Summary
Human antibodies to HIV-1 can neutralize a broad range of viral isolates in vitro and protect non-human primates against infection1,2. Previous work showed that antibodies exert selective pressure on the virus but escape variants emerge within a short period of time3,4. However, these experiments were performed before the recent discovery of more potent anti-HIV-1 antibodies and their improvement by structure-based design5-9. Here we re-examine passive antibody transfer as a therapeutic modality in HIV-1-infected humanized mice (hu-mice). Although HIV-1 can escape from antibody monotherapy, combinations of broadly neutralizing antibodies (bNAbs) can effectively control HIV-1 infection and suppress viral load to levels below detection. Moreover, in contrast to antiretroviral therapy (ART)10-12, the longer half-life of antibodies led to viremic control for an average of 60 days after cessation of therapy. Thus, combinations of potent monoclonal antibodies can effectively control HIV-1 replication in hu-mice, and should be re-examined as a therapeutic modality in HIV-1-infected individuals.
Treatment of HIV-1 infection was ineffective until antiretroviral drugs were applied in combination permitting sustained suppression of viremia13,14. Despite this resounding success, the burden of daily medication, side effects, and resistance to antiretroviral drugs necessitate a continuing search for additional complementary therapeutic modalities15.
To examine the potential of recently discovered antibodies to effectively control HIV-1 infection, we used non-obese diabetic (NOD) mice that carry targeted disruptions of the recombinase activating gene 1 (Rag1−/−) and interleukin receptor common gamma chain (IL2RγNULL) reconstituted with human fetal liver-derived CD34+ hematopoietic stem cells16,17. Hu-mice were preferred to nonhuman primates for these experiments because the latter produce anti-human antibodies that alter the bioavailability of the injected human antibodies after only one to two weeks.
Hu-mice were analyzed for engraftment (Supplementary Fig. 1) and infected intraperitoneally (i.p) with a CCR5-tropic HIV-1 isolate (NL4-3 carrying a YU2 envelope; HIV-1YU2)18. Viral load in serum was determined by quantitative RT-PCR with a limit of detection of 800 copiesiml (Supplementary Fig. 2). Viremia was established (geometric mean of 1.06×105 copies/ml) by 14-20 days, and was stable for 60 days before decreasing to a geometric mean of 1.9×104 copies/ml at 120 days after infection (Fig. 1a). Persistent viremia was associated with progressive reduction in CD4+ T cells as measured by decreasing CD4+/CD8+ T cell ratios (Supplementary Fig. 3).
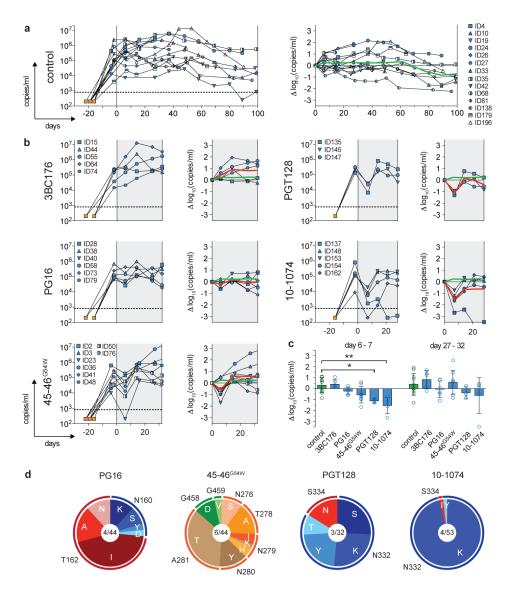
Figure 1. Monotherapy using broadly neutralizing antibodies in HIV-1YU2-infected hu-mice.
a, Left panel shows viral loads (RNA copies/ml, y-axis) measured over time (days, x-axis) in untreated HIV-1YU2-infected hu-mice (control group). Each line represents a single mouse and symbols reflect viral load measurements. Symbol characters correspond to individual mice as indicated (right). Hu-mice were infected with HIV-1YU2 (i.p.) between day −22 and −16 (orange square) and baseline viral loads were measured between day −4 and −2 (Supplementary Table 1). Dotted line represents limit of detection for viral load determination (800 copies/ml). Right panel shows changes in log10 (RNA copies/ml) from baseline (grey line) at day 0 with green line representing the average in viral load changes. b, Illustration of HIV-1 viral loads as in (a) but with single antibody treatment (shaded in grey) starting at day 0. Red line represents the average in viral load changes superimposed with averages of the control group (green line, a). c, Changes in log10 (RNA copies/ml) 6-7 days and 27-32 days after treatment initiation. Columns and error bars represent mean and standard deviation (SD), respectively. Significant statistical differences among groups were determined by performing a Kruskal-Wallis-test with Dunn-multiple comparison post-hoc test using GraphPad Prism version 5.0b for Mac OS X, GraphPad Software, San Diego California, USA. Significant differences among groups were detected at day 6-7 but not at the later time point (27-32 days). Asterisks (*, p≤0.05; **, p≤0.01) indicate a significant difference compared to the control group. d, Sequence analysis of HIV-1 gp120 after viral rebound while on therapy with single bNAbs (Supplementary Fig. 7). Pie charts show the dsitribution of amino acid changes at the antibodies’ respective target sites. A selection of amino acid substitutions was tested in vitro and confirmed antibody escape (Supplementary Fig. 8 and Table 2b). Mutations are relative to HIV-1YU2 and numbered according to HXBc2. Total number of analyzed mice/gp120 sequences is indicated in the center of the charts (Supplementary Fig. 7, Supplementary Table 2b).
To confirm that HIV-1YU2 infection in hu-mice is associated with viral diversification19 we cloned and sequenced 69 gp120 envelopes from 10 infected mice (Fig. 1a). After accounting for randomly introduced PCR errors (Supplementary Fig. 4a and b), we observed an average of 3.2 nucleotide substitutions per gp120 sequence, corresponding to a substitution rate of 2.2×10−3/bp (Supplementary Fig. 4b and c). We conclude that HIV-1YU2 infection is well established by 14-20 days in hu-mice, it persists for several months, and the virus mutates generating viral swarms18,19.
To examine the effects of bNAbs on established HIV-1 infection, we treated groups of 5-9 (3-8 analyzed) mice with antibody monotherapy using five different bNAbs. The antibodies were selected based on their potency and breadth in in vitro neutralization assays and because they target different epitopes. 45-46G54W is the most potent anti-CD4 binding site (CD4bs) antibody reported to date5, PG16 targets the V1/V2 loop region8,20, PGT128 is a glycan-dependent anti-V3 loop antibody7 and 10-1074 is a more potent variant of PGT1217,21 that has no measurable affinity for protein-free complex-type N-glycans in microarrays21. 3BC176 recognizes a conformational, yet to be defined epitope, and neutralizes HIV-1 strains that are resistant to potent CD4bs-antibodies22. Mice were treated subcutaneously with 0.5 mg of antibody/mouse (≅ 20 mg/kg) either once or twice a week based on the antibodies’ half-lives in mice, which varied from 0.7 to 6.3 days (Supplementary Fig. 5).
6-7 days after initiation of therapy, mice treated with PGT128 or 10-1074 showed an average decrease of 1.1 log10 (p≤0.05) and 1.5 log10 (p≤0.01) HIV-1 RNA copies/ml, respectively (Fig. 1b and c, Supplementary Table 1). The results for PG16 and 45-46G54W were variable, with an average decrease of 0.23 log10 and 0.56 log10 copies/ml (Fig. 1b and c, and Supplementary Table 1), while no effect was detected for 3BC176. However, with the exception of one mouse receiving 10-1074, viremia rebounded after 14-16 days and no significant differences between treated and control groups were detected thereafter (Fig. 1b and c). We conclude that monotherapy with PG16, 45-46G54W, PGT128 or 10-1074 results in an only transient decrease in the viral load in hu-mice infected with HIV-1YU2.
