Abstract
In the absence of an effective HIV-1 vaccine, passive immunization using broadly neutralizing Abs or Ab-like molecules could provide an alternative to the daily administration of oral antiretroviral agents that has recently shown promise as preexposure prophylaxis. Currently, no single broadly neutralizing Ab (bNAb) or combination of bNAbs neutralizes all HIV-1 strains at practically achievable concentrations in vivo. To address this problem, we created bispecific Abs that combine the HIV-1 inhibitory activity of ibalizumab (iMab), a humanized mAb directed to domain 2 of human CD4, with that of anti-gp120 bNAbs. These bispecific bNAbs (BibNAbs) exploit iMab’s potent anti–HIV-1 activity and demonstrated clinical efficacy and safety to anchor and thereby concentrate a second broadly neutralizing agent at the site of viral entry. Two BibNabs, PG9-iMab and PG16-iMab, exhibit exceptional breadth and potency, neutralizing 100% of the 118 viruses tested at low picomolar concentrations, including viruses resistant to both parental mAbs. The enhanced potency of these BibNAbs was entirely dependent on CD4 anchoring, not on membrane anchoring per se, and required optimal Ab geometry and linker length. We propose that iMab-based BibNAbs, such as PG9-iMab and PG16-iMab, are promising candidates for passive immunization to prevent HIV-1 infection.
Keywords: antiviral, neutralization, synergy
Despite decades of research, a sufficiently effective HIV-1 vaccine remains elusive. In the last several years, however, a number of broadly neutralizing Abs (bNAbs) have been isolated from HIV-1–infected individuals (1–6). Structural knowledge of such Abs in complex with the viral envelope (Env) glycoprotein gp120 (2, 3, 7–10) has raised the hope that immunogen design can be facilitated through a process of reverse vaccinology (11). In the meantime, alternative strategies for HIV-1 prevention are being pursued, including “treatment as prevention” (12), vaginal microbicides (13), and preexposure prophylaxis (PrEP) based on daily administration of antiretroviral drugs (14–16). In particular, PrEP using tenofovir with or without emtricitabine has demonstrated a protective efficacy of ∼40–70% in persons at high risk for HIV-1 infection; however, other studies using the same drugs have demonstrated no efficacy in similar populations. Higher efficacy would be expected if patients could better adhere to the prescribed regimen (14–16); however, the practical reality is that adherence on a daily basis is difficult, especially when patients are feeling well, regardless of the level of risk for HIV-1 infection.
PrEP using HIV-1–neutralizing Abs offers an attractive alternative to daily antiretroviral drugs. Abs are generally well tolerated and can be administered infrequently because of their longer half-life in vivo. Indeed, a recent multinational study of potential PrEP users showed that route of administration was the most important attribute regarding acceptability of a PrEP program, with a monthly or bimonthly injection preferred over daily or intermittent pills (17). In addition, numerous studies in macaques have demonstrated that passive immunization with bNAbs can prevent infection by hybrid simian-human immunodeficiency viruses (18–21). Until recently, however, such an approach was considered infeasible owing to the limited potency and/or breadth of the available bNAbs, such as 2F5, 4E10, and 2G12 (22, 23). The landscape has improved substantially with the advent of a collection of human anti-gp120 bNAbs, including PG9 and PG16 (5), VRC01 (4), 3BNC117 (6), PGT Abs (1), NIH45-46 and its engineered high-affinity variant NIH45-46G54W (9), and 10E8 (24). The impressive breadth and potency of these bNAbs against HIV-1 in vitro have energized the passive immunization field. Nevertheless, there remains a sizeable hole in the repertoire of these bNAbs, in that even at concentrations as high as 50 µg/mL, they cannot neutralize ∼2–30% of viral strains. Indeed, modeling a combination of 26 bNAbs provided only 89% viral coverage at clinically achievable concentrations (1).
In the present work, we sought to create Ab molecules that could neutralize 100% of the test strains of HIV-1 at clinically achievable concentrations. To this end, we constructed more than two dozen bispecific Abs, using the humanized anti-CD4 Ab ibalizumab (iMab) (25–27) as the scaffold onto which to engraft antigen-binding domains of select human anti-Env bNAbs. IMab was chosen because of its established safety record (well tolerated without serious adverse events) and anti–HIV-1 activity (>1 log reduction in viral load) in infected patients in phase 1a, 1b, 2a, and 2b clinical trials (25, 26, 28). This monoclonal Ab binds to domain 2 of human CD4, on a face opposite to the site of gp120 and MHC class II binding, and potently inhibits HIV-1 entry via a noncompetitive mechanism after virus attachment (29, 30). The effect of iMab on humoral responses to vaccination in vivo, as a surrogate of CD4–MHC II interactions, is currently under investigation in a clinical trial of HIV-uninfected individuals. The most notable constructs are bispecific fusion Abs PG9-iMab and PG16-iMab, which target both CD4 and the viral envelope and exhibit exceptional anti–HIV-1 profiles.
Results and Discussion
Construction and Biophysical Characterization of Ibalizumab-Based BibNAbs.
