Abstract
SUMO conjugation is known to occur in response to double-stranded DNA breaks in mammalian cells, but whether SUMO deconjugation has a role remains unclear. Here, we show that the SUMO/Sentrin/Smt3-specific peptidase, SENP7, interacts with the chromatin repressive KRAB-associated protein 1 (KAP1) through heterochromatin protein 1 alpha (HP1α). SENP7 promotes the removal of SUMO2/3 from KAP1 and regulates the interaction of the chromatin remodeler CHD3 with chromatin. Consequently, in the presence of CHD3, SENP7 is required for chromatin relaxation in response to DNA damage, for homologous recombination repair and for cellular resistance to DNA-damaging agents. Thus, deSUMOylation by SENP7 is required to promote a permissive chromatin environment for DNA repair.
Keywords: homologous recombination, HP1α, KAP1, SENP7, SUMO
INTRODUCTION
Mammalian double-strand break (DSB) repair is achieved by two main mechanisms; non-homologous end joining (NHEJ) or homologous recombination (HR). NHEJ ligates broken ends together, whereas HR requires a homologous template. Isoforms of the small ubiquitin-related modifiers (SUMOs, also called Sentrin/smt3) are covalently conjugated to various proteins in the mammalian DNA damage response. SUMO2 and 3 are 97% identical (referred to as SUMO2/3), whereas SUMO1 is more distantly related. SUMO2/3, but not SUMO1, readily forms polymers on substrates (polySUMO).
SUMOylation often acts to increase the interaction of modified proteins with those bearing SUMO interaction motifs (SIMs), having the consensus: V/I/L-V/I/L-x-V/I/L. SUMOylation can alter the function of individual proteins and groups of proteins, changing the efficacy of intracellular pathways [1]. In the response to DSBs, SUMOylation is essential for the proper recruitment of several repair factors [2].
Higher-order organization of chromatin also influences DNA repair. Chromatin-associated SUMO-modified proteins can recruit repressor complexes such as LSD1/CoREST1/HDAC [3] and the nucleosome remodelling and deacetylation (NuRD) complex [4]. Such repression must be released to promote relaxation and allow DNA repair [5, 6]. Both global and local chromatin structure is loosened following induction of a DNA lesion [5, 6], whereas in heterochromatic DNA, relaxation is necessary for HR proficiency [7].
In this study, we addressed whether a role for SUMO deconjugation exists in the repair of DSBs. The removal of SUMO from a target is performed by members of the SUMO/Sentrin/Smt3-specific peptidases (SENP) family of cysteine proteases, and by at least two non SENP proteases [8–10]. SENP1/2/3/5 deconjugate mono SUMO1/2/3, whereas SENP6/7 are specialized chain-editing enzymes [11, 12]. SENP1 and 2 have indirect roles in DNA repair by regulating the NF-κB pathway and p53 levels [13, 14]. SENP6 is reported to inhibit RPA70 SUMOylation [15], and SENP7 is a chromatin-associated protein that interacts with HP1α [16–18]. Here we report that SENP7 is specifically required for HR repair. We reveal that SENP7 regulates chromatin relaxation induced in response to DNA damage, through KRAB-associated protein 1 (KAP1)-associated chromatin remodellers.
RESULTS AND DISCUSSION
SENP enzymes are involved in DSB repair
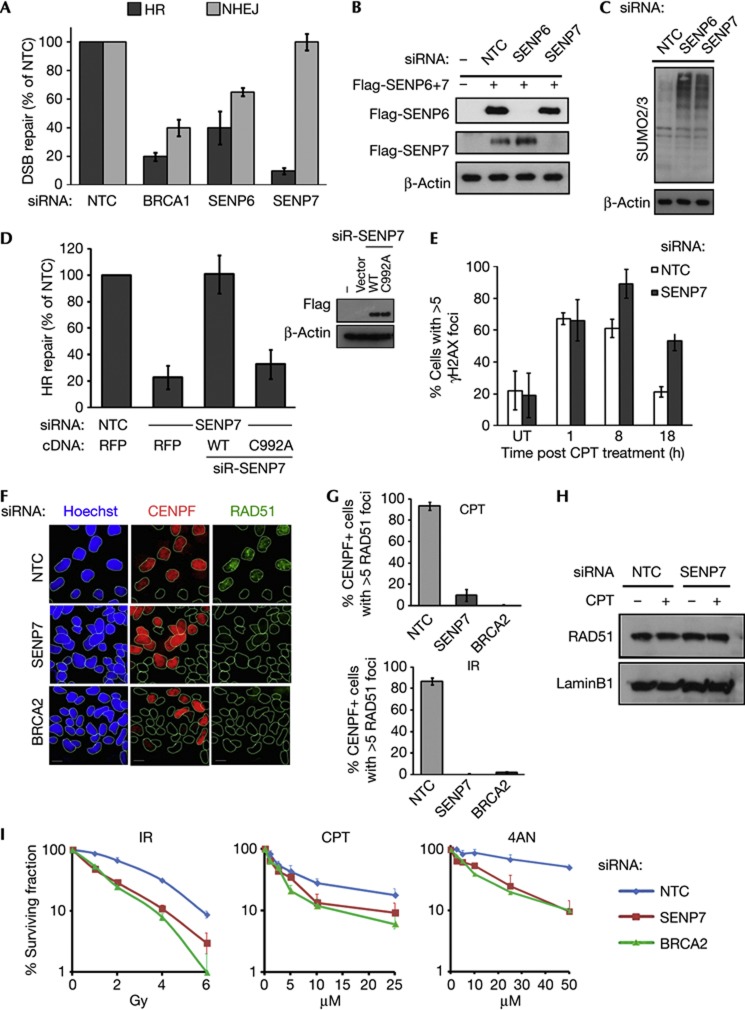
We first examined the effect of SUMO2 deconjugation mutants on HR repair. Despite the presence of endogenous SUMO2/3, expression of the deconjugating mutant (Q90P) or SENP6/7 binding mutant (N68A/D71H) [19, 20] impaired HR repair (supplementary Fig S1A–C online). This implicates deconjugation of SUMO in HR. Next, we measured HR and NHEJ DSB repair in cells depleted for each SENP. All but SENP3 and SENP5 small interfering RNA (siRNA) treatment reduced repair proficiency in these assays (Fig 1A,B, supplementary Fig S1D,E online). Given the effects of the N68A/D71H mutant and reduced repair efficiency in SENP6/7-depleted cells, we focused on the chain-editing SENPs. Perturbation of the cell cycle can indirectly influence repair. While SENP6 siRNA perturbed cell cycle kinetics, we observed little impact of SENP7 siRNA (supplementary