Abstract
Transcriptional enhancers are a primary mechanism by which tissue-specific gene expression is achieved. Despite the importance of these regulatory elements in development, responses to environmental stresses, and disease, testing enhancer activity in animals remains tedious, with a minority of enhancers having been characterized. Here, we have developed ‘enhancer-FACS-Seq’ (eFS) technology for highly parallel identification of active, tissue-specific enhancers in Drosophila embryos. Analysis of enhancers identified by eFS to be active in mesodermal tissues revealed enriched DNA binding site motifs of known and putative, novel mesodermal transcription factors (TFs). Naïve Bayes classifiers using TF binding site motifs accurately predicted mesodermal enhancer activity. Application of eFS to other cell types and organisms should accelerate the cataloging of enhancers and understanding how transcriptional regulation is encoded within them.
In metazoans, gene expression is regulated in a tissue-specific manner predominantly via noncoding genomic regions referred to as cis regulatory modules (CRMs) that regulate the expression of typically the nearby gene(s)1. CRMs contain one or more DNA binding sites for one or more sequence-specific transcription factors (TFs) that activate or repress gene expression. CRMs that activate gene expression are frequently referred to as transcriptional enhancers2.
The fruit fly Drosophila melanogaster has served as a powerful model organism for studies of transcriptional enhancers2. It has been estimated that there are ~50,000 enhancers in the D. melanogaster genome3, yet to date the tissue-specific activities of only ~1,800 are known4. Technology for identifying enhancers active in particular cell types would aid in defining functional cis regulatory elements and would facilitate computational identification of sequence features important for cell-type-specific enhancer activity. Currently, TF-occupied regions identified by chromatin immunoprecipitation (ChIP) are tested by low-throughput, traditional reporter assays5,6. Automated image analysis of reporter assays in embryos3,7 requires vast infrastructure and resources. Although highly parallel reporter assays have been developed recently8–13, none directly identify enhancer activity in a genomic context (i.e., integrated into the genome) in particular cell types of interest in a whole animal.
We have developed a new technology, termed ‘enhancer-FACS-Seq’ (eFS), for highly parallel identification of active, tissue-specific transcriptional enhancers in whole Drosophila embryos (Fig. 1a; Supplementary Fig. 1). As with traditional enhancer assays, each candidate CRM (cCRM) is cloned upstream of a reporter gene. Our key innovation is the replacement of microscopy to screen for tissue-specific enhancers with fluorescence activated cell sorting (FACS) of dissociated cells. In each fly, one marker (here, rat CD2 cell surface protein14) is used to label cells of a specific tissue for sorting by FACS, and the other marker (here, green fluorescent protein (GFP)) is used as a reporter of cCRM activity. Cells are sorted by tissue type and then by GFP fluorescence, allowing screening of hundreds of cCRMs in a time- and cost-efficient manner.
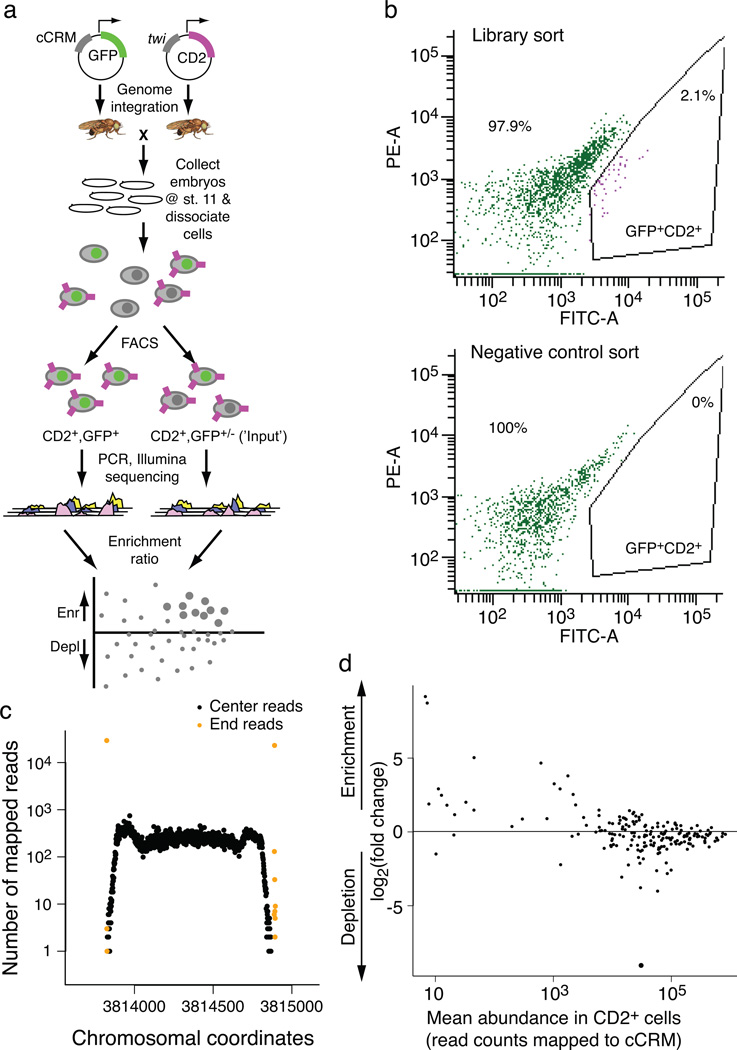
Fig. 1. Enhancer-FACS-Seq (eFS) methodology.
