Abstract
ABC transporters are a ubiquitous class of integral membrane proteins of immense clinical interest because of their strong association with human disease and pharmacology. To improve our understanding of these proteins, we used Membrane Yeast Two-Hybrid (MYTH) technology to map the protein interactome of all non-mitochondrial ABC transporters in the model organism Saccharomy cescerevisiae, and combined this data with previously reported yeast ABC transporter interactions in the BioGRID database to generate a comprehensive, integrated interactome. We show that ABC transporters physically associate with proteins involved in a surprisingly diverse range of functions. We specifically examine the importance of the physical interactions of ABC transporters in both the regulation of one another and in the modulation of proteins involved in zinc homeostasis. The interaction network presented here will be a powerful resource for increasing our fundamental understanding of the cellular role and regulation of ABC transporters.
Introduction
The ATP-binding cassette (ABC) superfamily is a major class of nucleotide-binding proteins found in all domains of life1. It is comprised of nine families, ABCA through ABCI, according to the classification scheme approved by the Human Genome Organization (HUGO), and its members are involved in many biological processes2–5.
While some ABC proteins are soluble and involved in processes such as DNA repair and chromosome segregation6, many are ATP-dependent, integral membrane ‘transporters’, that move a diverse range of compounds across cellular membranes7. These transporters are critical for maintaining cellular homeostasis via processes such as drug and metabolite export, lipid trafficking and nutrient uptake (among others)8.
ABC transporters have a conserved core structure of two cytosolic nucleotide binding domains (NBDs) and two transmembrane domains (TMDs). The NBDs are found in all ABC proteins and contain the regions responsible for ATP-binding and hydrolysis. The TMDs are comprised of multiple (usually 6) hydrophobic membrane spanning segments.
Mutations in ABC transporter genes are implicated in numerous inherited human diseases (see comprehensive reviews4,9). Overexpression of ABC transporters also plays a role in drug resistance of cancer cells and pathogenic microorganisms10,11. The study of ABC transporters is therefore of great clinical interest. However, despite considerable research to date, there is still much to learn about their function and regulation.
One powerful approach for studying protein function is via generation of protein interaction maps (‘interactomes’), which provide a broad-based look at the functional and regulatory associations within the cell. Therefore, to better understand ABC transporters, we set out to map the interactome of the members of this superfamily encoded within the genome of the budding yeast Saccharomyces cerevisiae using membrane yeast two-hybrid (MYTH) technology; a robust tool for studying interactions of full-length membrane proteins in their natural cellular environment12,13. S. cerevisiae encodes 30 ABC proteins, spanning five of the nine recognized ABC families. Of these, 22 are membrane bound transporters, many homologous to disease-linked human ABC proteins, making S. cerevisiae an ideal model organism for studying ABC transporter regulation and function5,14.
Herein, we describe the interactome mapping of all 19 non-mitochondrial ABC transporters encoded by the budding yeast genome. Our results reveal that ABC transporters physically associate with proteins involved in a variety of cellular processes, and provide novel insight into the cellular role and regulation of the members of this clinically relevant group of membrane transporters.
Results
Construction and Validation of MYTH Baits
Of the 22 ABC transporters in S. cerevisiae (Table 1), 19 are predicted to have cytosolic termini, a requirement for MYTH baits. The remaining 3 transporters, Atm1p, Mdl1p and Mdl2p, are localized to the inner mitochondrial membraneand were therefore omitted from the current study5.
Table 1. Saccharomyces cerevisiae ABC Transporters.
| ABC Transporter Family | ABC Transporter Name | Subcellular Localization |
|---|---|---|
| ABCB | Atm1p | Mitochondria |
| Mdl1p | Mitochondria | |
| Mdl2p | Mitochondria | |
| Ste6p | Plasma Membrane | |
| ABCC | Bpt1p | Vacuole |
| Nft1p | Vacuole | |
| Vmr1p | Vacuole | |
| Ybt1p | Vacuole | |
| Ycf1p | Vacuole | |
| Yor1p | Plasma Membrane | |
| ABCD | Pxa1p | Peroxisome |
| Pxa2p | Peroxisome | |
| ABCG | Adp1p | Vacuole |
| Aus1p | Plasma Membrane | |
| Snq2p | Plasma Membrane | |
| Pdr5p | Plasma Membrane | |
| Pdr10p | Plasma Membrane | |
| Pdr11p | Plasma Membrane | |
| Pdr12p | Plasma Membrane | |
| Pdr15p | Plasma Membrane | |
| Pdr18p | Plasma Membrane | |
| Yol075cp | Plasma Membrane |
We tagged all 19 non-mitochondrial transporters at their C-termini with the Cub-(YFP)-LexA-VP16 MYTH tag. Tagging was carried out endogenously (iMYTH approach) or, where the endogenously-tagged strains failed validation (see below), by cloning the target gene into the AMBV MYTH bait vector (tMYTH approach). Note that cloning of the NFT1 gene required correction of a nonsense mutation present in several laboratory strains15 (see Online Methods and Supplementary Results, Supplementary Fig. 1).
To validate tagged bait strains we used two approaches, the NubGI test and fluorescence microscopy13. The NubGI test ensured that the baits were properly expressed and did not non-specifically self-activate. In this test, we transformed bait strains with both positive (NubI) and negative (NubG) interaction control preys. To be suitable for screening, a bait strain needed to grow on interaction selection media when transformed with NubI, but not NubG, control prey (see example in Fig. 1a). We next used fluorescence microscopy on baits tagged with Cub-YFP-LexA-VP16, to ensure tagging did not disrupt proper subcellular localization, as shown for Pxa1p as an example (Fig. 1b).
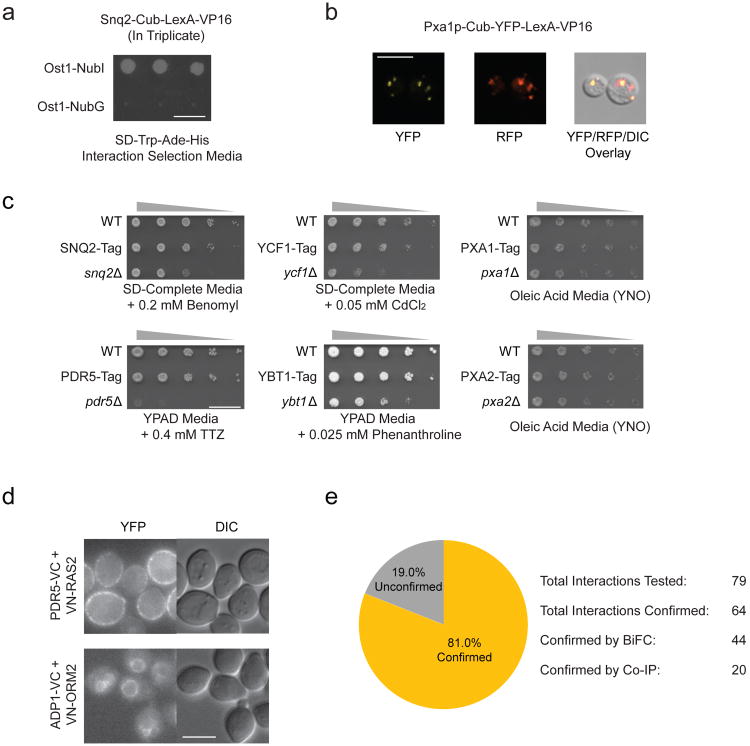
Figure 1.
