solution for 1 h at 20°C, except for cleaved caspase-3, which was incubated overnight at 4°C. For fluorescent actin staining in EPEC infected cells, 0.5 μg/mL of phalloidin conjugated to rhodamine (Sigma-Aldrich) was added during primary antibody incubation. 4′, 6-diamidino-2-phenylindole (DAPI; Invitrogen) was applied at 0.5 μg/mL in blocking solution for 5 min at 20°C post secondary antibody treatment. Secondary antibodies were all coupled to Alexa Fluor® dyes (Invitrogen, USA) applied to cells at 1:2000 in blocking solution for 1 h at 20°C. Coverslips were mounted onto microscope slides with Prolong Gold anti-fade reagent (Invitrogen). Images were acquired using a Zeiss confocal laser scanning microscope with a 100×/EC Epiplan-Apochromat oil immersion objective. Cleavage of caspase-8 or caspase-3 was quantified from at least three biological replicates performed in duplicate for both transfection and infection studies counting at least 100 cells per field.
Cell viability assays (MTT and propidium iodide staining)
HeLa cells were transfected in 24 well tissue culture plates for 18-24 h before being left untreated or treated with 20 ng/mL of Fcγ-FasL for a further 60 min. The cells were washed once with PBS and replaced with DMEM containing 0.1 μg/ml 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (Sigma) for 1 h, after which the medium was removed and 100 μL of dimethyl-sulfoxide (DMSO; Sigma) was added to each well. After thorough mixing on an orbital shaker for 1 min, the absorbance at 540 nm for each well was obtained using a FLUOstar Omega microplate reader (BMG Labtech, Germany). Results were obtained from at least three biological replicates performed in triplicate.
For analysis of cell viability by PI staining, HeLa cell monolayers were infected for 3 h before being incubated for 1 h in media supplemented with 50 μg/mL gentamycin with 20 ng/mL Fcγ-FasL. Propidium iodide (50 μg/ml) was added for the final 15 min of treatment. Cells were fixed and stained for confocal microscopy as previously mentioned. Phalloidin conjugated to FITC (Sigma-Aldrich) was used at 0.5 μg/mL to stain for actin. Duplicate coverslips were counted for PI positive cells, and results were obtained from at least three biological replicates counting at least 100 cells per field.
Animal experiments
All animal experimentation was approved by the University of Melbourne Animal Ethics Committee, applications 0808209 and 1112062. All genetically modified and spontaneous mutant mice are on a C57BL/6 background. Overnight cultures of bacterial strains were pelleted by centrifugation and resuspended in PBS. C57BL/6 (WT), Faslpr/lpr, Fasgld/gld and Bid−/− mutant mice (6 to 8 weeks old, male and female mice) were inoculated by oral gavage with 200 μL containing approximately 1 × 109 CFU of C. rodentium, or a lower dose of 1 × 108 CFU when testing for in prolonged infection (up to 33 days). The viable count of the inoculum was determined retrospectively by plating dilutions of the inoculum on plates with appropriate antibiotics. Animals were allocated to experimental groups to ensure even distribution of age, sex and weight. Investigators were not blinded to group allocation. Sample size was at least five animals per group for statistical power per biological replicate. Larger sample sizes (up to 12 mice per group) were used where mice were available. Infection experiments were repeated at least twice regardless of sample size to ensure reproducibility and results represent all experimental data combined from repeated infections. Mice were weighed every 2 days and faeces collected every 2 or 4 days for enumeration of CFU. The viable bacterial count per 100 mg of faeces was determined by plating serial dilutions in duplicate of faeces onto medium containing selective antibiotics.
Histological analysis and scoring of tissue pathology and diarrhoea
The increased intestinal pathology observed in CR-infected FAS-, FasL- and Bid-deficient mice revealed a function for the FAS apoptotic pathway in limiting the extent of colitis during gastrointestinal infection. This exacerbated disease compared to CR infection of WT mice likely arose from the difference between a complete block of FAS signalling in the gut epithelium of FAS-deficient mice as opposed to less complete inhibition of FAS signalling in the gut epithelium by NleB in WT mice, which depends on the timing and efficiency of CR attachment and NleB translocation. A grading system for diarrhoea was developed where a score of diarrhoea severity from 0-4 was recorded every 4 days. The scoring system can be interpreted as follows; 0 (no diarrhoea): solid stool with no sign of soiling around the anus. The stool is very firm when subjected to pressure with tweezers, 1 (very mild diarrhoea): formed stools that appear moist on the outside, and no sign of soiling around the anus. Stool is less firm when considerable pressure applied with tweezers, 2 (mild diarrhoea): formed stools that appear moist on the outside, and some signs of soiling around anus. Stools will easily submit to pressure applied with tweezers, 3 (diarrhoea): no formed stools with a mucous-like appearance. Considerable soiling around the anus and the fur around tail. Mouse takes a long time to pass stool, 4 (severe, watery diarrhoea): mostly clear or mucous-like liquid stool with very minimal solid present and considerable soiling around anus. Mouse may not be able to pass stool at all and may have a hunched appearance. Occasionally, blood may be observed in the stool.