Fig S1F online). Indirect impairment of the cellular response to DSBs might also occur as a consequence of trapping SUMO on substrates (Fig 1C) [11], potentially reducing free SUMO2/3 for de novo conjugation required in DNA damage response signalling. Therefore DNA repair assays in SENP6- or SENP7-depleted cells in which RFP-SUMO2 and SUMO3 were expressed were performed. This revealed that SUMO2/3 improved repair in SENP6/7-depleted cells, but did not fully restore HR in SENP7-depleted cells (supplementary Fig S1G and H online). This suggests the effects on HR following SENP7 depletion are not wholly caused by an indirect impact through SUMO homeostasis. To examine whether the enzymatic activity of SENP7 is required, we complemented knockdown with siRNA-resistant Flag-SENP7. Expression of WT, but not catalytic mutant (C992A), restored HR (Fig 1D), thus the impact of SENP7 siRNA on HR is through SENP7 protease loss.
Figure 1.
The SENP7 protease is required for HR repair. (A) Chain-editing SUMO proteases SENP6 and SENP7 are required for DSB repair. HeLa DR3-GFP (HR repair) and EJ5-GFP (NHEJ) reporters were treated with non-targeting control, BRCA1, SENP6 or SENP7 siRNA before transfection with Sce-I. (All bars throughout show standard error about the mean of three independent experiments). (B) Knockdown efficiency of SENP6/7. HEK293 expressing Flag-SENP6/SENP7 and treated with indicated siRNA. (C) Depletion of SENP6 and SENP7 increases SUMO2/3 conjugates. HEK293 transfected with indicated siRNA and lysates probed with SUMO2/3 antibody and anti-β-actin. (D) siRNA-resistant WT-SENP7, but not catalytic mutant, C992A, can compensate for SENP7 depletion in HR repair assays. HeLa DR3 depleted of endogenous SENP7 before transfection with Sce-I, RFP and siRNA-resistant Flag-SENP7. % HR repair relative to non-targeting control is shown. Inset shows WB of Flag-SENP7 expression and β-actin loading control. (E) Clearance of γH2AX foci after CPT treatment. U2OS depleted of SENP7 and treated with 1μM CPT (1 h), before washing and recovery. Cells stained against γH2AX. % cells with >5 foci were scored. (F–H) RAD51 foci are reduced in SENP7-depleted cells. U2OS transfected with siRNA before 2.5 Gy IR or 1 μM CPT/1 h. Cells were stained 4 h later for RAD51 and CENPF to identify G2-phase cells. % cells with >5 foci scored in CENPF-positive cells (G). Representative images of IR-treated cells are shown in (F). Immunoblot from parallel experiment lysed and blotted with RAD51 (H). (I) SENP7 depletion sensitizes cells to IR, CPT and PARP inhibition (4AN). Clonogenic assay of HeLa treated with indicated siRNA followed by doses of 4AN, IR or CPT. 4AN, 4-amino-1-8 naphthalimide; CPT, camptothecin; DSB, double-strand break repair; HR, homologous recombination; IR, ionizing radiation; NTC, non-targeting control; PARP, poly ADP ribose polymerase; siRNA, small interfering RNA; UT, untreated; WB, western blot; WT, wild type.
Poor HR repair slows clearance of γH2AX foci following DNA damage. After treatment with topoisomerase poison, camptothecin (CPT), γH2AX foci clearance was slowed in SENP7-depleted cells (Fig 1E). We next examined the ability of proteins required to signal and repair DSBs to recruit to γH2AX-decorated DNA. Recruitment of 53BP1, BRCA1 and the formation of RPA foci in G2 cells were similar in SENP7 siRNA and control-treated cells (supplementary Fig S1I online). However, RAD51 accumulation was severely restricted (Fig 1F–H). Therefore, early signalling events appear normal, but later steps such as RAD51 loading, invasion or homology search are impaired in SENP7-depleted cells. Similar findings have been noted in Drosophila heterochromatin and in human cell depletion of the p400, ATPase chromatin remodeller. Under these circumstances DSB signalling is not lost but inhibits RAD51-mediated repair [21, 22]. Consolidating these observations, we found that SENP7-depleted cells were sensitive to IR, CPT and the PARP inhibitor, 4-amino-1-8 naphthalimide (Fig 1I). These data show a requirement for the SUMO2/3 chain editor SENP7 in the response to DSBs and in HR repair.