To determine whether viral escape from monoclonal antibody therapy is linked to specific mutations we cloned and sequenced gp120 from 3-6 infected mice in each therapy group before (Supplementary Fig. 6) and after (Supplementary Fig. 7) viral rebound. In all cases, viral rebound was associated with recurring mutations (defined as substitutions within a range of 3 amino acids in a majority of gp120 clones in more than one mouse) in residues that map to the antibody target sites (Fig. 1d, Supplementary Fig. 7, Supplementary Table 2b). For example, escape from 45-46G54W, was linked to mutations in residues 276-281 or 458, 459, which are located in the CD4bs23. Likewise, escape from PG16 was associated with mutations at residues 160 or 162, which correspond to the potential N-linked glycosylation site (PNGS) in the V1/V2 loop8,20. Escape from PGT128 and 10-1074 was associated with mutations at residues 332 or 334, both of which abrogate the same PNGS (Fig. 1d; Supplementary Fig. 7, Supplementary Table 2b).
To verify that the putative escape mutations rendered HIV-1YU2 resistant to the respective antibodies, we produced selected HIV-1YU2 pseudoviruses carrying the mutations and tested them in neutralization assays in vitro. Recurrent mutations associated with escape were always associated with specific resistance to their respective antibodies (Supplementary Fig. 8). Passenger mutations, defined as occurring in only a single mouse, or in multiple mice but in fewer than 50% of gp120 sequences in those mice, had no apparent effect on viral sensitivity to the antibodies (Supplementary Fig. 8).
Although escape mutations were associated with antibody-targeted epitopes, some gp120 residues appeared to tolerate multiple alternative amino acid substitutions, while others were more restricted. For example, in hu-mice treated with 45-46G54W N279 mutated to H, K, or I, but A281, only became T (Fig. 1d; Supplementary Table 2b). Escape from PGT128, was associated with replacement of N332 with K, S, T, or Y, while it became K in 51 out of 53 sequences from 4 hu-mice treated with 10-1074 (Fig. 1d; Supplementary Fig. 7, Supplementary Table 2b). Consistent with this observation, N332K is resistant to both PGT128 and 10-1074, while N332Y is resistant to PGT128 but remains sensitive to high concentrations of 10-1074 (Supplementary Fig. 8).
Since the selected bNAbs target distinct epitopes on the HIV-1 spike, we asked whether treatment with a combination of three (tri-mix = 3BC176, PG16, 45-46G54W) or five (penta-mix = 3BC176, PG16, 45-46G54W, PGT128, and 10-1074) would alter the course of infection (Fig. 2, Supplementary Table 1). These combinations neutralize all but 2 (tri-mix 98.3%) and 1 (penta-mix 99.2%) of 119 mostly Tier 2 and 3 viruses from multiple clades with an IC80 (geometric mean) of 0.121 μg/ml and 0.046 μg/ml for tri-mix and penta-mix, respectively (Supplementary Fig. 9).
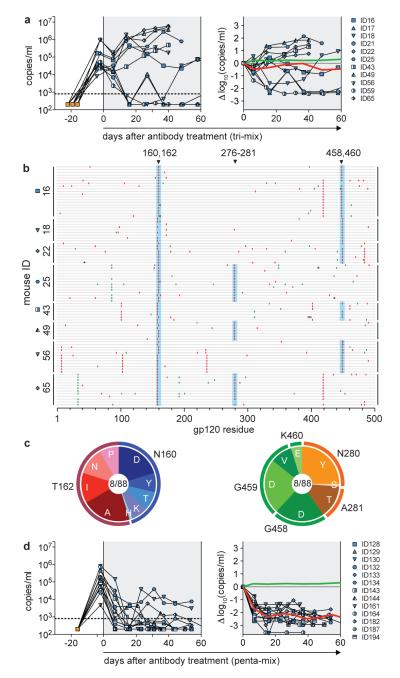
Figure 2. HIV therapy by a combination of three (tri-mix) or five (penta-mix) broadly neutralizing antibodies in HIV-1YU2-infected hu-mice.
a, Left panel shows viral loads (RNA copies/ml, y-axis) over time (days, x-axis) in HIV-1YU2-infected hu-mice treated with a combination of 3BC176, PG16 and 45-46G54W (tri-mix; grey shading). Each line represents a single mouse and symbols indicate viral load measurements (Supplementary Table 1). Infection and viral load determination was performed as in Fig. 1. Right panel shows changes in log10 (RNA copies/ml) from baseline at day 0. Red and green lines represent the average values in viral load change of tri-mix and control group (Fig. 1a), respectively. b, Individual gp120 envelope sequences cloned from single mice (y-axis) during tri-mix therapy after viral rebound. gp120 sequences are represented by horizontal gray bars with silent mutations indicated in green and amino acid replacements in red. Black asterisks represent mutations generating a stop codon and bold gray bars deletions. All mutations are relative to HIV-1YU2 and numbered according to HXBc2. Mutations at sites highlighted in blue can confer resistance to PG16- or 45-46G54W-mediated neutralization in vitro (Supplementary Fig. 8, Supplementary Table 3a). c, Pie charts as in Fig. 1d illustrating distribution of amino acid changes in gp120 (b) at PG16 (left) or 45-46G54W (right) respective target sites (Supplementary Table 3a). d, As in a, for HIV-1YU2-infected hu-mice treated with a combination of 3BC176, PG16, 45-46G54W, PGT128 and 10-1074 (penta-mix; grey shading).
A decline in the initial viral load was seen in 10 out of 11 tri-mix treated mice, but 7 rebounded to pre-treatment levels (Fig. 2a, Supplementary Table 1c, Supplementary Fig. 10). In contrast to monotherapy where mice almost never controlled viremia beyond 2 weeks of therapy, the tri-mix led to prolonged and effective HIV-1 control in 3 of 12 animals. In 2 of these mice viral load rebounded 20–40 days after cessation of therapy at a time when the YU2 gp120-reactive antibody concentration in serum decreased to a level below detection (Fig. 3a). In the third mouse (ID21), viremia was detected but remained low even in the absence of therapy for 60 days.
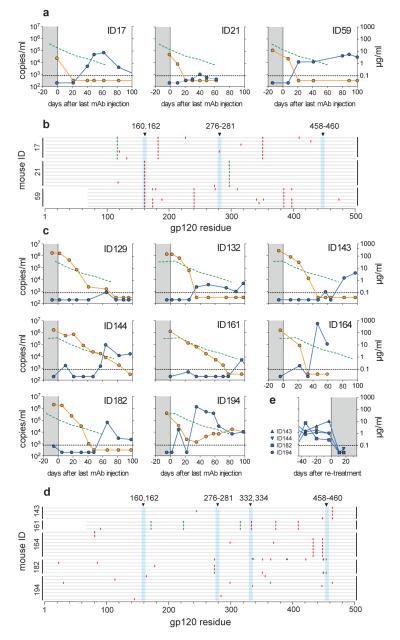
Figure 3. Viral rebound in HIV-1YU2-infected hu-mice after cessation of antibody therapy.
Viral load in RNA copies/ml (blue, left y-axis) and antibody concentration reactive to YU2 gp120 in μg/ml (orange, right y-axis) over time (x-axis) after the last antibody injection (day 0). Green dotted line indicates the viral load average of the control group (Fig. 1a). a, Viral load and YU2 gp120–reactive antibody concentration after stopping tri-mix therapy in mice that effectively controlled viremia below limit of detection. b, gp120 sequences illustrated as in Fig. 2b for viruses obtained after viral rebound from mice previously treated with tri-mix therapy. Vertical blue bars highlight sites in which mutations are able to confer resistance (Supplementary Fig. 2b, 3a). c, Viral load and YU2 gp120–reactive antibody concentration after stopping penta-mix therapy (Supplementary Fig. 3b). d, gp120 sequences for viruses obtained after rebound from mice previously treated with penta-mix therapy. e, Viral load of four mice re-treated with penta-mix therapy after viral rebound (c). Treatment period is highlighted in grey. Mouse numbers correspond to the mice in c.
