Abstract
Talin is a large adaptor protein that activates integrins and couples them to cytoskeletal actin. Talin contains an N-terminal FERM (band 4.1, ezrin, radixin, moesin) domain (the head) linked to a flexible rod comprised of 13 amphipathic helical bundles (R1–R13) that terminate in a C-terminal helix (DD) that forms an anti-parallel dimer. We derived a three-dimensional structural model of full-length talin at a resolution of approximately 2.5 nm using EM reconstruction of full-length talin and the known shapes of the individual domains and inter-domain angles as derived from small angle X-ray scattering. Talin adopts a compact conformation consistent with a dimer in which the two talin rods form a donut-shaped structure, with the two talin heads packed side by side occupying the hole at the center of this donut. In this configuration, the integrin binding site in the head domain and the actin-binding site at the carboxy-terminus of the rod are masked, implying that talin must unravel before it can support integrin activation and engage the actin cytoskeleton.
Keywords: Actin cytoskeleton, Electron microscopy, Focal adhesions, NMR, SAXS, Integrin
1. Introduction
The large adaptor protein talin (2 × 270 kDa) plays a key role in coupling the integrin family of cell adhesion molecules to the actin cytoskeleton (Critchley, 2009; Critchley and Gingras, 2008), and acts synergistically with kindlins to induce a conformational switch in integrins from a low to high affinity state (Moser et al., 2009; Shattil et al., 2010; Ye et al., 2011). There are two major talin isoforms (talin1 and talin2; 74% sequence identity) (Debrand et al., 2009) each made up of an N-terminal head (residues 1–400) linked to a large flexible rod (residues 482 to the C-terminus) by an unstructured linker that contains a calpain-II cleavage site (Fig. 1). The talin head comprises a FERM (band 4.1, ezrin, radixin, moesin) domain with F1, F2 and F3 domains, but it is atypical in that the ubiquitin-like F1 domain has a large unstructured insert (F1 loop), and is preceded by an additional domain F0 that also contains an ubiquitin-like fold (Goult et al., 2010b). Moreover, the crystal structure of the talin1 head shows that it adopts an extended structure (Elliott et al., 2010) rather than the clover-leafed conformation found in the ERM family of proteins (Bretscher et al., 2000). The talin head domain contains the primary integrin binding site, and details on how β-integrin tails are engaged by talin 1 or talin 2 F3-PTB-like FERM domains have now emerged (Anthis and Campbell, 2011; Anthis et al., 2009, 2010; Wegener et al., 2007). The talin rod comprises 62 amphipathic helices that are organized into a series of thirteen (R1–R13) 4- or 5-helix bundles as determined either by NMR or X-ray crystallography (Gingras et al., 2005, 2006, 2008, 2009, 2010; Goult et al., 2010a, 2013; Papagrigoriou et al., 2004). The talin rod is terminating at a C-terminal helix, referred to as the dimerization domain (DD) (Gingras et al., 2008) (Fig. 1). The rod contains a second integrin binding site of as yet uncertain function (Gingras et al., 2009; Moes et al., 2007), at least two actin binding sites (Hemmings et al., 1996) the best characterized of which is located in R13 (Gingras et al., 2008; Smith and McCann, 2007), and numerous binding sites for the cytoskeletal protein vinculin (Gingras et al., 2005), which itself can bind F-actin and many other ligands (Carisey and Ballestrem, 2010; Peng et al., 2011; Ziegler et al., 2006).
Fig. 1.
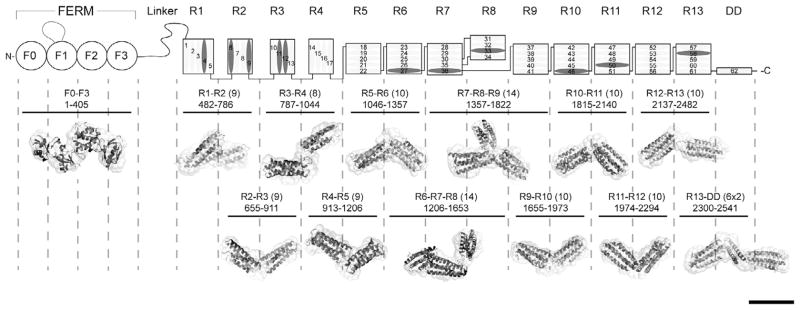
Domain structure of talin1 and SAXS analysis of talin1 polypeptides. The N-terminal FERM domain (residues 1–400), which is atypical in that it comprises 4 domains F0–F3 and a long unstructured loop in F1, is coupled to a flexible rod (residues 482–2541) by an unstructured linker. The 62 α-helices of the talin1 rod are organized into 13 amphipathic helical bundles (R1–R13) terminating with the C-terminal dimerization helix (DD). The rod comprises nine 5-helix bundles, and three of the four 4-helix bundles are arranged in tandem close to the N-terminus of the rod. Helices that support vinculin binding are in dark grey. The residue numbers of each polypeptide used in the study are shown (number of helices in brackets). The Small Angle X-ray Scattering (SAXS) envelope reconstruction of overlapping double and triple talin rod domain polypeptides is shown below the schematic diagram of talin1. The atomic resolution structures of the individual talin domains are fitted into the SAXS envelopes. The SAXS envelope of the talin FERM domain is also shown. Bar represents 5 nm.
The modular organization of talin is broadly consistent with that from negatively stained electron microscopy (EM) images of talin which showed about 12 globular regions (Winkler et al., 1997). Interestingly, sedimentation velocity, gel filtration and EM studies indicate that talin exists in both globular and extended conformations (Molony et al., 1987). However, the isolated talin rod was only found in a more extended form suggesting that an intra-molecular interaction between the head and rod is required for talin to adopt the globular conformation. Indeed, recent studies have identified an interaction between the F3 FERM domain and a region towards the C-terminus of the talin1 rod (Goksoy et al., 2008; Goult et al., 2009), and the NMR (Goult et al., 2009) and crystal (Song et al., 2012) structures of F3 bound to the R9 rod domain shows that the integrin-binding site in F3 is largely occluded. Mutations that compromise the F3/R9 interaction markedly accelerate the rate of focal adhesion assembly (Kopp et al., 2010) indicating that it is a major determinant of talin auto-inhibition. Recent three-dimensional super-resolution fluorescence microscopy studies on the orientation of talin in cell-extracellular matrix junction (focal adhesions) indicates that the talin head is located on the cytoplasmic face of the plasma membrane whereas the C-terminal region of the rod is co-located with actin filaments about 40 nm from the membrane (Kanchanawong et al., 2010). This suggests that engagement in focal adhesions renders full-length talin in an extended conformation with the rod at an angle of about 130° to the head.
The three-dimensional (3D) structure of the full-length talin remains elusive, although we have now determined the structures of all the domains in talin1 (Goult et al., 2013). Here, we employed electron microscopy (EM) and 3D image reconstruction to determine the overall conformation of full-length talin purified from turkey gizzard. We show that talin adopts a compact conformation, the volume of which fits that of a dimer. By combining the electron microscopy derived density map, shape information from the individual domains, and SAXS data on combinations of overlapping helical bundles spanning the entire rod, we provide a molecular model of full-length talin1. Independent NMR data on intra- and inter-molecular interactions were used here to further validate the derived model. In our model, the talin rod forms a donut-shaped structure that stacks against the rod of the adjacent subunit, with the ‘hole’ in the donut occupied by the talin heads. Because the integrin-binding site in the F3 FERM domain is masked by R9, we conclude that this represents an inactive form of the molecule, implying that talin must unravel before it can support integrin activation and engage the actin cytoskeleton.
2. Materials and methods
2.1. Protein purification and electron microscopy
Full-length talin1 was purified from turkey gizzard as described elsewhere (Schmidt et al., 1999). The isolated talin1 rod was prepared by cleaving talin1 with recombinant rat calpain 2 (Calbiochem) at a molar ratio of 1:300 (25 °C for 1 h) in 20 mM Tris buffer pH 7.0 containing 100 mM NaCl, 1 mM DTT and 2.5 mM CaCl2. Cleavage was terminated by adding a 10× molar excess of calpeptin (Calbiochem), and the talin1 rod purified from the talin1 head by gel filtration. Three independent talin1 preparations were used for EM studies. The talin concentrations were between 0.2 and 0.5 mg/ml. Fresh samples were diluted in 2 mM imidazole, pH 7.0, 50 mM KCl, 2 mM MgCl2, 1 mM EGTA, 0.2 mM DTT, 0.1 mM ATP, 0.02% azide, and applied to glow discharged, plasma cleaned (Solaris, Gatan) Quantifoil 1.2/1.3 holey carbon coated 400 mesh electron microscopy grids (Quantifoill, Micro Tools GmbH). Following 30 s incubation in a humidified chamber, a fresh drop of 4 μl protein was added, incubated for another minute, excess of liquid blotted and either stained with 2% aqueous uranyl acetate and air dried (negatively stained) or plunge frozen in liquid N2-cooled ethane (cryo data). Kodak Electron Image Film SO-163 was used to capture images of all samples suspended over holes. Images were acquired under low-dose conditions (~50 electrons/Angst2) using a Tecnai 12 G2 microscope (FEI company, Hillsboro, OR) equipped with a Denka Lab6 filament at 120 kV and a nominal magnification of 67,000×. The defocus was between 1 and 2 μm. The micrographs were digitized using a Photoscan Mark 2, Zeiss Intergraph Aerial Film Scanner (ZI Intergraph) at 7 μm raster and binned to a final pixel size of 0.55 nm.