To create PG9-iMab and PG16-iMab, we fused the single-chain variable fragment (scFv) of PG9 or PG16 (5) to the N terminus of the heavy chain of iMab via a flexible, 15-aa glycine-serine linker (Fig. 1A). PG9 and PG16, the other parental Abs, are well-characterized, potent, broadly neutralizing, somatically related Abs directed to quaternary epitopes on the envelope trimer that are V2- and glycan-dependent (8, 31). They neutralize ∼80% of HIV-1 strains with a median IC50 of 0.2 µg/mL, and are not autoreactive (5, 32). The resultant bispecific PG9-iMab and PG16-iMab have a molecular weight of 204 kDa, compared with 148 kDa for a native IgG.
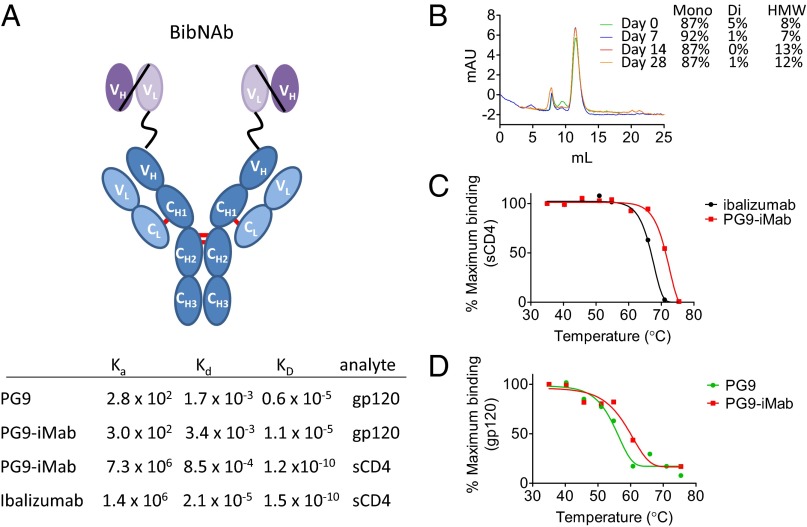
Fig. 1.
PG9-iMab bispecific Ab schema, binding kinetics, and biophysical characterization. (A) (Upper) Schematic depiction of the structure of PG9-iMab. The variable domains (VH and VL) of PG9 or PG16 were linked by a flexible glycine–serine linker to create a scFv (purple), which was linked to the N-terminal of the iMab heavy chain (dark blue). (Lower) Binding kinetics of PG9-iMab, iMab, and PG9 for sCD4 and monomeric gp120 (BaL strain), as determined by surface plasmon resonance. (B) Size-exclusion chromatogram of PG9-iMab (2 mg/mL in PBS) after incubation at 37 °C for up to 28 d. Mono, di, and HMW refer to monomeric, dimeric and high molecular weight forms, respectively. (C) Accelerated thermal stability profile of PG9-iMab and iMab for binding sCD4. (D) Accelerated thermal stability profile of PG9-iMab and PG9 for binding monomeric gp120 BaL.
The fusion of the scFvs to the iMab heavy chain was seemingly well tolerated. For example, yields of PG9-iMab and PG16-iMab from transient expression in 293A cells were comparable to those of iMab, suggesting no gross defects in protein folding. Furthermore, PG9-iMab was up to 95% monomeric after a single round of protein-A chromatography, as revealed by size-exclusion chromatography, even after a 28-d incubation at 37 °C (Fig. 1B) and following three freeze-thaw cycles. PG9-iMab also performed similarly to the parental mAbs in an accelerated thermal stability assay, which is often used to predict the stability of therapeutic proteins during long-term storage at 4 °C (Fig. 1 C and D). Importantly, the binding kinetics of PG9-iMab for both soluble CD4 and gp120, as determined by surface plasmon resonance, were similar to those of the parental iMab and PG9, respectively (Fig. 1A). Although the affinity of PG9-iMab (and PG9) for gp120 was low, this is likely an artifact of the use of gp120 monomer as an antigen, rather than a true reflection of the affinity of PG9 for its epitope presented in the context of native Env trimers. Indeed, binding of PG16 and PG16-iMab to monomeric gp120 was undetectable.
HIV-1 Neutralization Breadth and Potency: TZM-bl Assay.
To assess their breadth and potency against HIV-1, we first tested the neutralization activity of PG9-iMab and PG16-iMab and the parental Abs, using the TZM-bl/pseudovirus assay (33) against a panel of 118 diverse tier 2 and tier 3 viral strains in the Comprehensive Antibody-Vaccine Immunomonitoring Consortium (CA-VIMC) Antibody Core Lab of the Collaboration for AIDS Vaccine Discovery (CAVD) (34).
The addition of the PG9 or PG16 scFvs to iMab significantly improved both the breadth and potency of the parental mAbs (Fig. 2A). When tested at concentrations of up to 10 µg/mL, PG9-iMab and PG16-iMab each neutralized (as defined by >50% inhibition) 100% of the viruses tested, compared with 92% of viruses by iMab (P = 0.003, Fisher’s exact test), 81% by PG9 (P < 0.001, Fisher’s exact test), and 79% by PG16 (P < 0.001). Indeed, when using the more stringent >80% inhibition of infection for defining resistance, PG9-iMab and PG16-iMab neutralized 100% and 98% of viruses tested, respectively, compared with only 65% by iMab (P < 0.001, Fisher’s exact test), 71% by PG9 (P < 0.001, Fisher’s exact test), and 54% by PG16 (P < 0.001, Fisher’s exact test). We note, however, that two viruses required rather high concentrations of PG9-iMab (8.0 µg/mL and 10 µg/mL, respectively) to inhibit to the ≥80% level.