SENP7 SIMs and HP1α interaction promotes HR repair
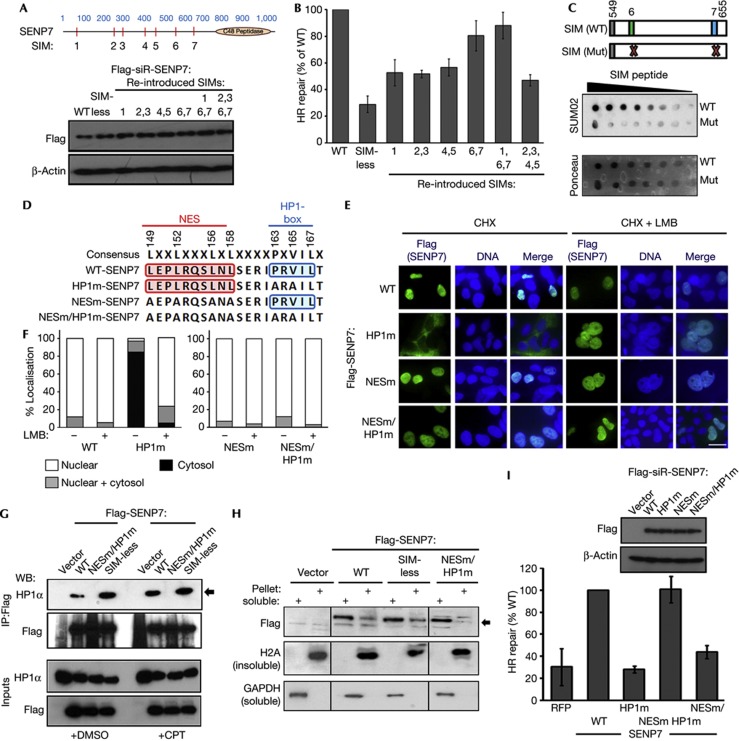
SENP7 contains seven potential SIMs outside the catalytic region. We mutated all of these to generate SIM-less-SENP7 and derivatives in which SIM sequences were re-introduced (Fig 2A; supplementary Fig S2A online). These changes had no impact on the subcellular localisation of the mutants, nor adversely affected expression (Fig 2A). SIM-less-SENP7 was unable to rescue HR repair (supplementary Fig S2B online), but inclusion of SIMs 6/7 restored the majority of the repair efficiency (Fig 2B). The fragment of SENP7 protein encompassing SIMs 6 and 7 interacted with polySUMO2 whereas the SIM mutant (supplementary Fig S2A online) did not (Fig 2C), indicating this region promotes interaction with SUMO2. Together these data suggest SUMO interaction contributes to the activity of SENP7 in HR repair.
Figure 2.
SENP7 SIMs and HP1-box are required to promote HR repair. (A) SIM mutants of SENP7. Location of each SIM (red lines) and the catalytic domain (yellow oval) are shown. Below, shows expression of Flag-SENP7 SIM mutants in HeLa. (B) SENP7 SIMs required for HR activity. HR rescue repair assays using Flag-siR-SIM mutants. % repair is measured relative to WT-SENP7. (C) SENP7 SIM 6 and 7 bind to polySUMO2. Cartoon of WT and SIM6 and 7 mutant fragments (aa 549–655). Spotted fragments were stained with Ponceau, incubated with polySUMO2, washed and probed for bound SUMO2. (D) SENP7 contains a NES adjacent to its HP1-box. NES and HP1-box consensus with the SENP7 NES and HP1 sequence and mutants made in this study. (E) Nuclear export of HP1-box mutant. U2OS expressing Flag-SENP7-WT and indicated mutants. 1 h before fixation and Flag staining, cells were treated with 10 μM cycloheximide and 6 nM Leptomycin-B or DMSO. Scale bar, 10 μm (F) Quantification of SENP7 localization from experiments described in (E) scored in 100 cells in three independent experiments (mean shown). (G) The NESm-HP1m mutant of SENP7 does not interact with endogenous HP1α. HEK293 expressing Flag-SENP7 constructs were untreated or exposed to CPT, lysed and immunoprecipitated with Flag. Eluates were blotted with Flag and HP1α. Lower panels=5% inputs. (H) SENP7 NESm-HP1m mutant is not localized with chromatin. HEK293 lysates expressing Flag-SENP7 constructs, separated into soluble and insoluble (pellet) fractions. Lysates were probed with Flag (SENP7), H2A (chromatin marker) and GAPDH (soluble marker). (I) HP1-box is required for HR repair. HR repair assays performed as for Fig 1C. % repair is shown relative to WT-SENP7. Inset image shows expression of mutants. CPT, camptothecin; DMSO, dimethyl sulphoxide; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HP1α, heterochromatin protein 1 alpha; HR, homologous recombination; NES, nuclear export sequence; WT, wild type.
SENP7 contains a conserved HP1-box (PxVxL) (Fig 2D) required for interaction with HP1 [16, 18]. To address the role of SENP7 chromatin localization and HP1 binding in HR, we mutated the sequence. However, mutant protein located to the cytoplasm (Fig 2E,F) and similarly siRNA depletion of heterochromatin protein 1 alpha (HP1α) resulted in a loss of endogenous SENP7 from the nucleus (supplementary Fig S2C online). Nuclear localization of the mutant could be restored by treatment with the CRM1 inhibitor, Leptomycin-B (LMB) or by substitution of residues in the nuclear export sequence (NES) adjacent to the HP1-box (Fig 2E,F) indicating that the interaction with CRM1 mediates SENP7 nuclear export when the HP1-box is mutated. Nuclear localization of the HP1-box mutant, whether by co-mutation of the NES or by LMB treatment, failed to restore interaction with HP1α (Fig 2G; supplementary Fig S2D online). In addition, the ability of SENP7 to co-purify with chromatin required its HP1-box, suggesting localisation through HP1α (Fig 2H). HR repair in SENP7-depleted cells could not be restored by expression of SENP7-HP1-box mutants (Fig 2I), suggesting HP1 interaction and chromatin localization of SENP7 are significant in its promotion of DNA repair.
