Abstract
Cohesin mediates sister chromatid cohesion and contributes to the organization of interphase chromatin through DNA looping. In vertebrate somatic cells, cohesin consists of Smc1, Smc3, Rad21, and either SA1 or SA2. Three additional factors Pds5, Wapl, and Sororin bind to cohesin and modulate its dynamic association with chromatin. There are two Pds5 proteins in vertebrates, Pds5A and Pds5B, but their functional specificity remains unclear. Here, we demonstrate that Pds5 proteins are essential for cohesion establishment by allowing Smc3 acetylation by the cohesin acetyl transferases (CoATs) Esco1/2 and binding of Sororin. While both proteins contribute to telomere and arm cohesion, Pds5B is specifically required for centromeric cohesion. Furthermore, reduced accumulation of Aurora B at the inner centromere region in cells lacking Pds5B impairs its error correction function, promoting chromosome mis-segregation and aneuploidy. Our work supports a model in which the composition and function of cohesin complexes differs between different chromosomal regions.
Keywords: aneuploidy, chromosome biorientation, cohesin, mouse model, pericentric heterochromatin
Introduction
Cohesin consists of a heterodimer of the Structural Maintenance of Chromosome (SMC) proteins Smc1 and Smc3, the alpha kleisin subunit Rad21/Scc1, and a protein known as Scc3 in yeast and stromal antigen (SA) in metazoa. Entrapment of the sister chromatids by cohesin allows faithful DNA repair by homologous recombination (HR) in G2 and accurate chromosome segregation during mitosis and meiosis (Nasmyth and Haering, 2009; Remeseiro and Losada, 2013). Cohesin’s ability to entrap two DNA segments can also act in cis to promote long-range DNA looping, which is crucial for transcriptional regulation, organization of replication factories, and locus rearrangement (Hadjur et al, 2009; Guillou et al, 2010; Kagey et al, 2010; Seitan et al, 2011). Mutations in cohesin and its regulators are present in at least two human syndromes, Cornelia de Lange (CdLS; Liu and Krantz, 2009) and Roberts/SC Phocomelia (RBS; Vega et al, 2005), and have also been associated with tumourigenesis (Barber et al, 2008; Solomon et al, 2011; Welch et al, 2012).
A number of proteins regulate the association of cohesin with chromatin throughout the cell cycle. Cohesin is loaded onto chromatin in G1 by the Scc2–Scc4 heterodimer (Ciosk et al, 2000; Gillespie and Hirano, 2004; Watrin et al, 2006). Live-cell analyses have indicated that chromatin-bound cohesin is constantly turning over (Gerlich et al, 2006; Bernard et al, 2008; Gause et al, 2010). Cohesin unloading depends on Wapl, whose function is particularly important during prophase in metazoa, when most cohesin is released from the condensing chromosomes (Gandhi et al, 2006; Kueng et al, 2006; Tedeschi et al, 2013). To counteract this Wapl-dependent unloading and stabilize cohesin to encircle the replicated sister chromatids, cohesin acetyl transferases (CoATs) must acetylate two lysine residues located in the head domain of Smc3 (Rolef Ben-Shahar et al, 2008; Unal et al, 2008; Zhang et al, 2008; Sutani et al, 2009; Feytout et al, 2011). In vertebrates, Smc3 acetylation is accompanied by binding of Sororin to cohesin (Lafont et al, 2010; Nishiyama et al, 2010). Some evidence suggests that the recruitment and/or function of Wapl and Sororin depend on the HEAT-repeat containing protein Pds5, although there are important differences among organisms. Pds5 provides an interaction surface for the FGF motifs present in Wapl and Sororin (Shintomi and Hirano, 2009; Nishiyama et al, 2010). The binding of Sororin to Pds5-bound cohesin after DNA replication and Smc3 acetylation has been proposed to displace Wapl, thereby stabilizing cohesin in human cells. In mitotic prophase, Sororin is released (Dreier et al, 2011; Liu et al, 2013) and Wapl promotes dissociation of most cohesin complexes, an action that is enhanced by cohesin phosphorylation (Hauf et al, 2005; Gandhi et al, 2006; Kueng et al, 2006).
In vertebrate cells, multiple players in cohesion have undergone gene duplication events and subsequent divergence. Recent reports suggest that distinct cohesin variants are preferred at different regions of the chromosome. For example, cohesin complexes in somatic cells can carry the SA1 or the SA2 subunit. Cohesin-SA1 mediates telomere cohesion, cohesin-SA2 supports centromere cohesion, and both contribute to chromosome arm cohesion (Canudas and Smith, 2009; Remeseiro et al, 2012a). Similarly, there are two CoATs in vertebrates; Esco1 and Esco2 (Hou and Zou, 2005). Bulk acetylation of Smc3 depends on both CoATs (Zhang et al, 2008; Nishiyama et al, 2010), whereas local acetylation at pericentric heterochromatin (PCH) depends specifically on Esco2 (Whelan et al, 2011). Finally, there are two versions of Pds5; Pds5A and Pds5B (Sumara et al, 2000; Losada et al, 2005). Both proteins are ∼1400 amino acids long with ∼72% sequence homology throughout most of the protein, including the two clusters of HEAT repeats (Neuwald and Hirano, 2000), but differing sequences in their C-terminal 300 amino acids. Currently, little is known about the functional specificity of the two Pds5 proteins. Both Pds5A and Pds5B can be found associated with either cohesin-SA1 or cohesin-SA2 (Losada et al, 2005). Previous studies have reported that knockout mice for either Pds5A or Pds5B die perinatally with several organ malformations reminiscent of CdLS, but no clear cohesion problems (Zhang et al, 2007, 2009). Here, we report the generation of distinct knockout alleles for Pds5A and Pds5B and the analysis of mouse embryonic fibroblasts completely lacking either protein. Contrary to previous results, we find that Pds5B is essential for centromeric cohesion. In cells lacking Pds5B, impaired function of Esco2 and binding of Sororin at PCH prevents proper cohesion establishment and maintenance in this region. Chromosome biorientation and segregation are defective in these cells, which often leads to aneuploidy.
Results
Non-redundant functions of Pds5A and Pds5B in embryonic development and cell proliferation
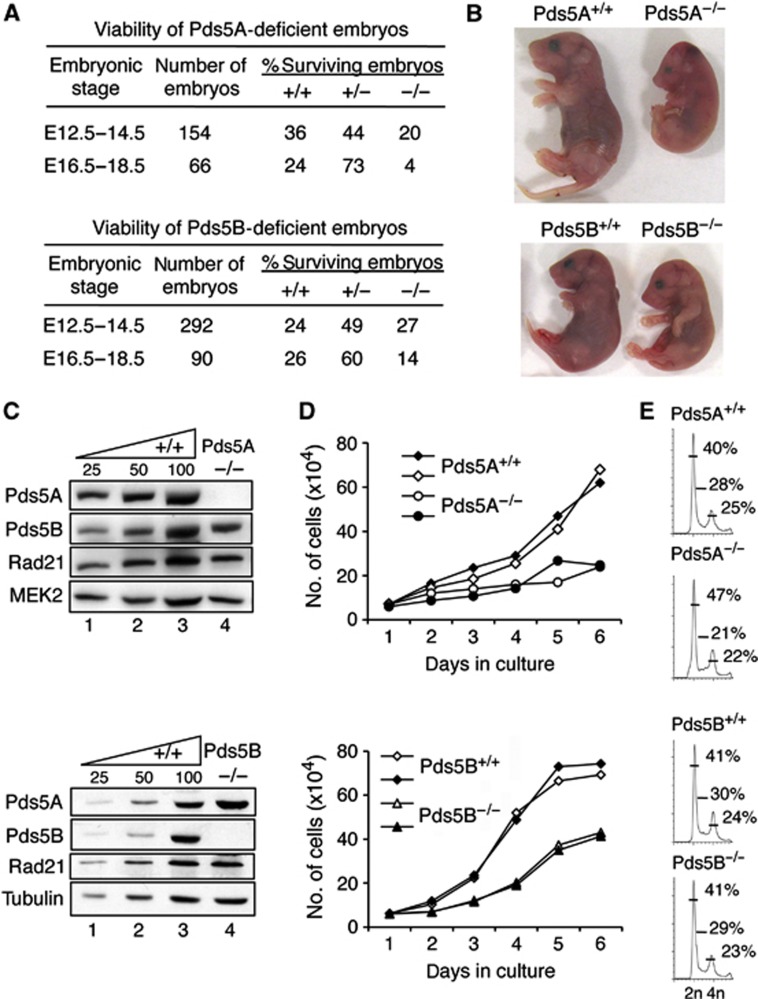
To explore the specific functions of the two Pds5 proteins, we generated mice carrying conditional knockout alleles for either Pds5A or Pds5B genes by insertion of loxP sites flanking either exon 6 in Pds5A or exons 4–5 in Pds5B (Supplementary Figure S1). Elimination of these exons leads to premature termination of translation and produces truncated proteins lacking HEAT repeats (176 out of 1332 amino acids for Pds5A and 124 out of 1442 amino acids in the case of Pds5B) that are unlikely to be functional. In both cases, crosses with mice ubiquitously expressing the Cre recombinase (CMV-Cre) generated heterozygous animals that were viable and fertile. However, homozygous progeny from subsequent backcrosses of heterozygous mice for either null allele was not obtained (out of >500 newborn mice), indicating that both genes are individually required for embryonic development. Embryonic lethality occurs at late post-implantation stages. Only 4% of Pds5A and 14% of Pds5B embryos from litters extracted between E16.5 and 18.5 carried homozygous null alleles compared to the expected 25% (Figure 1A). Pds5A-deficient embryos that survived to these late stages of development were noticeably smaller than their wild-type littermates (Figure 1B, top) and histological analysis revealed multiple anomalies in organogenesis (Supplementary Figure S2A and B). Late-stage Pds5B-deficient embryos were also smaller, but showed less severe defects (Figure 1B, bottom; Supplementary Figure S2C and D). Western blot analysis of fibroblasts obtained from E12.5 embryos homozygous for either knockout allele confirmed the absence of the corresponding protein and further revealed that lack of one Pds5 protein does not affect the levels of the other or of cohesin (Figure 1C). Finally, we observed that both Pds5A null and Pds5B null cells proliferate more slowly than wild-type cells, the defect being more pronounced for Pds5A (Figure 1D). Cell-cycle profiling reveals no major differences although asynchronous cultures of Pd5A null MEFs usually present a larger fraction of G1 cells and a smaller fraction of cells in S phase (Figure 1E). Taken together, our results indicate that both Pds5A and Pds5B are essential for proper cell proliferation and embryonic development.
