Abstract
RIG-I protects host cells against various RNA viruses by sensing viral RNAs in the cytoplasm. Crystal structures of RIG-I C-terminal domain bound to 5′-triphosphate dsRNA unveils how RIG-I recognizes the 5′-triphosphate moiety, a hallmark of viral RNAs (Lu et al., 2010).
The first line of defense against infections is mediated by innate pattern recognition receptors (PRRs), which include Toll-like receptors, RIG-I-like receptors (RLR), NOD-like receptors, and C-type lectin receptors (Takeuchi and Akira, 2010). Activation of these receptors leads to production of type I interferons and inflammatory cytokines to trigger the host antiviral program. RIG-I (retinoic acid inducible gene I) is the prototype of the RLR family that also include MDA5 and LGP2. In the cytoplasm, RIG-I and MDA5 detect a different set of RNA viruses, whereas LGP2 plays a regulatory role in the signaling pathway of RIG-I and MDA5 (Takeuchi and Akira, 2010). All RLR members share a central DExD/H-box helicase domain and a C-terminal domain (CTD) that detects viral RNAs. In addition, RIG-I and MDA5 have two CARD domains at the N-terminal region that are responsible for recruiting the downstream adaptor protein MAVS (also known as IPS-1, VISA, or CARDIF) (Takeuchi and Akira, 2010).
The identities of RNA ligands that activate RIG-I have been intensively debated (Schlee et al., 2009a). Recent studies suggested that only dsRNAs with 5′-triphosphate (5′-ppp) are capable of activating RIG-I and that previous confusions on the activity of 5′-ppp ssRNA may come from the use of in vitro transcribed RNAs with double-stranded byproducts (Schlee et al., 2009b; Schmidt et al., 2009). It should be noted that RIG-I sometimes plays a functional role in viruses (e.g., Reoviridae) that do not appear to generate 5′-ppp dsRNA (Yoneyama and Fujita, 2009). Structures of RIG-I CTD alone displayed a positively charged surface cleft, which was proposed to be the potential RNA binding site (Cui et al., 2008; Takahasi et al., 2008). However, because of the lack of a structure of RIG-I CTD bound to RNA, it is not clear how RIG-I is able to distinguish various RNA ligands and how the 5′-ppp moiety may facilitate the interaction.
In this issue of Structure, Pingwei Li’s group at Texas A&M University (Lu et al., 2010) reports structures of RIG-I CTD bound to a 14 bp GC-rich 5′-ppp dsRNA and a 12 bp AU-rich 5′-ppp dsRNA, respectively. These 5′-ppp dsRNAs contain palindromic sequences and were produced from in vitro transcription. In a parallel line of work, Dinshaw Patel’s group at the Memorial Sloan-Kettering Cancer Center (Wang et al., 2010) reported last month in Nature Structural Molecular Biology the structure of RIG-I CTD in complex with a chemically synthesized, 12 bp mixed content 5′-ppp dsRNA. Despite the different RNA contents and the apparent hydrolysis to 5′-pp in the Patel structure, the modes of RNA recognition in the three structures are remarkably similar, suggesting a conserved sequence independent recognition of 5′-ppp dsRNA.