SENP7 regulates SUMO2 modification of KAP1
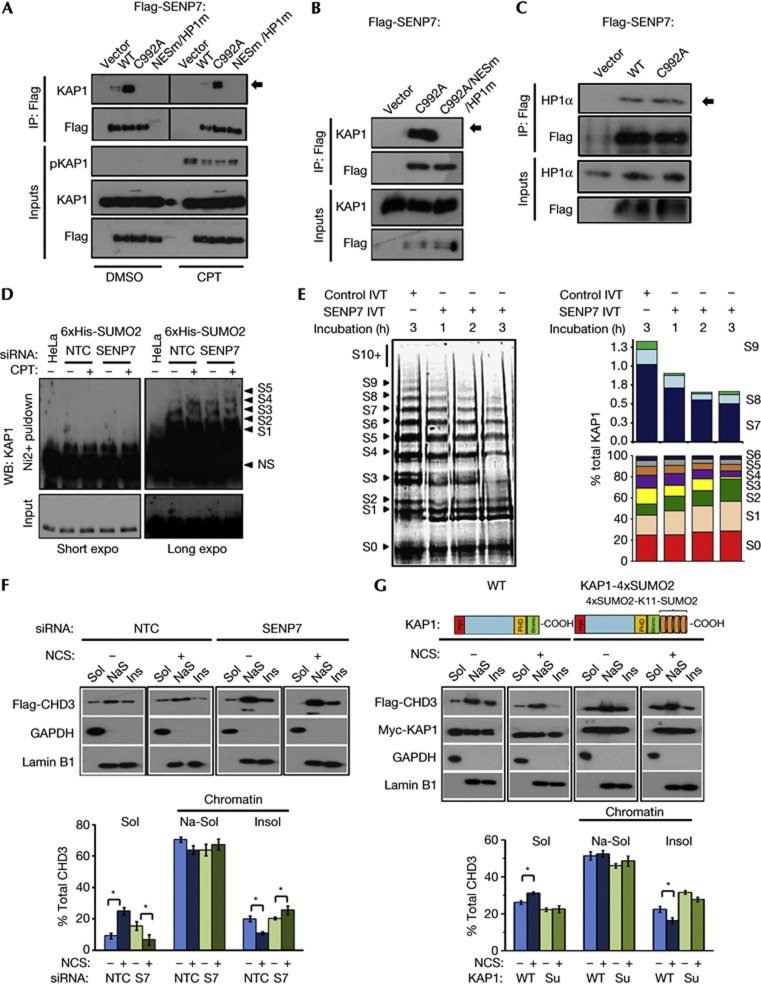
Despite several attempts, we were not able to detect changes in endogenous HP1α SUMOylation following SENP7 depletion. Therefore, we reasoned that SENP7 substrates might include proteins that are co-enriched with HP1α. KAP1 is modified by auto-SUMOylation, interacts with HP1α [4, 23] and can be both mono and polySUMOylated [24, 25]. Immunoprecipitation of WT-SENP7 revealed a weak interaction with endogenous KAP1, but no KAP1 was precipitated with NESm-HP1m-SENP7 (Fig 3A), suggesting SENP7–KAP1 interaction requires HP1 binding. Strikingly, SENP7-C992A interacted robustly with KAP1 in an HP1-box-dependent manner (Fig 3A,B). The fact that SENP7 C992A did not show increased interaction with HP1α suggests that the interaction with KAP1 is both through HP1α binding and secondarily through the SENP7 catalytic domain, consistent with an enzymatic trap (Fig 3C).
Figure 3.
SENP7 deSUMOylates KAP1 and regulates CHD3–chromatin association. (A) SENP7 interacts with KAP1. HEK293 expressing Flag-SENP7 was mock or CPT treated, lysed, immunoprecipitated and immunoblotted with Flag and KAP1. pSer824-KAP1 used as a marker of DNA damage. (B) SENP7–C992A interaction with KAP1 requires the HP1-box. HEK293 expressing Flag-C992A-SENP7 or Flag-C992A-NESm-HP1m-SENP7 immunoprecipitated and immunoblotted with Flag and KAP1. (C) HP1α interaction with WT and SENP7-C992A. HEK293 expressing Flag-WT and Flag-C992A-SENP7 were immunoprecipitated as above and blotted for HP1α. (D) KAP1 SUMO2ylation is increased on SENP7 depletion. 6xHis-SUMO2 expressing HeLa depleted with NTC or SENP7 siRNAs before CPT, lysed in 8 M urea and incubated with nickel-agarose. Eluates and 5% input were immunoblotted with KAP1. S=number of His-SUMO2 modifications.(E) DeSUMOylation of KAP1-polySUMO in vitro. Auto-SUMO2ylated GST-KAP1 incubated with full-length in vitro translated SENP7 for 1–3 h. Beads were washed, boiled and KAP1-SUMOylation detected on SPYRO ruby stained 6% gels. S0=unSUMOylated GST-KAP1. Quantification of SUMOylated KAP1, each SUMOylated species is calculated as % of total KAP1/lane. (F) SENP7 depletion increases CHD3–chromatin retention. HEK293 expressing WT Flag-CHD3 treated with indicated siRNA and NCS (200 ng/ml 1 h). Lysates were fractionated into soluble (Sol) and chromatin. The chromatin was subsequently fractionated into salt-soluble (NaS) and pellet (Ins) sub-fractions. Lysates were blotted with Flag and fraction markers. The proportions of Flag-CHD3 were quantified from three experiments below. Significance was assessed using Student’s t-test; *<0.05. (G) KAP1-4xSUMO2 increases CHD3 chromatin retention. HEK293 transfected with Flag-CHD3 and KAP1 or KAP1-4xSUMO2 (illustrated) and treated as for F. Quantification was as for F. Significance was assessed using Student’s t-test; *<0.05. CPT, camptothecin; GST, glutathione S-transferase; HP1α, heterochromatin protein 1 alpha; KAP1, KRAB-associated protein 1; NCS, neocarzinostatin; NTC, non-targeting control; siRNA, small interfering RNA; WT, wild type.