Figure 1.
Effects of Pds5A or Pds5B ablation on mouse embryonic development and cell proliferation. (A) Viability of Pds5A- and Pds5B-deficient embryos. Heterozygous animals carrying a null allele of Pds5A (top) and Pds5B (bottom) were mated. (B) Pictures of littermate E18.5 embryos of the indicated genotypes. (C) Western blot analysis of whole-cell extracts prepared from mouse embryo fibroblasts (MEFs) obtained from E12.5 embryos homozygous for each knockout allele. The Pds5 antibodies recognize the most C-terminal region of Pds5A and Pds5B. Rad21 is a cohesin subunit. MEK2 or tubulin is used as a loading control. (D) Growth curves of primary MEFs of the indicated genotypes (two clones each). (E) Representative cell-cycle profiles obtained by FACS analysis of primary MEFs of the indicated genotypes. The numbers indicate the percentage of cells in G1, S, and G2/M according to the Dean-Jet-Fox model.
Pds5 proteins are required for efficient cohesin unloading
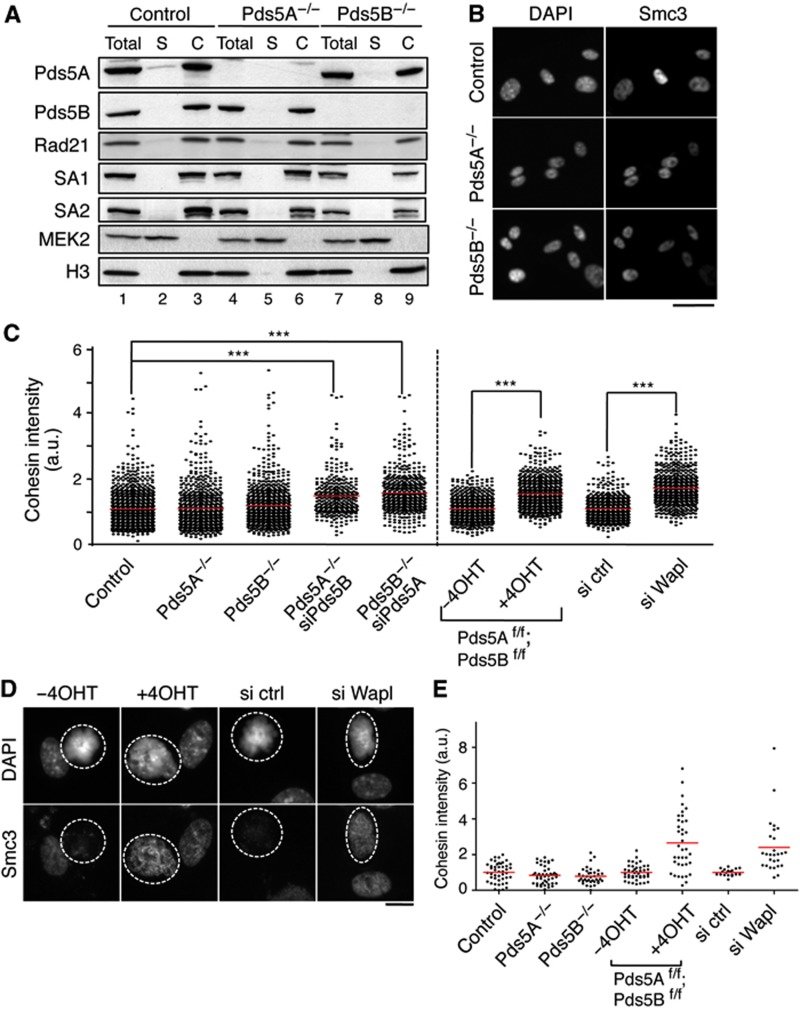
To determine how the absence of Pds5 proteins affects cohesin’s association with chromatin, chromatin bound levels of cohesin were analysed by chromatin fractionation (Figure 2A) and immunofluorescence (Figure 2B). Pds5A null and Pds5B null MEFs displayed similar levels of chromatin-bound cohesin to wild-type cells. We next employed two different strategies to simultaneously deplete both Pds5 proteins. One is transfection of siRNAs against Pds5A in Pds5B null cells or siRNAs against Pds5B in Pds5A null cells (Supplementary Figure S3A). The other is the generation of a Pds5 double knockout conditional (floxed, ‘f’) allele and a Cre-ERT2 transgene. Upon addition of 4-hydroxytamoxifen (4-OHT), activation of the Cre recombinase leads to decreased levels of both Pds5 proteins in MEFs carrying the double conditional knockout allele in homozygosis (Supplementary Figure S3B). A moderate but a significant increase in chromatin-bound cohesin could be detected by immunofluorescence in interphase cells when the levels of both Pds5 proteins were reduced (Figure 2C; Supplementary Figure S3C). As a control, we also measured cohesin intensity in MEFs in which Wapl was partially depleted using siRNA and obtained similar results (Figure 2C; Supplementary Figure S3C and D). We suggest that the increase in chromatin-bound cohesin likely reflects reduced cohesin unloading, consistent with a reduction in the presence of Wapl on chromatin in the absence of both Pds5A and Pds5B (see next section, Figure 3B). In mitosis, the absence of both Pds5 proteins also had a measurable effect on the efficiency of cohesin release by the prophase pathway (Figure 2D and E). We therefore conclude that Pds5 contributes to cohesin unloading both in interphase and in mitosis.
Figure 2.
Levels of chromatin-bound cohesin in the absence of the Pds5 proteins. (A) Chromatin fractionation of MEFs wild type (control henceforth), Pds5A−/−, and Pds5B−/−. Equivalent amounts of total cell extract (Total), soluble fraction (S), and chromatin-enriched fraction (C) were loaded. Rad21, SA1, and SA2 are cohesin subunits. The cytoplasmic kinase MEK2 and histone H3 are used as controls for the fractionation procedure. (B) Representative images of primary MEFs of the indicated genotypes pre-extracted with detergent before fixation to remove soluble proteins and stained with Smc3. Scale bar, 50 μm. (C) Box plot showing the quantification of cohesin staining (Smc1 or Smc3, in arbitrary units and normalized to the average value obtained in control cells) in interphase cells of the indicated genotypes and conditions. Western blot analyses to assess the efficiency of siRNA treatments are shown in Supplementary Figure S3. The following numbers of cells (n) from several clones (N) were measured: control, n=972, N=7; Pds5A−/− n=698, N=4; Pds5B−/−, n=742 cells, N=6; Pds5A−/− siPds5B, n=334, N=2; Pds5B−/− siPds5A, n=482, N=5; Pds5Af/f;Pds5Bf/f;Cre-ERT2 MEFs −4OHT and +4OHT, n=700 each, N=1; control and Wapl siRNA, n=600 each, N=1. Bonferroni’s multiple comparison test was used to assess significance. ***P<0.001. (D) Examples of primary MEFs in metaphase (surrounded by dotted line) treated with detergent before fixation and stained with Smc3. (E) Cohesin staining was measured in at least 35 metaphases for the indicated genotypes or condition except for control and si ctr and si Wapl in which n=19 and 26, respectively. Scale bar, 10 μm.