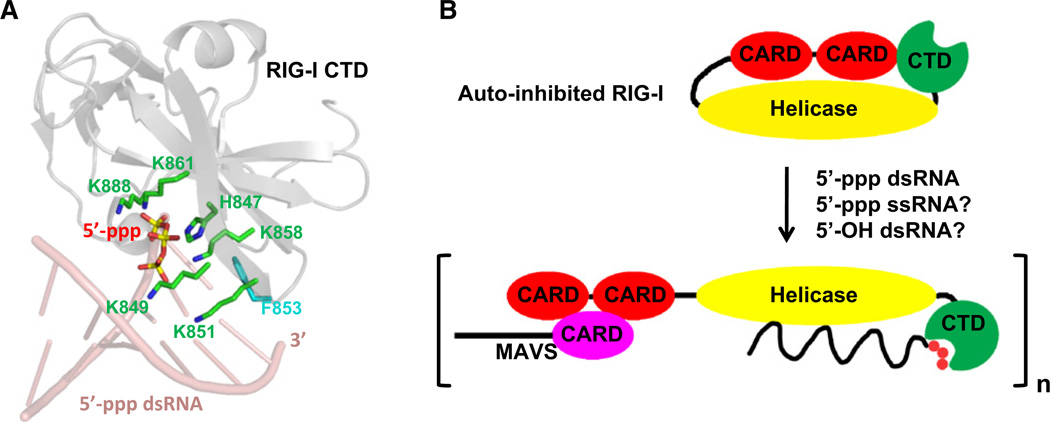
In all structures, each dsRNA adopts a standard A-form double helical structure and recruits two CTD molecules symmetrically. The contacts are made primarily through a few nucleotides at the 5′-end with the 5′-ppp moiety interacting extensively with RIG-I CTD at the positively charged cleft previously predicted. Most conspicuously, multiple Lys residues from noncontiguous segments of the CTD, including K888, K861, K858, K849, and K851, surround the 5′-ppp and hold it in place like “iron claws” (Figure 1A). Residue F853 of CTD stacks over the exposed terminal base pair. In addition to providing hydrophobic interaction energy, this interaction possibly acts as a torque to fix the orientation of the bound dsRNA to maximize the 5′-ppp interaction. The orientation differs by 30° to that of 5′-OH dsRNA in the complex with LGP2 (Li et al., 2009). Extensive structure-based mutagenesis experiments by both groups validated the importance of key contacting residues in RNA recognition and RIG-I signaling. In addition, using 5′-ppp RNA analogs containing 2′-OCH3 at positions 1–6 from the 5′-ppp end, Patel’s group (Wang et al., 2010) was able to demonstrate the relevance of the binding orientation of the dsRNA in the complex.
Figure 1. RNA Recognition and Activation of RIG-I.
(A) Recognition of 5′-ppp dsRNA by RIG-I CTD. RIG-I CTD (gray) and the first six pairs of the dsRNA (pink) are shown as ribbons. 5′-ppp and critical residues for 5′-ppp interaction are shown as stick models with atoms P in yellow, O in red, C in green, and N in blue. Residue F853 of RIG-I CTD, which stacks over the terminal base pair, is shown in cyan.
(B) A model of RNA-induced RIG-I activation. RIG-I may exist in an auto-inhibited conformation in the absence of viral RNAs. Upon RNA-binding, RIG-I may open up to allow oligomerization and recruitment of downstream signaling proteins such as MAVS.
Using surface plasmon resonance (SPR) and isothermal titration calorimetry (ITC), respectively, Li’s group and Patel’s group (Lu et al., 2010; Wang et al., 2010) determined that RIG-I CTD binds blunt-end 5′-ppp dsRNA with highest affinity, followed by 5′-OH dsRNA and 5′-ppp ssRNA. The structures presented by both groups explain the molecular basis for this selectivity of RIG-I CTD. While 5′-ppp ssRNA may preserve the interaction with the positively charged patch, it does not have the dsRNA conformation to optimize the interaction of other 5′-nucleotides with RIG-I or suffers significant entropic loss when in a dsRNA conformation. On the other hand, the 5′-OH dsRNA does not have the energetic contribution from the 5′-ppp moiety and may compensate by adopting an orientation similar to that in the LGP2:dsRNA complex. The high degree of salt-dependence of the interaction can be clearly anticipated by the electrostatic nature of the interaction.
One might predict that the highly electrostatic interaction between RIG-I CTD and 5′-ppp dsRNA would result in fast association in the interaction due to the long-range electrostatic attraction. However, SPR measurement by Li’s group showed that the interaction possesses slow association and slow dissociation. A slow association often indicates the requirement of conformational changes in the interaction. Indeed, comparison of the free and the bound forms of RIG-I CTD reveal local conformational adjustments, especially in a loop region that harbors the important Lys residues for 5′-ppp recognition. Perhaps the peculiarity that only Lys residues are involved in this process is partly due to its conformational flexibility, like individual claws that are perfect for “grabbing” the 5′-ppp moiety. Unlike the “kiss and run” nature of many enzymatic reactions, RIG-I signaling should require stabilization of a certain bound, and perhaps oligomerized, conformation for recruitment of downstream molecules. The unique slow dissociation kinetics of the interaction, with its half life in the range of minutes, could contribute to effective signaling.
