Abstract
A major risk factor for hepatocellular carcinoma (HCC) is hepatitis B virus (HBV), whose pathogenesis is exacerbated by the acquisition of mutations that accelerate carcinogenesis. We examined, with mass spectrometry, the temporality of an HBV 1762T/1764A double mutation in plasma and tumors. Initial studies found that 52 of 70 (74.3%) tumors from patients residing in Qidong, People's Republic of China, contained this HBV mutation. Paired plasma samples were available for six of the tumor specimens; four tumors had the HBV 1762T/1764A mutation, whereas three of the paired plasma samples were also positive. The potential predictive value of this biomarker was explored by using stored plasma samples from a study of 120 residents of Qidong who had been monitored for aflatoxin exposure and HBV infection. After 10 years of passive follow-up, there were six cases of major liver disease including HCC (four cases), hepatitis (one case), and cirrhosis (one case). All six cases had detectable levels of the HBV 1762T/1764A mutation up to 8 years before diagnosis. Finally, 15 liver cancers were selected from a prospective cohort of 1,638 high-risk individuals in Qidong on the basis of available plasma samples spanning the years before and after diagnosis. The HBV 1762T/1764A mutation was detected in 8 of the 15 cases (53.3%) before cancer. The persistence of detection of this mutation was statistically significant (P = 0.022, two-tailed). We therefore found that a prediagnosis biomarker of specific HBV mutations can be measured in plasma and suggest this marker for use as an intermediate endpoint in prevention and intervention trials.
Hepatocellular carcinoma (HCC) is a major cause of cancer morbidity and mortality in many parts of the world, including Asia and subSaharan Africa, where there are >500,000 new cases each year and >200,000 deaths annually in the People's Republic of China (P.R.C.) alone (1, 2). The major etiological factors associated with development of HCC in these regions are infection with hepatitis B virus (HBV) and/or hepatitis C virus (HCV) and lifetime exposure to high levels of aflatoxin B1 (AFB1) in the diet (3, 4). Detailed knowledge of the etiology of HCC has spurred many mechanistic studies to understand the pathogenesis of this nearly always fatal disease, and this knowledge is beginning to be translated into preventive interventions in high-risk populations.
HBV is a significant risk factor for HCC in the developing world, where there are >400 million viral carriers (5, 6). The biology, mode of transmission, and epidemiology of this virus continue to be actively investigated and have been recently reviewed (5, 6). The contribution of HBV to the pathogenesis of liver cancer is multifactorial and is complicated by the identification of mutant variants in HBV that modulate the carcinogenesis process (7-9). The HBV genome encodes its essential genes with overlapping ORFs; therefore, a mutation in the HBV genome can alter the expression of multiple proteins. In many cases of HCC in China and Africa, a double mutation in the HBV genome, an adenine-to-thymine transversion at nucleotide 1762 and a guanine-to-adenine transition at nucleotide 1764 (1762T/1764A), has been found in tumors (5, 10, 11). This segment of the HBV genome contains an overlapping sequence for the base core promoter and the HBV X gene; therefore, the double mutation in codons 130 and 131 of the HBV X gene reported in human HCC is identical to the 1762 and 1764 nucleotide changes (12, 13). The onset of these mutations has been also associated with the increasing severity of the HBV infection and cirrhosis (10, 11). Thus, the tracking of this polymorphism with disease outcomes makes it a candidate biomarker for early detection of HCC risk in individuals.
Several studies have now demonstrated that DNA isolated from serum and plasma of cancer patients contains the same genetic aberrations as DNA isolated from an individual's tumor (14-17). The process by which tumor DNA is released into circulating blood is unclear but may result from accelerated necrosis, apoptosis, or other processes (18). Recently, we have found that a specific codon 249 p53 mutation not only was detectable in plasma samples at the time of HCC diagnosis, but that it can be measured in some individuals at least 5 years before diagnosis (19). In the present study, we have extended the use of an electrospray ionization-MS-based method called short oligonucleotide mass analysis (SOMA) to measure the double 1762T/1764A mutation in the HBV genome in both tissue and plasma samples and examine the prevalance and temporality of this biomarker before and at the time of HCC diagnosis.
Methods
Case Materials. The tumor and plasma samples investigated in this report were obtained as part of three separate ongoing investigations of HCC and its risk factors in Qidong, P.R.C. First, the 70 tumor specimens were collected from surgical resections conducted on patients at the Qidong Liver Cancer Institute during the 1990s and archived at the Shanghai Cancer Institute. Second, plasma samples were collected during a longitudinal ecological study designed to assess HCC risk factors in 120 residents of Daxin Township, Qidong, P.R.C., as reported (20). Briefly, the 120 subjects were equally divided by gender and high levels of the S gene product (HBsAg) status with an age range of 19-73 years (mean age, 44.5 years) at the start of the study in 1993. The third study is an ongoing prospective cohort investigation started in 1992 where 852 healthy HBsAg-positive individuals (776 males and 76 females) and 786 HBsAg-negative individuals (723 males and 63 females) residing in Qidong were recruited (19). Their ages ranged from 20 to 65 years at the time of enrollment. In all cases, HCC diagnosis was made by one or more of the following means: (i) surgical biopsy; (ii) elevated serum α-fetoprotein (levels >100 ng/ml), with consistent clinical and radiological history; (iii) positive computerized axial tomography scan; and (iv) ultrasonography with consistent clinical history. These collaborations among the Shanghai Cancer Institute, the Qidong Liver Cancer Institute, and Johns Hopkins University and the respective consent forms and questionnaires have been approved by each respective Institutional Review Board for Human Research.