For histological analysis, mice inoculated with the higher dose of C. rodentium (1 × 109 CFU) were culled at day 10 or 12 post-infection and the distal portion of colon from the cecum to the rectum removed aseptically and weighed after removal of faecal pellets. Colons and spleens were homogenized mechanically in PBS using a Seward 80 stomacher, and the bacterial CFU per 100 mg of organ homogenate was enumerated by plating onto LB containing the appropriate antibiotics. Colons from a representative group of these mice were collected and fixed in 4% (wt/vol) paraformaldehyde (Sigma-Aldrich) and sectioned for H&E staining and assessment of gut pathology. A scoring system (0-3) was used by a veterinary pathologist to assess the extent of tissue damage: 0= no damage, 1= discrete lesion, 2= mucosal erosion, 3=extensive mucosal damage/ulceration (extending into muscularis and deeper).
Detection of C. rodentium and cleaved caspase-8 in mouse colon tissue by immunofluorescence microscopy
For immunofluorescence staining of mouse colons, the distal portion of colon from the caecum to the rectum was removed from representative groups of infected and uninfected C57BL/6 (WT) and Faslpr/lpr mice, gently flushed with a 1:1 mix of OCT™ compound (Tissue-Tek®) and PBS and frozen at −80°C in full strength OCT™ compound. 10 μm sections were cut on a Leica CM3050S cryostat and mounted onto Menzel-Gläser Superfrost® Plus slides (Thermo Fisher Scientific). Slide sections were dried for at least 2 h before fixing in absolute acetone at −20°C, followed by a 30 min block with Dako Protein-Free Block Serum (Dako). Sections were incubated with primary antibody; polyclonal rabbit anti-C. rodentium O152 (Statens Serum Institute, Copenhagen) diluted 1:100 or mouse specific monoclonal rabbit anti-cleaved caspase-8 (Asp387) (D5B2) XP® diluted 1:500 in 2.5% normal donkey serum (NDS) (Jackson Immunoresearch Laboratories Inc.) in PBS. Secondary antibody coupled to Alexa Fluor®-488 was applied at 1:1000 in 2.5% NDS followed by 2 min of exposure to Hoechst 33258 (Invitrogen) diluted at 1:2000 in 2.5% NDS. Slides were mounted with glass coverslips using Prolong Gold anti-fade reagent (Invitrogen) and images were acquired using a Zeiss LSM700 inverted Axio Observer with LED laser lines at 405 nm and 488 nm, and a Plan-Apochromat 20×/0.8 objective. To determine the average depth of penetration by C. rodentium in mouse intestinal crypts, Image J was used to measure the distance of bacterial staining from the epithelial surface on immunofluorescent images. Results are from five independent sections with at least three measurements taken from each section. To quantify the amount of cleaved caspase-8 in infected mouse colons, the relative fluorescence intensity of cleaved caspase-8 staining was expressed as a percentage of the relative fluorescence intensity of Hoechst staining from 5 independent infected fields of 3 mice for each bacterial strain.
Hyperplasia of colonic crypts (crypt height) was measured using MIRAX software on H&E stained sections at day 10 and 12 post-infection. Data are from 4 sections of distal colon measured at least 50 μm apart per animal from at least three individual mice per group.
Statistical analysis
All statistical analyses were performed using GraphPad Prism version 6.0. Statistical tests used were unpaired two-tailed Student’s t-test for pairwise comparisons between groups or One-way ANOVA with Dunnett’s Multiple Comparison Test for multiple comparisons where indicated. Variance was similar in all comparisons. Differences in faecal counts of CR from mice and diarrhoea scores were assessed using a Mann Whitney U test, where normal distribution was not assumed. P < 0.05 was considered to be significant.