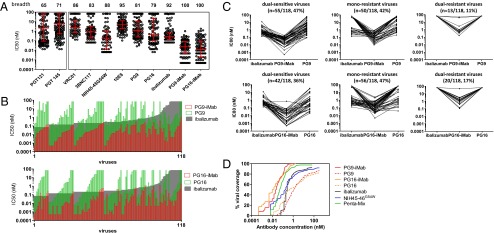
Fig. 2.
Neutralization breadth and potency of PG9-iMab, PG16-iMab, and parental mAbs against a diverse panel of 118 Env pseudoviruses. (A) Breadth and potency of PG9-iMab and PG16-iMab compared with a selection of other potent bNAbs. Numbers along the top indicate the breadth of each Ab, as defined by the percentage of viruses neutralized to >50%. Error bars represent median ± IQR. The PG9, PG16, 3BNC117, 10E8, iMab, PG9-iMab, and PG16-iMab data were generated against the same 118-virus panel. The VRC01 and NIH45-46G54W data were generated against an 82-virus subset of that panel. PGT121 and PGT145 data were generated against a different panel of 162 viruses and have been published previously (7). (B) (Upper) Paired IC50 (nM) values for PG9 (green open bars), iMab (gray closed bars), and PG9-iMab (red open bars) against a panel of 118 Env pseudoviruses in the TZM-bl assay. (Lower) Paired IC50 (nM) values for PG16 (green open bars), iMab (gray closed bars), and PG16-iMab (red open bars) against the 118-pseudovirus panel. Viruses are ordered by ascending iMab and PG9-iMab IC50 (Upper) and iMab and PG16-iMab IC50 (Lower). Maximum concentrations tested were 10 µg/mL (48 nM for BibNAbs and 67 nM for iMab, PG9, and PG16). (C) Paired IC80 (nM) of PG9-iMab, iMab, and PG9 (Upper) and paired IC80 (nM) of PG16-iMab, iMab, and PG16 (Lower) for dual-sensitive viruses (Left), monoresistant viruses (Center), and dual-resistant viruses (Right). (D) Percent viral coverage achieved by PG9-iMab and PG16-iMab. Cumulative frequency distribution of IC50 values of bNAbs tested against the 118-pseudovirus panel. NIH45-46G54W was tested against an 82-virus subset (9). The penta mix values have been published previously (36).
The BibNAbs also exhibited remarkable potency. Overall, on a molar basis, the geometric mean IC50 of PG9-iMab (19.3 pM; 4.0 ng/mL) and PG16-iMab (13.0 pM; 2.7 ng/mL) was 26-fold and 38-fold more potent than iMab (494.7 pM; 74.2 ng/mL), respectively, and 123-fold and 115-fold more potent than PG9 (2,378.1 pM; 356.7 ng/mL) and PG16 (1,494.7 pM; 224.2 ng/mL), respectively. Indeed, a majority of viruses were neutralized with an IC50 ≤70 pM (≤10 ng/mL) by PG9-iMab (74%) and PG16-iMab (72%), compared with 0%, 8%, and 25% for iMab, PG9 and PG16, respectively, at such low concentrations. Such improvements in Ab potency are highly synergistic according Chou and Talalay (35), whereby combination indexes <1 are considered synergistic. The combination indexes based on the IC50 and IC80 neutralization data were 0.11 ± 0.20 and 0.07 ± 0.17, respectively, for PG9-iMab and 0.02 ± 1.17 and 0.06 ± 0.37 for PG16-iMab. The improved breadth and potency of PG9-iMab and PG16-iMab are shown in Fig. 2 A–D.
The greater potency of PG9-iMab and PG16-iMab compared with the parental mAbs was not restricted to viruses resistant to one of the parental mAbs. Even dual-sensitive viruses were neutralized (IC80) 17-fold and 87-fold more potently by PG9-iMab than by iMab and PG9, respectively, and 43-fold and 161-fold more potently by PG16-iMab than by iMab and PG16, respectively (Fig. 2 B and C). This suggests that the enhanced potency was not due simply to the additive effects of two active agents. Remarkably, PG9-iMab and PG16-iMab neutralized viruses that were resistant to neutralization by both of the parental Abs (Fig. 2C, Center and Right). Perhaps the best demonstration of the improved activity of PG9-iMab and PG16-iMab are plots of their viral coverage with increasing Ab concentrations (Fig. 2D). In fact, PG9-iMab and PG16-iMab are also noticeably better NIH45-46G54W (9), an engineered high-affinity variant of a human mAb directed to the CD4-binding site on gp120, and even better than a combination of five highly potent bNAbs (Penta mix; NIH45-46G54W, PG16, 3BC176, PGT128, and 10-1074, a high-affinity variant of PGT121 (36) (Fig. 2D).
HIV-1 Neutralization Breadth and Potency: Peripheral Blood Mononuclear Cell Assay.