Mutation Detection by SOMA. DNA was isolated from tissue and plasma samples and SOMA performed as described (17, 19, 21). PCR was performed on this reaction mix by using the following primers: 5′-TTT GTT TAA AGA CTG GGA GGA CTG GAG GGA GGA GAT TAG GTT A-3′ (HBV-7 forward) and 5′-TGG TGC GCA GAC CAA TTT ATG CTG GAG GCC TCC TAG TAC AA-3′ (HBV-7 reverse). The thermocycling conditions were 95°C for 2 min, then 40 cycles of 94°C for 30 sec, 65°C for 30 sec, and 72°C for 30 sec, followed by a final extension of 72°C for 2 min. Negative controls (no DNA added) were included for each set of PCR. PCR product was purified by ethanol precipitation and digested with 8 units of BpmI (New England Biolabs) overnight at 37°C in a volume of 50 μl to release 8-bp internal fragments. A phenol-chloroform extraction followed by an ethanol precipitation in the presence of SeeDNA (Amersham Biosciences) was performed to purify samples for analysis by electrospray ionization-MS.
The digested fragments were resuspended in 10 μl of the HPLC mobile phase [70:30 (vol/vol)] solvent A/solvent B, where solvent A was 0.4 M 1,1,1,3,3,3-hexofluoro-2-propanol (pH 6.9), solvent B was 50:50 (vol/vol) 0.8 M 1,1,1,3,3,3-hexafluoro-2-propanol/methanol), and 8 μl was introduced into the HPLC coupled to the electrospray ionization-MS. HPLC was carried out at 30 μl/min by using a 1 × 150-mm Luna C18, 5-μl reversed phase column (Phenomenex, Torrance, CA) and Surveyor pumps (ThermoFinnigan, San Jose, CA). The gradient conditions were 70% A:30% B isocratic 1 min programmed to 100% B in 3 min, where it was held for 2.5 min followed by a return to 70% A:30% B in 1.5 min and isocratic elution for the remaining 32 min of the chromatography.
Mass spectra were obtained with a LCQ Deca ion-trap mass spectrometer (ThermoFinnigan) equipped with an electrospray ionization source operated in the negative ionization mode. The spray voltage was set at -4.0 kV, and the heated capillary was held at 240°C. Each of the oligonucleotide ions was isolated in turn and subjected to collision-induced dissociation at 30% collision energy. Full-scan spectra of the resultant fragment ions from m/z 600 to m/z 2,000 were acquired, and signals from up to three specific fragment ions were summed as a function of time for each of the oligonucleotides. The mass spectrometer was programmed to acquire data in the centroid mode (one scan, 200 msec; isolation width, 3 Da) by using scan events monitoring each [M-2H]2- oligonucleotide individually. Scan event 1, WT-s [5′-AAGGTCT-3′], m/z 1,099.20→750-2,000; scan event 2, WT-as [5′-ACCTTTA-3′], m/z 1,066.70→750-2,000; scan event 3, Mut-s [5′-ATGATCT-3′], m/z 1,086.70→750-2,000; scan event 4, Mut-as [5′-ATCATTA-3′], m/z 1,078.70→750-2,000). The fragment ions used for each oligonucleotide were WT-s, m/z 803.78 + 1,132.22 + 1,243.27; WT-as, m/z 910.07 + 1,531.29; Mut-s, m/z 914.30 + 1,227.29; and Mut-as, m/z 1,084.0 + 1,252.0. A sample was considered positive when fragments were observed in either or both sense and antisense channels for the mutant allele in at least three scans across the peak.
Data Analysis. All samples were coded before analysis to mask identity for both SOMA analysis and data interpretation. In addition, the coding of the samples was done to allow for randomization of the order of sample analysis and interspersion of control samples. For analysis of the prospective investigation, all of the mutant positive and negative data were dichotomized to the date of liver cancer diagnosis, resulting in scores for positive and negative mutant status before and after clinical diagnosis. The longitudinal data on each individual were summarized by the number of positive samples rB and rA out of the nB amplified samples before and nA amplified samples after the diagnosis, respectively. To not only provide a measure of the likelihood of mutation in HBV before and after HCC diagnosis but to also quantify the level of persistence, a β-binomial model was used. Specifically, if P denotes the probability of a sample showing an HBV double mutation, we modeled the belief about P as a β distribution with parameter Π(1-ρ)/ρ and (1-Π) (1-ρ)/ρ, and we modeled r given Π as binomial (n, Π), with n representing the number of amplified samples. It follows that the marginal distribution of r is β-binomial with mean n Π and variance n Π (1-Π)[1+(n-1)ρ]. The parameter Π represents p, the likelihood of mutation in HBV, and ρ is the correlation of the presence of mutation in within individual samples (i.e., persistence). Excess frequencies of low (e.g., =0) and/or high (e.g., =1) values of r/n are consonant with ρ > 0 (i.e., overdispersion caused by persistence). The inferences were summarized by providing the confidence intervals for Π and two-sided P values for testing the null hypothesis of lack of persistence (H0: ρ = 0). Maximum likelihood methods were implemented by using the egret statistical package (Cytel, Cambridge, MA). Methods used here to quantify persistence of mutation have previously proven useful for the assessment of persistence of human papilloma virus infection (22) and for the evaluation of clustering of inactive days in stays of patients in hospitals (23). These methods were also used to analyze p53 mutation data in a recent investigation (19).