Actin-talin-Rod arrays were grown for 24 h at 4 °C on positively charged lipid layers consisting of a 3:7 w/w solution of di-laurylphosphatidylinositol and didodecyldimethylammonium bromide dissolved in chloroform (Janssen et al., 2006; Taylor and Taylor, 1992; Ward et al., 1990). The lipid and surfactant mixture was layered over the polymerization buffer prior to the injection of G-actin (~0.35 μg/μl) for producing the arrays. The polymerization buffer contains 20 mM Na2PO4, 50 mM KCl, 1 mM ATP, 2 mM MgCl2, 1 mM EGTA, 1 mM DTT (pH 7.0) and ~0.06 μg/μl of talin rod. The monolayers were transferred to 200-mesh copper grids coated with lacey carbon films (EMS). Specimens were stained with 2% uranyl acetate and air-dried. Low-dose images were recorded with a Tecnai T12 G2 electron microscope (FEI Company, Hillsboro, OR) equipped with a Denka Lab6 filament at 120 kV at a nominal magnification of 52,000 and 1.5 μm defocus (electron dose 20 e−/Å2). Micrographs (Kodak Electron Image Film SO-163) were digitized with a Photoscan Mark 2, Zeiss Intergraph Aerial Film Scanner (ZI Intergraph) with a pixel size of 0.55 nm on the sample.
2.2. Image processing and model building
For the negatively stained data, in total 11800 images of individual particles, suspended over holes, were selected from 113 micrographs and used for the analysis. The particles were processed and analyzed using the EMAN (Ludtke et al., 1999), SPARX (Hohn et al., 2007) and CoAn (Volkmann and Hanein, 1999) software packages. During the processing, 4700 of the original particles were screened out and 7100 contributed to the final model. For the cryo conditions 9800 particles were selected for processing. All docking and modeling operations were performed using the CoAn suite (Volkmann and Hanein, 1999, 2003) and Chimera (Pettersen et al., 2004). The densities are deposited in the EMDB database. Docking of the atomic models of talin domains into the corresponding SAXS envelopes results in fits with a correlation of 0.93 or better for all domains (Fig. 1). Thus, “domain shape” can be regarded to refer interchangeably to low-resolution representations of the X-ray structures as well as to the SAXS data. To facilitate more accurate comparison of V angles between the SAXS models and the derived fits into the EM density, we used the SAXS representation for the modeling. Given the resolution of the EM data and the absence of high-resolution constraints, using atomic models for the fitting directly is highly underdetermined, and is prone to produce overfitting effects and spurious interpretations. The modeling was performed in an iterative manner starting with the DD dimer. The explicit constraints for placing the next domain into the EM density were (1) connectivity of the chain; (2) the low-resolution shape of the domains (SAXS and/or atomic models); (3) the approximate V-angles between domain pairs (SAXS); (4) absence of violations of the 2-fold symmetry; (5) absence of significant overlap with already placed domains or their symmetry mates, and (6) the quality of fit into the EM density of the complete assembly. This process was iterated until all domains were placed in the density. We only found a single configuration that fulfilled all these constraints simultaneously for all domains. All attempts to divert the placement of any of the domains significantly from this configuration had to be aborted after a few steps because one of the constraints became violated.
2.3. Expression of recombinant talin1 polypeptides and cell-based assays
Talin polypeptides were expressed in E. coli BL21 STAR (DE3) cultured in either LB for unlabelled protein, or in M9 minimal media for preparation of isotopically labeled samples for NMR. Recombinant His-tagged talin polypeptides were purified by nickel- affinity chromatography following standard procedures. The His-tag was removed by cleavage with AcTEV protease (Invitrogen), and the proteins were further purified by anion-exchange or cation-exchange depending on the construct. Protein concentrations were determined using the respective extinction coefficients at 280 nm based on absorption coefficients calculated from the aromatic content according to ProtParam (www.expasy.org). For the cell-based assays, human endothelial cells (HUVECs) were transfected with either scrambled (scrRNA) or talin1 siRNA pools and co-transfected with constructs encoding either GFP or mouse GFP-talin1 (WT) or mouse GFP-talinR2526G. After 72 h, cells were plated onto uncoated glass coverslips and analyzed 24 h later as described by Kopp et al., 2010.
2.4. NMR spectroscopy
Proteins were prepared in 20 mM sodium phosphate pH 6.5, 50 mM NaCl, 2 mM DTT, 10% (v/v) 2H2O. NMR spectra of all the proteins were obtained at 298 K using Bruker AVANCE DRX 600 or AVANCE AVII 800 spectrometers both equipped with CryoProbes. Proton chemical shifts were referenced to external DSS, and 15N and 13C chemical shifts were referenced indirectly using recommended gyromagnetic ratios (Wishart et al., 1995). Spectra were processed with TopSpin (Bruker) and analyzed using Analysis (Vranken et al., 2005). All NMR interaction studies were carried out in 20 mM phosphate pH 6.5, 50 mM NaCl, 2 mM DTT. A series of HSQC spectra were measured for the 15N-labeled protein alone (150 μM) and then in the presence of increasing amounts of talin polypeptide (0 μM, 150 μM or 450 μM) and the HSQC spectra analyzed for chemical shift and line-width changes (Table 2). The talin polypeptides indicated in Table 1, were grown in minimal media for preparation of 15N-labeled protein, purified as described above and characterized by 2D-HSQC NMR experiments.
Table 2.
NMR screen for intra-molecular interactions in talin1.
| Unlabeled/15N labeled | F0–F3 | R1–R3 | R4–R8 | R9–R12 | R13–DD |
|---|---|---|---|---|---|
| F0–F1 | nb | nb | nb | nb | nb |
| F2–F3 | b | B | nb | B | nb |
| F3–R1 | nb | nb | B | B | nb |
| R2–R3 | nb | nb | nb | nb | nb |
| R3–R4 | nb | nb | nb | B | nb |
| R5–R6 | nb | nb | nb | nb | nb |
| R7–R8 | nb | nb | b | nb | nb |
| R9–R10 | B | nb | b | nb | nb |
| R11–R12 | nb | nb | b | b | b |
The 15N-labeled talin polypeptides (left hand column) were incubated with the unlabeled talin fragments shown in the top row. The domain composition of the polypeptides is shown. nb = no binding, B = binding, b = broadening of the spectra suggestive of a possible interaction, but not conclusive.
Table 1.
SAXS data on the various talin polypeptides.
| Residues | Domains | Rg | Dmax | Angle (SAXS) | Angle (EM) |
|---|---|---|---|---|---|
| 482–786 | R1–R2 | 23.5 | 71 | 121 | 110 |
| 655–911 | R2–R3 | 27.4 | 87 | 124 | 128 |
| 787–1044 | R3–R4 | 29.7 | 97 | 146 | 145 |
| 913–1206 | R4–R5 | 26.9 | 90 | 132 | 139 |
| 1046–1357 | R5–R6 | 28.6 | 105 | 121 | 113 |
| 1206–1653 | R6–R7–R8 | 31.4 | 104 | 116 | 119 |
| 1357–1653 | R7–R8 | 27.8 | 85 | 125 | 111 |
| 1357–1822 | R7–R8–R9 | 33.2 | 110 | 126 | 109 |
| 1655–1973 | R9–R10 | 27.5 | 90 | 129 | 143 |
| 1815–2140 | R10–R11 | 28.1 | 85.5 | 124 | 137 |
| 1974–2294 | R11–R12 | 24.7 | 80 | 118 | 109 |
| 2137–2482 | R12–R13 | 33.1 | 120 | 109 | 95 |
| 2300–2541 | R13–DD | 37.2 | 124 | n/a | n/a |
Data obtained from SAXS analysis on the talin polypeptides listed. Rg is the radius of gyration, which gives an indication of the distribution of scattering mass of the polypeptide. Dmax is the maximum linear dimension of the polypeptide. The angle columns give the angle between the long principal axes of the respective domains in the SAXS and EM models respectively.
2.5. Solution X-ray scattering data collection and analysis
Small angle X-ray scattering (SAXS) experiments were carried out at three different stations; (i) the Synchrotron Radiation Source at Daresbury, UK, (ii) SOLEIL synchrotron, France, and (iii) the EMBL synchrotron in Hamburg, Germany. Measurements were performed at protein concentrations of 2 and 10 mg/ml in 20 mM sodium phosphate, 50 mM NaCl, 2 mM DTT, pH 6.5. Data were accumulated following the beamline recommendations, and before averaging, frames were inspected for X-ray induced damage or aggregation. No protein aggregation was detected and the linearity of the Guinier plots indicates that the protein solutions were homogeneous. The background was subtracted using the scattering from the buffer solution alone. Data reduction was carried out with software provided at the beamlines. Particle shapes were reconstructed ab initio with the bead modeling program GASBOR (Svergun et al., 2001) which represents the protein as a chain of dummy residues centered at the Cα positions. In addition the program BUNCH (Petoukhov and Svergun, 2005) was applied using the atomic coordinates of the individual domains reported here.