Overview of ABC transporters in Saccharomyces cerevisiae and validation of MYTH-tagged baits. (a) NubGI test of MYTH reporter strain expressing endogenously tagged Snq2p-Cub-LexA-VP16. The strain displays growth on interaction selection media only in the presence of the NubI-tagged positive interaction control prey, but not in the presence of the NubG-tagged negative interaction control prey. Scale bar is 10 mm. (b) Fluorescence microscopy images showing proper peroxisomal localization of P×a1p-Cub-YFP-LexA-VP16 expressed from MYTH reporter strain. P×a1 signal (yellow, YFP Channel) co-localizes with peroxisomal membrane-targeted control protein (red, RFP Channel). Scale bar is 6 μm. (c) Phenotypic challenge assay demonstrating normal function of MYTH-tagged ABC transporters. In all cases, strains expressing MYTH tagged ABC transporters display growth similar to that of the WT strain rather than to the reduced growth of strains carrying a deletion of the corresponding ABC transporter gene. TTZ = 2,3,5-Triphenyltetrazolium chloride. Scale bar is 15 mm.(d) Examples of BiFC images obtained for two of the interactions validated using BiFC. Scale bar is 5 μm. (e) Summary of the number of interactions tested by orthogonal assay which were positively confirmed by BiFC or Co-IP vs. the number of interactions which could not be confirmed.
Of the 19 ABC transporters, 17 passed both validation tests. For the 2 remaining transporters, Pdr11p and Pdr18p, bait strains were identified which passed the NubGI test, however YFP-tagged transporter was not expressed at levels suitable for detection by fluorescence microscopy. Since neither transporter was mislocalized (rather their specific localization could not be determined due to insufficient signal strength), we deemed both suitable for further screening. Of the 19 ABC baits selected for further analysis, 12 were endogenously tagged (iMYTH) baits and 7 were ectopically expressed (tMYTH) baits. Complete validation results are provided in Supplementary Figures 2 and 3, with the exception of those for Ycf1p, which have been reported previously16. Note that we analyzed Aus1p bait strains under anaerobic growth conditions, which cause a strong induction of Aus1p protein levels (Supplementary Fig. 4). Also note that Ste6p displays vacuolar luminal fluorescence, as previously published, due to its rapid recycling from the plasma membrane; Ste6p can only be detected on the plasma membrane when endocytosis is blocked17,18.
Finally, to ensure MYTH-tagging did not disrupt transporter function, we subjected a representative set of six baits, for which a suitable test condition could be identified, to transporter-specific phenotypic challenge. In all cases, bait strains grew comparably to wild-type, indicating normal transporter function (Fig. 1c).
MYTH Screening and Bait Dependency Testing
We carried out MYTH screening of ABC transporter baits using NubG-X cDNA and genomic prey libraries (where X is the DNA fragment expressing the prey) prepared from S. cerevisiae. Our initial results were assembled into a ‘preliminary’ interactome, which we further refined experimentally using the bait dependency test13 to help determine reproducibility and specificity of the interactions. All preys which passed this secondary testing were used in subsequent analysis and assembly of our final ABC transporter interactome (see Online Methods).
Our final dataset contained 285 unique interactions across 209 proteins (Supplementary Dataset 1). We found candidate interactors for all ABC transporter baits, except Pdr11p, which produced no screening results. Pdr11p could not be cloned for tMYTH screening, possibly due to toxicity of the overexpressed construct, and anaerobic conditions failed to significantly increase Pdr11p levels in the iMYTH strain (Supplementary Fig. 4). Pdr11p was found as an interacting prey, however, in screens using different ABC transporter baits and is therefore represented in the final interactome.
Validation by Orthogonal Assays
As a secondary validation of our results, we performed orthogonal testing using Bimolecular Fluorescence Complementation (BiFC)19 and co-immunoprecipitation (Co-IP) on a representative subset of interactions.
For BiFC, we endogenously tagged proteins at their N- or C-termini using the C-terminal half of YFP (VC) in yeast strain BY4742 (for ABC transporters) or the N-terminal half of YFP (VN) in yeast strain BY4741 (for interactors). Corresponding strains were mated, and we examined resultant diploids by fluorescence microscopy to identify cells producing YFP signal, indicating interaction of constructs (see example in Fig. 1d). Of the 79 interactions tested, 44 (55.6%) produced a positive BiFC signal (Supplementary Figs. 5 to 11).
We subjected the 35 interaction pairs which could not be confirmed by BiFC to Co-IP analysis using endogenously expressed TAP (tandem affinity purification) tagged baits20 and plasmids expressing full-length, HA (Human influenza hemagglutinin) tagged prey proteins21. Of these, 20 interactions (57%) produced a positive result, 11 interactions (32%) produced a negative result and 4 (11%) could not be tested due to technical issues with strain generation (Supplementary Figs. 12 and 13). The results of these two assays have an overall confirmation rate of 81.0% (Fig. 1e). Our failure to confirm a subset of the tested interactions may have resulted from altered expression and steric issues, since MYTH allows detection of both prey fragments and full-length preys, while our BiFC and Co-IP approaches use full-length preys exclusively. Overall, orthogonal testing confirms a strikingly high percentage of our MYTH interactions, providing excellent support for the quality of the ABC interactome.
Assembly of Integrated ABC Transporter Interactome
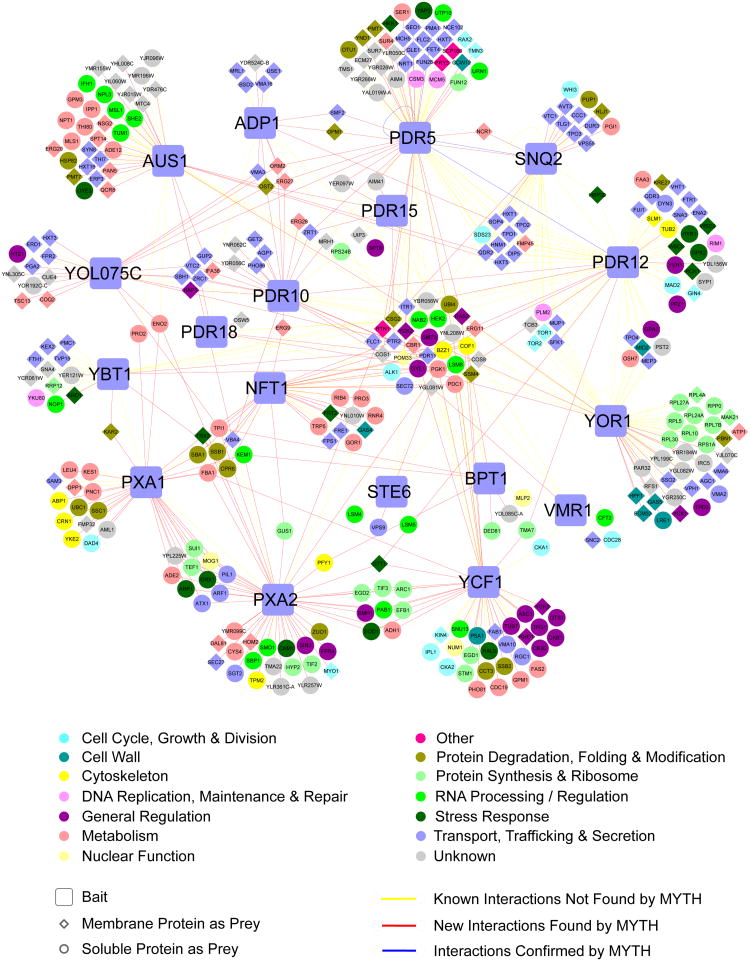
We integrated all MYTH screening results with the previously reported ABC transporter physical interactions available in the BioGRID database22, unambiguously compiling 537 unique binary interactions across 366 proteins. We then assigned each protein a functional classification, based upon manual inspection of Gene Ontology (GO) annotation and functional information available in the Saccharomyces Genome Database23. This annotated, integrated interactomeis presented in Figure 2. A variant of this interactome, showing protein conservation in humans and disease association is provided in Supplementary Figure 14 and Supplementary Dataset 2. An interactome showing only interactions identified in our MYTH screen is provided in Supplementary Figure 15.
Figure 2.
Integrated ABC transporter interactome. Novel interactions identified by MYTH screening (red lines) are shown alongside interactions previously reported in the BioGRID database (yellow lines). Previously reported interactions confirmed by MYTH are also shown, as nodes connected by blue lines. Individual nodes are coloured according to functional classification and are assigned a shape based on whether they are bait/prey and localized to the membrane or soluble fraction, as described in the legend.
