Abstract
Dengue virus cycles between mosquitoes and humans. Each host provides a different environment for viral replication, imposing different selective pressures. We identified a sequence in the dengue virus genome that is essential for viral replication in mosquito cells but not in mammalian cells. This sequence is located at the viral 3′ untranslated region and folds into a small hairpin structure. A systematic mutational analysis using dengue virus infectious clones and reporter viruses allowed the determination of two putative functions in this cis-acting RNA motif, one linked to the structure and the other linked to the nucleotide sequence. We found that single substitutions that did not alter the hairpin structure did not affect dengue virus replication in mammalian cells but abolished replication in mosquito cells. This is the first sequence identified in a flavivirus genome that is exclusively required for viral replication in insect cells.
TEXT
Dengue virus (DENV) is the most important mosquito-borne viral pathogen in humans. It represents an enormous public health problem around the world, with about 340 million infections per year. In nature, the virus cycles between humans and mosquitoes. The virus must use both cellular repertoires efficiently in order to succeed in infecting the two hosts. Fundamental differences in biological processes and biochemical machineries between human and mosquito cells, such as glycosylation, membrane composition and lipid metabolism, innate antiviral responses, and other processes subverted by the virus during infection, impose pressure on viral adaptation during host switching. Differences in DENV protein processing, encapsidation, and virion maturation in mosquito and mammalian cells have been reported (1–5). For example, the N terminus of the capsid protein is crucial for viral encapsidation in mammalian cells but not in mosquito cells (5). In addition, point mutations in NS4B have been reported to decrease viral replication in mosquito cells, while the same mutations have been shown to enhance replication in mammalian cells (2). Although it is evident that different interactions between the virus and the two hosts occur during the viral life cycle, the cellular processes and the host-specific underlying mechanisms involved are largely unknown. Here, we investigated RNA sequences present at the viral 3′ untranslated region (3′UTR) that were found to be essential for DENV replication in mosquito cells but dispensable in mammalian cells.
The DENV genome is an RNA molecule of about 11 kb that encodes a single open reading frame flanked by highly structured 5′- and 3′UTRs. The promoter for RNA synthesis, known as stem-loop A (SLA), is located at the 5′ end of the genome (6–8). This promoter binds and activates the polymerase NS5 that then initiates RNA synthesis at the 3′ end of a circularized viral genome. A great deal of information has been accumulated about the function of cis-acting RNA elements that enhance, silence, or promote DENV RNA synthesis. Based on numerous studies, key RNA elements present in the viral genome have been defined as essential for DENV replication (8–15). These elements include the SLA promoter, the cyclization elements present at both ends of the genome, the capsid-coding region hairpin element (cHP) located in the capsid protein-coding sequence, and the highly conserved 3′ stem-loop (3′SL). All of these elements were found to be necessary for viral replication in mammalian and mosquito cells.
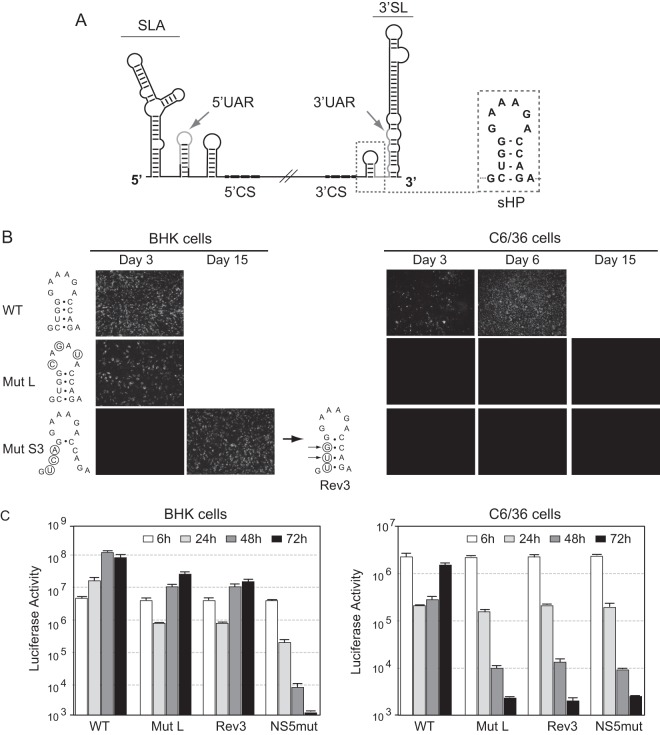