We next sought to assess the HIV-1–neutralizing activity of iMab-based BibNAbs using peripheral blood mononuclear cells (PBMCs) as target cells in the assay. This experiment was considered pertinent because TZM-bl cells express ∼2- to 10-fold more CD4 molecules than primary CD4+ T cells (37, 38), which could influence their activity. Thus, we tested PG9-iMab against a subset (n = 23) of the 118 strains previously tested in the TZM-bl/pseudovirus assay, using replication-competent reporter (Env-IMC-LucR) viruses in a PBMC neutralization assay (39).
This subset of viruses was shown to be representative of the full panel of HIV-1 strains (Fig. S1 A–D). The activity of PG9-iMab in the PBMC assay was quite similar to that observed in the TZM-bl assay. Except for one virus that was inhibited to only 80% in the TZM-bl assay, all viruses were inhibited to 100% maximum percent inhibition (MPI) in both assays. Furthermore, IC80 values were highly correlated between assays (Pearson’s r2 = 0.634, P < 0.001) (Fig. S2), and were only 1.1 ± 1.5-fold (median ± IQR) higher in the PBMC assay. These findings suggest that the enhanced activity of PG9-iMab is independent of the CD4 density on target cells and is not an artifact of the TZM-bl assay.
Mechanism of Synergistic Potency: Contribution of CD4 Anchoring.
We next addressed the mechanism behind the exceptional breadth and potency of iMab-based BibNabs. Because PG9 and PG16 are somatic mutants that recognize a similar, partially overlapping epitope and likely neutralize the virus via similar mechanisms, we focused our mechanistic studies on PG9-iMab. It is possible that the enhanced activity is due merely to the synergism of two active agents working in concert. Another possible explanation is that the longer reach of the anti-Env scFvs on the fusion molecule permits bivalent binding of two gp120 molecules on the virion surface, resulting in greater Ab avidity. It is also conceivable that iMab-based BibNabs anchor the active anti-Env moiety on cell surface CD4 and thereby concentrate their inhibitory activity at the precise location where it is needed.
To begin to discriminate among these competing possibilities, we constructed a PG9-iMab mutant (PG9-ΔiMab) by altering residue 33 (V–R) in CDR H1 and residue 102 (N–E) in CDR H3 of iMab. Based on the known structure of the complex of iMab Fab with human CD4 (29), these substitutions are expected to abrogate iMab binding to CD4. Indeed, binding of PG9-ΔiMab to CD4 molecules expressed on the surface of TZM-bl cells was undetectable by flow cytometry at concentrations up to 48 nM (10 µg/mL). When tested for neutralization against four HIV-1 strains exhibiting varying sensitivities to iMab and PG9, the potency of PG9-ΔiMab was indistinguishable from that of PG9, and the loss of CD4 binding was associated with a loss of enhanced activity (Fig. 3A). These findings not only rule out “longer reach” as an explanation for the potency of PG9-iMab, but also highlight the role of CD4 anchoring in the underlying mechanism.
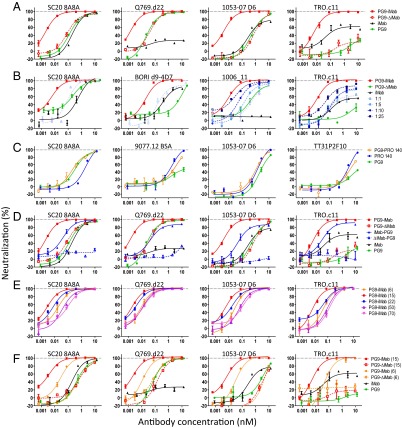
Fig. 3.
Neutralization data to elucidate the mechanism of PG9-iMab’s enhanced potency. (A) Abrogation of the CD4-binding function of PG9-iMab (PG9-ΔiMab) completely abolishes the enhanced potency of PG9-iMab. PG9-ΔiMab’s potency is indistinguishable from that of PG9. (B) Coadministration of iMab with PG9-ΔiMab at ratios of 1:1, 1:5, 1:10, and 1:25. (C) PG9-PRO 140 bispecific Ab exhibits only additive potency. (D) Neutralization activity of N-terminal and C-terminal BibNAbs. (E) Effects of reducing the length of the linker used to tether the PG9 scFv to iMab from 15 aa [PG9-iMab (15)] to 6 aa [PG9-iMab (6)] and of lengthening the linker to 20 aa [PG9-iMab (22)], 50 aa [PG9-iMab (50)], or 70 aa [PG9-iMab (70)]. (F) The reduced potency of PG9-iMab with a 6-aa linker [PG9-iMab (6)] compared with a 15-aa linker [PG9-iMab (15)] was not due to reduced activity of the PG9 scFv since the PG9-ΔiMab versions of PG9-iMab (15) [PG9-ΔiMab (15)] and PG9-iMab (6) [PG9-ΔiMab (6)] had similar potency, suggesting that the reduced potency of PG9-iMab (6) was related to the reduced “reach” of the PG9 scFv when bound to CD4.
To examine the question of synergism, we compared the neutralizing activity of PG9-iMab with that of a 1:1 mixture of iMab and PG9. As shown in Fig. 3B, this coadministration of iMab and PG9 yielded only additive effects, exhibiting virus-neutralization profiles far inferior to those observed for the fusion molecule. Increasing the iMab:PG9 ratio in the Ab mixture to 1:5, 1:10, and 1:25 produced a dose-dependent rise in potency; however, the activities remained well below that of PG9-iMab (Fig. 3B). These results dismiss the notion that the enhanced activity of PG9-iMab is related largely to the synergism of two agents working together. Instead, they point to the importance of the physical linkage of PG9 and iMab in mediating the activity of the bispecific Ab, likely by increasing the local concentration of PG9 at the target cell surface.