We noted higher-molecular weight species of KAP1 when the SENP7 catalytic mutant was overexpressed, potentially representing SUMO-KAP1 (Fig 3A inputs). To test the influence of endogenous SENP7 on KAP1-SUMOylation, we purified His-SUMO2 conjugates in denaturing conditions and probed for KAP1. Further SUMO2 modifications were evident in untreated and CPT-treated cells when SENP7 was depleted (Fig 3D). In addition polySUMOylated KAP1 could be deSUMOylated in vitro by SENP7 (Fig 3E), whereas mono-SUMOylated KAP1 could not (supplementary Fig S3A,B online). Together, these data indicate that the degree of polySUMO2-conjugated KAP1 is regulated by the associated protease, SENP7. We envisage that HP1α is responsible for tethering SENP7 to chromatin, and that SENP7 acts to restrain the levels of local SUMO2/3 conjugates on proteins such as KAP1.
SENP7 regulates CHD3 association with chromatin
SUMO1ylated KAP1 recruits the NuRD complex ATPase component CHD3 and the histone methyltransferase SET domain, bifurcated 1 (SETDB1) contributing to chromatin condensation [4, 6]. Condensed chromatin state is relieved on DNA damage by phosphorylation of KAP1 by ataxia telangiectasia mutated (ATM) protein kinase [5]. The phosphorylated KAP1 site is reported to reduce the influence of SUMO1-KAP1 by competing with the SIM of CHD3 for the interface with SUMO1, and in consequence reduce CHD3-KAP1 interaction and promote CHD3–NuRD complex dispersion and chromatin relaxation [6].
We tested whether the CHD3-SIM might also bind polySUMO2 using CHD3 peptides. The carboxy-terminal fragment bound to polySUMO2 in a manner dependent on its SIM motif (supplementary Fig S3C online), indicating binding to SUMO2 as well as SUMO1. Similarly, the CHD3 C-terminus was able to purify KAP1 from SENP7-depleted cells (supplementary Fig S3D online). We next examined whether chromatin association of CHD3 is regulated by SENP7 or polySUMO2-KAP1. In control cells, treatment with neocarzinostatin (NCS) increases the proportion of soluble CHD3, while decreasing the proportion on insoluble chromatin. In SENP7-depleted cells, this redistribution does not occur (Fig 3F). We fused 4xSUMO2 to C-terminal KAP1 to generate a mimic of polySUMO2-KAP1. In cells expressing this construct, the proportion of CHD3 in the soluble fraction was similarly unaffected by NCS treatment and the insoluble chromatin fraction did not decrease (Fig 3G). These findings correlate SENP7 activity and the restriction of KAP1-SUMO2 modification, with promoting reduced CHD3–chromatin interactions.
DNA damage-associated remodelling requires SENP7
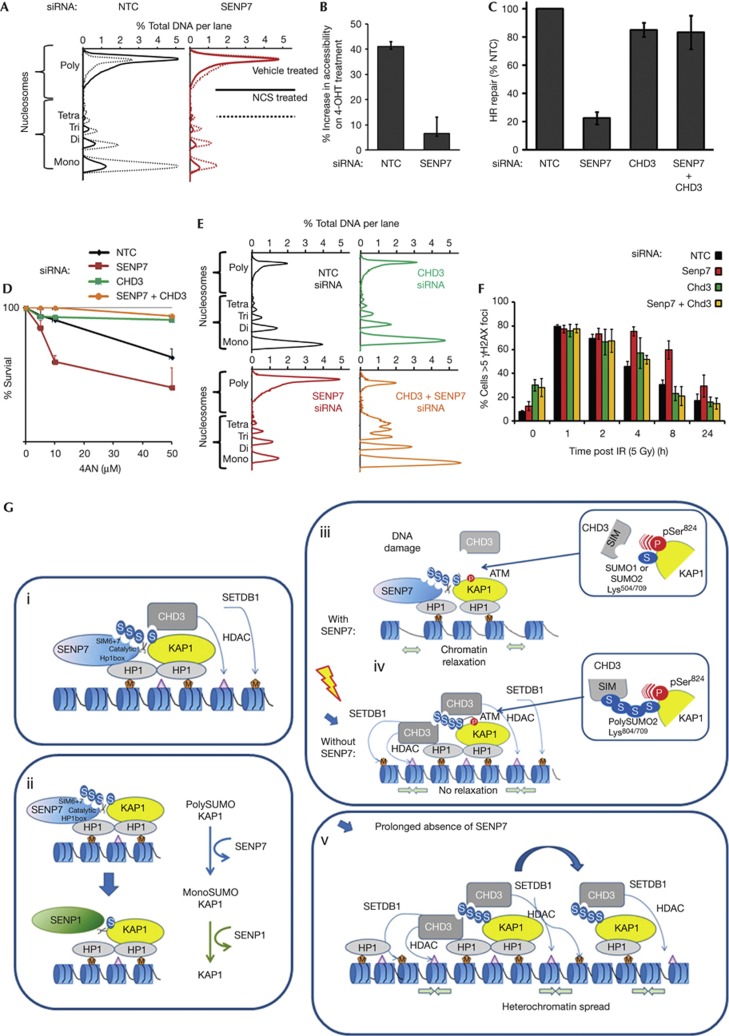
HDACs and the ATPase associated with the NuRD complex promote chromatin condensation [26], predicting that SENP7 is likely to regulate chromatin state. Consistent with this notion, prolonged SENP7 depletion has been reported to induce chromatin condensation [18]. We examined chromatin state after SENP7 depletion and found acute depletion did not have a gross effect (Fig 4A). However, after NCS treatment, or following a single euchromatic break, chromatin accessibility was reduced in SENP7-depleted cells (Fig 4A and B), indicating that SENP7 is required for chromatin relaxation in response to DNA damage.