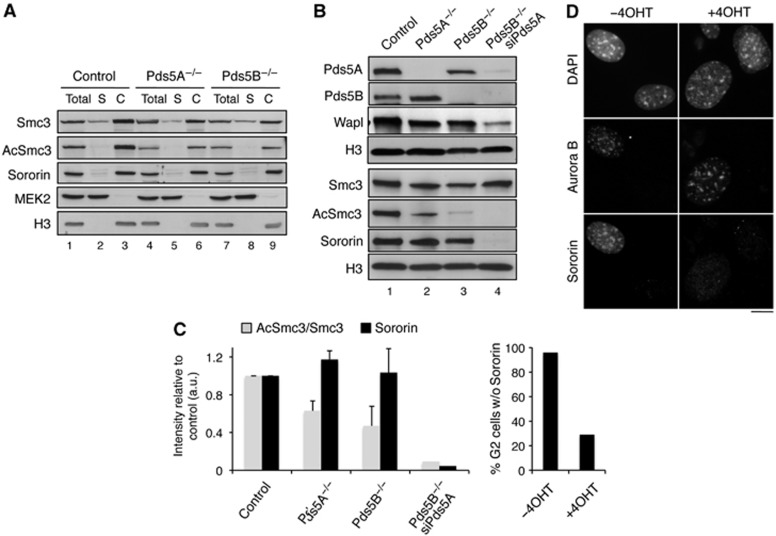
Figure 3.
Pds5A and Pds5B are required for full acetylation of cohesin and Sororin recruitment. (A) Western blot analysis of the chromatin fractionation shown in Figure 2A with additional antibodies. (B) Western blot analysis of the chromatin fraction of primary MEFs of the indicated genotypes untransfected or transfected with siRNAs against Pds5A (siPds5A). (C) Quantification of the indicated signals from the two experiments shown above and an additional one not shown. Bars represent mean+s.e.m. (D) G2 cells in asynchronous cultures of Pds5Af/f;Pds5Bf/f;Cre-ERT2 MEFs grown in the absence or presence of 4-OHT were stained with Aurora B and Sororin antibodies and counterstained with DAPI. The percentage of cells labelled by both Aurora B and Sororin was quantified among >160 G2 cells from two different clones and plotted in the graph below the images. Scale bar, 10 μm.
Pds5 proteins are required for Smc3 acetylation and Sororin recruitment
We next sought to test the contribution of Pds5 to cohesin acetylation and Sororin recruitment. Depletion of a single Pds5 protein leads to a partial reduction in acetylated cohesin based on the chromatin fractionation of asynchronous MEF cultures, but did not affect much Sororin recruitment (Figure 3A and C). In contrast, a strong reduction in both Smc3 acetylation and chromatin-bound Sororin could be observed when the levels of Pds5A were reduced by RNAi in Pds5B null MEFs (Figure 3B). Wapl binding to chromatin was also clearly decreased. Similar results were obtained using the Pds5 double knockout MEFs (Supplementary Figure S3E). However, FACS analysis and analysis of EdU incorporation in asynchronous cultures indicates a 2- to 3-fold reduction in the number of cells traversing S phase in the absence of both Pds5 proteins (Supplementary Figure S3F). Since Sororin recruitment occurs specifically during S phase, altered cell-cycle progression could explain, at least in part, the reduction of Sororin on chromatin. We therefore analysed the presence of Sororin by immunofluorescence in G2 cells. Pds5-depleted cells showed no Sororin staining (Figure 3D). Taken together, these results suggest that Pds5 proteins are required for cohesin acetylation in mammalian cells, as recently found in yeast (Vaur et al, 2012; Chan et al, 2013), as well as for recruitment of Sororin and Wapl.
Both Pds5A and Pds5B contribute to telomere and arm cohesion in mouse cells
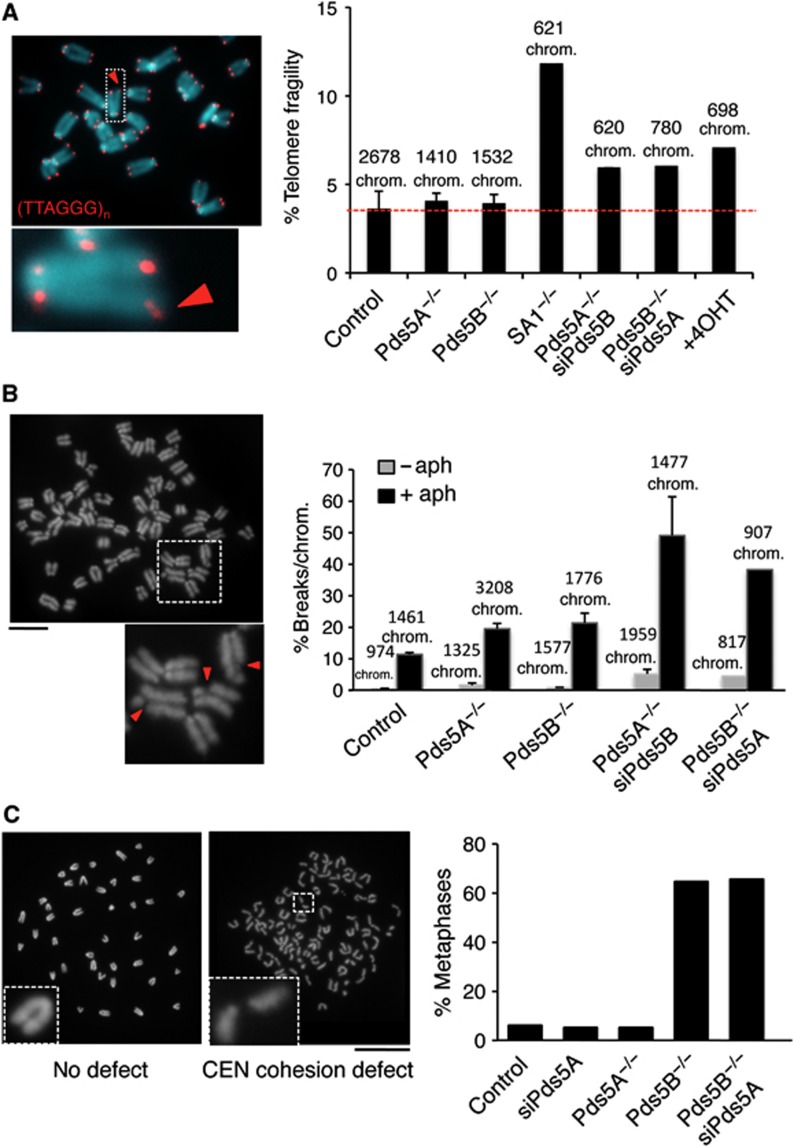
We next sought to investigate whether all populations of cohesin display a similar requirement for Pds5 activity by assessing cohesion at telomeres, arm, and centromeres. We performed fluorescence in situ hybridization (FISH) of interphase cells with two probes each from subtelomere and arm regions. Cohesion defects, revealed by the appearance of doublets, were not increased in Pds5A or Pds5B null cells compared to wild-type cells, suggesting that either Pds5 protein can mediate arm and telomere cohesion (Supplementary Figure S4). Cohesion facilitates HR-mediated DNA repair and recovery of stalled replication forks in regions difficult to replicate, like the telomeres or the fragile sites. Defective telomere replication results in irregularly shaped or multimeric signals upon hybridization with a telomeric repeat probe, a phenotype known as telomere fragility (Sfeir et al, 2009). Fragile sites generate breaks visible in mitotic chromosomes following exposure to a low dose of aphidicolin (Chan et al, 2009). We have previously shown that in the absence of cohesin or Sororin both telomere fragility and fragile site expression are increased (Remeseiro et al, 2012a). Thus, we used these two assays to evaluate telomere and arm cohesion more globally than with the FISH assay in the Pds5-deficient MEFs. Telomere fragility was similar in wild-type, Pds5A null, or Pds5B null MEFs but it increased in the absence of both Pds5 proteins, although to a lesser extent than in the SA1 null MEFS, which served as a positive control (Figure 4A). Similarly, a lack of Pds5A or Pds5B had a modest effect in the appearance of chromosome breaks in metaphase cells that had been treated with a low dose of aphidicolin, but the effect increased considerably when both Pds5 proteins were reduced (Figure 4B). From these results, we conclude that both Pds5A and Pds5B contribute to telomere and arm cohesion.
Figure 4.
Different requirements for Pds5A and Pds5B in telomere and centromere cohesion. (A) Metaphase chromosomes were stained with a telomeric repeat probe (red) and DAPI (blue). The image on the left corresponds to a wild-type cell. The fraction of fragile telomeres, that is, not round but elongated or double signals (red arrowhead) was measured for the indicated number of chromosomes from six clones control MEFs, three clones of Pds5A null or Pds5B null MEFs, two clones of SA1 null MEFs and one clone of the rest, and plotted (right). +4OHT refers to Pds5Af/f;Pds5Bf/f;Cre-ERT2 MEFs treated with 4-OHT for 5 days. (B) Quantification of breaks along the chromosome arms in metaphase chromosomes from cells either untreated (grey bars) or treated with 0.5 μM aphidicolin for 24 h (black bars). The images on the left show examples of the broken chromosomes (red arrowheads) in a Pds5B−/− cell. We counted the indicated number of chromosomes from two clones in all cases except Pds5B−/−siPds5A in which a single clone was used. (C) Representative images of metaphase spreads from control (left) and Pds5B−/− MEFs (right) showing proper and defective centromere cohesion, respectively. The graph below represents the percentage of cells showing more than five chromosomes with centromere cohesion defects among n⩾100 metaphase cells of each condition. We did not count metaphases showing only single chromatids, that is, in which pairs of sisters cannot be recognized. Scale bar, 20 μm.