Sequences obtained from mice that experienced viral rebound while on tri-mix therapy showed a combination of the mutations found in the PG16 and 45-46G54W monotherapy groups (Fig. 1d, 2b and c, Supplementary Table 2b, 3a). We verified that these mutations rendered HIV-1YU2 resistant to PG16 and 45-46G54W by producing the corresponding mutant pseudoviruses and testing them in neutralization assays in vitro (Supplementary Fig. 8). The pseudoviruses were not resistant to 3BC176 confirming that this antibody did not exert selective pressure on HIV-1YU2 and therefore just 2 of the 3 antibodies in the tri-mix were efficacious.
In contrast, sequences obtained from the mice that exhibited sustained viral control and rebounded after cessation of therapy either lacked any bNAb-associated mutation, or had a mutation mapped to the 45-46G54W (K282R) or PG16 (N162P) target site, but not both (Fig. 3 b, Supplementary Table 3a). In these mice, rebound viremia only occurred after YU2 gp120-reactive antibody levels decreased to below detection suggesting that the viruses that emerged were latent and remained susceptible to the tri-mix.
All 13 mice treated with the penta-mix showed a decrease in viral load 6-7 days after initiation of therapy (Fig. 2d, Supplementary Table 1d). However, in contrast to monotherapy and the tri-mix, all of the penta-mix treated mice remained below baseline during the entire treatment course (Fig. 2d, Supplementary Table 1d, Supplementary Fig. 10). Of the 13 mice, 11 had viral loads below or near the limit of detection. The two mice with the slowest reduction in viral load during treatment showed signs of severe graft versus host disease (GVHD). We conclude that penta-mix therapy reduces the viral load to levels below detection for up to 60 days in HIV-1YU2-infected hu-mice.
Penta-mix therapy was discontinued after 31–60 days and the mice were monitored for an additional 100 days (Fig. 3c). In 7 out of 8 mice that survived, viremia only rebounded after an average of 60 days (Fig. 3c). In contrast, mice treated with ART rebound after 10 days following discontinuation of therapy10. Viral rebound in penta-mix-treated mice was always correlated with decreased levels of the administered antibodies (Fig. 3c). Only one tri- (ID21) and one penta-mix (ID129) mouse failed to rebound. To determine their ability to support HIV-1 infection, these mice were re-infected with HIV-1YU2 (57.5 ng p24) and measured for viral load 2 weeks later. One of the two mice was infected but only at very low levels compared to the initial infection (Supplementary Fig. 11). Therefore prolonged control was primarily due to the long half-life of the injected antibodies.
We attempted to clone gp120 sequences from the plasma and cell-associated RNA of all penta-mix mice. Although we succeeded in obtaining sequences from 3 mice during the treatment period, every sequenced clone had at least one in-frame stop codon, all of which were consistent with signature APOBEC3G/F mutations (Supplementary Fig. 12, Supplementary Table 3b). In contrast, 27 of 28 gp120 sequences from viruses cloned after therapy was stopped and viral load rebounded did not have stop codons (Fig. 3d, Supplementary Table 3b). Furthermore, viruses that rebounded carried no or only one signature resistance mutation and remained susceptible to the penta-mix since viremia was controlled by re-treatment (Fig. 3e). Therefore, hu-mice treated with the penta-mix were unable to escape antibody pressure by way of envelope mutations, but the virus remained latent throughout the treatment period in at least 7 out of 8 mice.
HIV-1 infection in hu-mice differs from HIV-1 infection in humans in a number of important respects including a lower viral load, and a near absence of antibody-mediated immune responses, and therefore there is no pre-existing selective pressure on the envelope18 (Supplementary Fig. 13).
Previous antibody therapy experiments in hu-mice and humans concluded that treatment with combinations of antibodies had only limited effects against established HIV-1 infection4,24,25. However, the bNAbs used in those experiments (i.e. in mice b12, 2G12, 2F5; in humans 2G12, 4E10, 2F5) were orders of magnitude less potent than the ones used in this study. The difference in potency and the extended combination of broadly neutralizing antibodies is likely to account for the differences between our findings and earlier work.
Combination antibody therapy resembles antiretroviral or antimicrobial or anti-tumor combined therapy, in that escape requires the improbable appearance of multiple simultaneous mutations. However, antibodies differ from other therapeutic modalities for HIV in several respects. First, they can neutralize the pathogen directly; second, they have the potential to clear the virus and infected cells through engagement of innate effector responses26; third, immune complexes produced by the passively transferred antibodies may enhance immunity to HIV-127 and fourth antibodies have far longer half-lives than currently used antiretroviral drugs. Finally, anti-HIV-1 antibodies can be stably expressed in mammalian hosts for many months using adeno-associated viruses and therefore the potential exists to further prolong their bioavailability28,29. Although we have not combined antibodies and small molecule antiretroviral drugs, we speculate that such combinations may be especially effective because antibodies add a new modality to existing therapies. In addition, a combination of highly potent antibodies may be effective in suppressing viremia in individuals who do not tolerate anti-HIV medication.
This study establishes the principle that broadly neutralizing antibodies can suppress HIV-1 viremia to levels that are below detection in hu-mice for prolonged periods of time. Their efficacy as therapeutics and their long-term effects on HIV-1 infection in humans can only be evaluated in clinical trials.
Methods
Mice
NOD Rag1−/− IL2RγNULL (NOD.Cg-Ragltm1Mom Il2rgtm1Wjl/SzJ) mice were purchased from The Jackson Laboratory and bred and maintained at the Comparative Bioscience Center of the Rockefeller University according to guidelines established by the Institutional Animal Committee. All experiments were performed with authorization from the Institutional Review Board and the IACUC at the Rockefeller University.
Isolation of human hematopoietic stem cells (HSCs) and generation of humanized mice
Human fetal livers were procured from Advanced Bioscience Resources (ABR), Inc (Alameda) or the Human Fetal Tissue Repository (HFTR, Bronx, Ny). HSCs were isolated as previously described31. Briefly, human fetal liver was homogenized and incubated in digestion medium (HBSS with 0.1% collagenase IV (Sigma-Aldrich), 40mM HEPES, 2mM CaCl2 and 2U/ml DNAse I (Roche)) for 30 min at 37°C. Human CD34+ HSCs were isolated using a CD34+ HSC isolation kit (Stem Cell Technologies, Inc.) according to the manufacturers’ instructions. 1-5 day old NOD Rag1−/− IL2RγNULL (NRG) mice were irradiated with 100 cGy and 1.5-2×105 human CD34+ HSCs were injected intrahepatically 6 h after irradiation.
Flow cytometry
Flow cytometry was used to assess human hematopoiectic engraftment in mouse xenorecipients and to determine CD4+ to CD8+ T cell ratios. To this end, 200 μl of whole blood was collected from the superficial temporal vein of hu-mice and density gradient centrifugation (Ficoll-Paque™) was performed. Isolated peripheral blood mononuclear cells (PBMCs) were stained with anti-mouse CD45-PeCy7 (BioLegend), anti-human CD3-Pacific-Blue (BD Pharmingen), CD4-PE (BD), CD8-FITC (BD), CD16-Alexa700 (BD Pharmingen), CD19-APC (BD Pharmingen) and CD45-Pacific-Orange (Invitrogen) for 25 min at 4°C. Cells were washed and then fixed in 4% paraformaldehyde. To determine absolute numbers of CD4+ T cells in peripheral blood AccuCheck Counting Beads (Molecular Probes®) were added to the samples. FACS analysis was performed using a BD LSRFortessa cell analyzer (BD Biosciences) and data was analyzed using FlowJo Software (Tree Star).
HIV-1YU2 virus production
HIV-1YU2 was produced as previously described32 and levels of p24 in viral supernatant determined using Alliance HIV-1 p24 ANTIGEN ELISA Kit (PerkinElmer) according to the manufacturer’s instructions.