Figure 4.
SENP7 regulates chromatin decondensation required for HR repair. (A) SENP7 is required for chromatin relaxation in response to DNA damage. HEK293 transfected with siRNA before treatment with NCS. Nuclei were incubated with MNase (0.25 U/5 min). Data are represented as % signal/lane determined by densitometry. (B) SENP7 is required for localized euchromatin relaxation. Cells stably expressing oestrogen receptor-fused IPpoI transfected with siRNA and treated with vehicle or 4-OHT to induce DSB’s. Euchromatin accessible to digestion was assayed using EpiQ assay. Chromatin was digested and PCR amplification of the chromosome 1 site (DAB1 gene intron) adjacent to the IPpoI endonuclease target site was undertaken. Graph shows Qc (amplification efficiency) of (digested/undigested) for vehicle/4-OHT treatment. (C) CHD3 depletion restores HR repair in SENP7-depleted cells. HR repair assay performed as for Fig 1C. % GFP/RFP (HR repair) relative to GFP/RFP in NTC is shown. (D) CHD3 depletion confers PARP-inhibitor resistance of SENP7-depleted cells. Clonogenic assay of HeLa transfected with indicated siRNA followed by treatment with 4AN. (E) CHD3 depletion restores chromatin decondensation in SENP7-depleted cells. Cells transfected with indicated siRNAs before NCS treatment and MNase assay as in Fig 4A. (F) γH2AX clearance is restored by depletion of Chd3 in Senp7-depleted mouse cells. NIH3T3 transfected with NTC or Senp7 siRNA, exposed to 2.5 Gy IR and allowed to recover for indicated times before staining with antibody to γH2AX. Cells with >5 foci were counted (100 cells, in three independent experiments). (G). Model of SENP7 in promoting chromatin state in the presence and absence of DNA damage. (i) SENP7 and KAP1 are co-located on chromatin. SENP7 constitutively deSUMOylates KAP1 preventing formation of polySUMO2/3-KAP1. SUMO-KAP1 associates with NuRD subunit CHD3 and histone methyltransferase SETDB1 via SUMO interacting motifs. (ii) KAP1-SUMO homeostasis is a two-step process by which SENP7 deconjugates the polySUMO2/3 KAP1 and other SUMO proteases, such as SENP1 deconjugate the mono-SUMO2/3 KAP1. (iii) Following induction of DNA damage response, KAP1 is phosphorylated at Ser824 by ATM. KAP1 phosphorylation interferes with CHD3SIM–SUMO-KAP1 interaction, allowing dispersion of CHD3–NuRD complexes resulting in chromatin relaxation and repair of DNA. (iv) In the absence of SENP7, KAP1 is hyperSUMO2ylated. The increased SUMO2 conjugates in the vicinity of CHD3SIM negate the interference provided by the negatively charged KAP1-pSer824 and CHD3–NuRD is not released from chromatin and remodelling does not occur. (vi) Potentially prolonged absence of SENP7 promotes hyperSUMOylation of KAP1 leading to excessive remodeller accumulation and (through SETDB1) increased methylation of H3K9. This results in spread of heterochromatin factors and condensed chromatin. 4AN, 4-amino-1-8 naphthalimide; DSB, double-strand break repair; GFP, green fluorescence protein; HR, homologous recombination; IR, ionizing radiation; KAP1, KRAB-associated protein 1; NCS, neocarzinostatin; NTC, non-targeting control; NuRD, nucleosome remodelling and deacetylation; PARP, poly ADP ribose polymerase; PCR, polymerase chain reaction; RFP, red fluorescent protein; SETDB1, SET domain, bifurcated 1; siRNA, small interfering RNA.
To address whether SENP7 acts to promote DNA repair through its impact on chromatin remodeler recruitment, we co-depleted SENP7 and CHD3 before examining chromatin condensation, HR repair and resistance to PARP inhibitor. In each case, co-depletion of CHD3 bypassed the need for SENP7 (Fig 4C–E, supplementary Fig S3C online); therefore, SENP7 acts to counter chromatin condensation by the CHD3–NuRD complex and promote DSB repair.
Recruitment of the histone methyltransferase, SETDB1, also occurs via SUMO1-KAP1, [4] and we speculate SETDB1 interaction with chromatin might be similarly increased in the presence of polySUMO2-KAP1. Increased SETDB1 association would be an explanation for the enlarged HP1α chromatin deposits seen following SENP7 depletion in the human cells [18]. Thus SENP7 might act to prevent heterochromatin spread over time.
In contrast, murine Senp7 has been reported to maintain HP1α chromatin deposits [17]. Mindful of the potential for species differences, we examined if the role of SENP7 in DNA repair is conserved in mice. siRNA to Senp7 reduced clearance of γH2AX in murine NIH3T3 cells whereas γH2AX kinetics were normal in cells treated with Chd3 and Senp7 siRNA (Fig 4F). Thus the murine Senp7 also regulates DNA repair via Chd3, indicating functional conservation.
DNA repair in heterochromatin is dependent on chromatin remodelling. To specifically examine this compartment, we counted γH2AX foci in chromocentres in NIH3T3 cells. γH2AX foci were more prevalent within the chromocentres several hours after induction of damage in Senp7-depleted cells than in control-treated cells (supplementary Fig S3F online). This suggests a further slower rate of DNA repair, or reduced mobility of the damaged chromatin induced by Senp7 loss and suggests a role for Senp7 in heterochromatic DNA repair in addition to global and euchromatic environments.