(a) Overall design of enhancer-FACS-Seq (eFS). (b) FACS purification of GFP+CD2+ cells prepared from embryos resulting from a cross of Mef2-I-ED5:CD2 females to (upper panel) cCRM library transgenic males, and (lower panel) wild type (GFP-negative) males. In each panel, the plot shows yellow (“PE-A”) versus green (“FITC-A”) fluorescence for cells that pass the CD2+ gate out of 106 cells prepared from embryos. (c) Representative example of a cCRM, surrounding by native genomic flanking sequence, detected by eFS. (d) Enrichment ratios for cCRMs in twi:CD2− cells, as compared to twi:CD2+ cells. Large points: significantly enriched (P_adj < 0.1), small points: P_adj > 0.1.
RESULTS
Library of candidate cis regulatory modules (cCRMs)
We focused on embryonic mesoderm as our model system because it comprises a variety of cell types, the major regulatory factors governing mesoderm development are conserved between vertebrates and Drosophila15, and numerous data sets are available for genomic features associated with active enhancers. We created a plasmid library of hundreds of reporter constructs for ~1-kb cCRMs (see Online Methods; Supplementary Note 1; Supplementary Table 1) located next to mesodermally expressed genes and comprising: ChIP-CRMs6 bound by at least one of the somatic mesoderm TFs Twist (Twi), Tinman (Tin), or Myocyte enhancing factor 2 (Mef2); regions bound by the transcriptional coactivator CREB binding protein (CBP)16,17; regions containing DNase I hypersensitive sites (DHS)18; dense clusters of evolutionarily conserved motif occurrences for mesodermal TFs19; and additional regions surrounding mesodermal genes (see Supplementary Note 2).
Enhancer-FACS-Seq (eFS) experiments
Our cCRM plasmid library was injected into two batches of embryos. In the first batch, we injected ~3,500 embryos, and crossed transformant males to females from two different CD2 lines to identify enhancers active in distinct tissues: twi:CD2 for whole mesoderm, and Mef2-I-ED5:CD2 for a subset20 of largely fusion-competent myoblasts (FCMs). In the second batch, ~4,500 embryos were injected, from which transformant males were crossed to duf:CD2 females to identify activity in somatic mesoderm founder cells (FCs)21. Each resulting embryo has one GFP reporter under the control of one cCRM integrated at the same genomic site by the phiC31 integrase22. Use of a site-specific integrase avoids artifacts that would result if more than one cCRM were present in a cell and also avoids potential position effects on enhancer activity.
At developmental stages 11–12, embryos were dissociated and purified by FACS. From the twi:CD2 embryos, we collected ~315,000 GFP+CD2+ cells, ~198,000 GFP+CD2− cells and 1×106 mock-sorted cells (i.e., ‘input’) (see Online Methods; Fig. 1b; Supplementary Fig. 2; Supplementary Table 2). We collected fewer GFP+CD2+ cells from the Mef2-I-ED5:CD2 and duf:CD2 embryos (Supplementary Table 2) since the Mef2-I-ED5 enhancer is active in approximately 50-fold fewer cells than the twi enhancer, which is active in one-quarter to one-third of all cells at this stage, while the duf enhancer is active in the vast majority of the 660 FCs per embryo, nearly an order of magnitude fewer cells than for the Mef2-I-ED5 enhancer.