To further explore the role of cell surface anchoring, we fused the PG9 scFv via a 15-aa linker to the N terminus of PRO 140, a humanized mAb directed to human CCR5 with significant clinically proven antiviral activity against HIV-1 (40). The resultant bispecific Ab, PG9-PRO 140, was found to recognize both the viral envelope and human CCR5; however, its virus-neutralizing activity was no better than that of the more potent of the two parental Abs (Fig. 3C). There was no evidence of the enhanced potency seen for PG9-iMab. Thus, simply anchoring the scFv of PG9 to the cell surface, even when placed on the viral coreceptor, is insufficient to confer the enhanced antiviral potency.
Contribution of Ab Geometry and Linker Length.
In analyzing our data, we also noted highly significant direct correlations of the IC50 values for PG9 and PG9-iMab (Pearson’s r2 = 0.805, P < 0.001) and for PG16 and PG16-iMab (Pearson’s r2 = 0.828, P < 0.001) against the panel of 118 viruses tested (Fig. S3 A and B). These correlations are also present in the percent viral coverage profiles, where the profiles of PG9-iMab and PG16-iMab reflect the profiles of PG9 and PG16, respectively (Fig. S3C). This finding suggests that the high potency of PG9-iMab and PG16-iMab is specifically mediated by the gp120-binding activity of PG9 and PG16 scFvs. Taken together with previous findings, this conclusion led us to examine how the spatial relationship of this scFv to CD4, and hence to gp120, could affect the potency of PG9-iMab.
We first fused PG9 scFv to the C terminus of iMab heavy chain via a 15-aa linker, creating iMab-PG9. As shown in Fig. 3D, the virus-neutralizing potency of this new construct was more than 10-fold lower than that of PG9-iMab on average; however, when its CD4-binding property was abolished, the resultant ΔiMab-PG9 was substantially less active than PG9 or PG9-ΔiMab, indicating a major decrease in its affinity for gp120 when the scFv is placed in this orientation. Thus, although we were unable to draw a meaningful conclusion regarding spatial orientation from this experiment, these data highlight the remarkably enhanced activity of PG9 conferred by CD4 anchoring. Despite the >300-fold reduction in potency of PG9 scFv when fused to the C terminus of iMab heavy chain, iMab-PG9 was still more potent than PG9 (and iMab) for three of the four viruses examined (Fig. 3D).
This enhanced PG9 activity conferred by CD4 anchoring provides a mechanism by which PG9-iMab neutralizes dual iMab- and PG9-resistant viruses, such that mutations that reduce the affinity of the viral envelope for PG9 or PG9-like Abs to a sufficient degree to escape neutralization by PG9 are insufficient to escape the enhanced functional affinity of PG9 when concentrated at the site of viral entry, such as in the context of PG9-iMab, which binds to CD4 irrespective of the viral sensitivity to iMab.
We next assessed the impact of linker length in PG9-iMab on its antiviral potency. Reducing the linker to only six amino acids diminished the potency of PG9-iMab by sixfold (n = 4; P = 0.058, paired t test) (Fig. 3E). This drop in potency could be due either to a decreased “reach” or to constraints imposed by the shorter linker on the proper folding of PG9 scFv; however, the latter possibility is eliminated by the experimental results shown in Fig. 3F, demonstrating that PG9-ΔiMab with the 6-aa linker has comparable anti–HIV-1 activity to PG9. On the other hand, lengthening the linker to 22-, 50-, and 70-aa reduced the activity of PG9-iMab by 3-fold (n = 4; P = 0.151, paired t test), 12-fold (paired t test, n = 4; P = 0.046, paired t test), and 20-fold (n = 4; P = 0.041, paired t test), respectively (Fig. 3E). Collectively, these findings suggest that there is an optimal space into which to place the PG9 scFv in relation to CD4 receptors and gp120 trimers of the incoming HIV-1.
Model of Neutralization by Ibalizumab-Based BibNAbs.