Functional Analysis of ABC Transporter Interactome
The integrated interactome contains 14 functional groupings of varying size (Fig. 2 and Supplementary Fig. 16). The largest group (26%) corresponds to transport and related processes, consistent with the role of ABC transporters in the trafficking of cellular substances. The next largest group (16%) is proteins of unknown function, followed by those involved in metabolic processes (15%). The significant number of proteins of unknown function is intriguing, and investigation into these interactions may provide insight into new areas of ABC transporter regulation and function. The large number of metabolic proteins may be consistent with a role for ABC transporters in metabolic regulation and ‘quality control’, via the redirection, sequestration or export of key metabolites, with physical interactions providing immediate access to potential substrates, or allowing direct regulatory cross-talk between metabolic pathway constituents and the transporters. The remaining functional groups range in size from 1 to 7%, covering a range of diverse processes.
The association of ABC transporters with proteins of such a broad functional range suggests that their involvement in cellular function is more complex than previously demonstrated. Follow-up studies should help unlock this complexity and further elucidate the cellular role of these proteins. To this end, we carried out preliminary investigation of two interesting associations observed in our interactome – the physical interaction of ABC transporters with one another, and the interaction of ABC transporters with members of the zinc transport system.
Physical Interactions between ABC Transporters
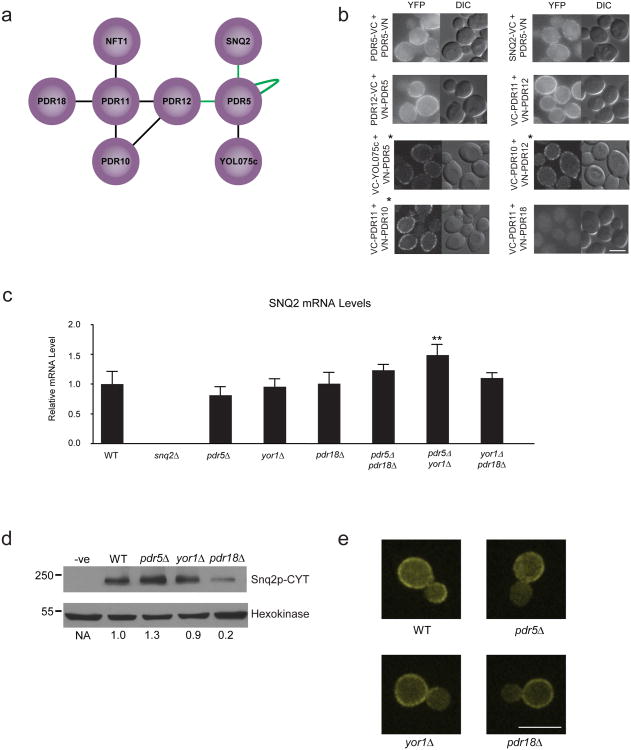
Examination of the interactome reveals a tendency of ABC transporters to interact with one another (Fig. 2). Our MYTH screen detected 6 novel physical associations between ABC transporters, and confirmed 3 previously reported interactions (Fig. 3a). We successfully validated these interactions using BiFC, with the exception of Pdr11p-Pdr18p, which failed to produce a signal, and Pdr11p-Nft1p, which could not be tested due to a nonsense mutation in the NFT1 gene (Fig. 3b). We did not detect six previously reported interactions (Pdr12p-Yor1p, Pdr12p-Snq2p, P×a1p-P×a2p, Ybt1p-P×a1p, Snq2p-Snq2p and Yor1p-Yor1p), possibly due to steric effects resulting from MYTH tagging.
Figure 3.
Investigation of interactions between full-length ABC transporters. (a) Interactions between full-length transporters detected in MYTH screening. Nodes connected by ‘green’ lines indicate interactions reported previously. (b) BiFC analysis of interactions between full-length ABC transporters detects clear signal for all interactor pairs, except Pdr11p-Pdr18p. Scale bar is 5 μm. Images marked with a ‘*’ were obtained via confocal microscopy as described in the Online Methods. (c) SNQ2 transcript level in BY4741 WT and ABC deletion strains as determined using qPCR. Values represent the average of three biological replicates (n=3) and are expressed relative to WT. Error bars indicate standard deviation. The pdr5Δyor1Δ double deletion (marked with an ‘**’) is the only strain with a significantly different level of SNQ2 transcript relative to WT (two-tailed t-test, p-value = 4.5×10−5). (d) Western blot analysis showing relative levels of Snq2p in BY4741 WT, pdr5Δ, yor1Δ and pdr18Δ strains. Hexokinase levels verify equal loading. Numbers below blot indicate ratio of Snq2p to Hexokinase, normalized to levels in WT strain. (e) Fluorescence microscopy visualization of Snq2p-CYT localization in BY4741 WT and ABC deletion strains. Snq2p-CYT displays identical peripheral localization in all strains, consistent with its predicted plasma membrane localization. Scale bar is 8 μm.
While half-transporters such as P×a1p and P×a2p associate to form full-size, functional ABC transporter molecules24, there has been little investigation into the significance of interactions between full-size ABC transporters, which may be is regulatory in nature. A recent study shows that deletion of the PDR5, SNQ2 or YOR1 genes produces compensatory activation of the remaining transporters, a result proposed to involve transcriptional changes25. Since we detected a clear, reciprocal physical interaction between Snq2p and Pdr5p (Fig. 3 a + b), we investigated if this compensatory activation might also involve effects at the protein level.
To this end we used BY4741 strains carrying deletions of the non-mitochondrial ABC transporter genes, obtained from the yeast deletion collection26. To measure transporter activity, we plated strains onto media containing benomyl (Snq2p substrate), cycloheximide (Pdr5p substrate) or 2,3,5-triphenyltetrazolium chloride (Pdr5p substrate)27.
As previously reported25, strains plated onto benomyl media displayed modest resistance in a yor1Δ background, and significantly increased resistance in pdr5Δ single deletion and pdr5Δ yor1Δ double deletion backgrounds (Supplementary Figs. 17 and 18). This effect disappeared in pdr5Δ snq2Δ and yor1Δ snq2Δ double deletions, consistent with it resulting from Snq2p activation (Supplementary Fig. 17). Compensatory activation was not evident in other ABC transporter single deletions (Supplementary Fig. 18), although a strong, unexpected sensitivity to benomyl was observed in the pdr18Δ strain (in addition to the predicted sensitivity of the snq2Δ strain), suggesting a previously unreported functional relationship between Snq2p and Pdr18p (Supplementary Figs. 17 and 18).
Unfortunately, we were unable to detect compensatory activation of Pdr5p under our test conditions (Supplementary Figs. 19, 20 and 21), likely a consequence of differences in strain background and growth conditions compared with earlier work25. We therefore focused on investigating the Snq2p phenotype, first examining the role of transcriptional changes using a quantitative real time PCR approach to measure relative SNQ2 transcript levels (Fig. 3c). Although we detected a modest increase of ∼1.5 fold in our pdr5Δ yor1Δ strain, we failed to find a significant change in SNQ2 transcript levels in other deletion strains. This result revealed that the compensatory activation of Snq2p in our strains was not correlated with transcript level, and therefore not primarily a result of transcriptional changes. We next compared levels of Snq2p protein in the single deletion strains to the levels in wild-type (Fig. 3d and Supplementary Fig. 22). No substantial increase in Snq2p protein levels was apparent in either the pdr5Δ or yor1Δ strains. We did, however, see a decrease in Snq2p protein level in the pdr18Δ strain, consistent with the increased benomyl sensitivity observed in this background. Additionally, we observed no gross changes in the plasma membrane localization of Snq2p in the deletion strains (Fig. 3e). These results suggest that, while the increased sensitivity to benomyl in the pdr18Δ deletion strain may be due to changes in protein level, the significant enhancement of Snq2p activity in the pdr5Δ and yor1Δ strains is not a consequence of changes in Snq2p expression or localization.