We extracted genomic DNA from the collected cells, amplified the cCRMs by PCR, and sequenced the resulting amplicons on the Illumina platform (see Online Methods). We mapped the sequencing reads (Fig. 1c; Supplementary Table 3) to the D. melanogaster genome using segemehl software23 (Supplementary Fig. 3). 213 and 400 cCRMs were detected (false discovery rate (FDR) < 5×10−5; see Online Methods) as having integrated into the fly genome from the first and second batches of injections, respectively. The greater number of cCRMs detected from the second batch was likely due to collection of transformant progeny from a larger number of injected embryos.
To evaluate the enhancer activity of the detected cCRMs, we calculated each cCRM’s enrichment in a particular cell population as compared to the corresponding ‘input’ sample (Fig. 1a) using DESeq software24. The input sample provides information on the baseline read counts due to cCRM representation within the embryo populations. In control experiments CD2+ and CD2− cells exhibited no significant differences in their cCRM content (Fig. 1d). Therefore, CD2+ cells were used as input sample for twi:CD2+GFP+, while for the rarer FCM and FC cell types CD2− cells were used as input (Supplementary Fig. 2).
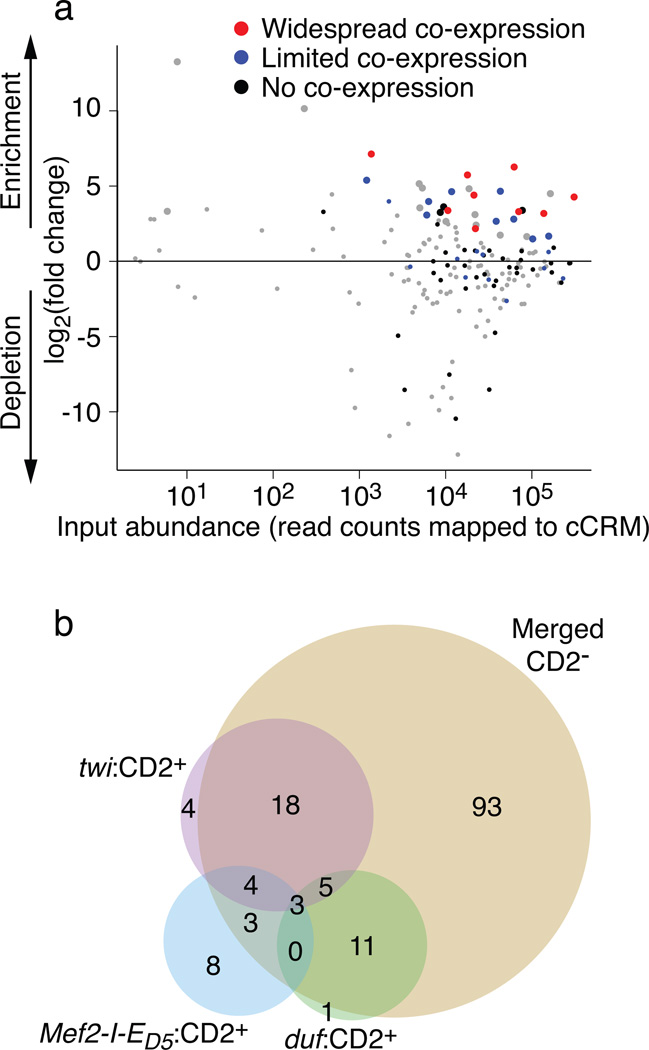
In total, 150 of the detected cCRMs were identified by eFS as being active enhancers (adjusted P-value (P_adj) < 0.1) in at least one cell population. Of these, 57 were active mesodermal enhancers: 34 in whole mesoderm (Fig. 2a), 18 in FCMs (Supplementary Fig. 4a), and 20 in FCs (Supplementary Fig. 4b). 12 of these 57 active mesodermal cCRMs overlap by at least 100 bp with a known mesodermal enhancer at an overlapping developmental timepoint in the REDfly database25 (Supplementary Table 4), while the remaining 45 represent putative novel mesodermal enhancers, including 16 in FCMs and 14 in FCs. Analysis of GFP+CD2− cells collected from twi:CD2, Mef2-I-ED5:CD2, and duf:CD2 embryos revealed 93 putative non-mesodermal enhancers (Fig. 2b; Supplementary Table 4). A recent study screened a genomic DNA library for enhancer activity in the S2 cell line and in cultured ovarian somatic cells13; only 13 of the 57 mesodermal enhancers and 11 of the 93 non-mesodermal enhancers identified by eFS overlap by at least 100 bp with enhancers found in that study. Indeed, this comparison highlights the value of eFS for identifying enhancers active in particular cell types of interest within whole embryos.