Supplementary Material
Extended Data Figure 1. NleB1 binds FADD, TRADD and RIPK1 and blocks extrinsic apoptosis. a, Co-immunoprecipitation of bacterially delivered NleB1-2HA with FADD-FLAG, TRADD-FLAG and RIPK1-FLAG in HeLa cells. Lane 1. HeLa cells infected with EPEC ΔnleB1 (pNleB1) (negative control). Lane 2, HeLa cells infected with EPEC ΔnleB1 (pNleB1-2HA). Actin; loading control. Representative immunoblot from at least three independent experiments b, GFP-Trap pull-down of FADD-FLAG or FADDΔDD-FLAG. NleB1 from EPEC E2348/69, EHEC O157:H7 EDL933 and C. rodentium (CR) ICC169 was used as indicated. Actin; loading control. Representative immunoblot from at least three independent experiments c, HeLa cells were infected with derivatives of EPEC as indicated for 4 h and left unstimulated (light grey bars) or stimulated with TNF for 8 h (dark grey bars). Results are the mean ± SEM of at least three independent experiments carried out in duplicate. *significantly greater than uninfected, unstimulated cells, P< 0.0001, one-way ANOVA with Dunnett’s Multiple Comparison Test. d, Apoptosis was induced by FasL and HeLa cells were stained with antibodies to cleaved caspase-8. UT, untransfected; Arrows indicate cells with cleaved caspase-8. Scale bar 5 μm. Representative images from at least three independent experiments e, Quantification of cleaved caspase-8 or cleaved caspase-3 by immunofluorescence microscopy in HeLa cells expressing GFP or GFP-NleB1. UT, untransfected; Results are the mean ± SEM percentage of cells with cleaved caspase-8 or cleaved caspase-3 from at least three independent experiments counting ~200 cells in triplicate. *P < 0.0001, unpaired, two-tailed t-test f, GlcNAcylation of FADD-FLAG detected using a monoclonal antibody to GlcNAc of immunoprecipitated lysate from HEK293T cells. Actin; loading control. Representative immunoblot from at least three independent experiments.
Extended Data Figure 4 GlcNAc modification of FADD by GST-NleB1. a, Intact protein mass spectrometry of FADD with R117A substitution incubated with GST-NleB1 and UDP-reveals no GlcNAc modification b, Intact protein mass spectrometry of FADD with S117A substitution incubated with GST-NleB1 and UDP-GlcNAc reveals a single GlcNAc modification c, In vitro assay for NleB1 GlcNAc modification of mutated forms of FADD as indicated. Representative immunoblot from at least three independent experiments.
Extended Data Figure 5. Comparison of FasL and TNF-mediated caspase-8 processing and analysis of NleB2 a, GFP-Trap pull-down of FADD-FLAG, TRADD-FLAG or RIPK1-FLAG with GFP-NleB2 in HEK293T cells. Actin; loading control. Representative immunoblot from at least three independent experiments b-d, Co-immunoprecipitation of bacterially delivered NleB2-2HA with FADD-FLAG, TRADD-FLAG and RIPK1-FLAG in HeLa cells using HA antibodies. Lane 1. HeLa cells infected with EPEC ΔnleB2 (pNleB2) (negative control). Lane 2, HeLa cells infected with EPEC ΔnleB2 (pNleB2-2HA). Actin; loading control. Representative immunoblots from at least three independent experiments e, Unprocessed and cleaved caspase-8 in HeLa cells infected for 3 h with derivatives of EPEC and left untreated or treated with Fcγ-FasL or TNF for 1 h as indicated. Representative immunoblot from at least three independent experiments f, Unprocessed and cleaved caspase-8 in HeLa cells infected for 45 min with derivatives of EPEC and left untreated or treated with TNF and CHX for 1 h as indicated. Actin; loading control. Representative immunoblot from at least three independent experiments g, Adherence of EPEC to HeLa cells. HeLa cells were infected with derivatives of EPEC for 3 h followed by treatment with FasL for 1 h. Number of recovered bacteria and the inoculum are shown for comparison. Mean ± SEM are indicated. Data are from three independent experiments performed in triplicate.