By acute depletion of SENP7 and examination of the cellular response to DNA damage, we have revealed a novel role for a SUMO chain-editing protease in promoting a permissive environment for HR repair. We propose a model in which chromatin-associated SENP7 acts to restrict the degree of polySUMO2/3ylation of local proteins, including KAP1, thereby allowing the regulation of chromatin remodellers bearing SIM motifs, such as CHD3, which in turn affect chromatin condensation state (Fig 4Gi and ii).
SENP enzymes represent an aspect of the SUMO pathway potentially accessible to small molecule inhibition, and these data suggest that targeting SENP7 might have therapeutic potential to enhance the efficacy of DNA-damaging agents. Further, as KAP1 has functions in transcription repression, particularly in epigenetic regulation and retroviral silencing [27], we anticipate a role for SENP7 in these processes.
METHODS
Statistics. All experiments were performed in triplicate, with error bars denoting standard error of mean. For IF counting, a minimum of 100 cells were counted per group in triplicate.
Plasmids and transfections. Full-length human SENP7 (CCDS2941) was subcloned into pCDNA5/FRT/TO with N-terminal Flag tag. Mutants were generated by site-directed mutagenesis unless otherwise stated and verified by sequencing. KAP1-4xSUMO2 encodes a cDNA with myc tag, full-length human KAP1 and 4 SUMO2 tandem sequences starting at codon 11 (Lys) and finishing with codon 98 (Gly) subcloned into pCDNA5/FRT/TO. The cDNA was generated by gene synthesis (GenScript) and included multiple silent mutations to reduce DNA sequence repetition. His-SENP7 SIM cDNA were generated by gene synthesis (GenScript) and cloned into pET15b vector (Novagen). Plasmid transfections were performed with Fugene6 (Promega) and siRNA with Dharmafect-1 (Dharmacon) according to the manufacturer’s instructions.
Cell culture and stable cell lines. All cells were maintained in DMEM under standard conditions. SENP7 stable cell lines were generated using the TRex-Flp-In HEK293 cell line system (Invitrogen).
HR/NHEJ repair assays, colony assays, MNase assays. HR/NHEJ repair assays and colony assays were performed essentially as for Butler et al [28]. For colony survival assays, cells were treated with indicated doses of CPT or 4-amino-1-8 naphthalimide for 1 h before re-plating. For MNase assays, cells were harvested and nuclei isolated in hypotonic buffer (10 mM Tris–HCl, pH 7.5, 2 mM MgCl2, 3 mM CaCl2, 320 mM sucrose, 1 mM DTT and complete EDTA-free protease inhibitor). Nuclei were digested at 25 °C with MNase (0.025 U/ml) in reaction buffer (10 mM Tris–HCl pH 7.5, 15 mM NaCl, 60 mM KCl, 1 mM CaCl2 and 250 mM sucrose). The reactions were stopped at 5 min with 20 mM EDTA and 2 mM EGTA. Genomic DNA was purified and separated on 1.2% agarose gel. Lanes of ethidium bromide-stained gels were scanned and profiles representing band intensity were obtained using GeneSnap software (SynGene USA). Peaks were quantified relative to the total signal from each lane.
Immunoprecipitations and pulldowns and peptide binding. Immunoprecipitation was carried out with M2 agarose beads (Sigma) or GFP-Trap (Chromotek) according to the manufacturer’s instructions. Bacterially expressed His-SENP7 fragments (WT and SIM6+7 m) were immobilized on PVDF, stained with Ponceau S and incubated with polySUMO2 (0–2 μg/ml) in PBS for 2 h at 25°C. Membranes were washed 5 × with PBST and immunoblotted against SUMO2/3. For CHD3, peptide pulldowns (peptide sequences in supplementary Information online), biotinylated CHD3 peptides (GenScript USA) were immobilized on streptavidin Dynabeads (Invitrogen). Peptide-conjugated beads were incubated with polySUMO2 (ENZO) for 2 h at 25 °C followed by three washes (10 mM Tris pH 7.5, 150 mM NaCl and 0.5 mM EDTA) and separated by SDS–PAGE on 4–20% gradient gel (NOVEX Invitrogen). Captured polySUMO2 were detected by immunoblot against SUMO2/3. For CHD3-KAP1 pulldowns 0.5 mg of lysates was incubated with 30 μl of peptide-conjugated streptavidin beads overnight at 4 °C, washed and blotted as for SUMO2/3.
Specific cut-site chromatin accessibility (EpiQ). HeLa–IPpoI cells [28] were transfected with siRNA 48 h before 16 h 4-OHT treatment to induce DSB in the DAB1 gene. EpiQ assay was performed per manufacturer’s instructions and digested DNA was PCR amplified with primers adjacent to the DAB1 DSB locus. Graph shows change in accessibility on 4-OHT treatment: Qc (amplification efficiency) of (digested/undigested) for vehicle/4-OHT treatment
Additional experimental details (primer sequences, antibodies and in vitro SUMOylation assay) can be found in supplemental data online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank J. Stark, (Duarte, CA) for pCBASce, M. Kastan, (Memphis, TN) for pBABE HA-ER-I-PpoI, OS Gabrielsen (Oslo) and P. Jeggo (Sussex) for CHD3 constructs, S. Jackson (Cambridge) for SUMO2+3-RFP WT, P Hemmerich (Jenna) for GFP-HP1α, R. Hay (Dundee) for HS2 cells and F Rauscher (Philadelphia) for GST-KAP1. This work was supported by the Breast Cancer Campaign: HRS, SAB-R and J.R.M. (2010NovPhD02, 2011NovSP004, 2006MaySF06), Cancer Research UK: R.M.D. (C8820/A15265), the Medical Research Council: DW (6900577). A.J.G., K.M.P. & K.J.L. are funded by MDS University of Birmingham.