Fig. 2. Active enhancers (P_adj < 0.1) identified from eFS data.
(a) Enrichment ratios for cCRMs in twi:CD2+GFP+ cells, as compared to twi:CD2 input cells. Large points: significantly enriched (P_adj < 0.1), small points: P_adj > 0.1. Results from traditional reporter assays revealed cCRMs whose GFP expression shows widespread (red), limited (blue), or no (black) co-expression with twi:CD2 expression. (b) Venn diagram of active enhancers (P_adj < 0.1) identified from different cell populations: twi:CD2+; Mef2-I-ED5:CD2+; duf:CD2+; nonredundant union of twi:CD2−, Mef2-I-ED5:CD2−, and duf:CD2− (“Merged CD2−”).
Validation of eFS results
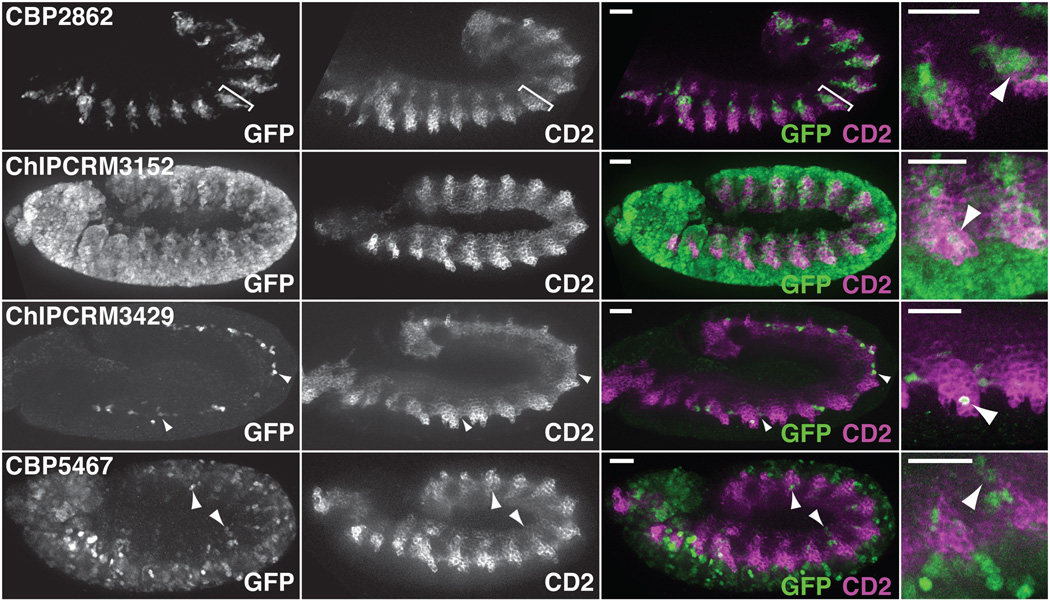
To validate our eFS results, we performed traditional reporter assays in whole Drosophila embryos (see Online Methods). For the twi:CD2+ eFS data, we tested 69 of the cCRMs, including: 21 putative active mesodermal enhancers (P_adj < 0.1) and 48 putative inactive cCRMs (P_adj > 0.1). The specificity of eFS was excellent among significantly enriched cCRMs: 18 of the 21 tested putative mesodermal enhancers drove expression in mesoderm at stage 11–12 (Fig. 3; Supplementary Fig. 5). eFS exhibited moderate sensitivity for significantly enriched enhancers that were active in relatively few mesodermal cells: 9 gave expression patterns that were manually assessed as ‘widespread co-expression’ (i.e., expression in a majority of strongly twi:CD2+ cells), while the other 9 drove ‘limited co-expression’ in smaller subsets of twi:CD2+ cells. 12 of the 48 putative inactive cCRMs drove ‘limited co-expression’ (Supplementary Fig. 5; Supplementary Table 4). Some of these eFS false negatives drove expression in cells that express low levels of CD2 and might have been missed by our relatively stringent FACS gate for collecting twi:CD2+ cells. In most cases, the observed expression domain was linked to an adjacent gene’s expression (Supplementary Table 5). Although the data are slightly noisier for FCM and FC enhancers (6 out of 9 tested putative FCM enhancers, and 9 out of 11 tested putative FC enhancers, drove mesodermal expression; Supplementary Fig. 5), likely because roughly 20-fold fewer CD2+GFP+ cells were collected from the more specific Mef2-I-ED5:CD2 and duf:CD2 lines, the results nevertheless demonstrate that eFS can successfully identify enhancers active in rarer cell types. In addition, the majority (35 of 47 tested) of cCRMs identified by eFS as active in any of the three CD2−GFP+ cell collections were indeed active at this developmental stage (Supplementary Table 6).