Quantitative reverse-transcriptase PCR assay
Blood was obtained from the superficial temporal vein and collected into EDTA-Microtubes (Sarstedt). After centrifugation, RNA was extracted from 100μl EDTA-plasma using a Qiagen MinElute Virus Spin Kit (Qiagen) and eluted in 50μl (2× 25μl elutions) RNAse free water (Qiagen). RNA samples were analyzed for HIV-1 RNA in a 50μl reaction mix using the 1× TaqMan® RT-PCR Mix, the 1× TaqMan® RT Enzyme Mix (TaqMan® RNA-to-Ct™ 1-Step kit, Applied Biosystems) and primers targeting a highly region conserved within the HIV-1 5′ long terminal repeat (FW 5′-GCCTCAATAAAGCTTGCCTTGA-3′; Rev 5′-GGCGCCACTGCTAGAGATTTT-3′)30. The internal probe (5′-AAGTAGTGTGTGC-CCGTCTGTTRTKTGACT-3′)30 contained a 5′ 6-carboxyfluorescein reporter and an internal/3′ ZEN-Iowa Black® FQ double-quencher (Integrated DNA Technologies, Inc.). As standard references (standard #1 and #2) two different dilutions from a single HIV-1NL4-3/BaL preparation were generated and quantified in duplicates by the Roche COBAS® TaqMan® HIV-1 Test, v.2.0 (Rolf Kaiser, University of Cologne, Germany). These reference aliquots were stored at −80°C and always extracted in parallel with mouse plasma samples. To generate a reference curve standard #1 was five-fold serially diluted from 173,500,000 to 4 HIV-1 RNA copies/ml (Supplementary Fig. 2). Standard #2 containing 2,600,000 copies/ml was used as a control for experimental variation: all sample Ct-values in a given experiment were adjusted by the difference between the experimental Ct-value for standard #2 and its expected Ct-value calculated using each experiment’s standard curve (generated using standard #1). Final viral load values were calculated using the control-adjusted Ct for each sample.
qRT-PCR assay, limit of detection
The lower limit of detection (LOD) for the assay used in this study was found at 800 copies/ml. HIV-1 culture supernatant of known RNA copy number was serially diluted two-fold to concentrations between 1600 and 200 copies/ml (corresponding to 64 to 8 copies per 20ul RNA extract). Each dilution was independently extracted and assayed at n=4 per experiment. The presence of a cycle threshold value for a given PCR reaction was scored as a detection event. Detection scores accumulated from five identical experiments were subjected to probit analysis, and the 85% LOD was found at 800 copies/ml for undiluted RNA extract (corresponding to 32 copies/PCR reaction). For RNA diluted five-fold in deionized nuclease free water, the 85% LOD was 640 copies/ml (corresponding to 5 copies/PCR reaction).
Antibody production, t1/2 estimation and administration
Antibodies were generated as previously described5,33,34. Briefly, HEK 293T/17 or HEK 293-6E cells were transiently transfected with equal amounts of immunoglobulin heavy and light chain expression vectors. After supernatant was harvested, purified IgG was obtained using HiTrap rProtein A FF columns (GE Healthcare) or Protein G sepharose 4 Fast Flow (GE Healthcare). Sterile filtration was performed for all antibodies shortly before injection by using Ultrafree-CL Centrifugal Filters (0.22 μm, Millipore). Antibody t1/2 was calculated by ln(2)/k where k is 1st order reaction rate constant and calculated as a slope of ln(antibody concentration) vs. time. Based on t1/2 of bNAbs HIV-1YU2-infected hu-mice were treated with 0.5 mg of each antibody injected subcutaneously once (3BC176) or twice (PG16, 45-46G54W, PGT128, 10-1074)/week. The tri-mix was composed of 3BC176, PG16 and 45-46G54W while the penta-mix included all five given antibodies. To determine antibody t1/2, 0.5 mg of antibody was administered intravenously by retro-orbital injection (Supplementary Fig. 5).
ELISA
Costar® 96-Well EIA/RIA Stripwell™ Plates were coated overnight with YU2 gp120 or goat Anti-Human IgG specific for Fcγ fragment at 250 and 150 ng/well, respectively. Plates were washed 3 × with distilled water and blocked for 1 h with PBST (0.1% Tween-20) containing 2% BSA and 1 μM EDTA. Standard IgG1 kappa (Sigma-Aldrich) and 10-1074 were used in duplicates as standard antibodies for quantifying total IgG and YU2 gp120-specific IgGs, respectively. After washing the plate, mouse plasma was applied in 1:3 serial dilutions with a 1:10 or 1:20 starting dilution and incubated for 2 h followed by washing the plate and 1 h incubation with Peroxidase-conjugated Goat Anti-human IgG (Jackson ImmunoResearch). Plates were washed and developed using ABTS single solution (Invitrogen). Optical density was measured at 405nm and IgG concentration determined by correlation to the standard curve of the reference antibodies. Of note, YU2 gp120 does not react with PG16 or 3BC176 and therefore these mAbs are not detected by the YU2 gp120 ELISA.
Neutralization
Single antibodies and tri- and penta-mix were tested against a panel of HIV-1 envelope-pseudoviruses as previously described6,35,36 or HIV-1YU2-mutant variants. Briefly, envelope-pseudoviruses were incubated with 5-fold serial dilutions of single antibodies or tri- and penta-mix samples and applied to TZM.bl cells that carry a luciferase-reporter gene. After 48 h cells were lysed and luminescence was measured. IC50 and IC80 reflect single antibody concentrations that caused a reduction in relative luminescence units (RLU) by 50% and 80%, respectively.
cDNA synthesis
Viral RNA was extracted as described above and subjected to first-strand cDNA synthesis using the SuperScript III reverse transcriptase kit (Invitrogen Life Technologies, Grand Island, Ny). For extraction of cellular RNA an RNA extraction kit was used (Qiagen). Sense and anti-sense primers used for cDNA synthesis were 5′-GGCTTAGGCATCTCCTATGGCAGGAAGAA-3′ and 5′-GGTGTGTAGTTCTGCCAATCAGGGAAGWAGCCTTGTG-3′, respectively.
PCR amplification and cloning
PCR amplifications were performed in ThermoGrid 96-well plates (Denville Scientific) using either the Clontech Advantage 2 PCR System (BD) or the Expand High Fidelity PCR System (Roche). All PCR amplifications were performed in a 50 μl reaction mix. The primers used for the first round of PCR were 5′-GGCTTAGGCATCTCCTATGGCAGGAAGAA-3′ and 5′-GGTGTGTAGTTCTGCCAATCAGGGAAGWAGCCTTGTG-3′. Primers for the second-round PCR were 5′-TAGAAA-GAGCAGAAGACAGTGGCAATGA-3′ and 5′-TCATCAATGGTGGTGATGATGATGTTTTTCTCTCTGCACCACTCTTCT-3′. Cycling parameters for 1st round PCR were 94°C for 2 min followed by 40 cycles of 94°C 30 sec, annealing temperature of 55-65°C for 45 s, 68°C 4 min, and a final extension at 68°C for 10 min, where the annealing temperature was 65°C for the first 3 cycles, 60°C for the next 11 cycles, and 55°C for the final 26 cycles. 3 μl of product from first-round was used as the template for the second-round of PCR. Cycling conditions for the second round PCR were 94°C for 2 min followed by 40 cycles of 94°C 30 sec, annealing temperature of 53-55°C for 45 sec, 68°C 2 min and 30 sec, and a final extension at 68°C for 10 min, where annealing temperature was 53°C for the first 9 cycles and 55°C for the next 31 cycles. Following the second-round PCR amplification, 0.5 μl Taq polymerase was added to each 50 μl reaction and an additional 72°C extension for 15 min was performed to add 3′ A overhangs for use in cloning. Gel purified PCR amplicons were ligated into pCR4-TOPO (Invitrogen) following the manufacturer’s instructions and transformed into One Shot TOP10 (Invitrogen) cells. Individual colonies were sequenced using M13F and M13R primers.