3. Results
3.1. Domain structure of talin1
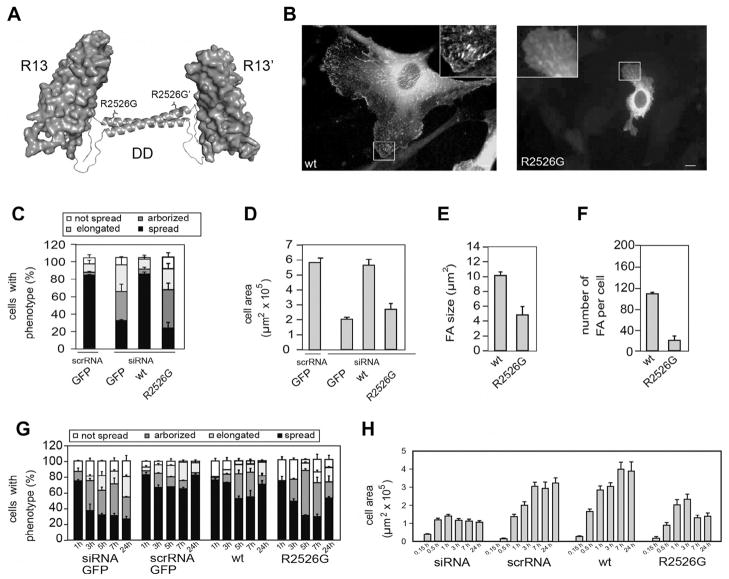
Talin1 is comprised of an atypical FERM domain which has an elongated structure (Elliott et al., 2010) joined to a long flexible rod by an ~80-residue unstructured linker (Fig. 1). We have recently mapped the boundaries of all the domains that make up the talin1 rod (Goult et al., 2013) and determined the structures of each domain or in some cases double domains either by NMR or X-ray crystallography (Gingras et al., 2005, 2006, 2008, 2009, 2010; Goult et al., 2010a, 2013; Papagrigoriou et al., 2004). The 62 amphipathic α-helices are organized into a modular assembly of 13 individual domains (R1–R13) comprised of either 4- or 5-helix bundles (Fig. 1). The rod terminates with a single helix that forms an anti-parallel coiled-coil dimer with the equivalent helix in another talin1 molecule (Gingras et al., 2008). Since all other rod domains are monomeric, we conclude that the last helix is responsible for assembly of the talin dimer. Importantly, mutation of a conserved residue (R2526G) at the interface between the anti-parallel helices abolishes dimerization (Gingras et al., 2008), and markedly reduces the ability of full-length talin1 to rescue cell spreading and focal adhesion assembly in talin1 knockdown endothelial cells (Fig. 2). These results indicate that talin functions as a dimer within the cell.
Fig. 2.
The ability of talin1 to support cell spreading and focal adhesion assembly is compromised by a R2526G mutation in the dimerization domain (DD). (A) The C-terminal actin-binding site of talin1 is comprised of rod domain R13 (a 5-helix bundle) and a C-terminal helix, the dimerization domain (DD). Mutation of R2526G abolishes dimerization of a R13-DD polypeptide and markedly reduces its affinity for filamentous actin (Gingras et al., 2008). (B) GFP-talin1 (left panel; WT) supported extensive cell spreading and was localized in abundant focal adhesions (FAs). In contrast, cells expressing the GFP-talin1 R2526G mutant were much less well spread and had far fewer FAs. (C) Cell morphology – cells were classified into four groups; not spread, elongated (cells 5× longer than wide), arborised (cells with >5 protrusions) or spread. (D) Cell area (E) FA size and (F) number per cell were quantified using ImageJ (NIH). Compared to cells expressing GFP-talin1, those expressing the GFP-talin1 R2526G mutant were more elongated or arborised, had a much reduced cell area, and FAs were reduced in number and size. (G, H) Time-course of cell spreading. Cells expressing GFP-talin1 R2526G managed to spread during the first 1 h after plating, but thereafter, the cells could not maintain a spread phenotype, and the spread cell area remained low.
3.2. Analysis of talin1 rod domain polypeptides in solution by SAXS
We have previously used a combination of SAXS and NMR data to characterize relative orientation of domains in multi-domain fragments from the talin head and rod region (Elliott et al., 2010; Gingras et al., 2008, 2010). The two methods provide complementary information, with SAXS ab initio reconstruction generating an averaged overall shape of the fragments and NMR demonstrating the relative dynamics of the domains that is related to the degree of motion and flexibility of the linker region.
SAXS shapes for 10 double and 2 triple overlapping talin1 rod fragments that cover the whole rod region are presented in Fig. 1. Each SAXS envelope has clearly identifiable globular elongated regions that fit well with the constituting domains. The correlation between density maps calculated from the beads defining the SAXS envelopes and density maps calculated from atomic structures of the docked domains at 2.5 nm resolution is larger than 0.93 in all cases, indicating that the SAXS shapes and the shapes calculated from the atomic models are interchangeable at this resolution. In all fragments, apart from R1–R2, the domains are connected in an end-to-end extended arrangement that shows a lack of inter-domain contacts apart from the regions adjacent to the linker between the domains. In the triple R7–R9 fragment the domains are well separated in a clover-leaf structure. This agrees with the unusual topology of the region, where R7 and R9 are directly connected by a linker, and R8 is inserted into the loop between the helices of R7 that is located close to the linker region (Gingras et al., 2010). The R1–R2 fragment has a more compact shape that fits well with the X-ray structure of the double R1–R2 frament that shows that the domains are stacked side-by-side via a hydrophobic interface (Papagrigoriou et al., 2004).
3.3. The talin rod domains are separated by short hinges
The SAXS envelopes reflect the average shapes of the molecule but provide no information on the variation of the relative domain orientation. Qualitatively, this information can be deduced from the comparison of the 1H,15N-HSQC spectra of the isolated domains and multi-domain fragments. Rigid structures corresponding to the SAXS profiles are expected to have highly anisotropic rotational diffusion and severely broadened NMR resonances. Relative motion of the domains would reduce this broadening. To have a significant effect, the amplitude of the motion has to be sufficiently large for a significant change in the orientation of the individual N–H vectors and fast on the relaxation timescale. In the extreme case where domains are separated by a long flexible linker, the resonances of the double-domain fragment will have similar linewidths to those of the isolated domains.
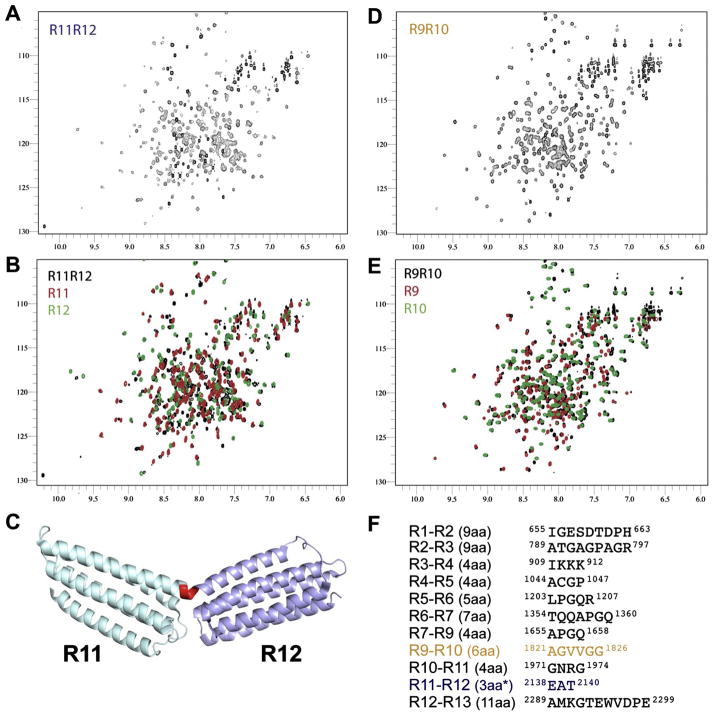
Despite the similarity in the SAXS shapes for the double-domain fragments we detected significant variation in the broadening of the resonances of the double domains compared to the resonances of the corresponding single domains (Supplementary Fig. 1 and Fig. 3). The largest broadening has been observed for the R11-R12 fragment (Fig. 3A, B). The spectrum of the double domain shows a large variation in the signal intensities with high-amplitude sharp signals observed for the unstructured N- and C-termini, while many signals from the folded regions are close to the noise level. In contrast, the signals of the isolated domains are uniform in intensities and have much smaller line-widths. The broadening and large variation in the intensity of the signals in double R11–R12 domain suggests anisotropy of the rotational diffusion and limited relative motion of the domains. This indicates a restricted relative motion of the domains. Correspondingly, the X-ray structure of the R11–R12 fragment has an open V-shape configuration with a well-defined short linker region that forms a helical turn that connects the helices from R11 and R12 into a continuous kinked helix (Fig. 3C). A very similar shape is observed in solution by SAXS, although with a sharper angle between the domains. This suggests that hinge-like motions of the individual domains around the linker region are limited.
Fig. 3.
The talin rod domains are separated by short hinges. NMR spectra (100 μM) of (A) R11–R12 (1974–2294), (B) overlaid with spectra of the R11 and R12 domains (red and green), (C) The structure of R11–R12 (PDB ID: 3DYJ) with R11 (magenta) and R12 (sky blue) attached via a continuous helix through the linker (red). (D) R9–R10 (1655–1973), (E) overlaid with the R9 and R10 domains (red and green) (F) The 11 rod domain linkers, the length of the linkers is shown in brackets. With the exceptions of R1–R2, R2–R3 and R11–R12 and R12–R13 the domains are connected via short 4–7aa linkers comprised of a high ratio of helix disrupting prolines and glycines.
Comparison of the chemical shifts for the R11–R12 and isolated domains support the elongated structure with a small contact area between the ends of the domains. The majority of the cross-peaks of the double domain are close or identical to the peaks detected in the spectra of the single domains, signifying a lack of structural changes in the domains. At the same time a distinct number of cross-peaks in the double domain spectrum are well separated from the signals of the isolated domains. These signals are expected to correspond to the linker region and the areas adjacent to it, with the chemical shift changes induced either by the structural changes in the linker region or by direct contacts between the ends of the domains.