Fig. 3. Validations of enhancers identified as active (P_adj < 0.1) by eFS.
Sample validations of eFS-identified enhancer activity. Constructs scored as driving “widespread co-expression” drove GFP specifically in a large fraction of the mesoderm (e.g., somatic mesoderm, bracketed, in CBP2862) or in mesodermal plus non-mesodermal cells (ChIPCRM3152). “Limited co-expression” generally described expression in isolated mesodermal cells (arrowheads, in ChIPCRM3429 and CBP5467) or in a specific mesodermal structure (Supplementary Fig. 5). Co-expression is observed as green and purple in the same cells, since the GFP in these embryos is nuclear, while CD2 is expressed on the cell surface. Assessment of co-expression was performed with the annotator being blind to the predicted activity of the cCRMs. Scale bars = 50 µm.
Comparisons of eFS data to other genomic data types
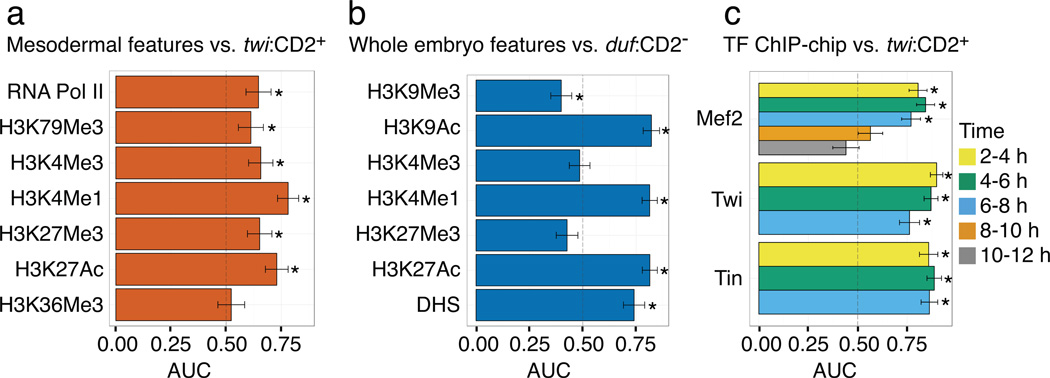
We examined the eFS-identified enhancers for enrichment of known enhancer-associated chromatin marks. Comparison to data from batch isolation of tissue-specific chromatin for immunoprecipitation (BiTS-ChIP) for mesodermal cells from stage 10–11 embryos26 showed that acetylation of histone H3 on lysine 27 (H3K27ac), monomethylation of histone H3 on lysine 4 (H3K4me1), H3K4 trimethylation (H3K4me3), H3K79me3 and RNA Pol II26–29, are enriched (area under receiver operating characteristic curve (AUC) ≥ 0.6, P < 0.05 by Wilcoxon-Mann-Whitney U-test) among enhancers found to be active in mesoderm by eFS (Fig. 4a). However, in contrast to a prior report that H3K27 trimethylation (H3K27me3) was depleted among active mesodermal enhancers26, we found H3K27me3 to be enriched among mesodermal enhancers. We also observed enrichment of H3K27Ac, H3K4me1 and H3K9Ac in comparisons of modENCODE data for 4–8 hr whole embryos17 to active enhancers identified by eFS in duf:CD2− cells, which approximate whole embryo samples (Fig. 4b, Supplementary Note 3). While H3K9Ac is known as a mark of active transcription start sites30, our observed enrichment of H3K9Ac among active enhancers supports the observation of H3K9Ac in the ‘strong enhancer’ chromatin state in human cells31.
Fig. 4. Enrichment of various genomic marks among eFS-identified enhancers.