Sequence Alignments and Mutation Analysis
Forward and reverse sequence reads from individual samples were assembled using Geneious Pro software version 5.5.6 (Biomatters Ltd) aligned to gp120YU2 (accession number M93258). Env sequences containing frameshift mutations or large deletions were excluded from further analysis. All sequences were analyzed for mutations relative to gp120YU2 using the Los Alamos Highlighter tool (http://www.hiv.lanl.gov/content/sequence/HIGHLIGHT/HIGHLIGHTXyPLOT/highlighter.html). and mutations were numbered using HXBc2 numbering as determined by the Los Alamos Sequence Locator tool (http://www.hiv.lanl.gov/content/sequence/LOCATE/locate.html). Mutations were considered recurrent when substitutions within a range of 3 amino acids occurred in the majority of gp120 clones in more than one mouse. The fidelity of the combined reverse transcription and nested PCR reactions was determined by amplifying monoclonal wild-type HIV-1YU2 by the same amplification procedures and identifying RT and PCR induced mutations (Supplementary Fig. 4).
Generation of HIV-1YU2 envelope mutants
Single, double and triple mutations were introduced into wildtype HIV-1YU2 envelope by site-directed mutagenesis using QuikChange (Multi-) Site-Directed Mutagenesis Kit (Agilent Technologies) according to the manufacturer’s specifications.
Statistical analysis
Krustal-Wallis-test with Dunn-multiple comparison or Mann-Whitney U post-hoc tests were performed using GraphPad Prism version 5.0b for Mac OS X, GraphPad Software, San Diego California USA, for comparisons of the viral load levels among different antibody treatment groups and control group (Fig. 1c, Supplementary Fig. 10) and for comparing gp120 mutation rates (Supplementary Fig. 4). Levels of significance are indicated by asterisks (*, p≤0.05; **, p≤0.01, ***, p≤0.001). Correlation between viral load (RNA copies/ml) and absolute numbers of CD4+ T cells in peripheral blood (Supplementary Fig. 2b) was analyzed by Spearman correlation coefficient (rho) using GraphPad Prism version 5.0b for Mac OS X. An unpaired student’s t-test was used to compare CD4+/CD8+ ratios of pre-infected mice in control and penta-mix group (Supplementary Fig. 3).
Supplementary Material
Supplementary Figure 1 Reconstitution analysis of humanized mice
Flow cytometry was used to analyze the level of chimerism before infection. a, Percentage of human CD45+ (hu-CD45+) cells of total CD45+ cells in peripheral blood of hu-mice (left). Percentage of human CD3+ and CD19+ cells of total hu-CD45+ cells (middle) and the distribution of human CD4+ and CD8+ T cells within the human CD3+ population (right). b, Plot shows viremia in copies/ml 14-20 days after infection with HIV-1YU2 (y-axis) and absolute numbers of CD4+ T cells/μl in peripheral blood (x-axis). Each dot represents a single mouse. Spearman correlation coefficient (rho) between viral load and CD4+ T cells was used to determine correlation (GraphPad Prism version 5.0b for Mac OS X). HIV-1YU2 infection of hu-mice with a higher number of CD4+ T cells in peripheral bolld correlates with a higher viral load after 14-20 days post infection. c, Reconstitution data for all HIV-1YU2-infected hu-mice.
Supplementary Figure 2. Standards used in qPCR assay to determine HIV-1 viral load
a, Averages of fifteen consecutive experiments with standard curve generated from five-fold serial dilutions of RNA extracted from 1.74 × 108 copies/ml HIV-1NL4-3/BaL. Error bars reflect standard deviation. b, Viral load values from two experiments containing three replicates per mouse (two replicates in one experiment (Exp. 1, gray) and the third replicate in a second experiment (Exp. 2, black)). Error bars for Exp. 1 reflect range between replicate values. c, Intra-assay and inter-assay variation were <log10 (0.5) units. d, Cycle threshold values are shown for two independent standards (Standard no. 1; 1.74 × 108 copies/ml; Standard no. 2; 2.6 × 106 copies/ml) extracted for each qPCR experiment. Cycle threshold variation between experiments similarly affected both standards.
Supplementary Figure 3. CD4+/CD8+ T cell ratio in HIV-1YU2-infected hu-mice
a, CD4+/CD8+ ratios in untreated control and penta-mix-treated mice prior to HIV-1YU2 infection. Both groups had similar baseline CD4+/CD8+ T cell ratios (P=0.8; unpaired Student’s t-test). ns, not significant. b, CD4+/CD8+ baseline levels are normalized to 1 and changes in ratio are measured over time for control and penta-mix-treated mice (day 0 reflects day of infection). In penta-mix-treated mice blue colored lines reflect the time from infection until therapy was stopped. Yellow line illustrates the changes of the CD4+/CD8+ T cell ratio after discontinuing penta-mix therapy.
Supplementary Figure 4. Quantification of PCR-induced mutations and detection of HIV-1 diversification
a, Each horizontal gray bar represents the sequence of a single gpl20 clone obtained by applying amplification and cloning techniques using monoclonal wild-type HIV-1YU2 as an RNA template. Synonymous nucleotide substitutions are indicated in green while non-synonymous substitutions are shown in red. Sequences are grouped by the polymerases used in PCR amplification (either Advantage 2 or Expand High Fidelity). Vertical black bars on the y-axis indicate groups of clones that came from the same starting HIV-1YU2 RNA template. Concentrations of the RNA template (copies/ml) are indicated. Overall, amplification with Advantage 2 Polymerase induced 38 substitutions over 37,275 base pairs, corresponding to an error rate of 1.02 × 10−3 per base. The amplification with Expand High Fidelity polymerase induced 24 substitutions over 38,766 base pairs, corresponding to an error rate of 6.19 × 10−4 per base. Of note, the starting concentration of the RNA template did not lead to a significant difference in PCR-induced nucleotide substitutions. b, Number of PCR-induced nucleotide substitutions within individual gpl20 clones is compared to nucleotide substitutions observed in HIV-1YU2-infected mice (sequences obtained from control group, Supplementary Fig. 6 and 7). While the majority of gp120 clones have 0 or 1 PCR-induced mutation, most of gp120 clones from infected mice carry multiple nucleotide substitutions. Mean is indicated in red. PCR-reactions were generally performed using Expand High Fidelity Polymerase. Significant statistical differences among groups were determined by using a Kruskal-Wallis test with Mann-Whitney U post-hoc test. Level of significance is indicated by asteriks (***P≤0.001). ns, not significant.
Supplementary Figure 5. t1/2 estimation of broadly neutralizing antibodies in mice
a, Antibody serum concentration (μg/ml) in NOD Ragl−/− IL2Rγnull mice after injection (i.v.) of 0.5 mg antibody (day 0, red arrow). Antibody concentrations were measured by ELISA and mean and standard deviation for the indicated number of mice are shown. b, t1/2 of the antibodies measured in NOD Ragl−/− IL2Rγnull mice. c, Neutralizing activity (IC50) of antibodies against YU2.DG and average t1/2-values of antibodies used in experiments. Neutralization activity is color-coded and IC50-values between 0.01 and 0.1 μg/ml are shown in red and IC50-values > 0.1 μg/ml in orange. d, Antibody concentrations in hu-mice injected on the schedule as in all treatment experiments. Green arrows indicate subcutaneous injections of 0.5 mg antibody. Based on its significantly longer t1/2, 3BC176 was injected only once a week. Shown are mean and standard deviation for the indicated number of mice. The absence of endogenous antibodies was confirmed in all mice used for t1/2 experiments.