Recently, we identified a small hairpin structure (sHP) preceding the large terminal 3′SL as a crucial element for dengue virus type 2 (DENV2) replication in mammalian cells (Fig. 1A) (16). Mutations altering base pairings of the sHP stem in the context of a full-length DENV clone were lethal, while mutations in the loop that did not alter the predicted structure were tolerated. We have previously observed that transfection of viral RNAs with mutations impairing sHP formation resulted in revertant and pseudorevertant viruses, rescued in cell culture, which restored both the sHP structure and viral replication. Here, we examined the requirement of this DENV cis-acting RNA element for replication in mosquito cells using the same methodology as previously described (16). Interestingly, when DENVs carrying nucleotide changes at the loop (called Mut L) or the stem (called Rev3) of the sHP obtained in mammalian cells (which maintain the sHP structure and replicate to high titers in BHK cells) were used to infect mosquito cells, no viral replication was detected (Fig. 1B). To confirm this observation and eliminate nucleotide changes in other regions of the genome, possibly acquired during the process of obtaining viral stocks in mammalian cells, transfections of RNAs instead of viral infections were performed. In vitro-transcribed RNA of infectious clones carrying three substitutions in the loop of the sHP (Mut L) or the substitutions within the sHP stem obtained in the revertant virus (Rev3) were used for transfections in mosquito cells. While the wild type (WT) had propagated throughout the complete monolayer at day 7 posttransfection, no sign of viral replication was observed with Mut L or Rev3. Transfected cells were passaged during 30 days to search for reversions but, unexpectedly, no immunofluorescence-positive cells were detected (data not shown).
Fig 1.
Differential roles of the sHP in viral replication in mosquito and mammalian cells. (A) Schematic representation of the DENV genome showing relevant cis-acting elements: the promoter stem-loop A (SLA), the capsid region hairpin (cHP), the cyclization sequences (CS) 5′ upstream AUG region (5′UAR) and 3′UAR and 5′- and 3′CS, and the 3′ stem-loop (3′SL). The nucleotide sequence corresponding to the sHP structure is indicated on the right. (B) Replication of DENVs carrying substitutions within the sHP. Replication of viruses carrying the indicated mutations in BHK cells was monitored by immunofluorescence assay at different times posttransfection. Supernatants from mammalian BHK cells were used to infect mosquito C6/36 cells. The nucleotide sequence of the revertant virus Rev3 obtained from BHK cells transfected with Mut S3 is indicated; the arrows show the spontaneous changes generated in cell culture. (C) Replication of sHP mutants in the context of a DENV carrying luciferase as reporter. Luciferase activity was measured as a function of time after transfection of RNAs corresponding to the WT, a replication negative control with a mutation in the catalytic site of the NS5 polymerase (NS5mut), the sHP mutant Mut L, and the revertant virus Rev3. Error bars show standard deviations.
To further analyze the defect of the mutant viruses, we incorporated the nucleotide changes in a DENV infectious clone carrying a luciferase reporter gene. This reporter system allows discrimination between DENV translation, RNA amplification, and viral particle formation by measuring luciferase activity as a function of time posttransfection and postinfection, as previously described (17). RNAs from the WT, a replication negative control with a mutation in the catalytic site of the NS5 polymerase, and the two sHP mutants, reporter Mut L and reporter Rev3, were transfected into BHK and C6/36 cells. All the RNAs transfected showed similar levels of luciferase activity at 6 h posttransfection, indicating efficient translation of the input genomes (Fig. 1C). In BHK cells, the replication of the sHP mutants was slightly delayed with respect to the replication of the WT, but both viruses showed high replication levels compared with that of the negative control NS5mut. In contrast, the RNA replication of reporter Mut L and reporter Rev3 in mosquito cells was impaired (Fig. 1C). The results suggest that the nucleotide sequence of the sHP appears to be important for DENV replication in mosquito cells.