ABC Transporters Are Involved in Zinc Homeostasis
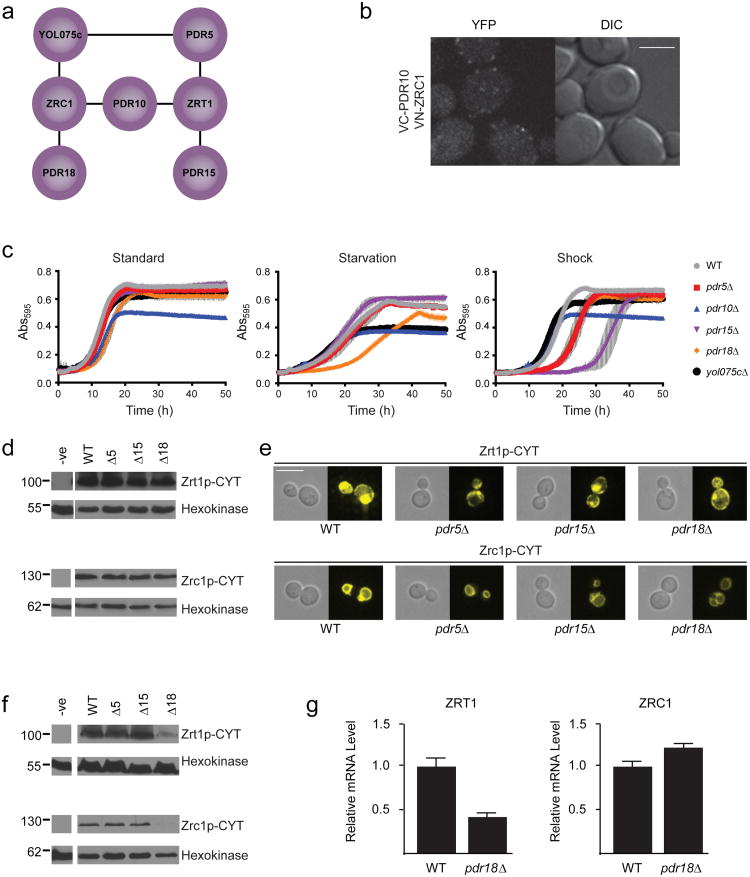
Another interesting observation is the association of several ABC transporters with two major yeast zinc transport proteins, Zrc1p (a vacuolar zinc uptake transporter responsible for zinc storage and detoxification28) and Zrt1p (a plasma membrane-localized, high affinity zinc uptake transporter induced under conditions of zinc limitation29). These interactions are summarized in Figure 4a.
Figure 4.
Investigating role of ABC transporters in zinc homeostasis. (a) Interactions between ABC transporters and the zinc transporters Zrc1p and Zrt1p. (b) BiFC signal observed for the Pdr10p-Zrc1p interaction pair. Scale bar is 5 μm. Image obtained using confocal microscopy as described in the Online Methods. (c) Growth of Y7092 WT and ABC deletion strains under standard, metal starvation and zinc shock conditions. (d) Western blot analysis of Zrt1p and Zrc1p levels in Y7092 WT and deletion strains grown under conditions of zinc limitation. Hexokinase levels verify equal loading. (e) Fluorescence microscopy analysis of localization of Zrt1p and Zrc1p inY7092 WT and deletion strains grown under conditions of zinc limitation. Scale bar is 6 μm. (f) Western blot analysis of Zrt1p and Zrc1p levels in Y7092 WT and deletion strains grown under zinc-replete conditions. Hexokinase levels verify equal loading.(g) ZRT1 and ZRC1 transcript levels in Y7092 WT and pdr18Δ deletion strains as determined using qPCR. Values represent the average of three biological replicates (n=3) and are expressed relative to WT. Error bars indicate standard deviation. Results show a significant reduction (two-tailed t-test, p-value = 7.9×10−4) of ZRT1 transcript in pdr18Δ cells relative to WT, but no change in ZRC1 transcript levels.
Considering the vacuolar localization of Zrc1p, its association with three plasma membrane localized ABC transporters is intriguing. We successfully validated the interaction between Pdr10p and Zrc1p using BiFC, observing a punctate signal located mainly around the cellular periphery (Fig 4b). This signal co-localized with CFP-tagged Cdc42p (which localizes to plasma, vacuolar and nuclear membranes, Supplementary Fig. 23a), while signal produced by mCherry-tagged Sec63p (which localizes to the endoplasmic reticulum), was more discretely separated (Supplementary Fig. 23b). This is consistent with the signal occurring in the plasma membrane rather than the cortical ER, although we also occasionally observed weak BiFC signal on the vacuolar membrane. Our results suggest that the Pdr10p-Zrc1p interaction occurs on the plasma membrane, with a small sub-population of (potentially irreversible) BiFC complex travelling to the vacuole, although the possibility of complex forming in the vacuole and moving to the plasma membrane cannot be definitively ruled out. Determining the exact nature and functional significance of this interaction will require further investigation.
We observed no signal when examining the interactions of Zrc1p with Pdr18p and Yol075cp by BiFC (Supplementary Figs. 8 and 9). Additionally, we could not test the interactions of ABC transporters with full-length Zrt1p using BiFC, due to the extracellular localization of both Zrt1p termini30. We were, however, able to confirm the Pdr18p-Zrc1p interaction and all three ABC transporter-Zrt1p interactions by co-immunoprecipitation (Supplementary Fig. 13). The Yol075cp-Zrc1p interaction was not successfully validated, possibly due to technical issues, although our data (see below) may implicate it in metal homeostasis.
To investigate the functional significance of these interactions, we compared the growth of Y7092 yeast cells deleted of these ABC transporters under conditions of zinc availability, limitation and shock (Fig. 4c). Deletion strains under ‘standard’ zinc replete conditions grew largely the same as wild-type, except pdr10Δ cells which had reduced absorbance at saturation. However, since we observed this phenotype in other media, it is unlikely related to zinc stress, and was not considered further. Under ‘starvation’ conditions (in media depleted of zinc and related metals by EDTA) pdr18Δ cells displayed a significantly increased lag phase, while yol075cΔ cells had a modest reduction in absorbance at saturation, suggesting possible roles of these transporters in metal homeostasis. We also investigated conditions of ‘zinc shock’, when cells are grown in low zinc media and then moved directly into conditions of high zinc availability. In low zinc, cells maximize zinc uptake, primarily via increased expression of Zrt1p. Upon transfer to high zinc conditions, cells rapidly take up zinc in detrimental amounts. To compensate, Zrc1p, whose expression is increased under low zinc conditions, rapidly sequesters zinc in the vacuole, thereby maintaining a safe cytosolic zinc concentration29,31. Strikingly, the pdr15Δ strain displayed a significant growth delay upon 2 mM zinc shock, while both pdr5Δ and pdr18Δ strains also showed modest sensitivity.
We decided to focus further investigation on Pdr5p, Pdr15p and Pdr18p, since the interactions of these proteins with Zrt1p or Zrc1p were confirmed by orthogonal assay, and the phenotypes of their corresponding deletions implicated them in zinc homeostasis.
We first examined Zrt1p and Zrc1p levels in Y7092 wild-type and deletion cells grown in zinc limited media, conditions identical to those experienced prior to zinc shock. Surprisingly, no change in protein level of either transporter occurred in deletion strains relative to wild-type (Fig. 4d and Supplementary Fig. 24). Gross localization of both transporters was also unchanged (Fig. 4e). When we examined zinc transport in these cells (Supplementary Fig. 25), however, we observed that the pdr15Δ strain had a significant increase in zinc uptake of ∼2.7 fold relative to wild-type, consistent with this strain's zinc shock sensitivity. This appeared to be Zrt1p-dependent, as the same increase was not evident in a pdr15Δ zrt1Δ double deletion. Interestingly, deletion of Zrt1p transporter alone did not lead to a reduction in zinc uptake, likely a result of compensatory activity of other zinc uptake systems, such as Fet4p and the low affinity zinc uptake transporter Zrt2p. Zinc uptake of the pdr5Δ and pdr18Δ cells was comparable to wild-type, indicating their modest sensitivity was not a result of greater zinc influx. Notably, however, pdr5Δ zrt1Δ and pdr18Δ zrt1Δ double deletions did experience significant decreases in zinc uptake, with levels ∼60% and 50% that of wild-type, respectively.