Comparison of the NMR data for the R11–R12 and R1–R2 fragments provides an additional reference for correlation of the spectral and structural changes. In R1–R2 the domains associate in a stable side-on staggered arrangement with a relatively large interface, which makes the overall structure more compact than the end-to-end structure of R11–R12. In agreement with a more compact structure the signals of R1–R2 have smaller line-width and more uniform intensity than for R11–R12, although they are noticeably broader than the signals of isolated R1 and R2 (Supplementary Fig. 1). The more extensive interface leads to larger number of chemical shift changes between the R1 and R2 double and single domains, compared to the R11–R12 region. Thus, for the double domains where X-ray structures are available we observe strong correlation between the NMR spectra, SAXS shapes and X-ray structures. This supports the conclusions derived from the combination of SAXS and NMR and provides the reference for the analysis of double domains where X-ray structures are not available.
The 1H,15N-HSQC spectrum of the R9–R10 fragment is distinctly different from the spectrum of the R11–R12 fragment despite their similarity in SAXS shape (Figs. 1 and 3D, E). Signals of R9–R10 have uniform intensities (Supplementary Fig. 2) and are broadened to a smaller degree compared to the corresponding isolated domains than R11–R12. The differences in the chemical shifts between R9–R10 and the isolated R9 and R10 domains are also much smaller than for R11–R12, and are localized in the immediate vicinity of the linker region (Supplementary Fig. 3). These results demonstrate that in the R9–R10 fragment, the domains have high relative mobility and small contact area. Notably, the intensities of the peaks corresponding to the linker region are very similar to the intensities of the signals of the domains, suggesting that the linker does not have increased dynamics with respect to the rest of the protein. This fits with the SAXS shape of close end-to-end contacts that would restrict the motion of the linker. We thus conclude that the linker region acts as a flexible hinge that allows the domains to change orientation relative to each other while at the same time preventing domain separation.
The differences in the inter-domain motions of the R9–R10 and R11–R12 fragments correlate with the length and amino acid composition of the linker regions (Fig. 3F). Strikingly, the linker length and composition is almost completely conserved between mouse and chicken talin1 indicating the importance of these flexible hinges (Supplementary Fig. 3). In R11–R12 the linker is composed of only three residues and does not include Gly or Pro residues that destabilize helical structures. In contrast, the R9–R10 linker is six residues long and contains three Gly residues, two of which are sequential. The linkers between the other talin1 rod domains are similar to the R9–R10 linker and most of them contain either Gly or Pro residues. As expected, the NMR spectra of the rest of the double domains are close in characteristics to the R9–R10 spectra, with limited resonance broadening and chemical shift changes (apart from the R2–R3 fragment) relative to the spectra of the corresponding isolated domains (Supplementary Fig. 1).
Interestingly, the R2–R3 fragment shows relatively extensive chemical shift changes compared to the isolated domains, despite the limited broadening and an elongated SAXS shape. All of the large chemical shift changes are related to the R3 domain, while the spectra of the R2 domain superimpose very well with the corresponding signals in the R2–R3 spectrum. Similarly, in the spectrum of the triple R2–R4 fragment both R2 and R4 signals align well, while many of the R3 signals are shifted significantly (Supplementary Fig. 4). This suggests that no structural changes occur within the R2 and R4 domains when they are part of larger fragments. The lack of interaction observed for R2 and R4, the relatively high mobility from limited broadening of the NMR signals, and the clear domain separation in the SAXS shapes suggests that the differences in the chemical shifts for the R3 domain are related to some conformational changes within the domain. These changes are expected to be limited, as the differences between the chemical shifts of R3 in the free form and within a larger fragment are significant, but relatively small (Supplementary Fig. 4D). The chemical shift changes are uniformly distributed through R3 and indicate overall rearrangement, most likely small changes in the relative positions of the helices due to the effect of end-to-end contact with other domains or end linker extensions. Our structure of the domain reveals a disruption in the internal hydrophobic core due to the presence of Thr residues (Goult et al., 2013). This could facilitate small rearrangements of the helices in the bundle without a change in the overall structure. Thus, despite the observed chemical shift differences, the R3 domain in the larger fragments behaves similarly to other domains.
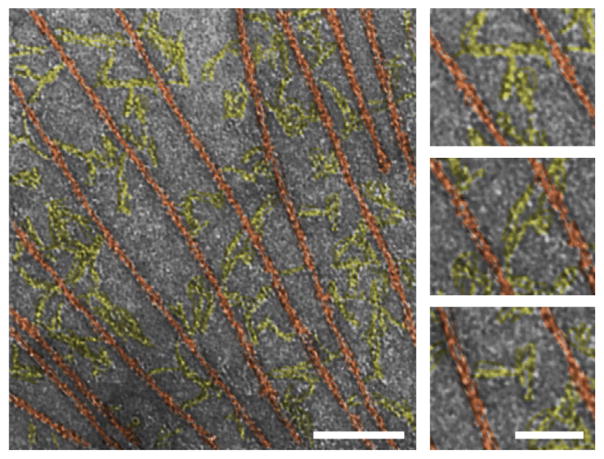
Overall, the combination of the elongated SAXS shapes and the NMR spectra indicates that all the domains in the talin1 rod are connected by hinge-like linkers that allow for a restricted relative domain motion without breaking end-to-end contacts. These topological features support an open chain-like structure for the talin1 rod that can adopt various extended configurations through segment- like motion of the individual domains. In electron micrographs of the isolated talin1 rod (residues 434–2541) in the presence of actin filaments (Fig. 4) the molecules form an intricate, flexible network connecting actin filaments at irregular distances, in line with the interpretation of hinge-like linkers between the rigid talin1 rod domains.
Fig. 4.
Representative electron micrograph of negatively stained talin rod in the presence of actin filaments. The talin1 rods (tinted yellow) are extended, and form a network connecting the actin filaments (tinted red). Bars represent 100 nm (left) and 50 nm (right).
3.4. Full-length talin1 forms a dimer, which adopts a compact conformation
To obtain three-dimensional (3D) information on full-length talin1, we generated 3D reconstructions of talin1 purified from turkey gizzard using electron microscopy (EM) and single particle reconstruction approaches (Fig. 5). Processing of data collected under cryo conditions resulted in a 3D reconstruction at 3.5 nm resolution. The inherent conformational variability within the sample appears to be a barrier in improving the resolution under cryo conditions, most likely owing to the low contrast. Indeed, the more efficient sorting and alignment protocols under the higher contrasted conditions in negative stain allowed us to generate higher resolution reconstructions at 2.5 nm. The high similarity of the reconstructions at the achievable cryo resolution of 3.5 nm under both conditions (correlation of 0.97 ± 0.02, Supplementary Fig. 5) assured us that the negatively stained data gives a valid, artifact-free representation of the talin1 structure. No preferred orientation of the particles with respect to the sample support was observed in negative stain or cryo and the distribution of orientations was even and complete (Fig. 5B–D). Prior to processing, a phase correction for the contrast transfer function was applied to all images. To generate an initial model, we first employed a two-dimensional, reference-free alignment procedure. Particles were sorted into 89 classes according to their mutual similarity without the use of an external reference. The averages of the respective classes represent unbiased characteristic views of the particles within the sample. The analysis revealed a number of compact, globular views and a smaller set of more extended-looking views, indicating a certain mix of conformations. Three different subsets where selected from the globular views and were used to generate initial references using a Fourier-based common-line approach. These three initial models were independently refined against the entire data set using an iterative refinement procedure. Convergence was achieved after 7–10 iterations. Both direct visualization and correlation-based symmetry determination indicated two-fold symmetry for all three initial reconstructions. Thus, refinement and reconstruction were repeated independently for all three starting models while imposing a two-fold symmetry constraint. Cross validation between the three resulting reconstructions verified model independence of the procedure, i.e. there was no significant difference between the three independent negatively stained reconstructions at the target resolution (3 nm) for this analysis according to the 0.5 Fourier shell correlation criterion (FSC0.5). We then proceeded to re-sort the data using the refined model and the initial models to increase homogeneity within the data set. This procedure is very effective in sorting out inhomogeneities in particle sets (Ludtke et al., 1999) and screened out approximately 40% of the initial data. We used the re-sorted data with the refined model from the previous step as a starting model to increase the resolution of the reconstruction. The final map is composed of a compact anti-parallel dimer of 12.5 nm × 11 nm × 9.5 nm dimension at a resolution of ~2.5 nm according to FSC0.5. The volume of this density is consistent with that estimated for a full-length talin dimer when calculated from its molecular weight (Fig. 5A) but is inconsistent with monomeric talin.
Fig. 5.
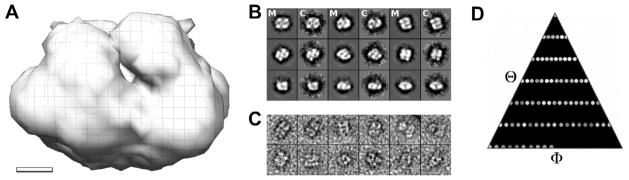
EM reconstruction of full-length talin. (A) Full-length talin1 adopts a compact shape with two-fold symmetry and volume consistent with two talin monomers. (B) Projections of the final, symmetrized reconstruction (M) correspond well to the corresponding non-symmetrized class average (C). (C) Representative raw particles, several of which show indications for 2-fold symmetry. (D) The distribution of particles in Euler angle space shows no preferred orientation. The bar in A represent 2.5 nm, the width of the boxes in B and C is 25 nm.