Enrichment of various genomic features (e.g., DHS, histone modifications, TF ChIP-binding) associated with active enhancers in (a, c) whole mesoderm (twi:CD2+) or (b) approximately whole embryos (duf:CD2−). AUC: area under receiver operator characteristic curve. * indicates P < 0.05 by Wilcoxon-Mann-Whitney U-test. Error bars indicate 1 standard deviation.
Our enhancer data allowed us to investigate which genomic data types6,16–18 provide the greatest utility in identifying enhancers. Occupancy by sequence-specific TFs (Twi, Tin, Mef2, Bagpipe (Bap), Biniou (Bin)) expressed specifically in the mesoderm was most enriched among active mesodermal enhancers (Fig. 4c; Supplementary Fig. 6). DHSs18 were nearly as enriched as enhancer-associated histone modifications (Fig. 4b, Supplementary Fig. 6).
Enrichment of transcription factor binding site motifs
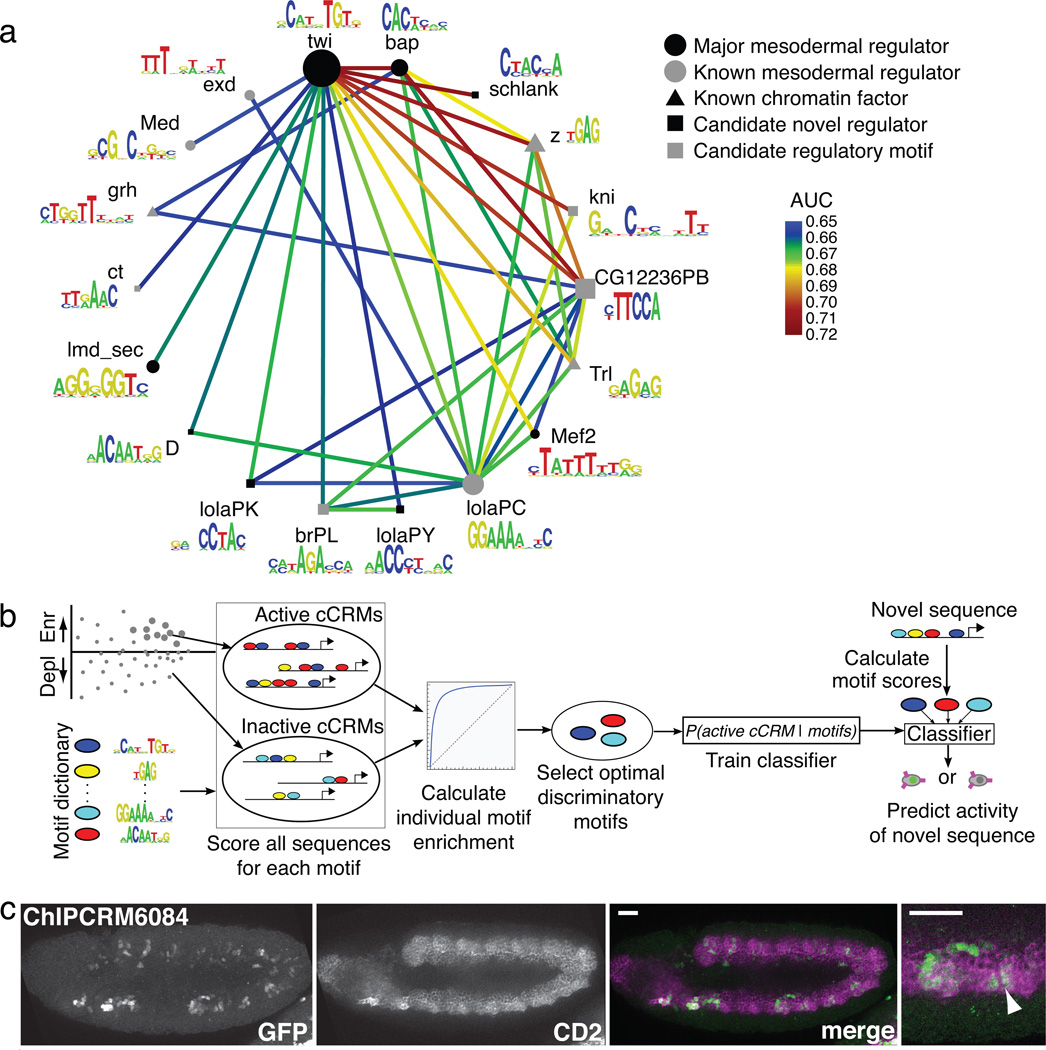
We separately analyzed each of the three sets of eFS-identified mesodermal enhancers (i.e., whole mesoderm, FCMs, or FCs) for over-represented motifs and pair-wise motif combinations that might be required for enhancer activity. We used the PhylCRM and Lever algorithms19 to determine enrichment of matches, scored according to their evolutionary conservation, to 567 publicly available Drosophila TF binding site motifs6,32–35 (see Online Methods). Numerous motifs were significantly enriched (AUC ≥ 0.65, FDR ≤ 0.1) either individually or in pair-wise combination (Fig. 5a; Supplementary Fig. 7, 8a; Supplementary Table 7) for the whole-mesoderm and FCM enhancers.