We also examined Zrt1p and Zrc1p levels during zinc replete growth, identical to the conditions experienced by cells in our starvation experiment prior to growth in metal-depleted media. While levels of Zrt1p and Zrc1p were unchanged in pdr5Δ and pdr15Δ cells, they were significantly reduced in pdr18Δ cells (Fig. 4f and Supplementary Fig. 24. This is consistent with the growth delay of pdr18Δ upon transfer to zinc limited media, since lower levels of these transporters (particularly Zrt1p) would initially reduce the capacity of the cells to take up zinc.
Using quantitative real time PCR, we observed that the lower Zrt1p levels correlated with reduced ZRT1 mRNA transcript, while the reduction of Zrc1p appeared to be independent of transcriptional changes (Fig. 4g). This suggests that a physical interaction with Pdr18p may be important for Zrc1p stability.
Discussion
Considering the ubiquitous nature of ABC transporters, and their major role in human disease, obtaining a better understanding of their regulation and function is of great clinical importance. To this end, we used the MYTH technology to successfully map the protein interactome of all 19 non-mitochondrial ABC transporters in the budding yeast S. cerevisiae. Combining the results of this screen with previously reported ABC transporter interactions in the BioGRID database allowed us to produce a comprehensive, integrated interactome, providing a detailed and unprecedented look at the cellular network involving these proteins.
The large number of functionally diverse interactors in our network suggests that ABC transporters have a more general involvement in cellular processes than previously suspected. Consistent with this is the large number of interactors of unknown function, demonstrating there is still much we do not understand about this protein class. Examination of the identifiable functional groups, however, does suggest significant involvement in transport and metabolic processes. Additionally, over 50% of the interactions involve a prey with an identifiable human ortholog, and of these, approximately 40% are associated with human disease, suggesting that further study of our interactome should provide information relevant to human disease states.
One interesting observation in our interactome is the tendency of full-size ABC transporters to interact with one another. While some of these interactions have been previously reported, our MYTH screening detected a number of previously unreported physical associations. Though the functional significance of these interactions has not been well studied, we suspect that they may play a regulatory role.
For instance, it is reported that deletion of PDR10 affects the activity of Pdr12p at the protein level, however the mechanism by which this occursisunknown32. Here we have shown that these two proteins physically interact, suggesting this association may be directly responsible for regulating Pdr12p activity. Such an interaction may be important in maintaining the stability and membrane microenvironment of Pdr12p, both of which were observed to be affected in an earlier study32. It is also known that deletion of the PDR5 gene leads to an increase in the activity of Snq2p25, two transporters between which we detected a robust physical interaction. Our investigation failed to detect changes in Snq2p localization, protein level or transcript amount in a pdr5Δ deletion, suggesting that the physical interaction between the proteins may directly lead to repression of Snq2p activity. We also observed that deletion of the PDR18 gene affects Snq2p activity, via a transcription-independent decrease in Snq2p protein level.
Together these results suggest that physical interactions between full-length transporters could serve as an important form of regulation and communication. Changes in the level of one transporter, for example in response to altered environmental conditions, would cause a shift in the binding equilibrium between transporters, acting as a means of rapidly altering transporter activity. While the full extent of such regulation is unknown, the detection of what appears to be a network of interconnected ABC transporter proteins suggests it may be fairly expansive. Consistent with this is our observation that Pdr18p, which is connected only indirectly to Snq2p through physical interactions with intermediate ABC transporters, appears to be important for the stability of Snq2p. Communication through such a network, however, may also involve factors other than direct physical interaction, such as alterations in membrane composition, a process in which Pdr18p and other ABC transporters have been implicated33–36. Further investigation will be necessary to validate and determine the exact nature of such a regulatory system.
Another interesting observation from our MYTH screen is the association between ABC transporters and the Zrc1/Zrt1p zinc transporters. Our experiments demonstrated the importance of PDR5, PDR15 and PDR18 for mounting a proper zinc shock response, although deletion of these genes was not associated with changes in the level or localization of Zrc1p or Zrt1p, which both play major roles in this response29,31. Interestingly, however, cells deleted of PDR15 experienced an apparent Zrt1p-dependent increase in zinc uptake, likely accounting for this strain's zinc shock sensitivity.
The source of the modest zinc shock sensitivity of the pdr5Δ and pdr18Δ deletions is less clear, and may reflect subtle effects on Zrc1p (which we found interacting with Pdr18p). A decrease in Zrc1p activity would cause an increased sensitivity to zinc shock, due to a reduced ability to sequester excess cytosolic zinc in the vacuole31. Such a reduction, and the consequent increase in cytosolic zinc concentration, might also be involved in the reduced uptake activity observed in the pdr5Δ zrt1Δ and pdr18Δ zrt1Δ double deletions, which are forced to rely on the lower affinity Zrt2p and Fet4p zinc transporters. Additional work should help determine the mechanisms involved and whether other zinc transporters play a role.
We also observed a significant drop in Zrt1p and Zrc1p levels in pdr18Δ cells grown under zinc replete conditions. The drop in Zrc1p level did not correlate with a transcriptional change, suggesting that loss of physical interaction with Pdr18p may decrease its stability. The drop in Zrt1p level did appear to result from reduced transcription, though this is likely a consequence of reduced Zrc1p activity. Lower Zrc1p activity would lead to increased cytosolic zinc, and a corresponding reduction in activity of the Zap1p transcription factor, responsible for activating the expression of Zrt1p37. The decreased level of these transporters is likely responsible for the delayed growth phenotype of pdr18Δ cells upon transfer from zinc-replete to zinc-limited conditions, possibly as a consequence of reduced vacuolar zinc stores (resulting from reduced Zrc1p activity during zinc replete growth) and/or reduced Zrt1p levels, whose activity is necessary for efficient uptake of zinc under conditions of zinc limitation29.
Intriguingly, the effects observed with Zrt1p and Zrc1p are similar to those observed between full-length ABC transporters. These regulatory effects are also reminiscent of the‘atypical’ role of the human ABC proteins Sur1 and Sur2, which serve as the regulatory components of ion channels38. It is tempting to speculate that ABC transporters may have a general role as functional regulators of membrane proteins, in addition to their known transport roles. Further study of the interactome should help determine if such regulation is a common occurrence and what regulatory mechanism is employed.
Our initial investigation of the integrated ABC transporter interactome has provided a number of novel insights into the cellular role of this important class of transport proteins. With the large number of other interactions still unexplored, further examination of the network promises to greatly increase our understanding ABC transporter function and regulation. Considering the high percentage of interactors with human orthologs, much of the information gained should also be directly applicable to higher organisms. We believe that the data presented here will serve as a comprehensive and powerful resource for further studies into this clinically relevant class of proteins, and be of great value in the development of novel therapeutics directed towards the treatment of diseases associated with ABC transporter dysfunction.
Online Methods
Construction of ABC Transporter MYTH Baits
iMYTH and tMYTH baits were generated as described previously13. In brief, iMYTH baits were generated by integration of Cub-(YFP)-LexA-VP16 KanMX cassette at the 3′ end of the ABC transporter gene of interest. tMYTH baits were generated using a ‘gap-repair’ approach39 into the MYTH bait plasmid AMBV (Dualsystems Biotech). For cloning of full-length NFT1 into AMBV4, two partially overlapping gene fragments, each correcting the nonsense mutation present in S288C strains, were amplified and used for cloning into AMBV4 plasmid (see Supplementary Figure 1). All baits were verified by PCR and sequencing. A list of primer sequences used for bait generation can be found in Supplementary Table 1.
Bait Localization by Fluorescence Microscopy
Subcellular localization of ABC transporter baits fused to Cub-YFP-LexA-VP16 MYTH tag was carried out using fluorescence microscopy. For all ABC transporters, with the exception of AUS1, cells expressing the ABC baits were grown to mid-log phase in SD-Complete (iMYTH bait strains) or SD-Leu (tMYTH bait strains) liquid medium at 30°C. Cells expressing AUS1 baits were grown anaerobically on solid SD-Complete media at 30°C for 3 days, in a sealed jar depleted of oxygen using BD GasPak Plus pouches. Cells from mid-log phase cultures or solid media (AUS1 bait) were examined at 630× magnification (11 z slices captured, 0.3 μm step size) using a Leica DMI 6000 B microscope with YFP, Texas Red and differential interference contrast (DIC) filter sets. Image capture and processing was performed using the Volocity software package (PerkinElmer).