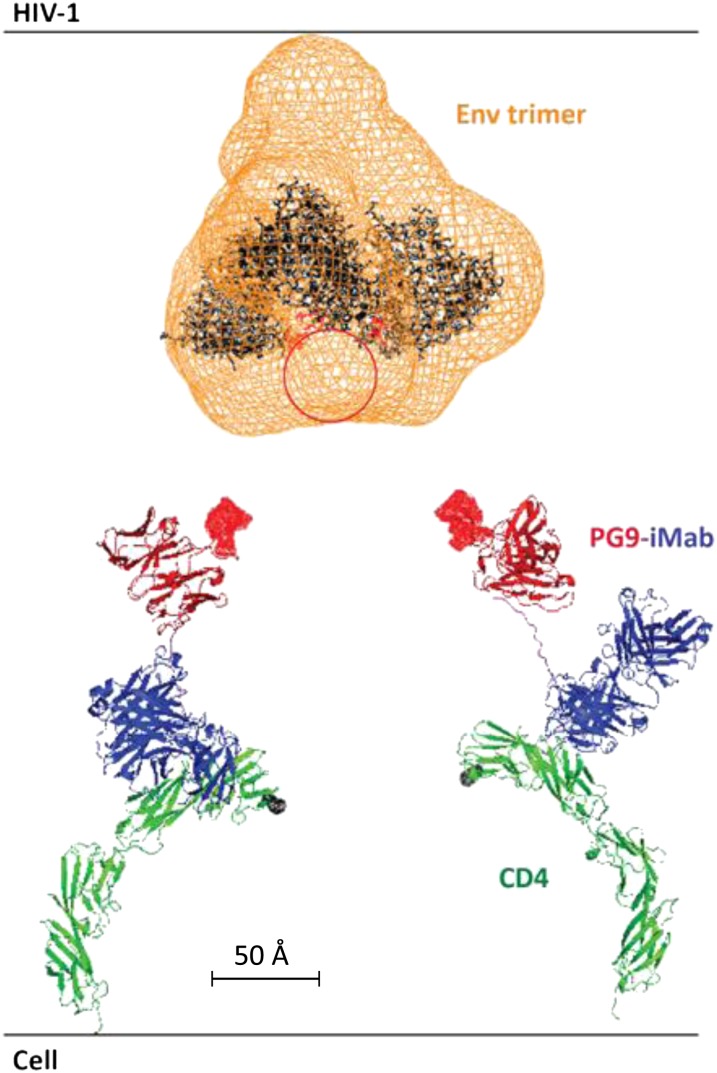
To further explore the enhanced potency of PG9-iMab, we modeled its spatial relationship to CD4 and the viral envelope trimer. Fig. 4 shows the gp120 trimer (41) together with a superimposition of the known CD4 structure (42) bound by iMab Fab (29), along with the known structure of PG9 scFv (8) connected by a 15-aa linker. The data shown in Fig. 3 A–F suggest that the enhanced potency of this bispecific Ab requires CD4 anchoring, which in turn greatly concentrates the PG9 scFv inside a space with a radius of 47 Å (length of the 15-aa linker) from the top of CD4. A radius of 17 Å (length of the 6-aa linker) is too short, and a radius greater than ∼70 Å (length of the 22-aa linker) is too long (Fig. 3E). Based on the results of the mAb mixture experiments (Fig. 3B), the degree of concentration of PG9 scFv within the critical space is likely more than 100-fold. It is apparent from Fig. 3E that PG9 scFv has easy access to its epitope at the outer tip of the envelope trimer (43). This Ab–antigen interaction appears to have a substantial spatial advantage over the binding between the CD4-binding sites of the envelope trimer and the first domain of CD4, the initial step in HIV-1 entry. Thus, we believe that the synergistic potency of PG9-iMab is the result of a large increase in PG9 scFv concentration at precisely the location where HIV-1 is trying to penetrate into its target cell, in addition to iMab’s activity in blocking a postbinding entry event.
Fig. 4.
Spatial relationship of PG9-iMab Fab, CD4, and the viral envelope trimer. A model of PG9-iMab Fab based on the crystal structures of PG9 (pdb3U4E, red), the iMab Fab and CD4 complex (pdb3O2D, blue and green, respectively), and an extended 20-aa linker (pink). The cyro-EM (orange mesh) of the Env trimer complex (41) is fitted with three copies of the gp120 crystal structure in black (pdb3DNN). The location of the missing V2 loops containing the PG9/16 epitope is approximated by the red circle, and the V2 loop binding residues on PG9 are highlighted as red surfaces. Modeling was performed using the PyMOL Molecular Graphics System, version 1.5.0.4 (Schrödinger).
Conclusion
IMab-based BibNAbs such as PG9-iMab and PG16-iMab have the antiviral breadth and potency to warrant serious consideration as potential agents for passive immunization or PrEP against HIV-1. They are the only Abs or Ab-like molecules described to date to neutralize all viruses tested, and to do so at extremely low concentrations, providing complete coverage against all HIV-1 strains tested at 1 nM and 2 nM, respectively (Fig. 2A), significantly better than that achieved by any Ab reported to date, including NIH45-46G54W (9). Although bispecific Abs have been successful in the clinic (44), we are mindful that their ultimate clinical utility will also depend on their pharmacokinetics, manufacturability, and lack of immunogenicity. Nevertheless, iMab-based BibNAbs such as PG9-iMab and PG16-iMab appear to be top candidates for passive immunization against HIV-1.
Materials and Methods
Bispecific Ab Cloning.
In-house generated iMab used for this study was generated by overlapping PCR of the iMab VH domain with a rhesus IgG1 Fc domain and cloned into a pcDNA3.1(+) vector (Invitrogen). To abrogate effector functions, L234A and L235A (LALA) mutations were introduced by site-directed mutagenesis (Stratagene). The genes encoding the scFv of PG9 and PG16 were chemically synthesized (GenScript) in the VH-VL orientation, linked together by a (Gly3Ser)4 linker. To create the bispecific Abs, the PG9 or PG16 scFvs were tethered to the iMab or PRO 140 heavy chain using a glycine-serine linker by overlapping PCR.
Ab Expression and Purification.
Ab expression and purification are described in detail in SI Materials and Methods.
Size-Exclusion Chromatography.
Size-exclusion fast-performance liquid chromatography is described in SI Materials and Methods.
Accelerated Thermal Stability Assay.