Supplementary Figure 6. Mutation analysis of gpl20 sequences within the first 8 days of antibody treatment
Individual gpl20 nucleotide sequences are represented by horizontal gray bars with silent mutations indicated in green and replacement mutations in red. Bold gray bars represent deletions and mutations that produced a stop codon are indicated by asterisks (*). All substitutions are relative to HIV-1YU2 (acc. number M93258) and numbered according to HXBc2. Beside the control sample (no treatment), all sequences were obtained 6-7 days after initiation of antibody therapy. Sequences are organized by the mouse from which the viral clone was obtained (y-axis). 6-7 days after initiation of therapy, i.e. before viral rebound, we found no recurrent mutations that mapped to known epitope targets in any of the treatment groups.
Supplementary Figure 7. Monotherapy with broadly neutralizing antibodies selects for viral escapes
a, Individual gpl20 envelope sequences are represented by horizontal gray bars with silent mutations indicated in green and amino acid replacements in red. Black asterisks represent mutations generating a stop codon and bold gray bars deletions. All mutations are relative to HIV-1YU2 (acc. number M93258) and numbered according to HXBc2. Sequences are organized by individual mice (mouse ID, y-axis) from which they were obtained. Highlighted sites refer to recurrent mutations as defined by amino acid substitutions within 3 amino acids in the majority of sequences in at least two mice (Supplementary Table 2). Blue highlighted areas are sites in which selected mutations confer resistance to respective antibodies (Supplementary Fig. 8). Gray highlighted areas in control group show recurrent mutations indicating a positive selection process (Supplementary Table 2a)19. Gray highlighted area in 3BC176 reflects a recurrent mutation that does not confer resistance to 3BC176 (Supplementary Fig. 8).
Supplementary Figure 8. Neutralization activity of bNAbs against YU2-mutants
IC50-values are shown for the indicated antibodies on YU2-mutant virions. Neutralization activity is color-coded and IC50-values < 0.1 μg/ml are shown in red, values between 0.1 and 1.0 μg/ml in orange, > 1 – 10 μg/ml in dark yellow, > 10 to 50 μg/ml in yellow and values > 50 μg/ml in white.
Supplementary Figure 9. Neutralization activity of tri- and penta-mix
a, Geometric mean IC80-values for all viruses grouped by clades. b, Individual IC50 and IC80-values for all tested viral strains. Neutralization activity is color-coded with values < 0.001 μg/ml highlighted in blue, between 0.001 - 0.01 μg/ml in red, > 0.01 - 0.1 μg/ml in orange, > 0.1 - 1 μg/ml in light orange, > 1 - 10 μg/ml in yellow and > 10 μg/ml in white.
Supplementary Figure 10. Comparing viral loads of tri- and penta-mix-treated mice
Changes in log10 (RNA copies/ml) 6-7 days and 27-32 days after treatment initiation with tri- and penta-mix. Columns and error bars represent mean and standard deviation, respectively. Kruskal-Wallis test with Dunn multiple comparison post-hoc test was performed using GraphPad Prism version 5.0b for Mac OS X, Graph-Pad Software, San Diego California USA. Asterisks (**p ≤ 0.01, ***p ≤ 0.001) indicate significant differences between groups.
Supplementary Figure 11. Re-infection of mouse ID21 and ID129
Viral load (copies/ml; blue line) and concentration of YU2 gpl20-reactive antibodies (grey line). Red arrows indicate the initial infection and re-infection with HIV-1YU2 (57.5 ng p24). Grey shaded area reflects antibody treatment (ID2l, tri-mix; ID129, penta-mix). Mouse ID2l has a significant slower increase in viral load compared to initial infection. Mouse ID129 died shortly after re-infection and viral load could not be confirmed.
Supplementary Figure 12. Sequence analysis during penta-mix treatment
Individual gpl20 envelope sequences cloned from single mice (y-axis) during penta-mix therapy in HIV-1YU2-infected hu-mice. Silent mutations are indicated in green and amino acid replacements in red. Black asterisks represent mutations generating a stop codon. All mutations are relative to HIV-1YU2 and numbered according to HXBc2. Sequences were only obtained from three mice and all of these sequences carried at least one stop codon (*) at residues that displayed an APOBEC3G/F motif (Supplementary Table 3b). Vertical blue bars highlight sites of recurrent mutations that were found in mono- (PG16, 45-46G54W, PGT128, 10-1074; Fig. lb, d) and tri-mix-therapy experiments (Fig. 2a, b, c).
Supplementary Figure 13. Natural antibody response to HIV-1YU2 infection in hu-mice
Figure shows antibody concentrations in μg/ml for total IgG for the indicated mice assayed (ELISA) on indicated days post infection (p.i.) as well as the IC50 neutralization titer determined in TZM-bl neutralization assay, a, control group; b, penta-mix group and c, uninfected controls. The values are color-coded as indicated in the legend (bottom right).
Acknowledgements
We thank Rolf Kaiser for analyzing viral loads of reference HIV-1 samples and Natalia N. Freund for producing aU2 gp120. We thank Brenna Flatley, Tamar Friling, Han Gao, Sabrina Sell and Susan Hinklein for assistance and technical support, Mayte Suarez-Farinas for advice on statistical analysis and Mariana Babayeva for helping with antibody t1/2 estimation. M.C.N. and F.K. have a pending patent application for the antibody 3BC176 and M.C.N., P.J.B. and H.M. for the antibody 10-1074 with the United States Patent and Trademark Office. These reagents are available with a Materials Transfer Agreement. F.K. (KL 2389/1-1), M.D. (DO 1450/1-1) and E.B. (BI 1422/1-1) were supported by the German Research Foundation (DFG). H.G., C.G. and R.-B. I. were supported by The German National Academic Foundation. M.S.S. was supported by the Bill and Melinda Gates Foundation’s Comprehensive Antibody Vaccine Immune Monitoring Consortium, grant number 1032144. A.P. is a recipient of a Liver Scholar Award from the American Liver Foundation. This work was supported in part by CAVD grant OPP1033115 from the Bill and Melinda Gates Foundation to M.C.N. and NIH grant AI081677 to M.C.N.. M.C.N., P.J.B. and P.D.B. are HHMI investigators.
Footnotes
Author Contributions: F.K., A.H.-S., J.A.H. planned and performed experiments and wrote the manuscript. H.G., J.F.H., S.B., H.M., L.A.S., R.D., A.A., T.Z., M.D., E.B., R.N.L., C.G., P.M., R.-B.I., T.R.E. and M.S. performed experiments. P.D.B., P.J.B., J.V.R., A.P. provided reagents, advise and edited the manuscript. M.C.N. planned experiments and wrote the manuscript.
Author information: The authors declare no competing financial interests.