Extended Data Figure 6. Infection of C57BL/6 and Faslpr/lpr mice with CR. Immunofluorescence staining of cleaved caspase-8 in different colonic sections from C57BL/6 mice infected with wild type CR and an nleB mutant as indicated, using antibodies to CR O-antigen (anti-O152, green) and cleaved caspase-8 (red). Intestinal tissue was visualised with Hoechst staining for DNA (blue). Arrows indicate cleaved caspase-8 positive cells sloughed into the gut lumen. Scale bar 100 μm Representative images from at least three separate sections of colon at least 100 μm apart (transverse or longitudinal), per animal from five individual mice per group. b, Time course of infection of C57BL/6 and Fas lpr mutant mice with CR. Each data point represents log10CFU/100 mg faeces per individual animal on the indicated days. Mean ± SEM are indicated, dotted line represents detection limit. CR, mice infected with wild type CR; ΔnleB mice infected with CR nleB mutant. *P < 0.05, **P < 0.01, ***P < 0.001. P values from Mann Whitney U. Dotted line represents the detection limit.
Extended Data Figure 7. Histological analysis of intestinal sections at day 10 from FAS-deficient mice infected with C. rodentium a, Bacterial load in the colon of mice infected with C. rodentium (CR). Each data point represents log10CFU/100 mg colon per individual animal on day 10 post-infection. CR, C57BL/6 (WT) mice infected with wild type CR; ΔnleB, C57BL/6 (WT) mice infected with CR nleB mutant; Faslpr/lpr CR, Faslpr/lpr mice infected with wild type CR, dotted line is detection limit. P = 0.0002, Mann Whitney U. Mean ± SEM are indicated b, Resected colon weights (between rectum and caecum) of individual animals on day 10 post-infection. CR, C57BL/6 (WT) mice infected with wild type CR; ΔnleB, C57BL/6 (WT) mice infected with CR nleB mutant; Faslpr/lpr CR, Faslpr/lpr mice infected with wild type CR. P = 0.0164, Mann Whitney U. Mean ± SEM are indicated. c, Mean ± SEM crypt height in μm in hematoxylin and eosin stained sections from C57BL/6 and Faslpr/lpr mice infected with wild type CR. UI, uninfected. Data are from 4 sections of colon measured at least 50 μm apart per animal from at least three individual mice per group d, Mean ± SEM tissue damage score in colon sections for individual mice infected with wild type CR. Scoring system is described in the Materials and Methods. UI, uninfected. P < 0.05, Mann Whitney U e, Hematoxylin and eosin stained sections of colon from C57BL/6 (WT) or Faslpr/lpr mice uninfected or infected with wild type CR (day 10 post-infection). UI, uninfected. Scale bar 100 μm a. mucosal inflammation b. neutrophil invasion of the muscularis mucosa c. erosion of the epithelium. Representative images from two sections of colon at least 100 μm apart (transverse or longitudinal) from at least three individual mice per group.
Extended Data Figure 8. Diarrhoea score and histological analysis of intestinal sections at day 12 from C57BL/6 (WT), Faslpr/lpr, Fasgld/gld and Bid−/− mice infected with CR. a, Bacterial load in the faeces of different mouse strains infected with CR as indicated. Each data point represents log10CFU/100 mg colon per individual animal. Dotted line is detection limit. Mean ± SEM are indicated b, Diarrhoea score at day 12 post-infection of different strains of mice infected with CR. Scoring system is described in the Methods. Mean ± SEM are indicated. P values from one-way ANOVA. c, Mean ± SEM crypt height in μm in haematoxylin and eosin stained sections from different mouse strains infected with wild type CR. UI, uninfected Data are from 4 sections of colon measured at least 50 μm apart per animal from at least three individual mice per group d, Haematoxylin and eosin stained sections of colon from different mouse strains infected with wild type CR. Scale bar 100 μm. Representative images from two sections of colon at least 100 μm apart (transverse or longitudinal) from at least three individual mice per group.
Extended Data Figure 9. Bacterial load in the faeces of Faslpr/lpr mice infected with wild type CR, an espI mutant and an espF mutant. a, Bacterial load in the faeces of mice during infection with derivatives of CR. Each data point represents log10CFU/100 mg faeces or colon as indicated per individual animal on days 4, 8, 10 and 12 post-infection. Mean ± SEM are indicated, dotted line represents detection limit. b, Diarrhoea score at day 4, 8, 10 and 12 post-infection. Scoring system is described in the Methods. Mean ± SEM are indicated. P values from one-way ANOVA c-d, Bacterial load in the faeces of mice infected with derivatives of CR. Each data point represents log10CFU/100 mg faeces per individual animal on day 8 post-infection. Mean ± SEM are indicated, dotted line represents detection limit. P values from Mann Whitney U.