Results
Detection of 1762T/1764A Mutation in HBV in Tumor and Plasma. Initial experiments were performed to validate the use of a MS technique, SOMA, for the measurement of a double 1762T/1764A mutation in the HBV genome. SOMA had been previously used for the detection of a number of other mutations, including a codon 249 mutation in p53 in plasma and tissues (17, 19). Following the design of the appropriate PCR primers, DNA was isolated from 70 liver tumor specimens from residents of Qidong, P.R.C., diagnosed between 1990 and 1998 and analyzed for the double HBV mutations as well as WT HBV integrated into the DNA. Shown in Table 1, 67 of 70 (95.7%) tumors contained detectable levels of either the HBV double mutation or integrated WT HBV DNA or both. Fifty-two of the specimens (74.3%) contained the 1762T/1764A HBV mutation. These findings are quantitatively similar to previous studies and consistent with the hypothesis that the double HBV mutation is acquired after WT integration (5, 10-13).
Table 1. Prevalence of HBV 1762T/1764A and WT HBV DNA in HCC tumors from Qidong, P.R.C.
| Year of diagnosis
|
Number of tumors
|
Integrated HBV DNA status 1762T/1764A mutation, WT
|
|||
|---|---|---|---|---|---|
| Positive/positive (%) | Positive/negative (%) | Negative/positive (%) | Negative/negative (%) | ||
| 1990 | 13 | 12 (92.3) | 0 (0) | 1 (7.7) | 0 (0) |
| 1996 | 17 | 11 (64.7) | 2 (11.8) | 4 (23.5) | 0 (0) |
| 1997 | 20 | 9 (45) | 4 (20) | 6 (30) | 1 (5) |
| 1998 | 20 | 6 (30) | 8 (40) | 4 (20) | 2 (10) |
| Total | 70 | 38 (54.3) | 14 (20) | 15 (21.4) | 3 (4.3) |
The results from the tumor DNA samples validated the MS method and established positive and negative DNA controls for measuring these fragments in plasma specimens. Paired plasma samples obtained at the time of surgery from six of the 70 liver cancer patients described above were available for study. Four of the six tumors contained the HBV 1762T/1764A double mutation. Of these four mutant positive tumors, there were detectable levels of the HBV 1762T/1764A double mutation in three of the plasma samples (75%). The two tumors that did not have the mutation were also negative for mutation in the paired plasma samples. Although all six of the tumors were positive for integrated WT HBV, only three of the paired plasma samples were positive (50%). These data demonstrate a relation between integrated viral DNA in a target tissue and its detection in plasma. Representative SOMA chromatograms of standards and samples obtained from tissue and plasma specimens are shown in Fig. 1.
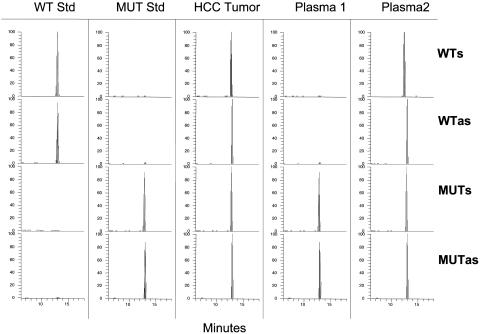
Fig. 1.
SOMA analysis of codon HBV 1762T/1764A mutations. WT Std and MUT Std represent standards for the WT sense and antisense and mutant sense and antisense signals. A representative chromatogram of a tumor sample containing integrated WT and tandem mutant HBV DNA is shown. The two plasma samples are from individuals scored as exhibiting medium levels of the HBV mutation. All scales are in relative abundance of 100% for the largest ion.
Prospective Measurement in Plasma of 1762T/1764A HBV Mutations. In 1993, 10 serial plasma samples from 120 residents of Daxin Township, Qidong, P.R.C., were collected over a 1-year period (20). These subjects were equally divided by gender and HBV status, and aflatoxin albumin biomarkers were assessed. During the year-long monitoring period, AFB-albumin adducts were detected in all but one of the samples tested. During the 10 years since the start of this study, the Qidong Liver Cancer Institute has continued to passively monitor all deaths and other serious illnesses, including HCC, in residents of Qidong. Within this 120-person cohort, who were all healthy at the time of enrollment, there have been 11 deaths and 3 additional serious diagnoses, including one case of hepatitis. The dates of the specific diagnoses and the determination of HBV 1762T/1764A mutation or WT HBV DNA status in the respective plasma samples are shown in Table 2. All six cases of major liver disease, including HCC (four cases), hepatitis (one case), and cirrhosis (one case), had detectable levels of the HBV 1762T/1764A up to 8 years before diagnosis. In contrast, only three of these six cases had detectable levels of the WT HBV DNA in plasma. None of the other eight cases of disease (Table 2), which were unrelated to liver function, had the HBV 1762T/1764A genotype. Thus, these findings supported the further exploration of the predictive power of the HBV mutation in plasma to presage the development of HCC.