3.5. The talin1 rod wraps around both talin heads in the dimer
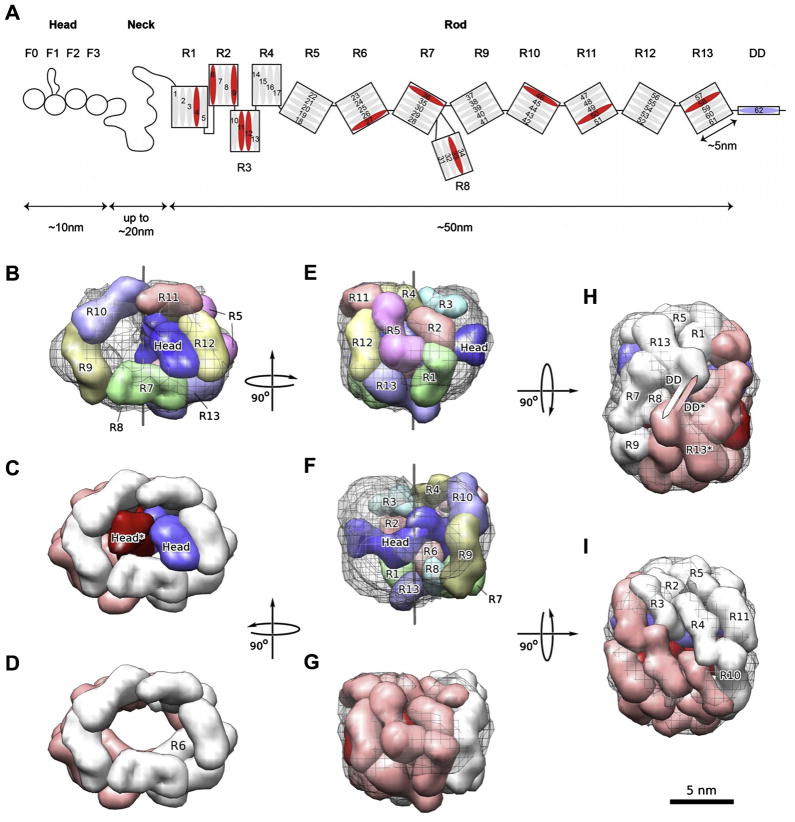
We then proceeded to generate a 3D model of the talin1-dimer domain organization by modeling the individual domain shapes using the V-angles as constraint by SAXS and NMR into the EM reconstruction (Fig. 6A–F). With the given constraints, this modeling task is akin to rolling a garden hose composed of segments that are connected with linkers of limited flexibility into a distinct, compact shape while also obeying a two-fold symmetry constraint. Thus, despite the relatively low resolution of the component models and the entire assembly, the configuration space for the modeling is extremely restricted. In order to obey the two-fold symmetry constraint of an anti-parallel dimer, the dimerization domain must be localized on the symmetry diad. In combination with the rather extended shape of the R13-DD dimer (Gingras et al., 2008) this constraint allowed us to unequivocally place this element into the reconstruction where the DD is located on the diad and the R13 domains follow a ridge in the density at the bottom of the reconstruction (Fig. 6F). With this configuration as a starting point, the shape of the helical bundle combined with the SAXS-determined V-angle between R12 and R13 were employed to place the next structural element, R12 into the density. A small adjustment of the V-angle (14°, see also Table 1) between R13 and R12 needed to be introduced to avoid major mismatches. This adjustment is justified by the hinge-like flexibility of the linker region. Larger angle variation, however, would be inconsistent with the limited linker flexibility observed by NMR. The same strategy was systematically applied to all of the helical bundles of the talin1 rod in descending order (down to R4). Because these bundles – with the exception of R8, which was handled as a unit with R7 (Gingras et al., 2010) – have their N- and C-termini on opposite ends, the placement could be achieved by using the location of the previous element and the inter-bundle V-angle as constraint. The N-terminal part of the rod required a modified approach. This region contains three sequential 4-helix domains R2–R3–R4 that have spatially close N- and C-termini. In addition, domains R1 and R2 make a side-to-side contact, resulting in a staggered domain arrangement. These constraints, in conjunction with those from their shapes and inter-domain angles lead to a reversal in direction for the inter-domain connections at R4 to match the remaining part of the outer layer of the EM shape (Supplementary Fig. 6).
Fig. 6.
Model Building. (A) Domain organization of talin1. (B) Front view of a single modeled talin1 monomer inside the EM density. The view is identical to that shown in Fig. 5a. The individually colored domain envelopes of the rod domains and the head domain are shown as solid surfaces. The EM reconstruction is shown as a black wire mesh. A thin black line marks the symmetry axis. (C) Front view of the entire talin1 dimer. The two monomers are colored white (rod)/blue (head) and pink (rod)/red (head) respectively. (D) Front view of talin1 dimer model with heads omitted. (E) Side view of talin1 monomer model. Color scheme follows (A). (F) View turned 180° around the symmetry axis from (E) for monomer model. Color scheme as in (A). (G) Side view of dimer model in the same orientation as (F). Color scheme follows (C). (H). Bottom view of dimer model along the symmetry axis showing the dimerization helices DD and DD* symmetric around the axis. Color scheme follows (C). (I) Top view of dimer model 180° from (H) along the symmetry axis. Color scheme follows (C). Bar represents 5 nm.
Overall, the inter-domain angles observed by SAXS fit well to the curvature of the EM shape and the whole chain of the domains corresponding to the talin1 rod can be fitted with only small adjustments of the inter-domain angles (<15° for all pairs, see Table 1). The restrictions imposed by the symmetry and the nature of the sequential connections between the domains leads to a unique fold of the talin1 rod into a compact arrangement corresponding to the EM shape (Fig. 6). In the resulting arrangement the two talin1 rods resemble a donut (Fig. 6D) with a ‘donut hole’ in the center that matches the shape and size of the SAXS envelopes of the two talin1 heads perfectly (Fig. 6B, C). In this model, the heads interact side by side and all helical bundles except for R6, which accommodates a groove in the head domain and is buried, contain some regions of accessible surface area. To test for distance constraints and accessibility, we modeled the unstructured F1 loop, which contains two major phosphorylation sites in platelet talin (Ratnikov et al., 2005), onto the EM structure as well (Supplementary Fig. 7).
3.6. Domain interactions mapped independently by NMR confirm the topology of the model
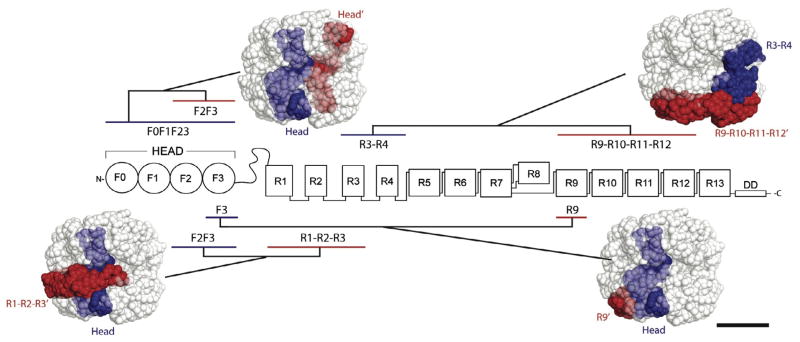
When generating the model of full-length talin1 we used only the knowledge of the inter-molecular interaction between the dimerization domains of two talin subunits (Gingras et al., 2008). The resulting arrangement has a large number of close contacts between domains that can be checked against the interactions between the domains reported in the literature and can be used to predict new interactions. The inactive state of talin is associated with the auto-inhibitory interaction of F3 with R9 (Goult et al., 2009) and F2F3 with R1–R2 (Banno et al., 2012). In the model, domains R1, R2 and R9 are next to each other (Fig. 6B) and are in direct contact with the F2F3 region of the talin head. The symmetry of the structure ensures that these contacts are fulfilled for both subunits. The proximity of the domains engaged in the auto-inhibitory interactions supports the model and suggests that the compact structure is stabilized by the interactions across the dimer.
To further test the model, we applied NMR-based approaches for detecting new interaction between talin1 domains. To reduce the number of combinations in the binding experiments we used nine 15N-labeled double domains – F0F1, F2F3, F3–R1, R1–R2, R3–R4, R5–R6, R7–R8, R9–R10, R11–R12 – and titrated these against 5 large unlabeled talin1 polypeptides – 1–482 i.e. F0–F1– F2–F3–linker), 482–911 (R1–R2–R3), 913–1653 (R4–R5–R6–R7–R8), 1659–2294 (R9–R10–R11–R12), 2300–2541 (R13-DD) – to yield a total of 45 titrations. For each experiment, the 15N-labeled sample (150 μM) was mixed with 0 μM, 150 μM or 450 μM unlabeled polypeptide, and the HSQC spectra analyzed for chemical shift and line-width changes (Table 2). For most of these experiments no significant spectral changes were observed following addition of the unlabeled fragment indicating no interaction. However, several combinations resulted in large spectral perturbations with chemical shift changes and increases in line-width (Supplementary Fig. 8). No precipitation was observed for any of the titrations and the intensity of the sharp peaks (from the C-terminal residues) remained constant throughout the experiments (corrected for the small dilution during the titrations) showing that the changes were not due to aggregation or precipitation. No changes in the spectra apart from small intensity reduction from the dilution were observed when equivalent amount of buffer was added to the 15N-labeled proteins in the control experiment. We therefore concluded that the changes are caused by the direct interactions between the domains. For most of the fragments large molecular weight of the resulting complexes prevented the detection of the signals in the bound state, apart from those corresponding to the unstructured N- and C-termini.