Extended Data Figure 2. Mass Spectra of peptides identifying Arg117 GlcNAc modification. Fragment ions of overlapping tryptic missed cleaved peptides confirm Arg117 of FADD as the modification site.
Extended Data Figure 3 Mass spectra obtained using ETD fragmentation on the Orbitrap Elite mass spectrometer. a, Red and blue spectra represent the c and z series ions that matches the peptide sequence DWKRLAR-HexNAc-ELKVSEAKM (addition of 203.0793 Da) produced from an AspN digest of FADD incubated with NleB1. Insert box shows the precursor ion present as a 4+ charge isotope series with a delta mass of 0.15 ppm between the measured and theoretical mass of the modified peptide. Xcorr score of 6.26 was obtained using the Sequest HT (V1.3) search engine. b, Total ion current of the AspN digest. c, Extracted ion chromatogram of mass 541.5449 belonging to the HexNAc modified peptide DWKRLARELKVSEAKM using isolation mass tolerance of 10ppm. d, ETD fragmentation spectra of the 541.55 precursor ion. Insert table shows the theoretical masses associated with the respective fragmentation ions.
Acknowledgments
We gratefully acknowledge Philippe Bouillet for the gift of Bid−/− mice and Thomas Cumming for assistance with animal work. This work was supported by the Australian National Health and Medical Research Council (Program Grant #606788 to E.L.H., Project Grants #637332, #1009145 to A.S., #1009145 to L.O.R., Australia Fellowship to A.S.), the Wellcome Trust to GF, the Juvenile Diabetes Foundation; the Leukaemi`a and the Lymphoma Society (New York; SCOR grant #7413) to AS. E.L.H was supported by an Australian Research Council Future Fellowship. J.S.P, M.K., T.W., C.G. and P.R. were supported by Australian Postgraduate Awards. This work was made possible through Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIISS.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
The authors declare no competing financial interests.
References
- 1.Wong AR, et al. Enteropathogenic and enterohaemorrhagic Escherichia coli: even more subversive elements. Mol Microbiol. 2011;80:1420–1438. doi: 10.1111/j.1365-2958.2011.07661.x. [DOI] [PubMed] [Google Scholar]
- 2.Srikanth CV, Mercado-Lubo R, Hallstrom K, McCormick BA. Salmonella effector proteins and host-cell responses. Cell Mol Life Sci. 2011;68:3687–3697. doi: 10.1007/s00018-011-0841-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashida H, et al. Shigella deploy multiple countermeasures against host innate immune responses. Curr Opin Microbiol. 2011;14:16–23. doi: 10.1016/j.mib.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 4.Park HH, et al. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu Rev Immunol. 2007;25:561–586. doi: 10.1146/annurev.immunol.25.022106.141656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strasser A, Jost PJ, Nagata S. The many roles of FAS receptor signaling in the immune system. Immunity. 2009;30:180–192. doi: 10.1016/j.immuni.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schneider-Brachert W, et al. Compartmentalization of TNF receptor 1 signaling: internalized TNF receptosomes as death signaling vesicles. Immunity. 2004;21:415–428. doi: 10.1016/j.immuni.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 7.Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- 8.Newton HJ, et al. The type III effectors NleE and NleB from enteropathogenic E. coli and OspZ from Shigella block nuclear translocation of NF-kappaB p65. PLoS Pathog. 2010;6:e1000898. doi: 10.1371/journal.ppat.1000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L, et al. Cysteine methylation disrupts ubiquitin-chain sensing in NF-kappaB activation. Nature. 2012;481:204–208. doi: 10.1038/nature10690. [DOI] [PubMed] [Google Scholar]
- 10.Gao X, et al. NleB, a bacterial effector with glycosyltransferase activity, targets GAPDH function to inhibit NF-kappaB activation. Cell Host Microbe. 2013;13:87–99. doi: 10.1016/j.chom.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imtiyaz HZ, Zhang Y, Zhang J. Structural requirements for signal-induced target binding of FADD determined by functional reconstitution of FADD deficiency. J Biol Chem. 2005;280:31360–31367. doi: 10.1074/jbc.M504138200. [DOI] [PubMed] [Google Scholar]