Table 2. HBV 1762T/1764A, WT HBV DNA in plasma from Qidong, P.R.C., in 1993.
| Diagnosis (year) | HBV 1762T/1764A mutation in plasma | HBV WT in plasma |
|---|---|---|
| HCC (1994) | Positive | Negative |
| HCC (2000) | Positive | Positive |
| HCC (2000) | Positive | Negative |
| HCC (2001) | Positive | Negative |
| Hepatitis (2000) | Positive | Positive |
| Bladder cancer and cirrhosis (2000) | Positive | Positive |
| Lung cancer (2001) | Negative | Negative |
| Esophageal cancer (1997) | Negative | Positive |
| Tuberculosis (1994) | Negative | Negative |
| Pancreatic cancer (2000) | Negative | Negative |
| Acute myocardial infarct (1997) | Negative | Negative |
| Cerebral hemorrhage (2002) | Negative | Negative |
| Cerebral hemorrhage (1997) | Negative | Negative |
| Dementia (1999) | Negative | Negative |
| Summary Case diagnosis | HBV 1762T/1764A mutations (%) | HBV WT (%) |
| HCC, hepatitis, cirrhosis ascites | 6/6 (100%) | 3/6 (50%) |
| Vascular disease, stroke, other cancers | 0/8 (0%) | 1/8 (12.5%) |
The examination of the temporality of the detection of the HBV mutation in plasma before and after the clinical diagnosis of HCC in the same patient was facilitated by the availability of longitudinally collected plasma samples from a cohort of 1,638 high-risk individuals in Qidong, P.R.C., that have been followed since 1992. Fifteen liver cancer cases diagnosed between 1997 and 2001 were selected for study on the basis of having available annual plasma samples with PCR amplifiable DNA that spanned the years around HCC diagnosis. Eighteen plasma samples from healthy U.S. adults were used as controls, and all samples were coded and randomized for analysis. Table 3 lists the details of the date of diagnosis, death, sample collection by year, and HBV mutation status. Of the 60 plasma samples analyzed, the HBV 1762T/1764A double mutation was found in 28 (46.7%) of the specimens. In contrast, only 5 of 60 (8.3%) plasma samples were positive for the WT HBV DNA. All of the plasma samples obtained from healthy U.S. controls were negative for HBV DNA.
Table 3. Plasma samples from liver cancer cases in Qidong, P.R.C.
| Individual | Date of diagnosis | Date of death | Year collected | Sample collected | HBV mutations |
|---|---|---|---|---|---|
| A | October 1999 | 1996 | Y | (+) | |
| 1997 | Y | (+) | |||
| 1998 | Y | (−) | |||
| 1999 | Y | (+) | |||
| 2000 | Y | (−) | |||
| B | March 2000 | 2000-4 | 1996 | Y | (+) |
| 1997 | Y | (+) | |||
| 1998 | Y | (+) | |||
| 1999 | Y | (−) | |||
| 2000 | N | / | |||
| C | April 1998 | 1996 | Y | (−) | |
| 1997 | N | / | |||
| 1998 | Y | (−) | |||
| 1999 | Y | (+) | |||
| 2000 | Y | (+) | |||
| D | October 1998 | 1996 | Y | (−) | |
| 1997 | N | / | |||
| 1998 | Y | (+) | |||
| 1999 | Y | (−) | |||
| 2000 | Y | (+) | |||
| E | September 1999 | 1999-11 | 1996 | Y | (−) |
| 1997 | Y | (−) | |||
| 1998 | Y | / | |||
| 1999 | Y | (−) | |||
| G | October 1999 | 1996 | Y | (−) | |
| 1997 | Y | (−) | |||
| 1998 | Y | (−) | |||
| 1999 | Y | (−) | |||
| 2000 | Y | (+) | |||
| H | October 1999 | 1996 | Y | (−) | |
| 1997 | Y | (−) | |||
| 1998 | N | / | |||
| 1999 | Y | (−) | |||
| 2000 | Y | (−) | |||
| K | April 1997 | 1998-10 | 1996 | Y | (+) |
| 1997 | Y | (+) | |||
| 1998 | Y | (+) | |||
| I | September 1997 | 2001-03 | 1996 | Y | (−) |
| 1997 | Y | (−) | |||
| 1998 | Y | (−) | |||
| 1999 | Y | (−) | |||
| 2000 | Y | (−) | |||
| J | October 1999 | 2001-4 | 1996 | Y | (+) |
| 1997 | Y | / | |||
| 1998 | Y | / | |||
| 1999 | Y | (+) | |||
| 2000 | N | / | |||
| L | May 2001 | 1996 | Y | (−) | |
| 1997 | N | / | |||
| 1998 | Y | (−) | |||
| 1999 | Y | (−) | |||
| 2000 | Y | (−) | |||
| M | October 1999 | 1996 | Y | (+) | |
| 1997 | Y | (+) | |||
| 1998 | Y | (+) | |||
| 1999 | Y | (+) | |||
| 2000 | N | / | |||
| N | October 2001 | 1996 | Y | (+) | |
| 1997 | Y | (−) | |||
| 1998 | Y | (−) | |||
| 1999 | Y | (+) | |||
| 2000 | Y | (−) | |||
| O | January 2001 | 1996 | Y | (+) | |
| 1997 | Y | (+) | |||
| 1998 | N | / | |||
| 1999 | Y | (+) | |||
| 2000 | N | / | |||
| P | October 2000 | 1996 | Y | (+) | |
| 1997 | Y | (−) | |||
| 1998 | Y | (+) | |||
| 1999 | Y | (−) | |||
| 2000 | Y | (+) |
Italics indicate data after diagnosis; (+), positive; (−), negative; /, no sample. Y, yes; N, no.