Bait Validation using NubGI Control Test
Suitability of generated baits for use in MYTH screening was assessed using the NubGI Control Test as previously described13. In brief, bait strains were transformed with Ost1p-NubI (positive interaction control) or Ost1p-NubG (negative interaction control) and spotted onto interaction selection media (SD-WAH for iMYTH strains or SD-WLAH for tMYTH strains). Plates were grown at 30°C for 2-3 days before analysis. Assays for AUS1 bait were done under anaerobic conditions to allow for proper induction of Aus1p.
Functionality Testing of Selected Baits
WT, MYTH-tagged (bait) and deletion strains were grown on YPAD plates for 2-3 days until single colonies appeared. Individual colonies were then diluted into 150 μL of sterile ddH20 and 10-fold serially diluted up to a maximal dilution of 10 000×, prior to spotting onto appropriate challenge media. Plates were grown at 30°C for 3-4 days prior to imaging.
MYTH Library Screening and Data Analysis
Library screening was carried out as previously described13, using a high efficiency yeast transformation protocol and both NubG-X cDNA and yeast genomic libraries. The cDNA library had a complexity of 2.2×106 independent clones and contained inserts ranging from 0.4 to 5 kb, with an average size of 0.7 kb (Dualsystems Biotech). The genomic library had an overall complexity of 4×107 and contained fragments ranging between 0.5 and 2 kb in size. Selection and transfer of colonies to liquid media and additional selection plates was carried out using a QP Interactor robotics system (Genetix). Interacting prey constructs obtained via MYTH screening were sequenced and identified using both automated analysis, consisting of a combination of in-house software and BLAST analysis tools, and subsequent manual inspection of individual sequences and chromatograms in order to properly identify prey candidates and eliminate spurious sequences. The resulting refined list of interactions comprised our initial MYTH interactome. This interactome was then further refined experimentally using the Bait Dependency Test, in which preys were transformed back into the original bait strain, as well as into a strain expressing artificial control bait (a minimal construct consisting of the Matα signal sequence, the single-pass transmembrane domain of the human T-cell surface glycoprotein CD4 and the Cub-LexA-VP16 tag). Preys that activated the reporter system in the original bait strain but not in the artificial control bait strain were deemed specific and used in subsequent analysis and assembly of the final ABC transporter interactome.
Bimolecular Fluorescence Complementation (BiFC) Analysis
BiFC analyses were carried out essentially as previously described40. Briefly, selected ABC transporter genes were endogenously tagged41 in strain BY4742 at either their 5′ or 3′ end with sequence encoding the C-terminal (‘VC’) fragment of YFP Venus42, while interactor genes were endogenously tagged in strain BY4741 at their 5′ or 3′ end with sequence encoding the N-terminal (‘VN’) fragment of YFP Venus. BY4741 and BY4742 strains were mated and resultant diploids examined for YFP signal by fluorescence microscopy. Image acquisition and processing were performed using Slidebook software (3i), as previously described43. For strains with relatively weak BiFC signals, we repeated imaging using a spinning disk confocal microscope (UltraView VoX; PerkinElmer Life and Analytical Sciences, Waltham, MA) equipped with a 100× oil NA 1.4 Plan-Apo Nikon objective lens, the EM CCD ImagEM (Back-Thinned Electron Multiplier CCD Camera), and a solid state 514-nm diode laser. A single optical section was captured for 1 sec exposure using 20% laser power (sensitivity 171) for a BiFC assay. To determine the localization of VC-PDR10/VN-ZRC1 interaction, the BiFC signal was captured in the presence of markers directed to the plasma membrane (pRS415-PMET-CFP-Cdc42) and endoplasmic reticulum (Sec63-mCherry). Image capture and processing were performed using the Volocity software package (PerkinElmer) and Image J software.
Generation of Integrated ABC Transporter Interactome
Yeast physical interactions were retrieved from BioGRID release 3.1.8122 and mapped onto our refined ABC gene list. Only interactions that contained at least one ABC gene were retained. By combining our novel interactions, we constructed an ABC interaction network containing 537 unique binary interactions among 366 proteins. Cytoscape was used to integrate and visualize the network44.
Gene Ontology analysis
GO∷TermFinder (v0.86) was deployed to carry out Gene Ontology (GO) annotation and evaluation45. Gene annotations were downloaded from SGD (as of November 23, 2011)23, while GO annotations were downloaded from the GO website (as of November 23, 2011)46.
Orthology mapping between yeast and human
To define a comprehensive orthology mapping table between yeast and human, we identified orthologous pairs using the clustered orthologous groups deposited in the InParanoid database (v7.0)47. For yeast genes without annotations in InParanoid, we manually checked their annotations in SGD23. Yeast YOR1 and PXA2 were found to be similar to human cystic fibrosis transmembrane receptor and ATP-binding cassette sub-family D member 1, and included in our orthology mapping table.
Compiling human disease-associated genes
To gain a high-confidence disease-gene annotation, we adopted the disease annotations deposited in McKusick's Online Mendelian Inheritance in Man (OMIM) database48 and the ‘morbid map’ table was downloaded from OMIM website (October 20, 2011). Disease-gene annotations were mapped to the genes in our interaction network.
Drug Challenge Assays
WT and ABC deletion strains (BY4741 background) were grown on YPAD +/appropriate antibiotic selection for 2-3. Individual colonies were diluted into 150 μL of sterile ddH2O and 10-fold serially diluted up to a maximal dilution of 1000×, prior to spotting onto SD-Complete media containing drug. Plates were grown at 30°C for 3-4 days prior to imaging.
Cell Growth for Protein and RNA Isolation
WT and deletion cells (BY4741 or Y7092 backgrounds) were inoculated from single colonies into SD-Complete (Standard, Metal Replete or Metal Limited) and grown overnight at 30°C to saturation. Overnight culture was diluted into fresh media and grown at 30°C for 4-5 hours. Cells were pelleted and stored at -20°C pending processing.
For analysis of aerobic vs. anaerobic induction of Aus1p-Cub-LexA-VP16 and Pdr11p-Cub-LexA-VP16, cells were grown on solid YPAD media at 30°C for 3 days either aerobically, or anaerobically in a sealed jar depleted of oxygen using BD GasPak Plus pouches. Cells were transferred from plates to microfuge tubes, rinsed briefly with SD-Complete media and stored at -20°C pending processing.
RNA Isolation and Quantitative Real-Time PCR Analysis
RNA was isolated from cells using the Masterpure Yeast RNA Purification Kit (Epicentre). cDNA synthesis was performed using Superscript II Reverse Transcriptase (Invitrogen). Quantitative real-time PCR amplification was performed using the Dynamo Flash SYBR Green qPCR Kit (Thermo Scientific) in a Light Cycler 480 II qPCR Machine (Roche) with the following amplification parameters: pre-incubation at 95°C for 7 min and 45 cycles of denaturation at 95°C for 10 seconds, annealing at 50°C for 35 seconds and extension at 72°C for 35 seconds. Primers used are listed in Supplementary Table 2. ZRT1, ZRC1 and SNQ2 transcript levels were expressed relative to levels of ACT1 housekeeping gene mRNA.
Protein Isolation and Western Blotting
Cells were resuspended in 500 μL 0.2M sodium hydroxide containing 0.2% β-mercaptoethanol and incubated on ice for 10 minutes. Proteins were precipitated by addition of 50 μL of 50% trichloroacetic acid, followed by a 10 minute incubation on ice. Precipitate was pelleted by centrifugation (18000 × g for 10 minutes at 4°C), resuspended in 50 μL 2× Sample Buffer (60 mM Tris, pH 6.8, 3% SDS, 10% glycerol) and protein concentration was determined using the Bio-Rad Protein Assay. Protein was resolved by SDS-PAGE using 10% gels and transferred to nitrocellulose membrane. MYTH-tagged proteins were detected using rabbit α-VP16 (Sigma) primary antibody and anti-rabbit IgG linked to horseradish-peroxidase (Cedarlane) as secondary antibody. Hexokinase was detected using α-hexokinase antibody coupled to horseradish peroxidase (Rockland Immunochemicals).