BibNabs and bNabs were incubated at temperatures ranging from 35 °C to 75 °C in 5-°C increments for 1 h in a thermal cycler. After incubation, samples were centrifuged at 10,600 × g to pellet insoluble aggregates, and then assessed by ELISA for binding of immobilized sCD4 or monomeric gp120 BaL.
Surface Plasmon Resonance.
Binding affinity analyses of mAbs and PG9-iMab for their respective ligands (sCD4 and monomeric gp120 BaL) were performed with the Biacore 3000 system, as described in SI Materials and Methods.
In Vitro Neutralization Assays.
Virus neutralization was assessed with a single-cycle assay using TZM-bl cells and HIV-1 pseudoviruses as described previously (33). Neutralization assays using HIV-1 Env pseudoviruses and TZM-bl cells as targets were performed by either the CAVD CA-VIMC’s Pre-Clinical Neutralizing Antibody Core Laboratory or our laboratory. The PBMC neutralization assay used infectious molecular clones expressing Renilla luciferase (Env-IMC-LucR) generated at the CA-VIMC’s Molecular Virology Core Laboratory and was performed there as described previously (39), except with PBMCs from four donors were pooled to reduce the contribution of PBMC donor variability. Nonlinear regression analysis was used to calculate IC50 and IC80 values and MPI.
Statistical Analyses.
Summary statistics are reported as median ± IQR. Differences in Ab potencies were assessed by Student’s paired t test and Fisher’s exact test of IC50 and IC80 and MPI as appropriate. Relationships of Ab activity were assessed by Spearman’s correlation analysis. Statistical significance was achieved at P ≤ 0.05. Statistics were generated by GraphPad Prism 5.03 software.
Supplementary Material
Acknowledgments
We thank Neal Padte, Yaoxing Huang, Moriya Tsuji, and members of the Ho laboratory for helpful discussions; Wendy Chen for figure preparation; and Dennis Burton and Wayne Koff for the PG9 neutralization data for the Collaboration for AIDS Vaccine Discovery (CAVD) virus panel. We also thank Francine McCutchan, Beatrice Hahn, David Montefiori, Michael Thomson, Ronald Swanstrom, Lynn Morris, Jerome Kim, Linqi Zhang, Dennis Ellenberger, and Carolyn Williamson for contributing HIV-1 envelope plasmids used in the CAVD virus panel, and Elise Zablowsky for performing neutralization assays. D.D.H. was supported by the Bill and Melinda Gates Foundation’s Collaboration for AIDS Vaccine Discovery (Grants OPP50714 and OPP1040732) and by the National Institutes of Health (Grant 1DP1DA033263-01). M.S.S. and C.O. were supported by the Bill and Melinda Gates Foundation’s Comprehensive Ab Vaccine Immune Monitoring Consortium (Grant 1032144).
Footnotes
Conflict of interest statement: D.D.H. is the scientific founder of TaiMed Biologics, Inc., which owns the commercial rights to ibalizumab. In this capacity, D.D.H. has equity in the company. C.S.P., R.S., and D.D.H. are holders of a patent describing fusion proteins, including PG9-iMab and PG16-iMab, for HIV-1 therapy.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304985110/-/DCSupplemental.
References
- 1.Walker LM, et al. Protocol G Principal Investigators Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477(7365):466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu X, et al. NISC Comparative Sequencing Program Focused evolution of HIV-1–neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333(6049):1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheid JF, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333(6049):1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu X, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329(5993):856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker LM, et al. Protocol G Principal Investigators Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326(5950):285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheid JF, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458(7238):636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 7.Pejchal R, et al. A potent and broad neutralizing Ab recognizes and penetrates the HIV glycan shield. Science. 2011;334(6059):1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McLellan JS, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing Ab PG9. Nature. 2011;480(7377):336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diskin R, et al. Increasing the potency and breadth of an HIV Ab by using structure-based rational design. Science. 2011;334(6060):1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou T, et al. Structural basis for broad and potent neutralization of HIV-1 by Ab VRC01. Science. 2010;329(5993):811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burton DR, Weiss RA. AIDS/HIV: A boost for HIV vaccine design. Science. 2010;329(5993):770–773. doi: 10.1126/science.1194693. [DOI] [PubMed] [Google Scholar]
- 12.Cohen MS, et al. HPTN 052 Study Team Prevention of HIV-1 infection with early antiretroviral therapy. N Engl J Med. 2011;365(6):493–505. doi: 10.1056/NEJMoa1105243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdool Karim Q, et al. CAPRISA 004 Trial Group Effectiveness and safety of tenofovir gel, an antiretroviral microbicide, for the prevention of HIV infection in women. Science. 2010;329(5996):1168–1174. doi: 10.1126/science.1193748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Celum C, Baeten JM. Tenofovir-based pre-exposure prophylaxis for HIV prevention: Evolving evidence. Curr Opin Infect Dis. 2012;25(1):51–57. doi: 10.1097/QCO.0b013e32834ef5ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Straten A, van Damme L, Haberer JE, Bangsberg DR. How well does PREP work? Unraveling the divergent results of PrEP trials for HIV prevention. AIDS. 2012;26(7):F13–F19. doi: 10.1097/QAD.0b013e3283522272. [DOI] [PubMed] [Google Scholar]