Table 4 summarizes the detection of the HBV 1762T/1764A double mutation in the pre- and postdiagnosis plasma samples. At the top of Table 4, the frequency of the mutation (i.e., 100 rA/nA and 100 rB/nB) in the nA and nB samples provided by individuals before and after diagnosis, respectively, is described. A majority of individuals did show mutation before the diagnosis, and the descriptive statistics at the top of Table 4 are consistent with overdispersion (particularly for samples prediagnosis) caused by persistence of the mutation for which β-binomial models are appropriate. Results from the analysis using a β-binomial distribution are shown at the bottom of Table 4. In the samples collected before diagnosis of liver cancer, 42.6% of the plasma samples had detectable levels of the HBV mutation, with a 95% confidence interval of 23.9-63.6%. The persistence of this prediagnosis marker was statistically significant (P = 0.022, two-tailed). These data indicate that 8 of 15 future patients with this marker can be detected at least 1 year and in some cases up to 5 years before cancer diagnosis. The HBV double mutation was detected in 52.1% of all plasma samples after the diagnosis of liver cancer with 95% confidence intervals from 29.8% to 73.5%. The persistence of this mutation after diagnosis was not statistically significant, which may reflect changes in the biology of late-stage HCC. Collectively, we have found that prediagnosis biomarkers of specific HBV mutations can be measured in plasma, and its occurrence reflects increased risk for the development of HCC.
Table 4. Detection of HBV 1762T/1764A mutations: pre- and post-liver cancer diagnosis.
| No. of individuals prediagnosis | No. of individuals postdiagnosis | |
|---|---|---|
| Percent of individuals with mutation | ||
| 0% | 7 (46.7%)* | 3 (27.3%)† |
| >0-50% | 2 (13.3%)* | 2 (18.2%)† |
| 51% to < 100% | 2 (13.3%)* | 2 (18.2%)† |
| 100% | 4 (26.7%)* | 4 (36.4%)† |
| Percent of mutant samples (95% confidence interval) | 42.6% (23.9, 63.6) | 52.1% (29.8, 73.5) |
| Persistence of mutation (P value) | 0.414 (P = 0.022) | 0.181 (P = 0.213) |
n = 15.
n = 11.
Discussion
The pathogenesis of HCC is complex, and the disease progresses through a multistage process with a majority of cases involving liver cirrhosis (4, 24). The worldwide geographic distribution of HCC illustrates that areas where viral hepatitis infection is endemic also have a high incidence of disease. The two major hepatitis viruses associated with HCC are HBV and HCV. HBV has a higher prevalence worldwide at this time, and there are >400 million HBV carriers worldwide (6). Because there is a <20% chronicity rate for HBV infection, >2 billion people are or have been exposed to this virus at some point during their lifetime. In hyperendemic areas, such as Korea and China, infection rates of HBV have exceeded 50% by age 30 (25). In these high-infection areas, perinatal transmissions account for 35-50% of HBV carriers (26). HCV appears to be associated with HCC in regions with relatively low prevalence of HBV infection. In China, HCV is found in <5% of HCC cases (27). Unfortunately, HCV infection results in a much higher incidence of chronicity and cirrhosis, which is beginning to contribute to the major rise in HCC in countries such as Japan and the U.S. (2, 7).
The etiology of HCC in some of the highest-risk regions is further complicated by the interplay between environmental chemical agents and HBV infection. Two major cohort studies have demonstrated the strong mutiplicative interaction between AFB1 exposure and HBV in the development of HCC. The first report culminated from monitoring and follow-up of >18,000 people in Shanghai (3, 28). A nested case-control study within this cohort revealed statistically significant increases in HCC for either aflatoxin exposure or HBV infection alone. For those people who had both aflatoxin and HBV exposure, there was a multiplicative interaction resulting in a relative risk for developing HCC of ≈60. A subsequent nested case-control study from a cohort of >15,000 people in Taiwan also found a strong interaction between aflatoxin and HBV (29). Collectively, these results strongly support a causal amplifying relationship between two major HCC risk factors. These findings have encouraged the development and validation of both aflatoxin and HBV biomarkers that can be used to identify high-risk individuals before HCC diagnosis.