Co-immunoprecipitation Experiments
Selected interaction test pairs were confirmed using endogenously expressed TAP (tandem affinity purification) tagged bait20 and inducible, overexpressed, full-length, HA (Human influenza hemagglutinin) tagged protein21 as prey. Plasmid overproducing the HA tagged protein was transformed into the TAP-tagged bait strain background. Positive transformants were selected on SD medium lacking uracil and successively grown at 30°C in the same liquid medium with 2% sucrose as carbon source. After sub-culturing and 4 hrs induction with yeast peptone and 2% galactose, cells were pelleted by high-speed centrifugation (4,000 × g for 20 min), resuspended in appropriate lysis buffer, and processed as described previously49. Immunoprecipitated samples were run on a 10% SDS-PAGE gel, transferred onto nitrocellulose membrane and probed with rabbit anti-HA polyclonal antisera (Pierce).
Tecan Growth Experiments
Overnight culture of WT and deletion strains (Y7092 background) grown in either ‘Metal Replete’ (Standard and Starvation conditions) or ‘Metal Limited’ (Shock condition) SD-Complete medium was diluted into fresh medium to an OD600 =∼ 0.15 and grown at 30°C for 4 to 5 hours. Cells were diluted to an OD600 =∼ 0.0625 in ‘Metal Limited’ medium and 95 μL volumes were transferred to the wells of a sterile 96-well plate (Greiner 655 180). For ‘Standard’ and ‘Shock’ conditions 5 μL of sterile 40 mM zinc chloride solution was added, to a final concentration of 2 mM zinc. For ‘Starvation’ conditions 5 μL sterile ddH2O was added. Plates were transferred to a Tecan GENios plate-reader where growth was monitored by measurement of OD595 every 15 minutes for 50 hours. GraphPad Prism 6 was used for data analysis.
Zinc Transport Assays
WT and deletion cells (Y7092 background) were grown to exponential phase in ‘Metal Limited’ SD-Complete medium and harvested. Pellets were washed three times and resuspended in fresh ‘Metal Limited’ SD-Complete media supplemented with 65Zn and incubated for 5 minutes. Cells were collected on glass fiber filters and washed with 10 mL ice-cold SSW (1 mM EDTA, 20 mM sodium citrate pH 4.2). Cell-associated radioactivity was measured by scintillation counter and reported as cpm. All results are normalized to WT.
Media Recipes
Media was prepared as previously described in detail13. Briefly, YPAD medium was made by dissolving 10g Yeast Extract, 20g Peptone, 20g D-Glucose, 40 mg Adenine Sulfate Dihydrate and 20g Agar (only for solid media) in a total volume of 1L ddH2O, and autoclaving. 2× YPAD was made in an identical manner but contained 20g Yeast extract, 40 g Peptone, 40g D-Glucose, 40 mg Adenine Sulfate Dihydrate and 20g Agar (only for solid media). SD-Complete and various drop-out media were made by dissolving 6.7g Yeast Nitrogen Base, 20 D-Glucose and 20g of Agar (only for solid media) in 900 mL of ddH2O followed by addition of appropriate 10× Nucleotide/Amino Acid solution (i.e. lacking whichever amino acids or nucleotides necessary for generation of the desired selective media) and autoclaving. ‘Metal Replete’ SD-Complete media was identical to SD-Complete media, but supplemented with 20 mM Sodium Citrate and adjusted to pH 4.2. ‘Metal Limited’ SD-Complete media was identical to ‘Metal Replete’ SD-Complete media, but contained 1 mM EDTA, pH 8.0. Oleic Acid (YNO) medium was made by dissolving 6.7 g Yeast Nitrogen Base and 20g of Agar in a total volume of 1L ddH2O supplemented with 10 mL BRIJ-35 (30% aqueous, Calbiochem), 1 mL Oleic Acid (>99%, Nu-Chek Prep Inc), 30 mL Glycerol and 100 mL 10× Nucleotide/Amino Acid solution (complete), followed by autoclaving. Additional antibiotics, drugs etc. were added to media where necessary at desired concentrations. 10× Nucleotide/Amino Acid solution was made by dissolving 400 mg Adenine Sulfate Dihydrate, 200 mg Arginine Hydrochloride, 200 mg Histidine Monohydrochloride, 300 mg Isoleucine, 1000 mg Leucine, 300 mg Lysine Hydrochloride, 1500 mg Methionine, 500 mg Phenylalanine, 2000 mg Threonine, 400 mg Tryptophan, 300 mg Tyrosine, 200 mg Uracil and 1500 mg Valine in a total volume of 1 L of ddH2O and autoclaving. Unless otherwise indicated above, all components used in media preparation were obtained from BioShop Inc.
Supplementary Material
Acknowledgments
We thank B. Andrews, C. Nislow, F. Vizeacoumar, C. Kurat and S. Alfred for providing reagents, assistance and equipment, L. Miller, S. Pagant, K. Kuchler and C. Klein for experimental assistance, and W.K. Huh for providing BiFC plasmids. We also thank V. Kanelis, K. Sokolinaand M. Ali for review of the manuscript. This work was supported by grants from the Canadian Institutes of Health Research, Canadian Foundation for Innovation, Natural Sciences and Engineering Research Council of Canada, Ontario Genomics Institute, Canadian Cystic Fibrosis Foundation, Canadian Cancer Society, University Health Network to I.S. and National Institutes of Health (R01-GM76375 to H.O.P. and R01-GM51508 to S.M.).
Footnotes
Author Contributions: I.S. (1, corresponding author) designed the project. J.S. was actively involved in all experiments, I.S. (1) and S.M. provided project guidance and assisted in manuscript preparation. K.J. and Z.Z. performed bioinformatics analysis. H.O.P. and M.E.L. performed BiFC experiments. A.R.Y. performed Westerns, phenotype assays and strain generation. A.H. carried out MYTH screening and zinc-related functional analysis. M.C., D.D., C.G., M.W., P.T. and V.W. carried out MYTH screening. S.L.S. and D.B. carried out MYTH screening of AUS1 under anaerobic conditions. K.D.D. carried out the PDR11 and AUS1 Western blots. C.G., M.J., J.F.G and M.B. performed co-immunoprecipitation experiments. C.B. and B.J.S.L. provided deletion strains. C.M.P. carried out zinc transport assays. H.O.P. and C.M.P. critically reviewed the manuscript. I.S. (2) was involved in bait strain generation and initial project design.
Competing Financial Interests Statement: The authors declare competing financial interests. Igor Stagljar is co-founder of Dualsystems Biotech, Switzerland.