To more rigorously examine the nucleotide sequence requirement of the sHP for DENV replication in mosquito cells, we designed mutants carrying single substitutions in each position of the loop and evaluated viral replication in both cell types. Mutants Mut L1 to L6 were designed as shown in Figure 2A in the context of the DENV reporter system. As we described previously, the sequence of the sHP folds locally in the linear form of the RNA, while the nucleotides of the sHP loop form an internal loop in the circular form of the genome (16). Therefore, for clarity, the location of each mutation is indicated in the two alternative structures (Fig. 2A). Single substitutions in mutants Mut L1 to L6 resulted in viruses that replicated efficiently in BHK cells (Fig. 2B). In contrast, transfection of the same viral RNAs into mosquito cells showed that mutants Mut L1, L2, L4, L5, and L6 were unable to replicate. The luciferase levels at 24, 48, and 72 h were similar to that observed with the replication-impaired control NS5mut (Fig. 2C). The only substitution that was tolerated was that of L3, whose replication was similar to that observed with the WT virus. These results provide strong evidence of different functions of the identified DENV sequence in mosquito and mammalian cells.
Fig 2.
The loop sequence of the sHP is essential for DENV replication in mosquito cells. (A) Schematic representation of linear and circular conformations of the DENV genome showing the sequence corresponding to the sHP and the internal loop, respectively. Substitutions in each position of the loop and the internal loop are indicated as L1 to L6. Internal loop mutations IL1 and IL2 at the 5′ end of the genome are also indicated. (B and C) Replication of DENV mutants in the luciferase reporter system in mammalian and mosquito cells. Luciferase activity was measured as a function of time after transfection of the RNAs corresponding to the controls and mutants Mut L1 to L6, IL1, and IL2. Error bars show standard deviations.
We also examined whether the nucleotide sequence of the internal loop predicted in the circular form of the genome plays a role during viral replication in mosquito or mammalian cells by mutating the 5′ end of the viral genome. RNAs with substitutions in the internal loop were constructed (Mut IL1 and IL2) (Fig. 2A). Transfections of the RNAs indicated that the two mutants (Mut IL1 and IL2) were able to replicate in both cell types (Fig. 2B and C). These results suggest that the internal loop sequence modified within the 5′ end of the genome is not critical for viral replication in mosquito or mammalian cells.
Sequence alignment of the four DENV serotypes indicates that the nucleotide sequence of the sHP is highly conserved (Fig. 3A). The only variation observed in nature is at position 3 of the loop, which was the only one that tolerated changes in our mutational analysis (Fig. 2, Mut L3). The sHP loop sequence, GAAAGA, provides high stability to the HP. This sequence belongs to the GNRA-like (GNNNRA) loops that have been previously described, in which N is any nucleotide and R is a purine (18). It is possible that the sequence of the loop provides stability for the sHP structure, rather than there being a role of the sequence itself. To examine whether the stability of the structure is critical for its function, we replaced the complete loop with two different loops, UNCG and GNRA. About 70% of the RNA tetraloop sequences identified in ribosomal RNAs from different organisms fall into either UNCG or GNRA families (19, 20). Hairpins with these loop sequences exhibit exceptionally high thermal stability. In addition, taking into account that the sHP loop resembles a GNRA-like loop, we compared the replication of viruses in which the G in the R position was replaced by A or U, which provided different stabilities. Four recombinant viruses with different loop sequences in the sHP (UUCG [UNCG], GAGA [GNRA], GAAAAA [GNNNRA], and GAAAUA [GNNNUA]) (Fig. 3B) were tested. All the RNAs transfected showed similar levels of luciferase activity at 6 h posttransfection, indicating efficient translation of the input genomes. Interestingly, complete replacement of the sHP loop sequence rendered RNAs that were competent for RNA replication in BHK cells (Fig. 3C). In contrast, none of the mutants were able to replicate in mosquito cells. In this case, the luciferase levels at 24, 48, and 72 h were similar to those observed with NS5mut. Because RNAs with sHP loops with different sequences and different predicted stabilities with respect to those of the WT were all competent for replication in mammalian cells but incompetent in mosquito cells, we conclude that altered structural stability is not the main cause of the great vulnerability to nucleotide changes observed for DENV replication in insect cells.
Fig 3.
Stability of the sHP structure is not the cause of the high vulnerability to nucleotide changes for DENV replication in mosquito cells. (A) Alignment showing high conservation of the sHP sequence in DENV types 1 to 4. The cyclization sequences 3′CS and 3′UAR are also indicated. (B) Schematic representation of mutants within the sHP that retain high stability. (C and D) Replication of DENV mutants in the luciferase reporter system in mammalian and mosquito cells as indicated in each case. Luciferase activity was measured as a function of time after transfection of viral RNAs corresponding to the controls and GAGA, UUCG, GAAAAA, and GAAAUA mutants. Error bars show standard deviations.