The development of new biomarkers for predicting an individual's risk for HCC after HBV infection is predicated on an understanding of the molecular pathways through which the virus mediates its effects (5, 7, 30). The pathobiology of HBV infection is also modulated through the selection and expression of a number of common viral mutants that affect a number of key viral proteins (8, 9, 31, 32). One of these common mutations is HBV 1762T/1764A, which affects the expression of both the HBV antigen, because the mutation lies in the basic core promoter, and the X gene (33). This double mutation induces an increased inflammatory response that becomes stronger as the progression of liver damage transits through chronic hepatitis and into a cirrhosis stage (34). The underlying mechanism of the effects of HBV antigen on the biology of inflammation and cirrhosis is still unclear, but there are substantial data that point to modulation of the immune surveillance system and immune tolerance in the presence and absence of this protein (33-35). The HBV 1762T/1764A double mutation also affects the amino acid sequence of the HBV X gene, because it resides in codons 130 and 131, thereby inducing lysine to methionine and valine to isoleucine alterations, respectively (36). The X gene protein has been found to have numerous biological activities, but its specific role and that of this mutant protein in the pathogenesis of liver cancer have yet to be elucidated (37). The 1762T/1764A double mutation occurs more frequently in people infected with the genotype C strains of HBV, which is the most common genotype found in East Asian patients (38-40).
The serology of HBV infection has been well established and validated in epidemiological studies. HBV is not cytotoxic for infected hepatocytes, and these cells synthesize and secrete HBsAg, which appears in blood before the onset of symptoms. Plasma HBsAg levels peak during the symptomatic phase and then decline to undetectable levels within 6 months of infection. HBV carrier status is then defined by HBsAg positivity in sequential samples obtained 6 months apart (6). HBeAg, HBV-DNA, and DNA polymerase also appear in the serum soon after HBsAg, and these are all biomarkers of active viral replication (41). Thus, these plasma HBV specific biomarkers reflect an intrinsic risk for the future development of HCC, but <10% of all exposed people will develop this cancer.
The use of a biomarker in blood for the early detection of HCC is well established using α-fetoprotein (42). Although the use of α-fetoprotein as a HCC diagnostic marker is widely used in high-risk areas because of its ease of use and low cost, this marker does suffer from low specificity because of its occurrence in diseases other than liver cancer (43, 44). This lack of specificity has contributed to the identification of other molecular biomarkers that are possibly more mechanistically associated with HCC development, including hypermethylation of the p16, p15 and GSTP1 promoter regions (16, 42, 45, 46). Results from investigations of p16, p15 and GSTP1 promoter hypermethylation indicate that these markers are prevalent in HCC, but there is as-of-yet limited information on the temporality of these genetic changes before clinical diagnosis. The recent report by Jackson et al. (19) also showed the potential use of specific p53 mutations in blood as a biomarker of HCC risk.
In this current investigation, we examined the temporality of detecting the HBV 1762T/1764A mutation and WT HBV in plasma and tumor specimens, both before and after the clinical diagnosis of HCC. This study was facilitated by the availability of prospectively collected tumor and plasma samples from high-risk individuals in Qidong, P.R.C. Similar to previous reports, almost all of the HCC tumors contained either the HBV tandem mutation or integrated HBV DNA and, in many cases, both (5, 10-13). To our knowledge for the first time, paired plasma tumor samples were examined for the HBV 1762T/1764A double mutation, and five of six plasma samples studied contained the same HBV mutational pattern as found in the tumor. This work was then extended to a cohort of 120 residents of Qidong who, after 10-year passive follow-up, contained four cases of HCC, one case of hepatitis, and one case of liver cirrhosis. All six cases had detectable levels of the HBV 1762T/1764A up to 8 years before diagnosis. The HBV mutation was explored in 15 HCC cases selected from a prospective cohort of 1,638 high-risk individuals in Qidong on the basis of available plasma samples spanning the years before and after diagnosis. The HBV 1762T/1764A mutation was detected in 8 of the 15 cases (53.3%) before cancer. The persistence of detection of this mutation was statistically significant (P = 0.022, two-tailed). Taken together, these data suggest the HBV 1762T/1764A double mutation in plasma could be a valuable predictive biomarker for HCC development. Further, the consistent finding of the HBV DNA in plasma reflecting the integrated DNA in the tumor suggests that shed DNA from tumor cells could be a significant source of this material (18).
The underlying mechanism and selection for the HBV 1762T/1764A double mutation are unclear at this time. Clearly inflammatory responses in cells produce large quantities of reactive oxygen species known to modify DNA and induce mutations (47). There is also a striking sequence specific binding and induction of mutations by aflatoxin at codon 249 of p53 in HCC (48-50). The WT sense strand sequence of p53 at codon 249 is AGG, and this codon is bracketed by two purines and two pyrimidines when reading from 5′ to 3′ resulting in a GGAGGCC sequence. Interestingly, the HBV 1762T/1764A double mutation lies in a sequence of AAAGGTC. The AGG is also bracketed by two purines and two pyrimidines when reading from 5′ to 3′. Thus, it is intriguing to speculate whether this common pattern shared by both p53 and HBV might be a mutational hot spot targeted by aflatoxin or some other chemical agent. Because both changes are induced mutations that contribute in some fraction to the pathogenesis of HCC, these biomarkers could be used in the early detection of disease. Thus, a combined use of exposure and genetic biomarkers might reveal the subset of high-risk people within a population who would most benefit from targeted mechanism-based interventions.
Acknowledgments
Financial support for this work was provided by the National Institute of Environmental Health Sciences through Grants P01 ES06052 and P30 ES03819.
Abbreviations: HCC, hepatocellular carcinoma; P.R.C., People's Republic of China; HBV, hepatitis B virus; HCV, hepatitis C virus; AFB1, aflatoxin B1; SOMA, short oligonucleotide mass analysis; HBsAg, high levels of the S gene product.