Pds5B ablation leads to centromeric cohesion defects
We next examined chromosome morphology in metaphase chromosome spreads. Pds5B null MEFs displayed pronounced centromeric cohesion defects with >60% of the metaphases from Pds5B null cells showing >5 chromosomes with clearly separated centromeres compared to <6% of the wild-type or Pds5A null cells (Figure 4C). Downregulation of Pds5A by RNAi in Pds5B null MEFs did not clearly enhance the defects caused by loss of Pds5B. The defect could also be observed in the absence of colcemide treatment, suggesting that is not a consequence of prolonged mitotic arrest (Supplementary Figure S5A). To confirm these data using another cell type, we prepared metaphase spreads from hepatocytes taken directly from E14.5 embryos. We observed centromeric cohesin defects in hepatocytes obtained from Pds5B null embryos but not in those from wild-type or Pds5A null embryos (Supplementary Figure S5B). Thus, we conclude that Pds5B is responsible for centromeric cohesion in mouse cells.
Pds5B recruits Sororin to PCH
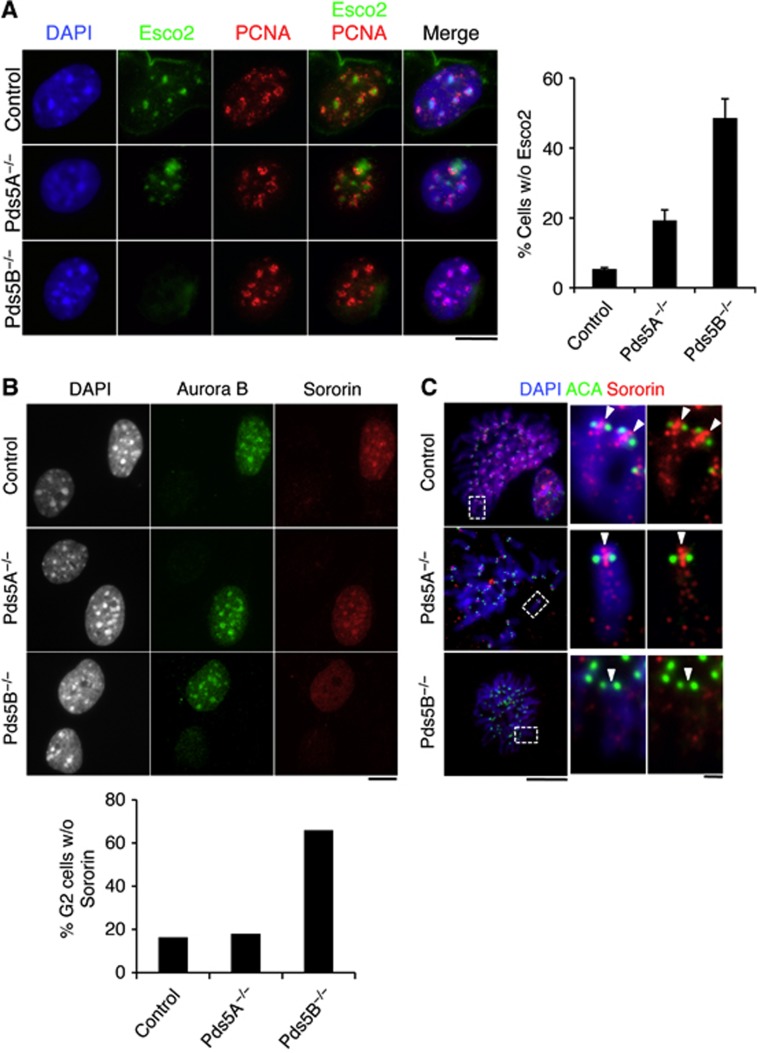
Cohesin acetylation at PCH occurs during mid-late S phase and depends on the Esco2 acetyl transferase (Whelan et al, 2011). Chromatin-bound levels of Esco2 are not consistently different between wild-type and Pds5B null cells (Supplementary Figure S3G) or cells depleted from both Pds5A and Pds5B (Supplementary Figure S3F), at least by immunoblotting. Thus, it is unlikely that Pds5 proteins recruit Esco2 to chromatin. During late S phase, wild-type MEFs show a characteristic horseshoe-shaped PCNA staining around DAPI-dense heterochromatin foci, with Esco2 staining inside (Figure 5A). In contrast, Esco2 was not visible in almost 50% of Pds5B null cells showing this PCNA staining pattern. This result suggests that Pds5B promotes Esco2 action on cohesin present at PCH in concert with DNA replication. Sororin accumulation at heterochromatin foci in G2 cells (Figure 5B) or at the inner centromere region of mitotic chromosomes (Figure 5C) was less evident in the absence of Pds5B. Taken together, these results indicate that Pds5B is required to promote cohesion establishment and maintenance near centromeres by facilitating cohesin acetylation by Esco2 and Sororin binding at PCH.
Figure 5.
Esco2 acetylation of cohesin and Sororin recruitment at PCH depend on Pds5B. (A) Esco2 (green) accumulates at the late replicating heterochromatin, labelled by PCNA (red), of control and Pds5A−/− MEFs, but not in Pds5B−/− MEFs. For the graph on the right, we counted the fraction of Esco2-negative/PCNA horseshoe-positive cells among cells (n) from several clones (N): control, n=168, N=2; Pds5A−/−, n=57; N=3; Pds5B−/− n=177, N=3. Scale bar, 10 μm. (B) G2 cells labelled by Aurora B (green) showing reduced accumulation of Sororin (red) at DAPI-dense foci of pericentric heterochromatin were scored among 300, 506, and 713 G2 cells in 2 clones each of control, Pds5A−/−, and Pds5B−/− MEFs. Scale bar, 10 μm. (C) Metaphase chromosomes prepared by cytospin were labelled with Sororin (red), ACA (anti-centromere antibody, green), and DAPI (blue). While Sororin stains the inner centromere region of control and Pds5A null chromosomes, it is absent from chromosomes lacking Pds5B. Scale bar, 10 μm; inset, 1 μm.
Centromeric cohesion defects lead to aneuploidy
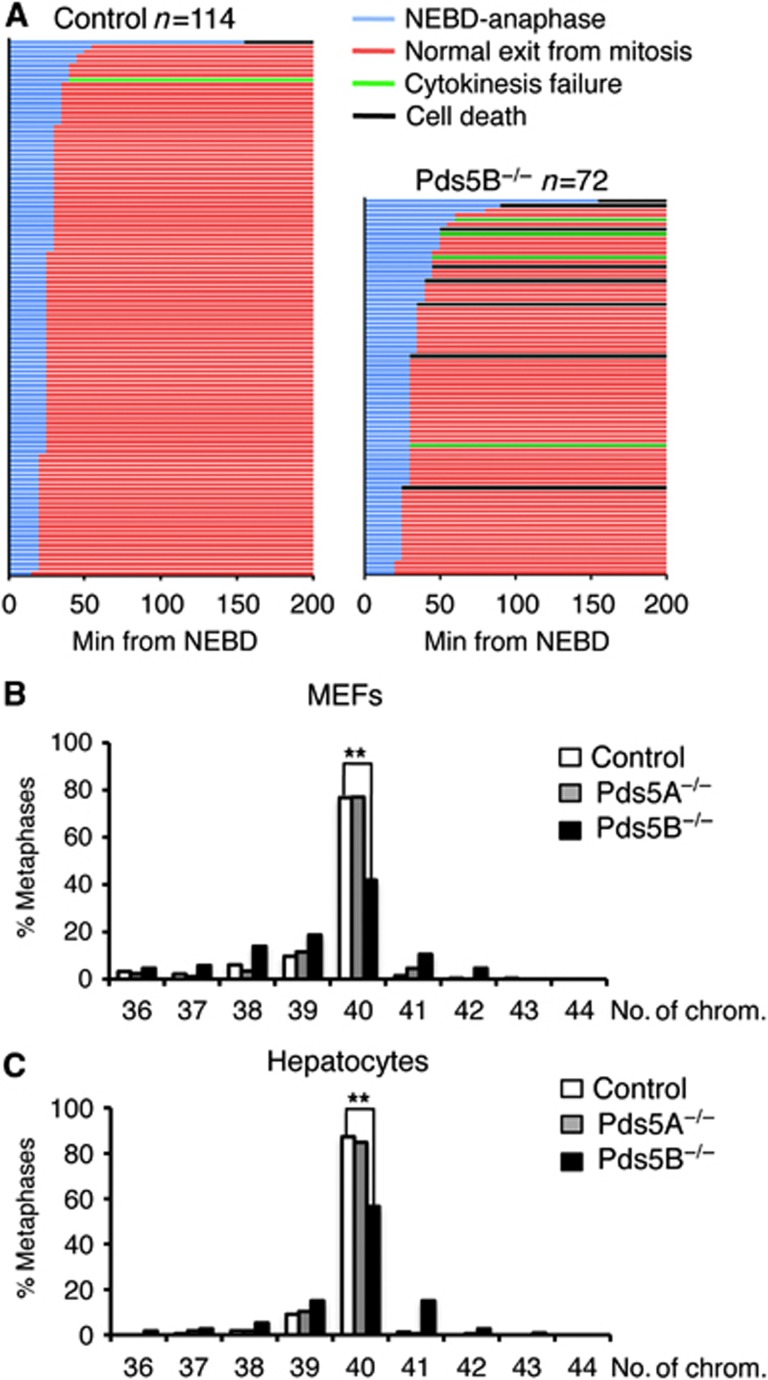
We next analysed mitotic progression in Pds5B null cells by live-cell imaging of primary MEFs transfected with histone H2B-mCherry (Figure 6A; Supplementary Figure S6). Despite defective centromeric cohesion, Pds5B null MEFs align their chromosomes and initiate anaphase with only a short delay compared to wild-type MEFs (30 and 25 min, respectively, from nuclear envelope breakdown (NEBD) to anaphase onset; Figure 6A, blue lines). Around 11% of Pds5B null cells died in mitosis (black lines) and 6% were unable to complete cytokinesis (green lines), most likely due to defective chromosome segregation during anaphase. Indeed, immunofluorescence of fixed cells revealed an increased incidence of lagging chromosomes and chromatin bridges in anaphase cells lacking Pds5B, as well as the presence of binucleated cells and cells with micronuclei (Supplementary Figure S7). Consistent with this, up to 58% of primary MEFs from Pds5B null embryos were aneuploid, compared to 23% of wild-type or Pds5A null MEFs (Figure 6B). This increase in aneuploidy in the absence of Pds5B was also observed in fetal hepatocytes with 43% aneuploid cells in Pds5B null embryos compared to 15% for Pds5A null embryos and 13% in wild-type embryos, Figure 6C).