We have demonstrated that the sequence of the loop of the sHP plays a crucial function in DENV replication only in mosquito cells. In addition, a possible role of nucleotides within the stem of the sHP was suggested by the lack of replication of Rev3 (Fig. 1B). To define whether the nucleotide sequence of the stem is required for viral replication in mosquito cells, we designed three different recombinant viruses with changes in one, two, or three nucleotides of the sHP stem, with the respective compensatory mutation(s) to maintain the structure (Rec 1, Rec 2, and Rec 3). The mutations were incorporated in both the reporter luciferase system and the DENV2 infectious clone. Transfection of reporter RNAs indicated that, while translation and RNA replication were efficient in BHK cells, in mosquito cells, the three RNAs showed luciferase kinetics similar to those of NS5mut (Fig. 4A and B). In addition, RNAs of the three mutants and the WT control were transfected into BHK and C6/36 cells, and immunofluorescence was followed as a function of time. In BHK cells, Rec1, Rec2, and Rec3 had propagated in the complete monolayer 3 days after transfection, while in mosquito cells, no viral replication was detected up to 15 days (Fig. 4C). Interestingly, no viral reversions were obtained even when the mosquito cells were passaged for an additional 2 weeks. Our studies conclusively identified a DENV sequence that is essential for viral replication in mosquito cells but not in mammalian cells.
Fig 4.
Substitutions within the stem of the sHP impair DENV replication in mosquito cells. (A and B) Replication of sHP mutants that change the sequence but maintain the stem structure in the context of the DENV luciferase reporter system. Luciferase activity in BHK and C6/36 cells was measured as a function of time after transfection of RNAs corresponding to WT and NS5Mut controls and mutants Rec 1, Rec 2, and Rec 3. Error bars show standard deviations. (C) Replication of DENV RNAs carrying substitutions, as indicated on the left, in mammalian BHK and mosquito C6/36 cells. Viral replication was monitored by immunofluorescence assay at different times posttransfection using specific anti-DENV antibodies.
Here, we found a nucleotide sequence in the DENV genome that is exclusively required for replication in mosquito cells. A remarkable effect on DENV replication was observed only in mosquito cells when single-nucleotide changes were introduced within the sHP. We propose that the identified sequence has at least two functions in viral replication. One function is linked to the secondary structure, which has previously been proposed to modulate the balance between the circular and linear forms of the genome, crucial for RNA synthesis (16), and the other function is linked to the nucleotide sequence required for viral replication in mosquito cells.
The mechanism by which this element participates in the viral process is unknown. According to previous reports, the sHP is considered part of the 3′SL, which contains the sHP and a large terminal HP (15). Although we have learnt a great deal about the function of RNA elements present in the flavivirus genome, the mechanisms of action of essential RNA elements, such as the 3′SL, are still enigmatic. In this regard, a recent report on West Nile virus (WNV) has demonstrated the generation of a microRNA derived from the 3′SL sequence in infected mosquito cells (21). It has been proposed that the accumulation of this small RNA enhances WNV replication. However, it remains to be determined whether this process also occurs during DENV replication. Interestingly, in flavivirus infections, the accumulation of subgenomic flavivirus RNAs (sfRNAs) derived from the 3′UTR have been reported to have a role in suppressing the host antiviral response both in mammalian and mosquito cells (22–24). In this regard, it will be interesting to investigate the impact of sHP mutations on the function of sfRNAs in the two hosts.
The finding of a strict requirement of the sHP sequence for viral replication in C6/36 cells is intriguing. It is possible that the sHP constitutes a binding site for a protein or a host-derived small RNA that is crucial for a viral process. Host proteins that interact with the DENV 3′UTR have been identified mainly in mammalian infected cells (25–28). Recently, specific binding of the host NF90 to the 3′SL and the host DDX6 helicase to the dumbbell structures present upstream from the sHP have been reported to play a role in DENV replication (29, 30). Studies using DENV-infected mosquito cells have also identified host proteins as binders of the viral 3′UTR (31, 32). However, the functions of these interactions during viral replication in mosquito cells have not been defined.
RNA cis-acting elements present at the DENV 5′ and 3′UTRs have previously been reported to enhance viral replication differentially in mosquito and mammalian hosts (1, 15, 33–35). However, the sHP sequence is the first one reported in the DENV genome that is essential for viral replication in mosquito cells and dispensable in mammalian cells. We believe that these results open new avenues for exploring mechanisms that regulate DENV replication in the insect host.