References
- 1.Wang, X. W., Hussain, P., Huo, T.-I., Wu, C.-G., Forgues, M., Hofseth, L. J., Brechot, C. & Harris C. C. (2002) Toxicology 181-182, 43-47. [DOI] [PubMed] [Google Scholar]
- 2.Kew, M. C. (2002) Toxicolology 181-182, 35-38. [Google Scholar]
- 3.Qian, G. S., Ross, R. K., Yu, M. C., Yuan, J. M., Gao, Y. T., Henderson, B. E., Wogan, G. N. & Groopman, J. D. (1994) Cancer Epidemiol. Biomarkers Prev. 3, 3-10. [PubMed] [Google Scholar]
- 4.Kensler, T. W., Qian, G.-S., Chen, J.-G. & Groopman, J. D. (2003) Nat. Cancer Rev. 3, 321-329. [DOI] [PubMed] [Google Scholar]
- 5.Arbuthnot, P. & Kew, M. (2001) Int. J. Exp. Pathol. 82, 77-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee, S. M. (1997) N. Engl. J. Med. 337, 1733-1745. [DOI] [PubMed] [Google Scholar]
- 7.Block, T. M., Mehta, A. S., Fimmel, C. J. & Jordan, R. (2003) Oncogene 22, 5093-5107. [DOI] [PubMed] [Google Scholar]
- 8.Locarnini, S., McMillan, J. & Bartholomeusz, A. (2003) Semin. Liver Dis. 23, 5-20. [DOI] [PubMed] [Google Scholar]
- 9.Okuda, K. (2002) J. Gastroenterol. Hepatol. 14, 401-405. [DOI] [PubMed] [Google Scholar]
- 10.Hou, J., Lau, G. K., Cheng, J., Cheng, C. C., Luo, K. & Carman, W. F. (1999) Liver 19, 411-417. [DOI] [PubMed] [Google Scholar]
- 11.Baptista, M., Kramvis, A. & Kew, M. C. (1999) Hepatology 29, 946-953. [DOI] [PubMed] [Google Scholar]
- 12.Hsia, C. C., Yuwan, H. & Tabor, E. (1996) Lancet 348, 625-626. [DOI] [PubMed] [Google Scholar]
- 13.Zhang, F., Zhu, Y. & Sun, Z. (1998) Chin. Oncol. 20, 18-21. [PubMed] [Google Scholar]
- 14.Nawroz, H., Koch, W., Anker, P., Stroun, M. & Sidransky, D. (1996) Nat. Med. 2, 1035-1037. [DOI] [PubMed] [Google Scholar]
- 15.Yamada, T., Nakamori, S., Ohzato, H., Oshima, S., Aoki, T., Higaki, N., Sugimoto, K., Akagi, K., Fujiwara, Y., Nishisho, I., et al. (1998) Clin. Cancer Res. 4, 1527-1532. [PubMed] [Google Scholar]
- 16.Wong, I. H., Lo, Y. M., Zhang, J., Liew, C. T., Ng, M. H., Wong, N., Lai, P. B., Lau, W. Y., Hjelm, N. M. & Johnson, P. J. (1999) Cancer Res. 59, 71-73. [PubMed] [Google Scholar]
- 17.Jackson, P. E., Qian, G. S., Friesen, M. D., Zhu, Y. R., Lu, P., Wang, J. B., Wu, Y., Kensler, T. W., Vogelstein, B. & Groopman, J. D. (2001) Cancer Res. 61, 33-35. [PubMed] [Google Scholar]
- 18.Anker, P., Mulcahy, H., Chen, X. Q. & Stroun, M. (1999) Cancer Metastasis Rev. 18, 65-73. [DOI] [PubMed] [Google Scholar]
- 19.Jackson, P. E., Kuang, S.-Y., Wang, J.-B., Strickland, P. T., Munoz, A., Kensler, T. W., Qian, G.-S. & Groopman, J. D. (2003) Carcinogenesis 24, 1-7. [DOI] [PubMed] [Google Scholar]
- 20.Wang, J.-S., Qian, G.-S., Zarba, A., He, X., Zhu, Y.-R., Zhang, B.-C., Jacobson, L., Gange, S. J., Muñoz, A., Kensler, T. W., et al. (1996) Cancer Epidemiol. Biomarkers Prev. 5, 253-261. [PubMed] [Google Scholar]
- 21.Laken, S. J., Jackson, P. E., Kinzler, K. W., Vogelstein., B., Strickland, P. T., Groopman, J. D. & Friesen, M. D. (1998) Nat. Biotechnol. 16, 1352-1356. [DOI] [PubMed] [Google Scholar]
- 22.Ahdieh, L., Muñoz, A., Vlahov, D., Trimble, C. L., Timpson, L. A. & Shah, K. (2000) Am. J. Epidemiol. 151, 1148-1157. [DOI] [PubMed] [Google Scholar]
- 23.Gange, S. J., Muñoz, A., Saez, M. & Alonso, J. (1996) Appl. Stat. 45, 371-382. [Google Scholar]
- 24.Thorgeirsson, S. S. & Grisham, J. W. (2002) Nat. Genet. 31, 339-346. [DOI] [PubMed] [Google Scholar]
- 25.Ding, X., Park, Y. N., Taltavull, T. C., Thung, S. N., Jin, X., Jin, Y., Trung, N. S., Edamoto, Y., Sata, T. & Abe, K. (2003) Jpn. J. Infect. Dis. 56, 12-18. [PubMed] [Google Scholar]