All fragments containing the F3-FERM domain and the R9 rod domain showed large changes in the HSQC spectra upon mixing, in agreement with the previously characterized interaction between these two domains (Supplementary Fig. 8A, B) (Goult et al., 2009) and validating our procedure. Other combinations where spectral changes were observed include significant broadening of the talin1 F2F3 spectra when titrated with the talin1 head, suggesting that the two heads in the dimer interact (Supplementary Fig. 8C). The interaction between F2F3 and the R1–R2–R3 region of the talin1 rod (Supplementary Fig. 8D), agrees with the previously reported interaction between F2F3 and the R1–R2 double domain (Banno et al., 2012). In addition, we detected interaction between R3–R4 and R9–R10–R11–R12. Larger broadening has been observed for R4 resonances, suggesting that this interaction is primarily mediated by this domain (Supplementary Fig. 8E). Finally, the F3–R1 construct interacts with the central portion (R4–R5–R6–R7–R8) of the talin1 rod (Supplementary Fig. 8F). The complex formation leads to the specific broadening of the F3 signals, with the linker region and the R1 domain having similar linewidths to that seen in the free form. Thus the interaction is limited to the F3 domain. Compared with the relatively tight binding of F3–R9 (Goult et al., 2009), disruption of which renders talin constitutively active (Kopp et al., 2010), the newly identified interactions are all relatively weak. However, it is clear from the chemical shift changes and strong line broadening that these interactions are specific and minimal or no changes were observed when the other fragments were used in the titrations.
All of the newly determined interactions are consistent with our EM model of full-length talin (Fig. 7), and provide further support for the arrangement of the talin domains. The head-head interaction is made possible through the close packing of the head region inside the compact structure, while the R3–R4 contacts R9–R10–R11–R12 in the outer layer. Significantly, all of the observed contacts can only be fulfilled in a compact, globular domain arrangement. Thus the interactions distributed along the whole talin molecule in combination with dimerization enable the folding of the free cytosolic talin into a globular inactive structure.
Fig. 7.
Intra- and inter-molecular interactions detected by NMR are consistent with the EM model for the compact form of talin1. 15N-labeled talin1 double domains; F0F1, F2F3, R1–R2, R3–R4, R5–R6, R7–R8, R9–R10, R11–R12 and R13–DD were screened against 5 large unlabeled talin1 polypeptides F0–F3, R1–R3, R4–R8, R9–R12 and R13–DD. The proximity of the interacting fragments is illustrated by highlighting the fragments on the model. Bar represents 5 nm.
4. Discussion
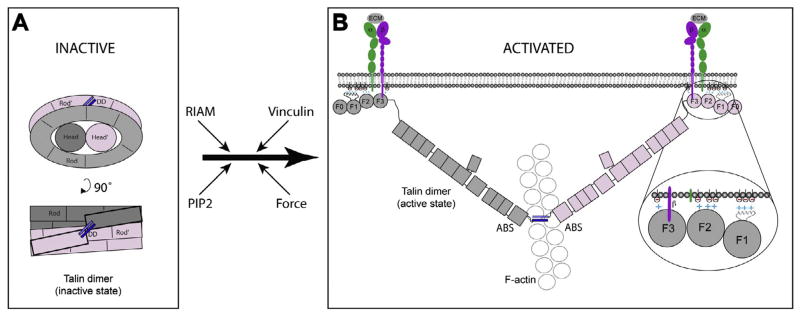
Based on EM and 3D image reconstruction of turkey gizzard talin1, a detailed knowledge of the structures of all the domains in talin1, and inter-bundle V-angle constraints derived from SAXS and NMR analyses of overlapping talin1 polypeptides, we have developed a model for the structure of full-length talin. The model shows that talin adopts a predominantly compact structure (12.5 nm × 11 nm × 9.5 nm), the volume of which fits that of a talin dimer. The major determinant of talin dimerization is the C-terminal helix, and its crystal structure shows that it forms an antiparallel coiled-coil (Gingras et al., 2008). Based on the two-fold symmetry constraints of an anti-parallel dimer, and the SAXS envelope of a talin1 polypeptide containing both R13 and the DD, we were able to unequivocally position these domains in the reconstruction. Importantly, the model reproduces the high affinity interaction between the F3-FERM domain and the R9 rod domain (Goult et al., 2009), without the latter being imposed as a restraint. The model is also consistent with the existence of several weaker intra-molecular interactions that we have detected by NMR, and which appear to make a significant contribution to the overall binding affinity of F3 to the rod. Thus, a Kd of 0.6 μM has been reported for binding of F3 to a talin rod polypeptide spanning R9–R12, while the Kd for binding to R9 alone was 3.6 μM, with weaker binding to R11–R12 (78 μM) (Goksoy et al., 2008). Overall, the data support a model in which the talin rod forms a donut-shaped structure that stacks against the rod of the other subunit, with the ‘hole’ in the donut occupied by both talin heads (Fig. 6). Given that the integrin-binding site in the F3-FERM domain is masked by the R9 rod domain (Goult et al., 2009), and the C-terminal actin-binding site in the R13-DD dimer faces towards the interior, it would seem likely that this represents an inactive form of talin.
Previous studies suggest that talin co-exists in several different conformations. Sedimentation equilibrium studies on gizzard talin point to a monomer–dimer equilibrium, though talin was found to be largely monomeric in physiological salt at concentrations below 0.7 mg of talin per ml (Molony et al., 1987). In contrast, equivalent studies on human platelet talin suggest that it is dimeric even at a concentration of 0.28 mg of talin per ml, and this was confirmed by cross-linking experiments (Goldmann et al., 1994). Whether these differences are due to the source of talin is unclear. Our own gel filtration studies on a recombinant mouse talin1 polypeptide containing R13 and the DD domain clearly establish that it forms a constitutive dimer, and that this is important for talin function (Gingras et al., 2008). Thus, introduction of a R2526G point mutation in the last helix (DD) inhibited dimerization and drastically reduced the activity of the C-terminal actin-binding site in vitro (Gingras et al., 2008). Moreover, we show here that introduction of this same mutation into full-length talin markedly reduced its ability to rescue the talin knockdown phenotype in cells, demonstrating that talin dimerization is also important in vivo.
Sedimentation velocity and gel filtration experiments indicate that gizzard talin is also more compact in low salt compared to 100 mM salt (Stokes radii of 6.5 nm and 7.3 nm respectively) (Molony et al., 1987). This was confirmed by EM of rotary shadowed gizzard talin which was globular in low salt, but appeared as a flexible elongated molecule ~60 nm in length in high salt. In contrast, the purified talin rod fragment was elongated under both conditions, suggesting that the talin head is required for talin to adopt a more compact structure (Molony et al., 1987). Similarly, 2D EM images of negatively stained gizzard talin confirmed that talin is more compact in low salt, whereas in 0.15 M KCl, the authors describe a U-shaped molecule 56 nm in length, although they also observed a small number of Y-shaped molecules (Winkler et al., 1997). In contrast, platelet talin has been described as a dumbbell- shaped molecule ~51 nm long with a globular region at both ends (Goldmann et al., 1994). Images of the more extended form of gizzard talin showed 10–12 globular “beads” ~3.8 nm in diameter, with variable center-to-center spacing (Winkler et al., 1997). Each “bead” had a calculated molecular mass of 24 kDa, which is broadly consistent with the mass of the structural domains found in talin. The isolated talin head contained two such “beads”, again consistent with the structure of the head in which the F0F1 and the F2F3 domains each pack together with a flexible interface between the two (Elliott et al., 2010). Interestingly, the “beads” in the isolated rod were less well defined than in full-length talin (Winkler et al., 1997) suggesting that the rod on its own is highly flexible, and that extended forms of talin are likely to exist in which the head still interacts with the rod restricting its mobility. The EM data and analysis presented here reveals that the predominant conformation of turkey gizzard talin in low salt is indeed a compact dimer. Interestingly, we observe that the isolated talin rod forms a network of extended ‘strings’ cross-linking adjacent actin filaments at low-salt concentrations, consistent with previous data showing that in the absence of the talin head, the rod adopts an extended conformation (Molony et al., 1987). The interaction surface between the talin F3-FERM domain and R9 is large, and is mediated by both hydrophobic and electrostatic interactions, although mutagenesis suggests that the latter provides most of the binding energy (Goult et al., 2009). This may explain why talin adopts an extended conformation at higher salt concentrations.
4.1. Mechanisms of talin regulation
Direct evidence that a compact inactive form of talin exists within the cell is currently lacking. However, much of the talin in cells is diffusely distributed in the cytoplasm, implying that the integrin and actin binding sites in talin are not exposed. This conclusion is supported by recent subcellular fractionation studies which show that the bulk of talin is in the cytosolic fraction, and that disruption of the intramolecular F3/R9 interaction enhances the amount of talin associated with the actin cytoskeleton (Banno et al., 2012). However, this was not sufficient to promote association of talin with the plasma membrane, which required disruption of the interaction between the F2F3 FERM and R1/R2 rod domains. The mechanisms that regulate the activity of talin are therefore the subject of much current interest (Fig. 8). Reconstitution of αIIbβ3 integrin activation in CHO cells has defined a pathway in which the small GTPase Rap1 and its effector RIAM lead to talin-dependent integrin activation, and recruitment of talin to the plasma membrane (Han et al., 2006; Lee et al., 2009; Watanabe et al., 2008). While talin binds directly to RIAM (Goult et al., 2013; Lee et al., 2009), whether the RIAM binding site(s) are exposed in the compact form of full-length talin has yet to be determined. It remains possible that other pathways synergize with RIAM to activate talin. For example, PIP2 has been reported to increase talin binding to integrins (Martel et al., 2001), and the talin head has been shown to bind acidic membrane phospholipids (Elliott et al., 2010; Goult et al., 2010b; Niggli et al., 1994). PIP2 levels increase in response to integrin-mediated adhesion (McNamee et al., 1993), and intriguingly, the type 1γ isoform of PIP kinase localizes to focal adhesions via high affinity binding to the talin F3 domain (Barsukov et al., 2003; de Pereda et al., 2005; Di Paolo et al., 2002; Ling et al., 2002). Moreover, deletion of the short talin binding sequence in PIP kinase type 1γ results in transient defects in β1-integrin-mediated adhesion to fibronectin, a reduction in the ability of integrins to exert force on the matrix, and markedly reduced rates of talin and vinculin recruitment to new adhesions (Legate et al., 2011). We have identified a series of basic residues in F1, F2 and F3 aligned on one surface of the talin1 FERM domain that bind PIP2 and are required for integrin activation (Elliott et al., 2010), including a cluster in the large unstructured F1 loop (Goult et al., 2010b). Modeling the ~35 amino acid long F1 loop suggests that it is large enough to protrude from the interior of the molecule (Supplementary Fig. 7). PIP2 induces the loop to form a helical structure in which basic residues are distributed along one surface of the helix. Perhaps helix formation and the consequent shortening of the loop brings talin closer to the membrane and facilitates PIP2 binding to F2 and F3. Moreover, the interaction between F3 and R9 is inhibited by PIP2 (Goksoy et al., 2008). Thus, PIP2 has the potential to play a key role in talin activation.