References
- 1.Hessell AJ, et al. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat Med. 2009;15:951–954. doi: 10.1038/nm.1974. doi:10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mascola JR, et al. Protection of Macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J Virol. 1999;73:4009–4018. doi: 10.1128/jvi.73.5.4009-4018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307312. doi: 10.1038/nature01470. doi:10.1038/nature01470nature01470 [pii] [DOI] [PubMed] [Google Scholar]
- 4.Poignard P, et al. Neutralizing antibodies have limited effects on the control of established HIV-1 infection in vivo. Immunity. 1999;10:431–438. doi: 10.1016/s1074-7613(00)80043-6. [DOI] [PubMed] [Google Scholar]
- 5.Diskin R, et al. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science. 2011;334:1289–1293. doi: 10.1126/science.1213782. doi:science.1213782 [pii]10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheid JF, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. doi:10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker LM, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. doi:10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker LM, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. doi:1178746 [pii]10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu X, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. doi:science.1187659 [pii]10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choudhary SK, et al. Suppression of human immunodeficiency virus type 1 (HIV-1) viremia with reverse transcriptase and integrase inhibitors, CD4+ T-cell recovery, and viral rebound upon interruption of therapy in a new model for HIV treatment in the humanized Rag2−/−{gamma}c−/− mouse. J Virol. 2009;83:8254–8258. doi: 10.1128/JVI.00580-09. doi:10.1128/JVI.00580-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nischang M, et al. Humanized Mice Recapitulate Key Features of HIV-1 Infection: A Novel Concept Using Long-Acting Anti-Retroviral Drugs for Treating HIV-1. PLoS One. 2012;7:e38853. doi: 10.1371/journal.pone.0038853. doi:10.1371/journal.pone.0038853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denton PW, et al. Generation of HIV latency in humanized BLT mice. J Virol. 2012;86:630–634. doi: 10.1128/JVI.06120-11. doi:10.1128/JVI.06120-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weiss RA. Special anniversary review: twenty-five years of human immunodeficiency virus research: successes and challenges. Clinical and experimental immunology. 2008;152:201–210. doi: 10.1111/j.1365-2249.2008.03645.x. doi:10.1111/j.1365-2249.2008.03645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finzi D, Siliciano RF. Viral dynamics in HIV-1 infection. Cell. 1998;93:665–671. doi: 10.1016/s0092-8674(00)81427-0. [DOI] [PubMed] [Google Scholar]
- 15.Arts EJ, Hazuda DJ. HIV-1 Antiretroviral Drug Therapy. Cold Spring Harbor perspectives in medicine. 2012;2:a007161. doi: 10.1101/cshperspect.a007161. doi:10.1101/cshperspect.a007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Traggiai E, et al. Development of a human adaptive immune system in cord blood cell-transplanted mice. Science. 2004;304:104–107. doi: 10.1126/science.1093933. doi:10.1126/science.1093933. [DOI] [PubMed] [Google Scholar]
- 17.Brehm MA, et al. Parameters for establishing humanized mouse models to study human immunity: analysis of human hematopoietic stem cell engraftment in three immunodeficient strains of mice bearing the IL2rgamma(null) mutation. Clin Immunol. 2010;135:84–98. doi: 10.1016/j.clim.2009.12.008. doi:10.1016/j.clim.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baenziger S, et al. Disseminated and sustained HIV infection in CD34+ cord blood cell-transplanted Rag2−/−gamma c−/− mice. Proc Natl Acad Sci U S A. 2006;103:15951–15956. doi: 10.1073/pnas.0604493103. doi:10.1073/pnas.0604493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ince WL, et al. Evolution of the HIV-1 env gene in the Rag2−/− gammaC−/−humanized mouse model. J Virol. 2010;84:2740–2752. doi: 10.1128/JVI.02180-09. doi:10.1128/JVI.02180-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLellan JS, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. doi:10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mouquet H. Complex-type N-glycan recognition by potent broadly-neutralizing HIV antibodies. 2012. submitted. [DOI] [PMC free article] [PubMed]
- 22.Klein F, et al. Broad neutralization by a combination of antibodies recognizing the CD4 binding site and a new conformational epitope on the HIV-1 envelope protein. J Exp Med. 2012;209:1469–1479. doi: 10.1084/jem.20120423. doi:10.1084/jem.20120423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwong PD, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. doi:10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mehandru S, et al. Adjunctive passive immunotherapy in human immunodeficiency virus type 1-infected individuals treated with antiviral therapy during acute and early infection. J Virol. 2007;81:11016–11031. doi: 10.1128/JVI.01340-07. doi:10.1128/JVI.01340-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trkola A, et al. Delay of HIV-1 rebound after cessation of antiretroviral therapy through passive transfer of human neutralizing antibodies. Nat Med. 2005;11:615–622. doi: 10.1038/nm1244. doi:nm1244 [pii]10.1038/nm1244. [DOI] [PubMed] [Google Scholar]
- 26.Nimmerjahn F, Ravetch JV. Antibody-mediated modulation of immune responses. Immunol Rev. 2010;236:265–275. doi: 10.1111/j.1600-065X.2010.00910.x. doi:IMR910 [pii]10.1111/j.1600-065X.2010.00910.x. [DOI] [PubMed] [Google Scholar]
- 27.Ng CT, et al. Passive neutralizing antibody controls SHIV viremia and enhances B cell responses in infant macaques. Nat Med. 2010;16:1117–1119. doi: 10.1038/nm.2233. doi:10.1038/nm.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson PR, et al. Vector-mediated gene transfer engenders long-lived neutralizing activity and protection against SIV infection in monkeys. Nat Med. 2009;15:901–906. doi: 10.1038/nm.1967. doi:10.1038/nm.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balazs AB, et al. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature. 2012;481:81–84. doi: 10.1038/nature10660. doi:10.1038/nature10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rouet F, et al. Transfer and evaluation of an automated, low-cost real-time reverse transcription-PCR test for diagnosis and monitoring of human immunodeficiency virus type 1 infection in a West African resource-limited setting. Journal of clinical microbiology. 2005;43:2709–2717. doi: 10.1128/JCM.43.6.2709-2717.2005. doi:10.1128/JCM.43.6.2709-2717.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Billerbeck E, et al. Development of human CD4+FoxP3+ regulatory T cells in human stem cell factor-, granulocyte-macrophage colony-stimulating factor-, and interleukin-3-expressing NOD-SCID IL2Rgamma(null) humanized mice. Blood. 2011;117:3076–3086. doi: 10.1182/blood-2010-08-301507. doi:10.1182/blood-2010-08-301507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang YJ, et al. Envelope-dependent, cyclophilin-independent effects of glycosaminoglycans on human immunodeficiency virus type 1 attachment and infection. J Virol. 2002;76:6332–6343. doi: 10.1128/JVI.76.12.6332-6343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheid JF, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. doi:nature07930 [pii]10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 34.Tiller T, et al. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. Journal of immunological methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. doi:10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montefiori DC. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr Protoc Immunol. 2005 doi: 10.1002/0471142735.im1211s64. Chapter 12, Unit 12 11, doi:10.1002/0471142735.im1211s64. [DOI] [PubMed] [Google Scholar]
- 36.Li M, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. doi:10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 Reconstitution analysis of humanized mice
Flow cytometry was used to analyze the level of chimerism before infection. a, Percentage of human CD45+ (hu-CD45+) cells of total CD45+ cells in peripheral blood of hu-mice (left). Percentage of human CD3+ and CD19+ cells of total hu-CD45+ cells (middle) and the distribution of human CD4+ and CD8+ T cells within the human CD3+ population (right). b, Plot shows viremia in copies/ml 14-20 days after infection with HIV-1YU2 (y-axis) and absolute numbers of CD4+ T cells/μl in peripheral blood (x-axis). Each dot represents a single mouse. Spearman correlation coefficient (rho) between viral load and CD4+ T cells was used to determine correlation (GraphPad Prism version 5.0b for Mac OS X). HIV-1YU2 infection of hu-mice with a higher number of CD4+ T cells in peripheral bolld correlates with a higher viral load after 14-20 days post infection. c, Reconstitution data for all HIV-1YU2-infected hu-mice.
Supplementary Figure 2. Standards used in qPCR assay to determine HIV-1 viral load
a, Averages of fifteen consecutive experiments with standard curve generated from five-fold serial dilutions of RNA extracted from 1.74 × 108 copies/ml HIV-1NL4-3/BaL. Error bars reflect standard deviation. b, Viral load values from two experiments containing three replicates per mouse (two replicates in one experiment (Exp. 1, gray) and the third replicate in a second experiment (Exp. 2, black)). Error bars for Exp. 1 reflect range between replicate values. c, Intra-assay and inter-assay variation were <log10 (0.5) units. d, Cycle threshold values are shown for two independent standards (Standard no. 1; 1.74 × 108 copies/ml; Standard no. 2; 2.6 × 106 copies/ml) extracted for each qPCR experiment. Cycle threshold variation between experiments similarly affected both standards.
Supplementary Figure 3. CD4+/CD8+ T cell ratio in HIV-1YU2-infected hu-mice
a, CD4+/CD8+ ratios in untreated control and penta-mix-treated mice prior to HIV-1YU2 infection. Both groups had similar baseline CD4+/CD8+ T cell ratios (P=0.8; unpaired Student’s t-test). ns, not significant. b, CD4+/CD8+ baseline levels are normalized to 1 and changes in ratio are measured over time for control and penta-mix-treated mice (day 0 reflects day of infection). In penta-mix-treated mice blue colored lines reflect the time from infection until therapy was stopped. Yellow line illustrates the changes of the CD4+/CD8+ T cell ratio after discontinuing penta-mix therapy.