Author contributions: AJG and JRM performed the experiments, designed the research and wrote the paper. All other authors performed experiments, contributed to discussions and edited the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Psakhye I & Jentsch S (2012) Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151: 807–820 [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S & Mailand N (2011) The ubiquitin- and SUMO-dependent signaling response to DNA double-strand breaks. FEBS Lett 585: 2914–2919 [DOI] [PubMed] [Google Scholar]
- Ouyang J Shi Y Valin A Xuan Y & Gill G (2009) Direct binding of CoREST1 to SUMO-2/3 contributes to gene-specific repression by the LSD1/CoREST1/HDAC complex. Mol Cell 34: 145–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov AV et al. (2007) PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell 28: 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y Bielopolski D Galanty Y Lukas C Taya Y Schultz DC Lukas J Bekker-Jensen S Bartek J & Shiloh Y (2006) Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol 8: 870–876 [DOI] [PubMed] [Google Scholar]
- Goodarzi AA Kurka T & Jeggo PA (2011) KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat Struct Mol Biol 18: 831–839 [DOI] [PubMed] [Google Scholar]
- Goodarzi AA Noon AT & Jeggo PA (2009) The impact of heterochromatin on DSB repair. Biochem Soc Trans 37: 569–576 [DOI] [PubMed] [Google Scholar]
- Hickey CM Wilson NR & Hochstrasser M (2012) Function and regulation of SUMO proteases. Nat Rev Mol Cell Biol 13: 755–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz S et al. (2012) Ubiquitin-specific protease-like 1 (USPL1) is a SUMO isopeptidase with essential, non-catalytic functions. Embo Rep 13: 930–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin EJ Shin HM Nam E Kim WS Kim JH Oh BH & Yun Y (2012) DeSUMOylating isopeptidase: a second class of SUMO protease. Embo Rep 13: 339–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen LN Geoffroy MC Jaffray EG & Hay RT (2009) Characterization of SENP7, a SUMO-2/3-specific isopeptidase. Biochem J 421: 223–230 [DOI] [PubMed] [Google Scholar]
- Lima CD & Reverter D (2008) Structure of the human SENP7 catalytic domain and poly-SUMO deconjugation activities for SENP6 and SENP7. J Biol Chem 283: 32045–32055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M Chiu SY & Hsu W (2011) SUMO-specific protease 2 in Mdm2-mediated regulation of p53. Cell Death Differ 18: 1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH Mabb AM Gill GB Yeh ET & Miyamoto S (2011) NF-kappaB induction of the SUMO protease SENP2: a negative feedback loop to attenuate cell survival response to genotoxic stress. Mol Cell 43: 180–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou H Huang C Singh M Carpenter PB & Yeh ET (2010) Regulation of DNA repair through deSUMOylation and SUMOylation of replication protein A complex. Mol Cell 39: 333–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa RS Nagao K Masuda HT Iwasaki O Hirota T Nozaki N Kimura H & Obuse C (2010) Human POGZ modulates dissociation of HP1alpha from mitotic chromosome arms through Aurora B activation. Nat Cell Biol 12: 719–727 [DOI] [PubMed] [Google Scholar]
- Maison C Romeo K Bailly D Dubarry M Quivy JP & Almouzni G (2012) The SUMO protease SENP7 is a critical component to ensure HP1 enrichment at pericentric heterochromatin. Nat Struct Mol Biol 19: 458–460 [DOI] [PubMed] [Google Scholar]
- Bawa-Khalfe T Lu LS Zuo Y Huang C Dere R Lin FM & Yeh ET (2012) Differential expression of SUMO-specific protease 7 variants regulates epithelial-mesenchymal transition. Proc Natl Acad Sci USA 109: 17466–17471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekes M Prudden J Srikumar T Raught B Boddy MN & Salvesen GS (2011) The dynamics and mechanism of SUMO chain deconjugation by SUMO-specific proteases. J Biol Chem 286: 10238–10247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alegre KO & Reverter D (2011) Swapping small ubiquitin-like modifier (SUMO) isoform specificity of SUMO proteases SENP6 and SENP7. J Biol Chem 286: 36142–36151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiolo I Minoda A Colmenares SU Polyzos A Costes SV & Karpen GH (2011) Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell 144: 732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courilleau C Chailleux C Jauneau A Grimal F Briois S Boutet-Robinet E Boudsocq F Trouche D & Canitrot Y (2012) The chromatin remodeler p400 ATPase facilitates Rad51-mediated repair of DNA double-strand breaks. J Cell Biol 199: 1067–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White DE Negorev D Peng H Ivanov AV Maul GG & Rauscher FJ 3rd (2006) KAP1, a novel substrate for PIKK family members, colocalizes with numerous damage response factors at DNA lesions. Cancer Res 66: 11594–11599 [DOI] [PubMed] [Google Scholar]
- Bruderer R Tatham MH Plechanovova A Matic I Garg AK & Hay RT (2011) Purification and identification of endogenous polySUMO conjugates. Embo Rep 12: 142–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golebiowski F Matic I Tatham MH Cole C Yin Y Nakamura A Cox J Barton GJ Mann M & Hay RT (2009) System-wide changes to SUMO modifications in response to heat shock. Sci Signal 2: ra24. [DOI] [PubMed] [Google Scholar]
- Bowen NJ Fujita N Kajita M & Wade PA (2004) Mi-2/NuRD: multiple complexes for many purposes. Biochimica et biophysica acta 1677: 52–57 [DOI] [PubMed] [Google Scholar]
- Iyengar S & Farnham PJ (2011) KAP1 protein: an enigmatic master regulator of the genome. J Biol Chem 286: 26267–26276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler LR Densham RM Jia J Garvin AJ Stone HR Shah V Weekes D Festy F Beesley J & Morris JR (2012) The proteasomal de-ubiquitinating enzyme POH1 promotes the double-strand DNA break response. EMBO J 31: 3918–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.