Is 5′-ppp absolutely required for RIG-I activation and generation of interferon response? Li’s group (Lu et al., 2010) showed that transfection of 5′-ppp dsRNA, 5′-ppp ssRNA, or 5′-OH dsRNA all led to IFN-β reporter activity, while Patel’s group (Wang et al., 2010) showed that only 5′-ppp dsNRA is able to activate RIG-I signaling. Since 5′ OH-dsRNA and 5′-ppp ssRNA bind to RIG-I CTD at lower affinities in comparison with 5′-ppp dsRNA, a higher effective concentration may be required to activate RIG-I. Li’s group (Lu et al., 2010) used an RNA concentration of 50 nM in their transfection assay, whereas Patel’s group (Wanget al., 2010) used 5 nM, which may explain the different observations. The observation that RNA ligands other than 5′-ppp dsRNA are able to activate RIG-I signaling is in agreement with some previous studies but different from others (Cui et al., 2008; Schlee et al., 2009b; Schmidt et al., 2009; Takahasi et al., 2008). It appears then that one must understand both the chemical natures of the endogenous viral ligands and their cellular concentrations to predict whether they could potently activate RIG-I.
How does the current structure help to understand the mechanistic basis of RIG-I activation? The RIG-I CTD was also known as the regulatory domain (RD), which has been shown to possess the ability to interact with other regions of RIG-I, leading to RIG-I auto-inhibition in the absence of RNA ligands (Figure 1B). It is believed that once RIG-I CTD senses viral RNAs, conformational changes are induced, leading to an open-up of the structure. Subsequently, RIG-I oligomerizes and recruits the adaptor protein MAVS to activate the downstream signaling events (Schlee et al., 2009a; Yoneyama and Fujita, 2009) (Figure 1B). While the RIG-I CARD domains are important for recruitment of MAVS through CARD:CARD interactions, the role of the helicase domain is far less clear. It has been noted that RIG-I CTD is structurally related to a GDP exchange factor of Rab GTPases, which raises the possibility that the CTD activates the helicase domain by structural modulation of the ATP-binding site (Cui et al., 2008). It has also been proposed that the ATPase activity of RIG-I, rather than its RNA unwinding ability, is required for the conformational changes that promote downstream signaling (Yoneyama and Fujita, 2009). Structural and biochemical studies of full length RIG-I, both alone and in complex with RNAs, will give more insights into the process of RIG-I activation.
REFERENCES
- Cui S, Eisenacher K, Kirchhofer A, Brzozka K, Lammens A, Lammens K, Fujita T, Conzelmann KK, Krug A, Hopfner KP. Mol. Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Li X, Ranjith-Kumar CT, Brooks MT, Dharmaiah S, Herr AB, Kao C, Li P. J. Biol. Chem. 2009;284:13881–13891. doi: 10.1074/jbc.M900818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Hu H, Ranjith-Kumar CT, Brooks MT, Hou TY, Hu F, Herr AB, Strong RK, Kao CC, Li P. Structure. 2010;18(this issue):1032–1043. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M, Hartmann E, Coch C, Wimmenauer V, Janke M, Barchet W, Hartmann G. Immunol. Rev. 2009a;227:66–74. doi: 10.1111/j.1600-065X.2008.00724.x. [DOI] [PubMed] [Google Scholar]
- Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, et al. Immunity. 2009b;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Schwerd T, Hamm W, Hellmuth JC, Cui S, Wenzel M, Hoffmann FS, Michallet MC, Besch R, Hopfner KP, et al. Proc. Natl. Acad. Sci. USA. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahasi K, Yoneyama M, Nishihori T, Hirai R, Kumeta H, Narita R, Gale M, Jr, Inagaki F, Fujita T. Mol. Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ludwig J, Schuberth C, Goldeck M, Schlee M, Li H, Juranek S, Sheng G, Micura R, Tuschl T, et al. Nat. Struct. Mol. Biol. 2010;17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Fujita T. Immunol. Rev. 2009;227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]