- 26.Broderick, A. L. & Jonas, M. M. (2003) Semin. Liver Dis. 23, 59-68. [DOI] [PubMed] [Google Scholar]
- 27.Merican, I., Guan, R., Amarapapuka, D., Alexander, M. J., Chutaputti, A., Chien, R. N., Hasnian, S. S., Leung, N., Lesmana, L., Phiert, P. H., et al. (2000) J. Gastroenerol. Hepatol. 15, 1356-1361. [DOI] [PubMed] [Google Scholar]
- 28.Ross, R. K., Yuan, J.-M., Yu, M., Wogan, G. N., Qian, G.-S., Tu, J.-T., Groopman, J. D., Gao, Y.-T. & Henderson, B. E. (1992) Lancet 339, 943-946. [DOI] [PubMed] [Google Scholar]
- 29.Wang, L. Y., Hatch, M., Chen, C. J., Levin, B., You, S. L., Lu, S. N., Wu, M. H., Wu, W. P., Wang, L. W., Wang, Q., et al. (1996) Int. J. Cancer 67, 620-625. [DOI] [PubMed] [Google Scholar]
- 30.Fattovich, G. (2003) Sem. Liver Dis., 23, 47-58. [DOI] [PubMed] [Google Scholar]
- 31.Francois, G., Kew, M., Damme, P. V., Mphahlele, M. J. & Meheus, A. (2001) Vaccine 19, 3799-3815. [DOI] [PubMed] [Google Scholar]
- 32.Hannoun, C., Horal, P. & Lindh, M. (2000) J. Gen. Virol. 81, 75-83. [DOI] [PubMed] [Google Scholar]
- 33.Hadziyannis, S. J. & Vassilopoulos, D. (2001) Hepatology 34, 617-624. [DOI] [PubMed] [Google Scholar]
- 34.Yotsuyanagi, H., Hino, K., Tomita, E., Toyoda, J., Yasuda, K. & Iino, S. (2002) J. Hepatol. 37, 355-363. [DOI] [PubMed] [Google Scholar]
- 35.Parekh, S., Zoulim, F., Ahn, S. H., Tsai, A., Li, J., Kawai, S., Khan, N., Trepo, C., Wangs, J. & Tong, S. (2003) J. Virol. 77, 6601-6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, J., Buckwold, V. E., Hon, M.-W. & Ou, J.-H. (1999) J. Virol. 73, 1239-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arbuthnot, P., Capovilla, A. & Kew, M. (2000) J. Gastroenterol. Hepatol. 15, 357-368. [DOI] [PubMed] [Google Scholar]
- 38.Lindh, M., Hannoun, C., Dhillon, A. P., Norkans, G. & Horal, P. (1999) J. Infect. Dis. 179, 775-782. [DOI] [PubMed] [Google Scholar]
- 39.Yuen, M.-F., Sablon, E., Yuan, H.-J., Wong, D. K.-H., Hui, C.-K., Wong, B. C.-Y., Chan, A. O.-O. & Lai, C. L. (2003) Hepatology 37, 562-567. [DOI] [PubMed] [Google Scholar]
- 40.Cho, S. W., Shin, Y. J., Hahm, K. B., Jin, J. H., Kim, Y. S., Kim, J. H. & Kim, H. J. (1999) J. Korean Med. Sci. 14, 424-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seeger, C. & Mason, W. S. (2000) Microbiol. Mol. Biol. Rev. 64, 51-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong, I. H., Lo, Y. M. & Johnson, P. J. (2001) Ann. N.Y. Acad. Sci. 945, 36-50. [PubMed] [Google Scholar]
- 43.Johnson, P. J. (1999) J. Gastroenterol. Hepatol. 14, S32—S36. [DOI] [PubMed] [Google Scholar]
- 44.Johnson, P. J. (2001) Clin. Liver Dis. 5, 145-159. [DOI] [PubMed] [Google Scholar]
- 45.Wong, I. H., Lo, Y. M., Lai, P. B. & Johnson, P. J. (2000) Clin. Chem. 46, 1420-1422. [PubMed] [Google Scholar]
- 46.Zhong, S., Tang, M. W., Yeo, W., Liu, C., Lo, Y. M. & Johnson, P. J. (2002) Clin. Cancer Res. 8, 1087-1092. [PubMed] [Google Scholar]
- 47.Cooke, M. S., Evans, M. D., Dizdaroglu, M. & Lunec, J. (2003) FASEB J. 17, 1195-1214. [DOI] [PubMed] [Google Scholar]
- 48.Aguilar, F., Hussain, S. P. & Cerutti, P. (1993) Proc. Natl. Acad. Sci. USA 90, 8586-8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smela, M. E., Currier, S. S., Bailey, E. A. & Essigmann, J. M. (2001) Carcinogenesis 22, 535-545. [DOI] [PubMed] [Google Scholar]
- 50.Smela, M. E., Hamm, M. L., Henderson, P. T., Harris, C. M., Harris, T. M. & Essigmann, J. M. (2002) Proc. Natl. Acad. Sci. USA 99, 6655-6660. [DOI] [PMC free article] [PubMed] [Google Scholar]