- 16.Grant RM, et al. iPrEx Study Team Preexposure chemoprophylaxis for HIV prevention in men who have sex with men. N Engl J Med. 2010;363(27):2587–2599. doi: 10.1056/NEJMoa1011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisingerich AB, et al. Attitudes and acceptance of oral and parenteral HIV preexposure prophylaxis among potential user groups: A multinational study. PLoS ONE. 2012;7(1):e28238. doi: 10.1371/journal.pone.0028238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hessell AJ, et al. Broadly neutralizing monoclonal antibodies 2F5 and 4E10 directed against the human immunodeficiency virus type 1 gp41 membrane-proximal external region protect against mucosal challenge by simian-human immunodeficiency virus SHIVBa-L. J Virol. 2010;84(3):1302–1313. doi: 10.1128/JVI.01272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hessell AJ, et al. Effective, low-titer Ab protection against low-dose repeated mucosal SHIV challenge in macaques. Nat Med. 2009;15(8):951–954. doi: 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mascola JR, et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000;6(2):207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 21.Mascola JR, et al. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J Virol. 1999;73(5):4009–4018. doi: 10.1128/jvi.73.5.4009-4018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehandru S, et al. Adjunctive passive immunotherapy in human immunodeficiency virus type 1-infected individuals treated with antiviral therapy during acute and early infection. J Virol. 2007;81(20):11016–11031. doi: 10.1128/JVI.01340-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trkola A, et al. Delay of HIV-1 rebound after cessation of antiretroviral therapy through passive transfer of human neutralizing antibodies. Nat Med. 2005;11(6):615–622. doi: 10.1038/nm1244. [DOI] [PubMed] [Google Scholar]
- 24.Huang J, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human Ab. Nature. 2012;491(7424):406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobson JM, et al. Safety, pharmacokinetics, and antiretroviral activity of multiple doses of ibalizumab (formerly TNX-355), an anti-CD4 monoclonal Ab, in human immunodeficiency virus type 1-infected adults. Antimicrob Agents Chemother. 2009;53(2):450–457. doi: 10.1128/AAC.00942-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuritzkes DR, et al. Antiretroviral activity of the anti-CD4 monoclonal Ab TNX-355 in patients infected with HIV type 1. J Infect Dis. 2004;189(2):286–291. doi: 10.1086/380802. [DOI] [PubMed] [Google Scholar]
- 27.Burkly LC, et al. Inhibition of HIV infection by a novel CD4 domain 2-specific monoclonal Ab: Dissecting the basis for its inhibitory effect on HIV-induced cell fusion. J Immunol. 1992;149(5):1779–1787. [PubMed] [Google Scholar]
- 28.Fessel WJ, et al. The efficacy of an anti-CD4 monoclonal Ab for HIV-1 treatment. Antiviral Res. 2011;92(3):484–487. doi: 10.1016/j.antiviral.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freeman MM, et al. Crystal structure of HIV-1 primary receptor CD4 in complex with a potent antiviral Ab. Structure. 2010;18(12):1632–1641. doi: 10.1016/j.str.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song R, et al. Epitope mapping of ibalizumab, a humanized anti-CD4 monoclonal Ab with anti-HIV-1 activity in infected patients. J Virol. 2010;84(14):6935–6942. doi: 10.1128/JVI.00453-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doores KJ, Burton DR. Variable loop glycan dependency of the broad and potent HIV-1–neutralizing antibodies PG9 and PG16. J Virol. 2010;84(20):10510–10521. doi: 10.1128/JVI.00552-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonsignori M, et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol. 2011;85(19):9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei X, et al. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother. 2002;46(6):1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seaman MS, et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol. 2010;84(3):1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: The combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 36.Klein F, et al. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature. 2012;492(7427):118–122. doi: 10.1038/nature11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nokta MA, et al. Chemokine/CD4 receptor density ratios correlate with HIV replication in lymph node and peripheral blood of HIV-infected individuals. AIDS. 2001;15(2):161–169. doi: 10.1097/00002030-200101260-00004. [DOI] [PubMed] [Google Scholar]
- 38.Lee B, Sharron M, Montaner LJ, Weissman D, Doms RW. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc Natl Acad Sci USA. 1999;96(9):5215–5220. doi: 10.1073/pnas.96.9.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edmonds TG, et al. Replication competent molecular clones of HIV-1 expressing Renilla luciferase facilitate the analysis of Ab inhibition in PBMC. Virology. 2010;408(1):1–13. doi: 10.1016/j.virol.2010.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jacobson JM, et al. Phase 2a study of the CCR5 monoclonal Ab PRO 140 administered intravenously to HIV-infected adults. Antimicrob Agents Chemother. 2010;54(10):4137–4142. doi: 10.1128/AAC.00086-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455(7209):109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu H, Kwong PD, Hendrickson WA. Dimeric association and segmental variability in the structure of human CD4. Nature. 1997;387(6632):527–530. doi: 10.1038/387527a0. [DOI] [PubMed] [Google Scholar]
- 43.Mao Y, et al. Subunit organization of the membrane-bound HIV-1 envelope glycoprotein trimer. Nat Struct Mol Biol. 2012;19(9):893–899. doi: 10.1038/nsmb.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bargou R, et al. Tumor regression in cancer patients by very low doses of a T cell-engaging Ab. Science. 2008;321(5891):974–977. doi: 10.1126/science.1158545. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.