- 12.Wang L, et al. The Fas-FADD death domain complex structure reveals the basis of DISC assembly and disease mutations. Nat Struct Mol Biol. 2010;17:1324–1329. doi: 10.1038/nsmb.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strasser A, Harris AW, Huang DC, Krammer PH, Cory S. Bcl-2 and Fas/APO-1 regulate distinct pathways to lymphocyte apoptosis. EMBO J. 1995;14:6136–6147. doi: 10.1002/j.1460-2075.1995.tb00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly M, et al. Essential role of the type III secretion system effector NleB in colonization of mice by Citrobacter rodentium. Infect Immun. 2006;74:2328–2337. doi: 10.1128/IAI.74.4.2328-2337.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jost PJ, et al. XIAP discriminates between type I and type II FAS-induced apoptosis. Nature. 2009;460:1035–1039. doi: 10.1038/nature08229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaufmann T, et al. The BH3-only protein bid is dispensable for DNA damage- and replicative stress-induced apoptosis or cell-cycle arrest. Cell. 2007;129:423–433. doi: 10.1016/j.cell.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 17.Mundy R, et al. Identification of a novel Citrobacter rodentium type III secreted protein, EspI, and roles of this and other secreted proteins in infection. Infect Immun. 2004;72:2288–2302. doi: 10.1128/IAI.72.4.2288-2302.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng W, et al. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc Natl Acad Sci U S A. 2004;101:3597–3602. doi: 10.1073/pnas.0400326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wickham ME, Brown NF, Boyle EC, Coombes BK, Finlay BB. Virulence is positively selected by transmission success between mammalian hosts. Curr Biol. 2007;17:783–788. doi: 10.1016/j.cub.2007.03.067. [DOI] [PubMed] [Google Scholar]
- 20.Wickham ME, et al. Bacterial genetic determinants of non-O157 STEC outbreaks and hemolytic-uremic syndrome after infection. J Infectious Dis. 2006;194:819–827. doi: 10.1086/506620. [DOI] [PubMed] [Google Scholar]
- 21.Karmali MA, et al. Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J Clin Microbiol. 2003;41:4930–4940. doi: 10.1128/JCM.41.11.4930-4940.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isono T. O-GlcNAc-specific antibody CTD110.6 cross-reacts with N-GlcNAc2-modified proteins induced under glucose deprivation. PLoS One. 2011;6:e18959. doi: 10.1371/journal.pone.0018959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levine MM, et al. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet. 1978;1:1119–1122. doi: 10.1016/s0140-6736(78)90299-4. [DOI] [PubMed] [Google Scholar]
- 26.Perna NT, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 27.Zurawski DV, et al. The NleE/OspZ family of effector proteins is required for polymorphonuclear transepithelial migration, a characteristic shared by enteropathogenic Escherichia coli and Shigella flexneri infections. Infect Immun. 2008;76:369–379. doi: 10.1128/IAI.00684-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garmendia J, et al. TccP is an enterohaemorrhagic Escherichia coli O157:H7 type III effector protein that couples Tir to the actin-cytoskeleton. Cell Microbiol. 2004;6:1167–1183. doi: 10.1111/j.1462-5822.2004.00459.x. [DOI] [PubMed] [Google Scholar]
- 29.Mundy R, et al. Identification of a novel type IV pilus gene cluster required for gastrointestinal colonization of Citrobacter rodentium. Mol Microbiol. 48:795–809. doi: 10.1046/j.1365-2958.2003.03470.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Extended Data Figure 1. NleB1 binds FADD, TRADD and RIPK1 and blocks extrinsic apoptosis. a, Co-immunoprecipitation of bacterially delivered NleB1-2HA with FADD-FLAG, TRADD-FLAG and RIPK1-FLAG in HeLa cells. Lane 1. HeLa cells infected with EPEC ΔnleB1 (pNleB1) (negative control). Lane 2, HeLa cells infected with EPEC ΔnleB1 (pNleB1-2HA). Actin; loading control. Representative immunoblot from at least three independent experiments b, GFP-Trap pull-down of FADD-FLAG or FADDΔDD-FLAG. NleB1 from EPEC E2348/69, EHEC O157:H7 EDL933 and C. rodentium (CR) ICC169 was used as indicated. Actin; loading control. Representative immunoblot from at least three independent experiments c, HeLa cells were infected with derivatives of EPEC as indicated for 4 h and left unstimulated (light grey bars) or stimulated with TNF for 8 h (dark grey bars). Results are the mean ± SEM of at least three independent experiments carried out in duplicate. *significantly greater than uninfected, unstimulated cells, P< 0.0001, one-way ANOVA with Dunnett’s Multiple Comparison Test. d, Apoptosis was induced by FasL and HeLa cells were stained with antibodies to cleaved caspase-8. UT, untransfected; Arrows indicate cells with cleaved caspase-8. Scale bar 5 μm. Representative images from at least three independent experiments e, Quantification of cleaved caspase-8 or cleaved caspase-3 by immunofluorescence microscopy in HeLa cells expressing GFP or GFP-NleB1. UT, untransfected; Results are the mean ± SEM percentage of cells with cleaved caspase-8 or cleaved caspase-3 from at least three independent experiments counting ~200 cells in triplicate. *P < 0.0001, unpaired, two-tailed t-test f, GlcNAcylation of FADD-FLAG detected using a monoclonal antibody to GlcNAc of immunoprecipitated lysate from HEK293T cells. Actin; loading control. Representative immunoblot from at least three independent experiments.