Fig. 8.
Schematic diagram of inactive and activated talin. (A) Compact inactive form of the talin dimer. The talin rod forms a donut-shaped structure with the talin heads occupying the hole in the center. The talin subunits are colored pink and grey and the dimerization helix is in blue. (B) When activated, the talin dimer adopts an extended structure in which the F3 FERM domains bind to the cytoplasmic domains of β-integrin subunits (purple), while basic residues on the F1, F2 and F3 FERM domains engage acidic membrane phospholipids (inset). Both interactions are required for integrin activation and binding to the extracellular matrix (ECM) (Anthis et al., 2009). The C-terminal actin-binding site (as a dimer) binds to a single actin filament (Gingras et al., 2008). Possible pathways to talin activation are indicated. Force exerted on talin is thought to facilitate vinculin binding to the talin rod, stabilizing integrin/talin/actin complexes, but this is not shown in the diagram.
Vinculin has also been implicated in talin activation since expression of the vinculin Vd1 domain that binds talin has been shown to promote a focal adhesion enlargement via a mechanism that relies on talin binding, and is independent of actomyosin contraction (Humphries et al., 2007). The puzzle is that the vinculin- binding sites in the talin rod appear to be buried within the hydrophobic core of the amphipathic helical bundles that make up the rod (Gingras et al., 2005, 2006; Papagrigoriou et al., 2004; Patel et al., 2006), and force exerted on an integrin/talin/actin complex is thought to be required for exposure of the these sites (del Rio et al., 2009). However, if as seems likely, talin exists in a number of different conformations, it is possible that vinculin might shift the equilibrium towards a more extended form. An interaction between F2F3-FERM and the R1/R2 rod domains inhibits membrane localization of talin, and it is possible that vinculin binding to R2 relieves this constraint (Banno et al., 2012). Finally, the possibility that phosphorylation plays a role in talin activation is raised by the observation that in platelets, the thrombin-induced redistribution of talin from the cytosol to the membrane is associated with a 4-fold increase in talin phosphorylation (Bertagnolli et al., 1993). Interestingly, the two major phosphorylation sites (T144/T150) in platelet talin are in the large unstructured F1 loop (Ratnikov et al., 2005).
In summary, we derived a structural model of the auto-inhibited form of talin. The model consists of two interacting talin monomers in which each talin rod forms half of a “double donut”, with the talin heads side by side occupying the hole at the center of the donut. The model indicates that the structural basis for auto-inhibition is rooted in the fact that the integrin binding site in the head and the actin-binding site at the C-terminus of the rod are masked. As a consequence, talin must unravel before it can support integrin activation and engage the actin cytoskeleton. While many of the details of activation remain to be determined, the model reported here lays the foundation for a better understanding of the mechanisms involved in talin activation.
Supplementary Material
Acknowledgments
DH thanks Larnele Hazelwood for assisting with the EM sample preparations. This work was initially supported by the NIH Cell Migration Consortium (CMC) Grant U54 GM64346 from the National Institute of General Medical Sciences (NIGMS) to DRC, DH and NV with additional support for the modeling from NIH grant R01 GM76503 to NV. Following the sunset of the CMC, the work was supported by NIGMS grant U01 GM094663 to NV and DH and Wellcome Trust and Cancer Research UK grants to DRC.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jsb.2013.05.014.
References
- Anthis NJ, Campbell ID. The tail of integrin activation. Trends Biochem Sci. 2011;36:191–198. doi: 10.1016/j.tibs.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthis NJ, Wegener KL, Critchley DR, Campbell ID. Structural diversity in integrin/talin interactions. Structure. 2010;18:1654–1666. doi: 10.1016/j.str.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthis NJ, Wegener KL, Ye F, Kim C, Goult BT, et al. The structure of an integrin/talin complex reveals the basis of inside-out signal transduction. EMBO J. 2009;28:3623–3632. doi: 10.1038/emboj.2009.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banno A, Goult BT, Lee H, Bate N, Critchley DR, et al. Subcellular localization of talin is regulated by inter-domain interactions. J Biol Chem. 2012;287:13799–13812. doi: 10.1074/jbc.M112.341214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsukov IL, Prescot A, Bate N, Patel B, Floyd DN, et al. Phosphatidylinositol phosphate kinase type 1gamma and beta1-integrin cytoplasmic domain bind to the same region in the talin FERM domain. J Biol Chem. 2003;278:31202–31209. doi: 10.1074/jbc.M303850200. [DOI] [PubMed] [Google Scholar]
- Bertagnolli ME, Locke SJ, Hensler ME, Bray PF, Beckerle MC. Talin distribution and phosphorylation in thrombin-activated platelets. J Cell Sci. 1993;106:1189–1199. doi: 10.1242/jcs.106.4.1189. [DOI] [PubMed] [Google Scholar]
- Bretscher A, Chambers D, Nguyen R, Reczek D. ERM-Merlin and EBP50 protein families in plasma membrane organisation and function. Annu Rev Cell Dev Biol. 2000;16:113–143. doi: 10.1146/annurev.cellbio.16.1.113. [DOI] [PubMed] [Google Scholar]
- Carisey A, Ballestrem C. Vinculin, an adapter protein in control of cell adhesion signalling. Eur J Cell Biol. 2010;90:157–163. doi: 10.1016/j.ejcb.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchley DR. Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Annu Rev Biophys. 2009;38:235–254. doi: 10.1146/annurev.biophys.050708.133744. [DOI] [PubMed] [Google Scholar]
- Critchley DR, Gingras AR. Talin at a glance. J Cell Sci. 2008;121:1345–1347. doi: 10.1242/jcs.018085. [DOI] [PubMed] [Google Scholar]
- de Pereda JM, Wegener KL, Santelli E, Bate N, Ginsberg MH, et al. Structural basis for phosphatidylinositol phosphate kinase type Igamma binding to talin at focal adhesions. J Biol Chem. 2005;280:8381–8386. doi: 10.1074/jbc.M413180200. [DOI] [PubMed] [Google Scholar]
- Debrand E, El Jai Y, Spence L, Bate N, Praekelt U, et al. Talin 2 is a large and complex gene encoding multiple transcripts and protein isoforms. FEBS J. 2009;276:1610–1628. doi: 10.1111/j.1742-4658.2009.06893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, et al. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G, Pellegrini L, Letinic K, Cestra G, Zoncu R, et al. Recruitment and regulation of phosphatidylinositol phosphate kinase type 1 gamma by the FERM domain of talin. Nature. 2002;420:85–89. doi: 10.1038/nature01147. [DOI] [PubMed] [Google Scholar]
- Elliott PR, Goult BT, Kopp PM, Bate N, Grossmann JG. The Structure of the talin head reveals a novel extended conformation of the FERM domain. Structure. 2010;18:1289–1299. doi: 10.1016/j.str.2010.07.011. (London, England: 1993) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AR, Ziegler WH, Frank R, Barsukov IL, Roberts GC, et al. Mapping and consensus sequence identification for multiple vinculin binding sites within the talin rod. J Biol Chem. 2005;280:37217–37224. doi: 10.1074/jbc.M508060200. [DOI] [PubMed] [Google Scholar]
- Gingras AR, Vogel KP, Steinhoff HJ, Ziegler WH, Patel B, et al. Structural and dynamic characterization of a vinculin binding site in the talin rod. Biochemistry. 2006;45:1805–1817. doi: 10.1021/bi052136l. [DOI] [PubMed] [Google Scholar]
- Gingras AR, Bate N, Goult BT, Patel B, Kopp PM, et al. Central region of talin has a unique fold that binds vinculin and actin. J Biol Chem. 2010;285:29577–29587. doi: 10.1074/jbc.M109.095455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AR, Bate N, Goult BT, Hazelwood L, Canestrelli I, et al. The structure of the C-terminal actin-binding domain of talin. EMBO J. 2008;27:458–469. doi: 10.1038/sj.emboj.7601965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AR, Ziegler WH, Bobkov AA, Joyce MG, Fasci D, et al. Structural determinants of integrin binding to the talin rod. J Biol Chem. 2009;284:8866–8876. doi: 10.1074/jbc.M805937200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goksoy E, Ma YQ, Wang X, Kong X, Perera D, et al. Structural basis for the autoinhibition of talin in regulating integrin activation. Mol Cell. 2008;31:124–133. doi: 10.1016/j.molcel.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann WH, Bremer A, Haner M, Aebi U, Isenberg G. Native talin is a dumbbell-shaped homodimer when it interacts with actin. J Struct Biol. 1994;112 (1):3–10. doi: 10.1006/jsbi.1994.1002. [DOI] [PubMed] [Google Scholar]