Figure 6.
Chromosome segregation defects and aneuploidy in the absence of Pds5B. (A) Graphical summary of the progression through mitosis of control (n=114) and Pds5B−/− MEFs (n=72), as observed by live-cell imaging after transfection with H2B-mCherry (red) to label chromatin. Each line represents the progression through mitosis of a single cell and it is coloured according to the legend shown above the graph. Nuclear envelope breakdown (NEBD) is considered as t=0. Still images from some of the movies can be found in Supplementary Figure S6. (B) Graph showing the distribution in the number of chromosomes of at least 80 metaphases from 4 clones of control, 2 clones Pds5A−/− and 5 clones of Pds5B−/− primary MEFs. **P<0.005 (unpaired t-test). (C) Same analysis in cells from hepatocytes taken from E14.5 embryos. We examined 165 metaphases from three wild-type embryos, 165 metaphases from 2 Pds5A null embryos, and 109 metaphases from 4 Pds5B null embryos. **P<0.005 (unpaired t-test).
Impaired localization and activation of Aurora B in Pds5B-deficient cells
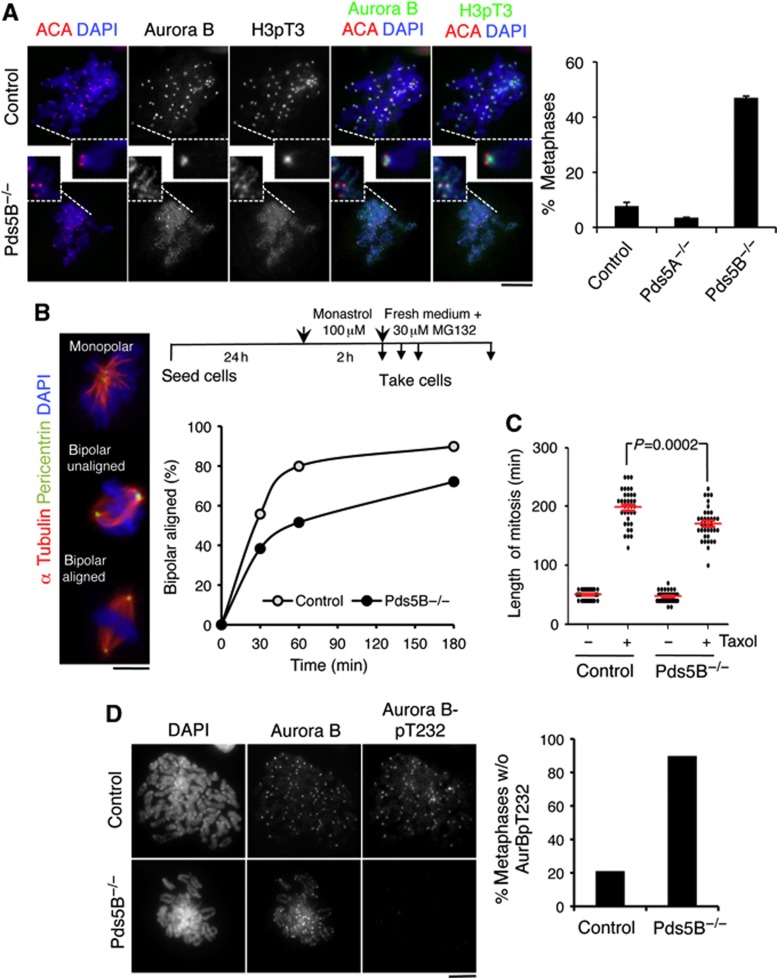
Centromeric cohesion generates tension by opposing the pulling forces of microtubules on kinetochores. In the absence of tension, Aurora B destabilizes kinetochore-microtubule attachments and the ‘empty’ kinetochores generate a signal that activates the spindle assembly checkpoint (SAC). Proper localization of Aurora B is essential for this function and depends on Bub1-mediated phosphorylation of H2A at Threonine 120 and Haspin-mediated phosphorylation of histone H3 on Threonine 3 (H3pT3) at the inner centromere region (Kelly et al, 2010; Wang et al, 2010, 2012; De Antoni et al, 2012). Bub1 was properly localized at centromeres independently of Pds5 proteins (Supplementary Figure S8). In contrast, accumulation of H3pT3 and Aurora B at the inner centromere was clearly reduced in cells lacking Pds5B (Figure 7A; Supplementary Figure S9). On the basis of this, Pds5B is likely to be required for recruitment of Haspin to centromeres, as has recently been reported in fission yeast (Yamagishi et al, 2010). To assess the downstream consequences of impaired Aurora B localization, we treated MEFs with the Eg5-inhibitor monastrol, which arrests cell in prometaphase with monopolar spindles and syntelic attachments (Kapoor et al, 2000). Correction of these syntelic attachments to form bipolar spindles, which depends on Aurora B activity, was delayed in the Pds5B null cells (Figure 7B). Moreover, live-cell imaging of MEFs treated with the microtubule stabilizing drug taxol, which activates the SAC in an Aurora B-dependent manner (Vader et al, 2007), indicated that cells exit mitosis earlier in the absence of Pds5B. This suggests that Pds5B null cells cannot maintain the mitotic arrest due to reduced local activity of Aurora B (Figure 7C). Indeed, a dramatic reduction in the levels of active Aurora B, as judged by staining with an antibody against Aurora B phospho-Threonine 232, was observed in metaphase cells lacking Pds5B (Figure 7D). Thus, Pds5B is important not only for centromeric cohesion, but also for proper accumulation of active Aurora B at centromeres.
Figure 7.
Pds5B is required for proper localization and activation of Aurora B. (A) Metaphase chromosomes prepared by cytospin were labelled with Aurora B, H3pT3, ACA, and DAPI. In control cells, and also in Pds5A null cells, Aurora B accumulates between the sister kinetochores labelled by ACA (top). In Pds5B−/− cells, Aurora B and H3pT3 signals appear all along chromosome arms and their centromeric enrichment is dramatically reduced (bottom). Scale bar, 10 μm. For the graph on the right, at least 85 metaphases from four different clones were scored for each genotype. Data represent mean+s.e.m. (B) Experimental design for the monastrol washout experiment (top) and representative images of the spindles observed after staining with alpha tubulin (red), pericentrin (green), and DAPI (blue). Between 65 and 175 spindles were counted in each time point for each genotype. (C) Box plot showing the duration of mitosis (as measured by live-cell imaging from NEBD to chromosome decondensation) in control and Pds5B−/− MEFs treated or not with taxol. Thirty-six cells were scored for each condition. (D) Metaphase chromosomes from control and Pds5B−/− MEFs prepared by cytospin were labelled with Aurora B and phospho(T232)Aurora B. We scored the staining of 58 metaphases from 2 control clones and 45 metaphases from 2 Pds5B−/− clones. Scale bar, 10 μm.