Extended Data Figure 4 GlcNAc modification of FADD by GST-NleB1. a, Intact protein mass spectrometry of FADD with R117A substitution incubated with GST-NleB1 and UDP-reveals no GlcNAc modification b, Intact protein mass spectrometry of FADD with S117A substitution incubated with GST-NleB1 and UDP-GlcNAc reveals a single GlcNAc modification c, In vitro assay for NleB1 GlcNAc modification of mutated forms of FADD as indicated. Representative immunoblot from at least three independent experiments.
Extended Data Figure 5. Comparison of FasL and TNF-mediated caspase-8 processing and analysis of NleB2 a, GFP-Trap pull-down of FADD-FLAG, TRADD-FLAG or RIPK1-FLAG with GFP-NleB2 in HEK293T cells. Actin; loading control. Representative immunoblot from at least three independent experiments b-d, Co-immunoprecipitation of bacterially delivered NleB2-2HA with FADD-FLAG, TRADD-FLAG and RIPK1-FLAG in HeLa cells using HA antibodies. Lane 1. HeLa cells infected with EPEC ΔnleB2 (pNleB2) (negative control). Lane 2, HeLa cells infected with EPEC ΔnleB2 (pNleB2-2HA). Actin; loading control. Representative immunoblots from at least three independent experiments e, Unprocessed and cleaved caspase-8 in HeLa cells infected for 3 h with derivatives of EPEC and left untreated or treated with Fcγ-FasL or TNF for 1 h as indicated. Representative immunoblot from at least three independent experiments f, Unprocessed and cleaved caspase-8 in HeLa cells infected for 45 min with derivatives of EPEC and left untreated or treated with TNF and CHX for 1 h as indicated. Actin; loading control. Representative immunoblot from at least three independent experiments g, Adherence of EPEC to HeLa cells. HeLa cells were infected with derivatives of EPEC for 3 h followed by treatment with FasL for 1 h. Number of recovered bacteria and the inoculum are shown for comparison. Mean ± SEM are indicated. Data are from three independent experiments performed in triplicate.
Extended Data Figure 6. Infection of C57BL/6 and Faslpr/lpr mice with CR. Immunofluorescence staining of cleaved caspase-8 in different colonic sections from C57BL/6 mice infected with wild type CR and an nleB mutant as indicated, using antibodies to CR O-antigen (anti-O152, green) and cleaved caspase-8 (red). Intestinal tissue was visualised with Hoechst staining for DNA (blue). Arrows indicate cleaved caspase-8 positive cells sloughed into the gut lumen. Scale bar 100 μm Representative images from at least three separate sections of colon at least 100 μm apart (transverse or longitudinal), per animal from five individual mice per group. b, Time course of infection of C57BL/6 and Fas lpr mutant mice with CR. Each data point represents log10CFU/100 mg faeces per individual animal on the indicated days. Mean ± SEM are indicated, dotted line represents detection limit. CR, mice infected with wild type CR; ΔnleB mice infected with CR nleB mutant. *P < 0.05, **P < 0.01, ***P < 0.001. P values from Mann Whitney U. Dotted line represents the detection limit.