References
- 1.Lewis VG, Ween MP, McDevitt CA. The role of ATP-binding cassette transporters in bacterial pathogenicity. Protoplasma. 2012;249:919–42. doi: 10.1007/s00709-011-0360-8. [DOI] [PubMed] [Google Scholar]
- 2.Verrier PJ, et al. Plant ABC proteins--a unified nomenclature and updated inventory. Trends Plant Sci. 2008;13:151–9. doi: 10.1016/j.tplants.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Dean M, Rzhetsky A, Allikmets R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 2001;11:1156–66. doi: 10.1101/gr.184901. [DOI] [PubMed] [Google Scholar]
- 4.Dean M, Annilo T. Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu Rev Genomics Hum Genet. 2005;6:123–42. doi: 10.1146/annurev.genom.6.080604.162122. [DOI] [PubMed] [Google Scholar]
- 5.Paumi CM, Chuk M, Snider J, Stagljar I, Michaelis S. ABC transporters in Saccharomyces cerevisiae and their interactors: new technology advances the biology of the ABCC (MRP) subfamily. Microbiol Mol Biol R. 2009;73:577–93. doi: 10.1128/MMBR.00020-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hopfner K. Rad50/SMC proteins and ABC transporters: unifying concepts from high-resolution structures. Curr Opin Struc Biol. 2003;13:249–255. doi: 10.1016/s0959-440x(03)00037-x. [DOI] [PubMed] [Google Scholar]
- 7.Jones PM, O’Mara ML, George AM. ABC transporters: a riddle wrapped in a mystery inside an enigma. Trends Biochem Sci. 2009;34:520–31. doi: 10.1016/j.tibs.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Jones PM, George AM. The ABC transporter structure and mechanism: perspectives on recent research. Cell Mol Life Sci. 2004;61:682–99. doi: 10.1007/s00018-003-3336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottesman MM, Ambudkar SV. Overview: ABC transporters and human disease. J Bioenerg Biomembr. 2001;33:453–8. doi: 10.1023/a:1012866803188. [DOI] [PubMed] [Google Scholar]
- 10.Cannon RD, et al. Efflux-mediated antifungal drug resistance. Clin Microbiol Rev. 2009;22:291–321. doi: 10.1128/CMR.00051-08. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szakacs G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5:219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 12.Stagljar I, Korostensky C, Johnsson N, Te Heesen S. A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc Natl Acad Sci USA. 1998;95:5187–92. doi: 10.1073/pnas.95.9.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Snider J, et al. Detecting interactions with membrane proteins using a membrane two-hybrid assay in yeast. Nat Protoc. 2010;5:1281–93. doi: 10.1038/nprot.2010.83. [DOI] [PubMed] [Google Scholar]
- 14.Kovalchuk A, Driessen AJM. Phylogenetic analysis of fungal ABC transporters. BMC Genomics. 2010;11:177. doi: 10.1186/1471-2164-11-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mason DL, Mallampalli MP, Huyer G, Michaelis S. A region within a lumenal loop of Saccharomyces cerevisiae Ycf1p directs proteolytic processing and substrate specificity. Eukaryot Cell. 2003;2:588–98. doi: 10.1128/EC.2.3.588-598.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paumi CM, et al. Mapping protein-protein interactions for the yeast ABC transporter Ycf1p by integrated split-ubiquitin membrane yeast two-hybrid analysis. Mol Cell. 2007;26:15–25. doi: 10.1016/j.molcel.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 17.Kelm KB, Huyer G, Huang JC, Michaelis S. The internalization of yeast Ste6p follows an ordered series of events involving phosphorylation, ubiquitination, recognition and endocytosis. Traffic. 2004;5:165–80. doi: 10.1111/j.1600-0854.2004.00168.x. [DOI] [PubMed] [Google Scholar]
- 18.Kölling R, Hollenberg CP. The ABC-transporter Ste6 accumulates in the plasma membrane in a ubiquitinated form in endocytosis mutants. EMBO J. 1994;13:3261–71. doi: 10.1002/j.1460-2075.1994.tb06627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9:789–98. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 20.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–41. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 21.Gelperin DM, et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005;19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stark C, et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2011;39:D698–704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cherry JM, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–5. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shani N, Valle D. A Saccharomyces cerevisiae homolog of the human adrenoleukodystrophy transporter is a heterodimer of two half ATP-binding cassette transporters. Proc Natl Acad Sci USA. 1996;93:11901–6. doi: 10.1073/pnas.93.21.11901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolaczkowska A, Kolaczkowski M, Goffeau A, Moye-Rowley WS. Compensatory activation of the multidrug transporters Pdr5p, Snq2p, and Yor1p by Pdr1p in Saccharomyces cerevisiae. FEBS Lett. 2008;582:977–83. doi: 10.1016/j.febslet.2008.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–91. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 27.Rogers B, et al. The pleitropic drug ABC transporters from Saccharomyces cerevisiae. J Mol Microbiol Biotechnol. 2001;3:207–14. [PubMed] [Google Scholar]
- 28.MacDiarmid CW, Milanick MA, Eide DJ. Biochemical properties of vacuolar zinc transport systems of Saccharomyces cerevisiae. J Biol Chem. 2002;277:39187–94. doi: 10.1074/jbc.M205052200. [DOI] [PubMed] [Google Scholar]
- 29.Zhao H, Eide D. The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc Natl Acad Sci USA. 1996;93:2454–8. doi: 10.1073/pnas.93.6.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gitan RS, Shababi M, Kramer M, Eide DJ. A cytosolic domain of the yeast Zrt1 zinc transporter is required for its post-translational inactivation in response to zinc and cadmium. J Biol Chem. 2003;278:39558–64. doi: 10.1074/jbc.M302760200. [DOI] [PubMed] [Google Scholar]
- 31.MacDiarmid CW, Milanick MA, Eide DJ. Induction of the ZRC1 metal tolerance gene in zinc-limited yeast confers resistance to zinc shock. J Biol Chem. 2003;278:15065–72. doi: 10.1074/jbc.M300568200. [DOI] [PubMed] [Google Scholar]
- 32.Rockwell NC, Wolfger H, Kuchler K, Thorner J. ABC transporter Pdr10 regulates the membrane microenvironment of Pdr12 in Saccharomyces cerevisiae. J Membr Biol. 2009;229:27–52. doi: 10.1007/s00232-009-9173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cabrito TR, Teixeira MC, Singh A, Prasad R, Sá-Correia I. The yeast ABC transporter Pdr18 (ORF YNR070w) controls plasma membrane sterol composition, playing a role in multidrug resistance. Biochem J. 2011;440:195–202. doi: 10.1042/BJ20110876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pomorski T, et al. Drs2p-related P-type ATPases Dnf1p and Dnf2p Are Required for Phospholipid Translocation across the Yeast Plasma Membrane and Serve a Role in Endocytosis. Mol Biol Cell. 2003;14:1240–1254. doi: 10.1091/mbc.E02-08-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Prinz WA. ATP-binding cassette (ABC) transporters mediate nonvesicular, raft-modulated sterol movement from the plasma membrane to the endoplasmic reticulum. J Biol Chem. 2004;279:45226–34. doi: 10.1074/jbc.M407600200. [DOI] [PubMed] [Google Scholar]
- 36.Wilcox LJ, et al. Transcriptional profiling identifies two members of the ATP-binding cassette transporter superfamily required for sterol uptake in yeast. J Biol Chem. 2002;277:32466–72. doi: 10.1074/jbc.M204707200. [DOI] [PubMed] [Google Scholar]
- 37.Eide DJ. Homeostatic and adaptive responses to zinc deficiency in Saccharomyces cerevisiae. J Biol Chem. 2009;284:18565–9. doi: 10.1074/jbc.R900014200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burke MA, Mutharasan RK, Ardehali H. The sulfonylurea receptor, an atypical ATP-binding cassette protein, and its regulation of the KATP channel. Circ Res. 2008;102:164–76. doi: 10.1161/CIRCRESAHA.107.165324. [DOI] [PubMed] [Google Scholar]
- 39.Degryse E, Dumas B, Dietrich M, Laruelle L, Achstetter T. In vivo cloning by homologous recombination in yeast using a two-plasmid-based system. Yeast. 1995;11:629–40. doi: 10.1002/yea.320110704. [DOI] [PubMed] [Google Scholar]
- 40.Sung M, Huh W. Bimolecular fluorescence complementation analysis system for in vivo detection of protein – protein interaction in Saccharomyces cerevisiae. Yeast. 2007;24:767–775. doi: 10.1002/yea.1504. [DOI] [PubMed] [Google Scholar]
- 41.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–61. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 42.Nagai T, et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 43.Lee ME, et al. The Rho1 GTPase acts together with a vacuolar glutathione S-conjugate transporter to protect yeast cells from oxidative stress. Genetics. 2011;188:859–70. doi: 10.1534/genetics.111.130724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shannon P, et al. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boyle EI, et al. GO∷TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ashburner M, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ostlund G, et al. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 2010;38:D196–203. doi: 10.1093/nar/gkp931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick's Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2009;37:D793–6. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Babu M, Krogan NJ, Awrey DE, Emili A, Greenblatt JF. Systematic characterization of the protein interaction network and protein complexes in Saccharomyces cerevisiae using tandem affinity purification and mass spectrometry. Methods Mol Biol. 2009;548:187–207. doi: 10.1007/978-1-59745-540-4_11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.