Discussion
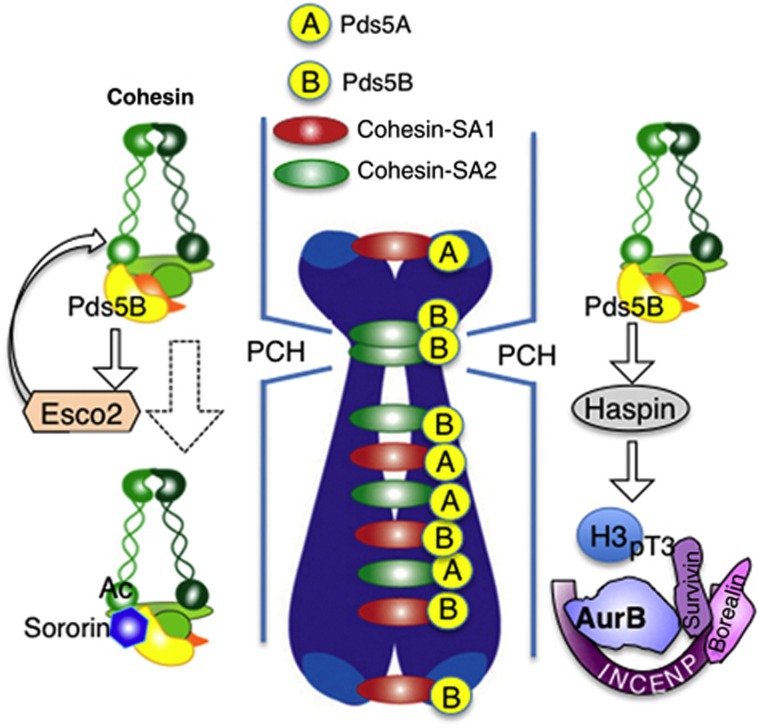
To assess the specific roles of the two Pds5 proteins in vertebrates, we have generated loss-of-function alleles for Pds5A and Pds5B in mice. Previous reports suggested that these genes were redundant because homozygous knockout mice for either gene could survive until birth and presented similar developmental defects (Zhang et al, 2007, 2009). Moreover, no cohesion defects were detected in this previous work. Here, by generating independent knockout alleles, we demonstrate that both Pds5A and Pds5B are essential to fulfill embryonic development. Analysis of primary MEFs lacking Pds5A, Pds5B, or both proteins revealed that both Pds5A and Pds5B contribute to telomere and arm cohesion whereas Pds5B is specifically required for centromeric cohesion. We do not know the reason for the discrepancy with the abovementioned studies regarding detection of centromere cohesion defects in metaphase spreads. In our hands, this phenotype is observed not only in Pds5B knockout MEFs but also in hepatocytes from Pds5B knockout embryos. Also in HeLa cells, downregulation of Pds5B levels results in centromeric cohesion defects (Losada et al, 2005). Taken together with previous analysis of SA1 and SA2 in mouse (Remeseiro et al, 2012a) and human cells (Canudas and Smith, 2009), it is likely that cohesin-SA1-Pds5A and cohesin-SA1-Pds5B act at telomeres, cohesin-SA2-Pds5B functions preferentially at centromeres, and these three complexes as well as cohesin-SA2-Pds5A perform cohesion along chromosome arms (Figure 8). The molecular determinants that direct these specificities remain to be identified. It is also unclear why telomeric and centromeric cohesin complexes should be different. We speculate that cohesion serves different purposes at different chromosomal regions. While telomere cohesion ensures telomere replication, centromere cohesion promotes chromosome biorientation by providing tension and proper localization of the chromosomal passenger complex (CPC).
Figure 8.
Specific cohesin complexes function at different chromosomal regions. A schematic representation of the specificity of different cohesin complexes (with SA1 or SA2 and Pds5A or Pds5B) to perform cohesion in different regions of the chromosome. Whether this functional distribution reflects a specific localization of the corresponding complexes is not known. The presence of Pds5B at pericentric heterochromatin (PCH) promotes cohesin acetylation by Esco2 during DNA replication and Sororin recruitment. Pds5B is also important for proper localization of Aurora B at the inner centromere region of mitotic chromosomes, maybe by promoting Haspin recruitment in mitosis.
Centromeric cohesion is viewed as being the most critical for proper chromosome segregation and is the likely reason for cohesin accumulation at the centromeres of mitotic chromosomes (McGuinness et al, 2005; Watanabe and Kitajima, 2005). Pds5B null cells have major defects in centromeric cohesion as judged by metaphase chromosome spreads, but nevertheless are capable of forming a metaphase plate and initiating anaphase with only a short delay compared with wild-type cells. We envision that cohesion mediated by cohesin bound to Pds5A along chromosome arms, as well as DNA catenation, provides sufficient linkages between the sister chromatids to allow chromosome alignment in Pds5B null cells. Even though inter-kinetochore stretch may be reduced in the absence of cohesin-mediated centromere cohesion, the residual intra-kinetochore stretch may be sufficient to satisfy the SAC (Maresca and Salmon, 2009; Uchida et al, 2009). However, we suspect that Pds5B null cells begin anaphase without having achieved proper biorientation, which leads to chromosome segregation defects and, eventually, to aneuploidy (Kapoor et al, 2006). We have shown that the accumulation of Aurora B at the inner centromere is defective in Pds5B null cells, possibly through the impaired recruitment of Haspin, which may further prevent the correction of improper attachments and a cell-cycle delay (Cimini et al, 2006; Vader et al, 2007; Santaguida et al, 2011). In S. pombe, Pds5 is also required for proper localization of Haspin (Hrk1) and a physical interaction between the two proteins has been detected (Yamagishi et al, 2010).
Results from different model organisms have suggested that Pds5 proteins act as both positive and negative regulators of cohesion, functioning for the establishment and maintenance of cohesion, but also for its destabilization (Hartman et al, 2000; Panizza et al, 2000; Tanaka et al, 2001; Dorsett et al, 2005; Losada et al, 2005; Rowland et al, 2009; Shintomi and Hirano, 2009; Sutani et al, 2009; Vaur et al, 2012). Our analysis indicates that Pds5A and Pds5B are indeed critical for cohesion establishment and maintenance in vertebrates, as these proteins are required for full Smc3 acetylation and Sororin binding. A role for Pds5 in promoting the association of Sororin with chromatin in Xenopus egg extracts had been already reported but this role was thought to derive from the fact that Sororin binds cohesin through Pds5 (Nishiyama et al, 2010). We here show that Pds5 proteins are also necessary for the acetylation of Smc3, although the molecular mechanisms underlying this requirement remain to be identified. A recent study proposes that S. cerevisae Pds5 both promotes Smc3 acetylation by Eco1 and prevents its deacetylation by Hos1 (Chan et al, 2013). Consistent with previous results in Xenopus showing that Esco2 recruitment does not depend on cohesin and, therefore, on Pds5 (Higashi et al, 2012), we have not seen decreased binding of Esco2 to chromatin even when the two Pds5 proteins are absent. However, a cross-talk must exist between the CoATs and Pds5 proteins, which is essential for Smc3 acetylation and cohesion establishment and maintenance. In particular, we observe the reduced presence of Esco2 at replicating PCH foci in Pds5B null cells, which correlates with reduced Sororin at PCH and centromere cohesion defects. A lack of centromeric cohesion has been proposed to underlie the pathology of the Roberts syndrome (Vega et al, 2005; Monnich et al, 2011; Whelan et al, 2011). From the results presented here, one could expect Pds5B mutations to be causative of this disease. However, all cases reported to date display homozygous mutations in Esco2. Loss of a single Pds5B allele does not lead to centromeric cohesion defects, at least in MEFs, whereas loss of both alleles impairs embryonic development and results in prenatal or perinatal death (Zhang et al, 2007, 2009). The same may be true in humans.
Decreased proliferation of Pds5B null cells could be explained by mitotic cell death and aneuploidy. Cells lacking Pds5A have a stronger proliferation defect and Pds5A null embryos present an earlier lethality, but in this case cells display correct ploidy and no mitotic defects. Previous results indicate that Pds5 regulates transcription in Drosophila (Dorsett et al, 2005; Gause et al, 2010). We speculate that the Pds5A null phenotypes may be related to altered transcription. Future experiments will have to address the genome-wide distribution of Pds5A and Pds5B and the effects of their ablation on gene expression during development.
Materials and methods
Generation of knockout alleles for PdsA and Pds5B
A Pds5A targeting vector containing 6.4-kb (right) and 5.6-kb (left) homology arms, an FRT-flanked neomycin cassette and two loxP sites flanking this cassette and exon 6 was generated by Ozgene. We generated a Pds5B targeting vector in pDELBOY-3X with 5.3-kb (right) and 4.6-kb (left) homology arms, two loxP sites flanking exons 4–5 and an FRT-flanked neomycin cassette. These vectors were electroporated into the 129/SvPas embryonic stem (ES) cells and neomycin-resistant clones were screened by Southern blotting and PCR. Verified ES cell clones were microinjected into C57BL/6J blastocysts and germline transmission was assessed. Animals carrying the Pds5A lox frt allele were mated with Tg-CMV-Cre mice to generate mice with the Pds5A null allele. For the Pds5B null allele, animals carrying the Pds5B lox frt allele were mated first with Tg-pCAG-Flpe mice and then with Tg-CMV-Cre mice. Mice carrying both the Pds5A lox frt and Pds5B lox alleles in the same chromosome as well as a transgene encoding a Cre recombinase fused to oestrogen receptor (Cre-ETR2) were also generated. Mice were housed in a pathogen-free animal facility following the animal care standards of the institution. The following primers were used for genotyping by PCR: Pds5A-fw: 5′-GGACACTTTAGCAGTTACCTCAGC-3′; Pds5A-rv1: 5′-ACCCTAAGTCCCAATGCACC-3′; Pds5A-rv2: 5′-GGCGGAAAGAACCATCTAGC-3′; Pds5B-fw: 5′-GCCCTTCTTTCATTGTTTAC-3′; Pds5B-Rv: 5′-GGTTTGCAGAGAGTTCTAGC-3′ (see Supplementary Figure S1 for localization).