- Goult BT, Gingras AR, Bate N, Barsukov IL, Critchley DR, et al. The domain structure of talin: residues 1815–1973 form a five-helix bundle containing a cryptic vinculin-binding site. FEBS Lett. 2010a;584:2237–2241. doi: 10.1016/j.febslet.2010.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goult BT, Bate N, Anthis NJ, Wegener KL, Gingras AR, et al. The structure of an interdomain complex that regulates talin activity. J Biol Chem. 2009;284:15097–15106. doi: 10.1074/jbc.M900078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goult BT, Bouaouina M, Elliott PR, Bate N, Patel B, et al. Structure of a double ubiquitin-like domain in the talin head: a role in integrin activation. EMBO J. 2010b;29:1069–1080. doi: 10.1038/emboj.2010.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goult BT, Zacharchenko T, Bate N, Tsang R, Hey F, et al. RIAM and vinculin binding to talin are mutually exclusive and regulate adhesion assembly and turnover. J Biol Chem. 2013;288:8238–8249. doi: 10.1074/jbc.M112.438119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lim CJ, Watanabe N, Soriani A, Ratnikov B, et al. Reconstructing and deconstructing agonist-induced activation of integrin alphaIIbbeta3. Curr Biol. 2006;16:1796–1806. doi: 10.1016/j.cub.2006.08.035. [DOI] [PubMed] [Google Scholar]
- Hemmings L, Rees DJG, Ohanian V, Bolton SJ, Gilmore AP, et al. Talin contains three actin-binding sites each of which is adjacent to a vinculin-binding site. J Cell Sci. 1996;109:2715–2726. doi: 10.1242/jcs.109.11.2715. [DOI] [PubMed] [Google Scholar]
- Hohn M, Tang G, Goodyear G, Baldwin PR, Huang Z, et al. SPARX, a new environment for Cryo-EM image processing. J Struct Biol. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Humphries JD, Wang P, Streuli C, Geiger B, Humphries MJ, et al. Vinculin controls focal adhesion formation by direct interactions with talin and actin. J Cell Biol. 2007;179:1043–1057. doi: 10.1083/jcb.200703036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen ME, Kim E, Liu H, Fujimoto LM, Bobkov A, et al. Three-dimensional structure of vinculin bound to actin filaments. Mol Cell. 2006;21:271–281. doi: 10.1016/j.molcel.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, et al. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468:580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp PM, Bate N, Hansen TM, Brindle NP, Praekelt U, et al. Studies on the morphology and spreading of human endothelial cells define key inter- and intramolecular interactions for talin1. Eur J Cell Biol. 2010;89:661–673. doi: 10.1016/j.ejcb.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Lim CJ, Puzon-McLaughlin W, Shattil SJ, Ginsberg MH. RIAM activates integrins by linking talin to ras GTPase membrane-targeting sequences. J Biol Chem. 2009;284:5119–5127. doi: 10.1074/jbc.M807117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legate KR, Takahashi S, Bonakdar N, Fabry B, Boettiger D, et al. Integrin adhesion and force coupling are independently regulated by localized PtdIns(4,5)2 synthesis. EMBO J. 2011;30:4539–4553. doi: 10.1038/emboj.2011.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling K, Doughman RL, Firestone AJ, Bunce MW. Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature. 2002;420:89–93. doi: 10.1038/nature01082. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Martel V, Racaud-Sultan C, Dupe S, Marie C, Paulhe F, et al. Conformation, localization, and integrin binding of talin depend on its interaction with phosphoinositides. J Biol Chem. 2001;276:21217–21227. doi: 10.1074/jbc.M102373200. [DOI] [PubMed] [Google Scholar]
- McNamee HP, Ingber DE, Schwartz MA. Adhesion to fibronectin stimulates inositol lipid synthesis and enhances PDGF-induced inositol lipid breakdown. J Cell Biol. 1993;121:673–678. doi: 10.1083/jcb.121.3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moes M, Rodius S, Coleman SJ, Monkley SJ, Goormaghtigh E, et al. The integrin binding site 2 (IBS2) in the talin rod domain is essential for linking integrin beta subunits to the cytoskeleton. J Biol Chem. 2007;282:17280–17288. doi: 10.1074/jbc.M611846200. [DOI] [PubMed] [Google Scholar]
- Molony L, Mccaslin D, Abernethy J, Paschal B, Burridge K. Properties of talin from chicken gizzard smooth-muscle. J Biol Chem. 1987;262:7790–7795. [PubMed] [Google Scholar]
- Moser M, Legate KR, Zent R, Fassler R. The tail of integrins, talin, and kindlins. Science. 2009;324:895–899. doi: 10.1126/science.1163865. [DOI] [PubMed] [Google Scholar]
- Niggli V, Kaufmann S, Goldmann WH, Weber T, Isenberg G. Identification of functional domains in the cytoskeletal protein talin. Eur J Biochem. 1994;224 (3):951–957. doi: 10.1111/j.1432-1033.1994.00951.x. [DOI] [PubMed] [Google Scholar]
- Papagrigoriou E, Gingras AR, Barsukov IL, Bate N, Fillingham IJ, et al. Activation of a vinculin-binding site in the talin rod involves rearrangement of a five-helix bundle. EMBO J. 2004;23:2942–2951. doi: 10.1038/sj.emboj.7600285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel B, Gingras AR, Bobkov AA, Fujimoto LM, Zhang M, et al. The activity of the vinculin binding sites in talin is influenced by the stability of the helical bundles that make up the talin rod. J Biol Chem. 2006;281:7458–7467. doi: 10.1074/jbc.M508058200. [DOI] [PubMed] [Google Scholar]
- Peng X, Nelson ES, Maiers JL, DeMali KA. New insights into vinculin function and regulation. Int Rev Cell Mol Biol. 2011;287:191–231. doi: 10.1016/B978-0-12-386043-9.00005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, et al. UCSF Chimera – a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Ratnikov B, Ptak C, Han J, Shabanowitz J, Hunt DF, et al. Talin phosphorylation sites mapped by mass spectrometry. J Cell Sci. 2005;118:4921–4923. doi: 10.1242/jcs.02682. [DOI] [PubMed] [Google Scholar]
- Schmidt JM, Zhang J, Lee HS, Stromer MH, Robson RM. Interaction of talin with actin: sensitive modulation of filament crosslinking activity. Arch Biochem Biophys. 1999;366:139–150. doi: 10.1006/abbi.1999.1204. [DOI] [PubMed] [Google Scholar]
- Shattil SJ, Kim C, Ginsberg MH. The final steps of integrin activation: the end game. Nat Rev Mol Cell Biol. 2010;11:288–300. doi: 10.1038/nrm2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SJ, McCann RO. A C-Terminal dimerization motif is required for focal adhesion targeting of talin1 and the interaction of the talin1 I/LWEQ module with F-actin. Biochemistry. 2007;46:10886–10898. doi: 10.1021/bi700637a. [DOI] [PubMed] [Google Scholar]
- Song X, Yang J, Hirbawi J, Ye S, Perera HD, et al. A novel membrane-dependent on/off switch mechanism of talin FERM domain at sites of cell adhesion. Cell Res. 2012;22:1533–1545. doi: 10.1038/cr.2012.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from X-ray solution scattering. Biophys J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KA, Taylor DW. Formation of 2-D paracrystals of F-actin on phospholipid layers mixed with quaternary ammonium surfactants. J Struct Biol. 1992;108:140–147. doi: 10.1016/1047-8477(92)90013-z. [DOI] [PubMed] [Google Scholar]
- Volkmann N, Hanein D. Quantitative fitting of atomic models into observed densities derived by electron microscopy. J Struct Biol. 1999;125:176–184. doi: 10.1006/jsbi.1998.4074. [DOI] [PubMed] [Google Scholar]
- Volkmann N, Hanein D. Docking of atomic models into reconstructions from electron microscopy. Methods Enzymol. 2003;374:204–225. doi: 10.1016/S0076-6879(03)74010-5. [DOI] [PubMed] [Google Scholar]
- Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Ward RJ, Menetret JF, Pattus F, Leonard K. Method for forming two-dimensional paracrystals of biological filaments on lipid monolayers. J Electron Microsc Tech. 1990;14:335–341. doi: 10.1002/jemt.1060140408. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Bodin L, Pandey M, Krause M, Coughlin S, et al. Mechanisms and consequences of agonist-induced talin recruitment to platelet integrin alphaIIbbeta3. J Cell Biol. 2008;181:1211–1222. doi: 10.1083/jcb.200803094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener KL, Partridge AW, Han J, Pickford AR, Liddington RC, et al. Structural basis of integrin activation by talin. Cell. 2007;128:171–182. doi: 10.1016/j.cell.2006.10.048. [DOI] [PubMed] [Google Scholar]
- Winkler J, Lunsdorf H, Jockusch BM. Energy-filtered electron microscopy reveals that talin is a highly flexible protein composed of a series of globular domains. Eur J Biochem. 1997;243:430–436. doi: 10.1111/j.1432-1033.1997.0430a.x. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, et al. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- Ye F, Kim C, Ginsberg MH. Molecular mechanism of inside-out integrin regulation. J Thromb Haemost: JTH. 2011;9 (Suppl 1):20–25. doi: 10.1111/j.1538-7836.2011.04355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler WH, Liddington RC, Critchley DR. The structure and regulation of vinculin. Trends Cell Biol. 2006;16:453–460. doi: 10.1016/j.tcb.2006.07.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.