Supplementary Figure 4. Quantification of PCR-induced mutations and detection of HIV-1 diversification
a, Each horizontal gray bar represents the sequence of a single gpl20 clone obtained by applying amplification and cloning techniques using monoclonal wild-type HIV-1YU2 as an RNA template. Synonymous nucleotide substitutions are indicated in green while non-synonymous substitutions are shown in red. Sequences are grouped by the polymerases used in PCR amplification (either Advantage 2 or Expand High Fidelity). Vertical black bars on the y-axis indicate groups of clones that came from the same starting HIV-1YU2 RNA template. Concentrations of the RNA template (copies/ml) are indicated. Overall, amplification with Advantage 2 Polymerase induced 38 substitutions over 37,275 base pairs, corresponding to an error rate of 1.02 × 10−3 per base. The amplification with Expand High Fidelity polymerase induced 24 substitutions over 38,766 base pairs, corresponding to an error rate of 6.19 × 10−4 per base. Of note, the starting concentration of the RNA template did not lead to a significant difference in PCR-induced nucleotide substitutions. b, Number of PCR-induced nucleotide substitutions within individual gpl20 clones is compared to nucleotide substitutions observed in HIV-1YU2-infected mice (sequences obtained from control group, Supplementary Fig. 6 and 7). While the majority of gp120 clones have 0 or 1 PCR-induced mutation, most of gp120 clones from infected mice carry multiple nucleotide substitutions. Mean is indicated in red. PCR-reactions were generally performed using Expand High Fidelity Polymerase. Significant statistical differences among groups were determined by using a Kruskal-Wallis test with Mann-Whitney U post-hoc test. Level of significance is indicated by asteriks (***P≤0.001). ns, not significant.
Supplementary Figure 5. t1/2 estimation of broadly neutralizing antibodies in mice
a, Antibody serum concentration (μg/ml) in NOD Ragl−/− IL2Rγnull mice after injection (i.v.) of 0.5 mg antibody (day 0, red arrow). Antibody concentrations were measured by ELISA and mean and standard deviation for the indicated number of mice are shown. b, t1/2 of the antibodies measured in NOD Ragl−/− IL2Rγnull mice. c, Neutralizing activity (IC50) of antibodies against YU2.DG and average t1/2-values of antibodies used in experiments. Neutralization activity is color-coded and IC50-values between 0.01 and 0.1 μg/ml are shown in red and IC50-values > 0.1 μg/ml in orange. d, Antibody concentrations in hu-mice injected on the schedule as in all treatment experiments. Green arrows indicate subcutaneous injections of 0.5 mg antibody. Based on its significantly longer t1/2, 3BC176 was injected only once a week. Shown are mean and standard deviation for the indicated number of mice. The absence of endogenous antibodies was confirmed in all mice used for t1/2 experiments.
Supplementary Figure 6. Mutation analysis of gpl20 sequences within the first 8 days of antibody treatment
Individual gpl20 nucleotide sequences are represented by horizontal gray bars with silent mutations indicated in green and replacement mutations in red. Bold gray bars represent deletions and mutations that produced a stop codon are indicated by asterisks (*). All substitutions are relative to HIV-1YU2 (acc. number M93258) and numbered according to HXBc2. Beside the control sample (no treatment), all sequences were obtained 6-7 days after initiation of antibody therapy. Sequences are organized by the mouse from which the viral clone was obtained (y-axis). 6-7 days after initiation of therapy, i.e. before viral rebound, we found no recurrent mutations that mapped to known epitope targets in any of the treatment groups.
Supplementary Figure 7. Monotherapy with broadly neutralizing antibodies selects for viral escapes
a, Individual gpl20 envelope sequences are represented by horizontal gray bars with silent mutations indicated in green and amino acid replacements in red. Black asterisks represent mutations generating a stop codon and bold gray bars deletions. All mutations are relative to HIV-1YU2 (acc. number M93258) and numbered according to HXBc2. Sequences are organized by individual mice (mouse ID, y-axis) from which they were obtained. Highlighted sites refer to recurrent mutations as defined by amino acid substitutions within 3 amino acids in the majority of sequences in at least two mice (Supplementary Table 2). Blue highlighted areas are sites in which selected mutations confer resistance to respective antibodies (Supplementary Fig. 8). Gray highlighted areas in control group show recurrent mutations indicating a positive selection process (Supplementary Table 2a)19. Gray highlighted area in 3BC176 reflects a recurrent mutation that does not confer resistance to 3BC176 (Supplementary Fig. 8).
Supplementary Figure 8. Neutralization activity of bNAbs against YU2-mutants
IC50-values are shown for the indicated antibodies on YU2-mutant virions. Neutralization activity is color-coded and IC50-values < 0.1 μg/ml are shown in red, values between 0.1 and 1.0 μg/ml in orange, > 1 – 10 μg/ml in dark yellow, > 10 to 50 μg/ml in yellow and values > 50 μg/ml in white.
Supplementary Figure 9. Neutralization activity of tri- and penta-mix
a, Geometric mean IC80-values for all viruses grouped by clades. b, Individual IC50 and IC80-values for all tested viral strains. Neutralization activity is color-coded with values < 0.001 μg/ml highlighted in blue, between 0.001 - 0.01 μg/ml in red, > 0.01 - 0.1 μg/ml in orange, > 0.1 - 1 μg/ml in light orange, > 1 - 10 μg/ml in yellow and > 10 μg/ml in white.
Supplementary Figure 10. Comparing viral loads of tri- and penta-mix-treated mice
Changes in log10 (RNA copies/ml) 6-7 days and 27-32 days after treatment initiation with tri- and penta-mix. Columns and error bars represent mean and standard deviation, respectively. Kruskal-Wallis test with Dunn multiple comparison post-hoc test was performed using GraphPad Prism version 5.0b for Mac OS X, Graph-Pad Software, San Diego California USA. Asterisks (**p ≤ 0.01, ***p ≤ 0.001) indicate significant differences between groups.
Supplementary Figure 11. Re-infection of mouse ID21 and ID129
Viral load (copies/ml; blue line) and concentration of YU2 gpl20-reactive antibodies (grey line). Red arrows indicate the initial infection and re-infection with HIV-1YU2 (57.5 ng p24). Grey shaded area reflects antibody treatment (ID2l, tri-mix; ID129, penta-mix). Mouse ID2l has a significant slower increase in viral load compared to initial infection. Mouse ID129 died shortly after re-infection and viral load could not be confirmed.
Supplementary Figure 12. Sequence analysis during penta-mix treatment
Individual gpl20 envelope sequences cloned from single mice (y-axis) during penta-mix therapy in HIV-1YU2-infected hu-mice. Silent mutations are indicated in green and amino acid replacements in red. Black asterisks represent mutations generating a stop codon. All mutations are relative to HIV-1YU2 and numbered according to HXBc2. Sequences were only obtained from three mice and all of these sequences carried at least one stop codon (*) at residues that displayed an APOBEC3G/F motif (Supplementary Table 3b). Vertical blue bars highlight sites of recurrent mutations that were found in mono- (PG16, 45-46G54W, PGT128, 10-1074; Fig. lb, d) and tri-mix-therapy experiments (Fig. 2a, b, c).
Supplementary Figure 13. Natural antibody response to HIV-1YU2 infection in hu-mice
Figure shows antibody concentrations in μg/ml for total IgG for the indicated mice assayed (ELISA) on indicated days post infection (p.i.) as well as the IC50 neutralization titer determined in TZM-bl neutralization assay, a, control group; b, penta-mix group and c, uninfected controls. The values are color-coded as indicated in the legend (bottom right).