ACKNOWLEDGMENTS
We are grateful to Richard Kinney for the dengue virus cDNA clone. We also thank the members of Andrea Gamarnik's laboratory for helpful discussions and ideas about this work.
A.V.G. is a member of the Argentinean Council of Investigation (CONICET). This work was supported by grants from the Agencia Argentina de Promoción Científica y Tecnológica (PICT-2010), the NIH (1R01AI095175-01), and DARPA (grant HR001111C0094).
Footnotes
Published ahead of print 12 June 2013
REFERENCES
- 1. Groat-Carmona AM, Orozco S, Friebe P, Payne A, Kramer L, Harris E. 2012. A novel coding-region RNA element modulates infectious dengue virus particle production in both mammalian and mosquito cells and regulates viral replication in Aedes aegypti mosquitoes. Virology 432:511–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hanley KA, Manlucu LR, Gilmore LE, Blaney JE, Jr, Hanson CT, Murphy BR, Whitehead SS. 2003. A trade-off in replication in mosquito versus mammalian systems conferred by a point mutation in the NS4B protein of dengue virus type 4. Virology 312:222–232 [DOI] [PubMed] [Google Scholar]
- 3. Helt AM, Harris E. 2005. S-phase-dependent enhancement of dengue virus 2 replication in mosquito cells, but not in human cells. J. Virol. 79:13218–13230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mondotte JA, Lozach PY, Amara A, Gamarnik AV. 2007. Essential role of dengue virus envelope protein N glycosylation at asparagine-67 during viral propagation. J. Virol. 81:7136–7148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Samsa MM, Mondotte JA, Caramelo JJ, Gamarnik AV. 2012. Uncoupling cis-acting RNA elements from coding sequences revealed a requirement of the N-terminal region of dengue virus capsid protein in virus particle formation. J. Virol. 86:1046–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Filomatori CV, Iglesias NG, Villordo SM, Alvarez DE, Gamarnik AV. 2011. RNA sequences and structures required for the recruitment and activity of the dengue virus polymerase. J. Biol. Chem. 286:6929–6939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Filomatori CV, Lodeiro MF, Alvarez DE, Samsa MM, Pietrasanta L, Gamarnik AV. 2006. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 20:2238–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lodeiro MF, Filomatori CV, Gamarnik AV. 2009. Structural and functional studies of the promoter element for dengue virus RNA replication. J. Virol. 83:993–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alvarez DE, Filomatori CV, Gamarnik AV. 2008. Functional analysis of dengue virus cyclization sequences located at the 5′ and 3′UTRs. Virology 375:223–235 [DOI] [PubMed] [Google Scholar]
- 10. Alvarez DE, Lodeiro MF, Luduena SJ, Pietrasanta LI, Gamarnik AV. 2005. Long-range RNA-RNA interactions circularize the dengue virus genome. J. Virol. 79:6631–6643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clyde K, Barrera J, Harris E. 2008. The capsid-coding region hairpin element (cHP) is a critical determinant of dengue virus and West Nile virus RNA synthesis. Virology 379:314–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Khromykh AA, Meka H, Guyatt KJ, Westaway EG. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 75:6719–6728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. You S, Falgout B, Markoff L, Padmanabhan R. 2001. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J. Biol. Chem. 276:15581–15591 [DOI] [PubMed] [Google Scholar]
- 14. Yu L, Markoff L. 2005. The topology of bulges in the long stem of the flavivirus 3′ stem-loop is a major determinant of RNA replication competence. J. Virol. 79:2309–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zeng L, Falgout B, Markoff L. 1998. Identification of specific nucleotide sequences within the conserved 3′-SL in the dengue type 2 virus genome required for replication. J. Virol. 72:7510–7522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Villordo SM, Alvarez DE, Gamarnik AV. 2010. A balance between circular and linear forms of the dengue virus genome is crucial for viral replication. RNA 16:2325–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Samsa MM, Mondotte JA, Iglesias NG, Assuncao-Miranda I, Barbosa-Lima G, Da Poian AT, Bozza PT, Gamarnik AV. 2009. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 5:e1000632. 10.1371/journal.ppat.1000632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moody EM, Feerrar JC, Bevilacqua PC. 2004. Evidence that folding of an RNA tetraloop hairpin is less cooperative than its DNA counterpart. Biochemistry 43:7992–7998 [DOI] [PubMed] [Google Scholar]