MEF isolation and cell culture, RNA interference, and chromatin fractionation
Primary MEFs were isolated from E12.5 embryos and cultured in DMEM supplemented with 20% FBS and antibiotics under 90% humidity and 5% CO2. For proliferation assays, 60 000–80 000 cells were seeded in six wells, and each day the cells in one well were trypsinized and counted. For RNA interference, siGENOME SMARTpool siRNAs from Dharmacon (M-054017-01 for Pds5A; M-058400-01 for Pds5B; M-047528-01 for Wapl) were introduced in MEFs at a final concentration of 100 nM with the Neon transfection system (Invitrogen). Knock-down efficiencies were assessed 72 h after transfection by western blot analysis of whole-cell extracts. Double conditional knockout primary MEFs (Pds5A f/f; Pds5B f/f; Cre-ERT2) were maintained in medium with 1 μM 4-OHT for 4–6 days and the efficiency of elimination of Pds5A and Pds5B protein was assessed by western blotting. Whole-cell extracts from MEFs for western blot analysis were prepared as in Remeseiro et al (2012a). For chromatin fractionation, we followed the protocol from Mendez and Stillman (2000).
Metaphase spreads, FISH, telomere fragility assay, and cytospin
Chromosome spreads from MEFs and fetal livers, FISH of interphase nuclei, as well as the telomere fragility assay were performed as previously described (Remeseiro et al, 2012a). In the indicated cases, cells were cultured in 0.5 μM aphidicolin for 24 h before harvesting. For cytospin, primary MEFs were swollen in 75 mM KCl for 20 min at 37°C and spun onto poly-L-lysine coated coverslips at 1000, r.p.m. for 10 min using a Cytospin-3 centrifuge (Shandon). Cells were fixed in 4% paraformaldehyde (PFA) in PBS and processed for immunofluorescence.
Live-cell imaging and monastrol washout assay
Primary MEFs at p2 were transfected with H2B-mCherry and seeded on chamber slides. Time-lapse live-cell imaging was performed using the Delta Vision system (Applied Precision). Phase-contrast and fluorescent images were acquired with a × 40 objective every 5 min for 24 h. For the experiment in Figure 7C, 1.5 μM Taxol was added to the cells right before starting recording. The videos were analysed using the ImageJ 1.43 software. For the monastrol recovery assay, MEFs growing on coverslips were treated with 100 μM monastrol for 2 h, washed five times with PBS and incubated with media containing 30 μM MG132 for up to 3 h before fixation in 4% PFA and analysis by immunofluorescence.
Immunofluorescence and image analysis
MEFs grown on coverslips were fixed with 4% PFA for 20 min and permeabilized in 0.25% Triton X-100 in PBS for 5 min on ice. When required, cells were pre-extracted with 0.5% Triton X-100 in CSK buffer (10 mM Pipes pH 7.0, 100 mM NaCl, 3 mM MgCl2 and 300 mM sucrose) for 5 min before fixation. Cells were routinely blocked with 3% BSA, 0.05% Tween-20 in PBS for 30 min. Primary and secondary antibodies were diluted in blocking solution and incubated for 1 h each. Detection of Sororin required overnight incubation and including 10% normal donkey serum in the blocking solution. DNA was counterstained with 1 μg/ml DAPI. A Leica DM6000 microscope was used to obtain greyscale images, which were later pseudo-coloured and merged using Adobe Photoshop, and analysed in Image J (http://rsb.info.nih.gov/ij) or Definiens software.
Antibodies
Rabbit polyclonal antibodies against Rad21 and Pds5A were generated using peptides as immunogens (Rad21, GGDQDQEERRWNKRTQQMLC; Pds5A, CKKAVPAERQIDLQR) and affinity purified. Other custom-made antibodies have been described: Smc1, Smc3, SA1, SA2, Sororin (Remeseiro et al, 2012a; Remeseiro et al, 2012b); Pds5B (Losada et al, 2005); acetylated Smc3 (a gift from K Shirahige; Nishiyama et al, 2010); Esco2 (a gift from G Whelan and G Eichele; Whelan et al, 2011); Wapl (a gift from JM Peters; Tedeschi et al, 2013); Bub1 (a gift from S Taylor; Taylor et al, 2001). Additional antibodies from commercial sources are α-tubulin (Sigma, DM1A); ACA (Antibodies Inc., 15-235); Aurora B (AIM-1, BD Transduction Laboratories); Aurora B-pT232 (Rockland, 600-401-677); histone H3 (Abcam, ab1791); Mek2 (BD, M24520); H3pThr3 (Millipore 07-424); PCNA (Santa Cruz, PC10).
Statistical analysis
Statistical analysis was performed using the GraphPad Prism 5 software. Unpaired two-tailed Student’s t-test was applied for Figures 6B, C and 7C. For Figure 2C, Bonferroni’s multiple comparison test was used. Numbers of clones (N) and cells (n) are indicated in figure legends. Data are shown as mean±s.e.m. (standard error of the mean) when at least three clones/conditions/experiments were analysed. P-values were considered as statistically not significant when P>0.05 and were not depicted in the figures.
Supplementary Material
Acknowledgments
We are grateful to G Eichele, JM Peters, K Shirahige, and S Taylor for providing antibodies; to JC Cigudosa and members of his group for help with FISH; to M Cañamero for the histopathological analyses; to S Ortega and the Transgenic Mice Unit for generation of the Pds5B mice; to M Malumbres for advise; to S Remeseiro and A Cuadrado for help with additional experimental procedures. We also acknowledge IM Cheeseman and T Hirota for helpful discussions and critically reading the manuscript. This research has been supported by the Spanish Ministry of Economy and Competitiveness (grants BFU2007-66627, SAF-2010-21517 and CSD2007-00015 INESGEN to AL; FPI fellowship to MR-T).
Author contributions: AL designed and supervised the study. MC and MR-T performed and analysed most of the experiments with contributions from MR-C. IB designed and made the Pds5B targeting construct. AL wrote the manuscript with ideas and comments from the rest of the authors.
Footnotes
The authors declare that they have no conflict of interest.
References
- Barber TD, McManus K, Yuen KW, Reis M, Parmigiani G, Shen D, Barrett I, Nouhi Y, Spencer F, Markowitz S, Velculescu VE, Kinzler KW, Vogelstein B, Lengauer C, Hieter P (2008) Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci USA 105: 3443–3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Schmidt CK, Vaur S, Dheur S, Drogat J, Genier S, Ekwall K, Uhlmann F, Javerzat JP (2008) Cell-cycle regulation of cohesin stability along fission yeast chromosomes. EMBO J 27: 111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canudas S, Smith S (2009) Differential regulation of telomere and centromere cohesion by the Scc3 homologues SA1 and SA2, respectively, in human cells. J Cell Biol 187: 165–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Gligoris T, Upcher W, Kato Y, Shirahige K, Nasmyth K, Beckouet F (2013) Pds5 promotes and protects cohesin acetylation. Proc Natl Acad Sci USA 110: 13020–13025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying S, Hickson ID (2009) Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol 11: 753–760 [DOI] [PubMed] [Google Scholar]
- Cimini D, Wan X, Hirel CB, Salmon ED (2006) Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr Biol 16: 1711–1718 [DOI] [PubMed] [Google Scholar]
- Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K (2000) Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell 5: 243–254 [DOI] [PubMed] [Google Scholar]
- De Antoni A, Maffini S, Knapp S, Musacchio A, Santaguida S (2012) A small-molecule inhibitor of Haspin alters the kinetochore functions of Aurora B. J Cell Biol 199: 269–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D, Eissenberg JC, Misulovin Z, Martens A, Redding B, McKim K (2005) Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila. Development 132: 4743–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreier MR, Bekier ME 2nd, Taylor WR (2011) Regulation of sororin by Cdk1-mediated phosphorylation. J Cell Sci 124: 2976–2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feytout A, Vaur S, Genier S, Vazquez S, Javerzat JP (2011) Psm3 acetylation on conserved lysine residues is dispensable for viability in fission yeast but contributes to Eso1-mediated sister chromatid cohesion by antagonizing Wpl1. Mol Cell Biol 31: 1771–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi R, Gillespie PJ, Hirano T (2006) Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol 16: 2406–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gause M, Misulovin Z, Bilyeu A, Dorsett D (2010) Dosage-sensitive regulation of cohesin chromosome binding and dynamics by Nipped-B, Pds5, and Wapl. Mol Cell Biol 30: 4940–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich D, Koch B, Dupeux F, Peters JM, Ellenberg J (2006) Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Curr Biol 16: 1571–1578 [DOI] [PubMed] [Google Scholar]
- Gillespie PJ, Hirano T (2004) Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr Biol 14: 1598–1603 [DOI] [PubMed] [Google Scholar]
- Guillou E, Ibarra A, Coulon V, Casado-Vela J, Rico D, Casal I, Schwob E, Losada A, Mendez J (2010) Cohesin organizes chromatin loops at DNA replication factories. Genes Dev 24: 2812–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M (2009) Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460: 410–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman T, Stead K, Koshland D, Guacci V (2000) Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J Cell Biol 151: 613–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters JM (2005) Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol 3: e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi TL, Ikeda M, Tanaka H, Nakagawa T, Bando M, Shirahige K, Kubota Y, Takisawa H, Masukata H, Takahashi TS (2012) The prereplication complex recruits XEco2 to chromatin to promote cohesin acetylation in xenopus egg extracts. Curr Biol 22: 977–988 [DOI] [PubMed] [Google Scholar]