Extended Data Figure 7. Histological analysis of intestinal sections at day 10 from FAS-deficient mice infected with C. rodentium a, Bacterial load in the colon of mice infected with C. rodentium (CR). Each data point represents log10CFU/100 mg colon per individual animal on day 10 post-infection. CR, C57BL/6 (WT) mice infected with wild type CR; ΔnleB, C57BL/6 (WT) mice infected with CR nleB mutant; Faslpr/lpr CR, Faslpr/lpr mice infected with wild type CR, dotted line is detection limit. P = 0.0002, Mann Whitney U. Mean ± SEM are indicated b, Resected colon weights (between rectum and caecum) of individual animals on day 10 post-infection. CR, C57BL/6 (WT) mice infected with wild type CR; ΔnleB, C57BL/6 (WT) mice infected with CR nleB mutant; Faslpr/lpr CR, Faslpr/lpr mice infected with wild type CR. P = 0.0164, Mann Whitney U. Mean ± SEM are indicated. c, Mean ± SEM crypt height in μm in hematoxylin and eosin stained sections from C57BL/6 and Faslpr/lpr mice infected with wild type CR. UI, uninfected. Data are from 4 sections of colon measured at least 50 μm apart per animal from at least three individual mice per group d, Mean ± SEM tissue damage score in colon sections for individual mice infected with wild type CR. Scoring system is described in the Materials and Methods. UI, uninfected. P < 0.05, Mann Whitney U e, Hematoxylin and eosin stained sections of colon from C57BL/6 (WT) or Faslpr/lpr mice uninfected or infected with wild type CR (day 10 post-infection). UI, uninfected. Scale bar 100 μm a. mucosal inflammation b. neutrophil invasion of the muscularis mucosa c. erosion of the epithelium. Representative images from two sections of colon at least 100 μm apart (transverse or longitudinal) from at least three individual mice per group.
Extended Data Figure 8. Diarrhoea score and histological analysis of intestinal sections at day 12 from C57BL/6 (WT), Faslpr/lpr, Fasgld/gld and Bid−/− mice infected with CR. a, Bacterial load in the faeces of different mouse strains infected with CR as indicated. Each data point represents log10CFU/100 mg colon per individual animal. Dotted line is detection limit. Mean ± SEM are indicated b, Diarrhoea score at day 12 post-infection of different strains of mice infected with CR. Scoring system is described in the Methods. Mean ± SEM are indicated. P values from one-way ANOVA. c, Mean ± SEM crypt height in μm in haematoxylin and eosin stained sections from different mouse strains infected with wild type CR. UI, uninfected Data are from 4 sections of colon measured at least 50 μm apart per animal from at least three individual mice per group d, Haematoxylin and eosin stained sections of colon from different mouse strains infected with wild type CR. Scale bar 100 μm. Representative images from two sections of colon at least 100 μm apart (transverse or longitudinal) from at least three individual mice per group.
Extended Data Figure 9. Bacterial load in the faeces of Faslpr/lpr mice infected with wild type CR, an espI mutant and an espF mutant. a, Bacterial load in the faeces of mice during infection with derivatives of CR. Each data point represents log10CFU/100 mg faeces or colon as indicated per individual animal on days 4, 8, 10 and 12 post-infection. Mean ± SEM are indicated, dotted line represents detection limit. b, Diarrhoea score at day 4, 8, 10 and 12 post-infection. Scoring system is described in the Methods. Mean ± SEM are indicated. P values from one-way ANOVA c-d, Bacterial load in the faeces of mice infected with derivatives of CR. Each data point represents log10CFU/100 mg faeces per individual animal on day 8 post-infection. Mean ± SEM are indicated, dotted line represents detection limit. P values from Mann Whitney U.
Extended Data Figure 2. Mass Spectra of peptides identifying Arg117 GlcNAc modification. Fragment ions of overlapping tryptic missed cleaved peptides confirm Arg117 of FADD as the modification site.
Extended Data Figure 3 Mass spectra obtained using ETD fragmentation on the Orbitrap Elite mass spectrometer. a, Red and blue spectra represent the c and z series ions that matches the peptide sequence DWKRLAR-HexNAc-ELKVSEAKM (addition of 203.0793 Da) produced from an AspN digest of FADD incubated with NleB1. Insert box shows the precursor ion present as a 4+ charge isotope series with a delta mass of 0.15 ppm between the measured and theoretical mass of the modified peptide. Xcorr score of 6.26 was obtained using the Sequest HT (V1.3) search engine. b, Total ion current of the AspN digest. c, Extracted ion chromatogram of mass 541.5449 belonging to the HexNAc modified peptide DWKRLARELKVSEAKM using isolation mass tolerance of 10ppm. d, ETD fragmentation spectra of the 541.55 precursor ion. Insert table shows the theoretical masses associated with the respective fragmentation ions.