- 19. Antao VP, Lai SY, Tinoco I., Jr 1991. A thermodynamic study of unusually stable RNA and DNA hairpins. Nucleic Acids Res. 19:5901–5905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Woese CR, Winker S, Gutell RR. 1990. Architecture of ribosomal RNA: constraints on the sequence of “tetra-loops”. Proc. Natl. Acad. Sci. U. S. A. 87:8467–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hussain M, Torres S, Schnettler E, Funk A, Grundhoff A, Pijlman GP, Khromykh AA, Asgari S. 2012. West Nile virus encodes a microRNA-like small RNA in the 3′ untranslated region which up-regulates GATA4 mRNA and facilitates virus replication in mosquito cells. Nucleic Acids Res. 40:2210–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, van der Aa L, Liu WJ, Palmenberg AC, Shi PY, Hall RA, Khromykh AA. 2008. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 4:579–591 [DOI] [PubMed] [Google Scholar]
- 23. Schnettler E, Sterken MG, Leung JY, Metz SW, Geertsema C, Goldbach RW, Vlak JM, Kohl A, Khromykh AA, Pijlman GP. 2012. Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and mammalian cells. J. Virol. 86:13486–13500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schuessler A, Funk A, Lazear HM, Cooper DA, Torres S, Daffis S, Jha BK, Kumagai Y, Takeuchi O, Hertzog P, Silverman R, Akira S, Barton DJ, Diamond MS, Khromykh AA. 2012. West Nile virus noncoding subgenomic RNA contributes to viral evasion of the type I interferon-mediated antiviral response. J. Virol. 86:5708–5718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lei Y, Huang Y, Zhang H, Yu L, Zhang M, Dayton A. 2011. Functional interaction between cellular p100 and the dengue virus 3′ UTR. J. Gen. Virol. 92:796–806 [DOI] [PubMed] [Google Scholar]
- 26. Paranjape SM, Harris E. 2007. Y box-binding protein-1 binds to the dengue virus 3′-untranslated region and mediates antiviral effects. J. Biol. Chem. 282:30497–30508 [DOI] [PubMed] [Google Scholar]
- 27. Polacek C, Friebe P, Harris E. 2009. Poly(A)-binding protein binds to the non-polyadenylated 3′ untranslated region of dengue virus and modulates translation efficiency. J. Gen. Virol. 90:687–692 [DOI] [PubMed] [Google Scholar]
- 28. Yocupicio-Monroy RM, Medina F, Reyes-del Valle J, del Angel RM. 2003. Cellular proteins from human monocytes bind to dengue 4 virus minus-strand 3′ untranslated region RNA. J. Virol. 77:3067–3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gomila RC, Martin GW, Gehrke L. 2011. NF90 binds the dengue virus RNA 3′ terminus and is a positive regulator of dengue virus replication. PLoS One 6:e16687. 10.1371/journal.pone.0016687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ward AM, Bidet K, Yinglin A, Ler SG, Hogue K, Blackstock W, Gunaratne J, Garcia-Blanco MA. 2011. Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3′ UTR structures. RNA Biol. 8:1173–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. De Nova-Ocampo M, Villegas-Sepulveda N, del Angel RM. 2002. Translation elongation factor-1alpha, La, and PTB interact with the 3′ untranslated region of dengue 4 virus RNA. Virology 295:337–347 [DOI] [PubMed] [Google Scholar]
- 32. Yocupicio-Monroy M, Padmanabhan R, Medina F, del Angel RM. 2007. Mosquito La protein binds to the 3′ untranslated region of the positive and negative polarity dengue virus RNAs and relocates to the cytoplasm of infected cells. Virology 357:29–40 [DOI] [PubMed] [Google Scholar]
- 33. Alvarez DE, De Lella Ezcurra AL, Fucito S, Gamarnik AV. 2005. Role of RNA structures present at the 3′UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology 339:200–212 [DOI] [PubMed] [Google Scholar]
- 34. Cologna R, Rico-Hesse R. 2003. American genotype structures decrease dengue virus output from human monocytes and dendritic cells. J. Virol. 77:3929–3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Men R, Bray M, Clark D, Chanock RM, Lai CJ. 1996. Dengue type 4 virus mutants containing deletions in the 3′ noncoding region of the RNA genome: analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J. Virol. 70:3930–3937 [DOI] [PMC free article] [PubMed] [Google Scholar]