- Hou F, Zou H (2005) Two human orthologues of Eco1/Ctf7 acetyltransferases are both required for proper sister-chromatid cohesion. Mol Biol Cell 16: 3908–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA (2010) Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor TM, Lampson MA, Hergert P, Cameron L, Cimini D, Salmon ED, McEwen BF, Khodjakov A (2006) Chromosomes can congress to the metaphase plate before biorientation. Science 311: 388–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ (2000) Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol 150: 975–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AE, Ghenoiu C, Xue JZ, Zierhut C, Kimura H, Funabiki H (2010) Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science 330: 235–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters JM (2006) Wapl controls the dynamic association of cohesin with chromatin. Cell 127: 955–967 [DOI] [PubMed] [Google Scholar]
- Lafont AL, Song J, Rankin S (2010) Sororin cooperates with the acetyltransferase Eco2 to ensure DNA replication-dependent sister chromatid cohesion. Proc Natl Acad Sci USA 107: 20364–20369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Rankin S, Yu H (2013) Phosphorylation-enabled binding of SGO1-PP2A to cohesin protects sororin and centromeric cohesion during mitosis. Nat Cell Biol 15: 40–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Krantz ID (2009) Cornelia de Lange syndrome, cohesin, and beyond. Clin Genet 76: 303–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Hirano T (2005) Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J Cell Sci 118: 2133–2141 [DOI] [PubMed] [Google Scholar]
- Maresca TJ, Salmon ED (2009) Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J Cell Biol 184: 373–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuinness BE, Hirota T, Kudo NR, Peters JM, Nasmyth K (2005) Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol 3: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez J, Stillman B (2000) Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol 20: 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnich M, Kuriger Z, Print CG, Horsfield JA (2011) A zebrafish model of Roberts syndrome reveals that Esco2 depletion interferes with development by disrupting the cell cycle. PLoS One 6: e20051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH (2009) Cohesin: its roles and mechanisms. Annu Rev Genet 43: 525–558 [DOI] [PubMed] [Google Scholar]
- Neuwald AF, Hirano T (2000) HEAT repeats associated with condensins, cohesins, and other complexes involved in chromosome-related functions. Genome Res 10: 1445–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Ladurner R, Schmitz J, Kreidl E, Schleiffer A, Bhaskara V, Bando M, Shirahige K, Hyman AA, Mechtler K, Peters JM (2010) Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell 143: 737–749 [DOI] [PubMed] [Google Scholar]
- Panizza S, Tanaka T, Hochwagen A, Eisenhaber F, Nasmyth K (2000) Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr Biol 10: 1557–1564 [DOI] [PubMed] [Google Scholar]
- Remeseiro S, Cuadrado A, Carretero M, Martínez P, Drosopoulos WC, Cañamero M, Schildkraut CL, Blasco MA, Losada A (2012a) Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J 31: 2076–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remeseiro S, Cuadrado A, Gómez-López G, Pisano DG, Losada A (2012b) A unique role of cohesin-SA1 in gene regulation and development. EMBO J 31: 2090–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remeseiro S, Losada A (2013) Cohesin, a chromatin engagement ring. Curr Opin Cell Biol 25: 63–71 [DOI] [PubMed] [Google Scholar]
- Rolef Ben-Shahar T, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F (2008) Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science 321: 563–566 [DOI] [PubMed] [Google Scholar]
- Rowland BD, Roig MB, Nishino T, Kurze A, Uluocak P, Mishra A, Beckouet F, Underwood P, Metson J, Imre R, Mechtler K, Katis VL, Nasmyth K (2009) Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity. Mol Cell 33: 763–774 [DOI] [PubMed] [Google Scholar]
- Santaguida S, Vernieri C, Villa F, Ciliberto A, Musacchio A (2011) Evidence that Aurora B is implicated in spindle checkpoint signalling independently of error correction. EMBO J 30: 1508–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE, Teng G, Carroll T, Terry A, Horan K, Marks H, Adams DJ, Schatz DG, Aragon L, Fisher AG, Krangel MS, Nasmyth K, Merkenschlager M (2011) A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature 476: 467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T (2009) Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintomi K, Hirano T (2009) Releasing cohesin from chromosome arms in early mitosis: opposing actions of Wapl-Pds5 and Sgo1. Genes Dev 23: 2224–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon DA, Kim T, Diaz-Martinez LA, Fair J, Elkahloun AG, Harris BT, Toretsky JA, Rosenberg SA, Shukla N, Ladanyi M, Samuels Y, James CD, Yu H, Kim JS, Waldman T (2011) Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science 333: 1039–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM (2000) Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol 151: 749–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutani T, Kawaguchi T, Kanno R, Itoh T, Shirahige K (2009) Budding yeast Wpl1(Rad61)-Pds5 complex counteracts sister chromatid cohesion-establishing reaction. Curr Biol 19: 492–497 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Hao Z, Kai M, Okayama H (2001) Establishment and maintenance of sister chromatid cohesion in fission yeast by a unique mechanism. EMBO J 20: 5779–5790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, Hussein D, Wang Y, Elderkin S, Morrow CJ (2001) Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J Cell Sci 114: 4385–4395 [DOI] [PubMed] [Google Scholar]
- Tedeschi A, Wutz G, Huet S, Jaritz M, Wuensche A, Schirghuber E, Davidson IF, Tang W, Cisneros DA, Bhaskara V, Nishiyama T, Vaziri A, Wutz A, Ellenberg J, Peters JM (2013) Wapl is an essential regulator of chromatin structure and chromosome segregation. Nature 501: 564–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida KS, Takagaki K, Kumada K, Hirayama Y, Noda T, Hirota T (2009) Kinetochore stretching inactivates the spindle assembly checkpoint. J Cell Biol 184: 383–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE (2008) A molecular determinant for the establishment of sister chromatid cohesion. Science 321: 566–569 [DOI] [PubMed] [Google Scholar]
- Vader G, Cruijsen CW, van Harn T, Vromans MJ, Medema RH, Lens SM (2007) The chromosomal passenger complex controls spindle checkpoint function independent from its role in correcting microtubule kinetochore interactions. Mol Biol Cell 18: 4553–4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaur S, Feytout A, Vazquez S, Javerzat JP (2012) Pds5 promotes cohesin acetylation and stable cohesin-chromosome interaction. EMBO Rep 13: 645–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega H, Waisfisz Q, Gordillo M, Sakai N, Yanagihara I, Yamada M, van Gosliga D, Kayserili H, Xu C, Ozono K, Jabs EW, Inui K, Joenje H (2005) Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat Genet 37: 468–470 [DOI] [PubMed] [Google Scholar]
- Wang F, Dai J, Daum JR, Niedzialkowska E, Banerjee B, Stukenberg PT, Gorbsky GJ, Higgins JM (2010) Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science 330: 231–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Ulyanova NP, Daum JR, Patnaik D, Kateneva AV, Gorbsky GJ, Higgins JM (2012) Haspin inhibitors reveal centromeric functions of Aurora B in chromosome segregation. J Cell Biol 199: 251–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Kitajima TS (2005) Shugoshin protects cohesin complexes at centromeres. Philos Trans R Soc Lond B Biol Sci 360: 515–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrin E, Schleiffer A, Tanaka K, Eisenhaber F, Nasmyth K, Peters JM (2006) Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr Biol 16: 863–874 [DOI] [PubMed] [Google Scholar]
- Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, Wartman LD, Lamprecht TL, Liu F, Xia J, Kandoth C, Fulton RS, McLellan MD, Dooling DJ, Wallis JW, Chen K, Harris CC, Schmidt HK, Kalicki-Veizer JM, Lu C et al. (2012) The origin and evolution of mutations in acute myeloid leukemia. Cell 150: 264–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan G, Kreidl E, Wutz G, Egner A, Peters JM, Eichele G (2011) Cohesin acetyltransferase Esco2 is a cell viability factor and is required for cohesion in pericentric heterochromatin. EMBO J 31: 71–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y, Honda T, Tanno Y, Watanabe Y (2010) Two histone marks establish the inner centromere and chromosome bi-orientation. Science 330: 239–243 [DOI] [PubMed] [Google Scholar]
- Zhang B, Chang J, Fu M, Huang J, Kashyap R, Salavaggione E, Jain S, Kulkarni S, Deardorff MA, Uzielli ML, Dorsett D, Beebe DC, Jay PY, Heuckeroth RO, Krantz I, Milbrandt J (2009) Dosage effects of cohesin regulatory factor PDS5 on mammalian development: implications for cohesinopathies. PLoS One 4: e5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Jain S, Song H, Fu M, Heuckeroth RO, Erlich JM, Jay PY, Milbrandt J (2007) Mice lacking sister chromatid cohesion protein PDS5B exhibit developmental abnormalities reminiscent of Cornelia de Lange syndrome. Development 134: 3191–3201 [DOI] [PubMed] [Google Scholar]
- Zhang J, Shi X, Li Y, Kim BJ, Jia J, Huang Z, Yang T, Fu X, Jung SY, Wang Y, Zhang P, Kim ST, Pan X, Qin J (2008) Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol Cell 31: 143–151 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.