Fig. 5. Computational motif analysis of eFS-identified active enhancers.
(a) TF binding site motifs or motif combinations significantly enriched (AUC ≥ 0.65, FDR ≤ 0.1) among eFS-identified active enhancers in twi:CD2+ cells. Nodes represent motifs for sequence-specific DNA-binding proteins that target chromatin-modifying PcG and trxG complexes to DNA (triangles), major mesodermal regulators (black circles), other factors known to have a role in mesodermal gene expression (gray circles), putative novel regulators (black squares); putative regulatory motifs for which the representative factors shown are not expressed in the embryonic mesoderm at the appropriate time (gray squares) and may be recognized by other trans-acting factors. Edges represent significant pair-wise AND combinations. Node diameter is proportional to (AUC-0.5)2 considering the Lever AUC for the individual motif. (b) Schema of classifier analysis. (c) Maximum Intensity Projection of GFP expression driven by ChIPCRM6084, correctly predicted to drive co-expression with twi:CD2. Co-expression is observed and was assessed as described in Fig. 3. Scale bars = 50 µm.
For each of these two sets of enhancers, we observed strong enrichment of the primary, known master regulator of that cell population: Twi for whole mesoderm36, and Lmd for FCMs20,37. Motifs for other known mesodermal regulators were found in enriched combinations, including Bap, Lola-PC, and Mef2 in whole mesoderm, and Twi and Mef2 in FCMs. We also saw strong enrichment of motifs for sequence-specific DNA-binding proteins – z, grh, and Trl (also known as GAGA Factor (GAF)) – that participate in recruitment of chromatin-modifying PcG and trxG proteins38, supporting prior findings of the enrichment of the z and/or Trl motifs among regions bound by Mef2, Twi, or Tin in ChIP-chip39. For the eFS-identified FC enhancers, no individual motifs or combinations thereof met our statistical significance criteria, although a few combinations for known and candidate mesodermal regulators narrowly missed our thresholds (Supplementary Table 7).
FCM enhancers show enrichment for a variety of motifs (e.g., Twi and Trl) in combination with a Lmd motif, supporting the previously observed enrichment of these motifs in Lmd ChIP-Seq peaks35. We also observed numerous significantly enriched motif combinations (e.g., many involving the uncharacterized zinc finger protein CG7928) not found in the Lmd ChIP-Seq study35. Since eFS data are not constrained by occupancy by a particular TF, they allow for more unbiased identification of regulatory motifs. We also observed enrichment of numerous motif combinations comprising a master regulator and a factor with either ubiquitous or mesoderm-specific expression at the appropriate stage but no known role in mesoderm development (e.g., schlank, Lola-PK), suggesting novel regulators of mesodermal expression (square nodes in Fig. 5a and Supplementary Fig. 7).
Machine learning classifier to predict mesodermal enhancer activity
We developed a machine learning approach to model whether cCRMs will be active or inactive in mesoderm or specifically in FCMs. We selected the mesodermal TF binding site motifs6,32, independently in 10-fold cross-validation, that were most discriminatory in distinguishing active versus inactive cCRMs (see Online Methods). We then trained a Naïve Bayes classifier40 (Fig. 5b) based on the number and quality of matches to the discriminatory motifs, independently for whole mesoderm, FCMs, and FCs.
We assessed the accuracy of our models by 10-fold cross-validation. The whole mesoderm model achieved an AUC of 0.74 (P = 3.9 × 10−4, Wilcoxon-Mann-Whitney test) using 12 discriminatory motifs, while the FCM-specific model achieved an AUC of 0.93 (P = 1.2 × 10−6, Wilcoxon-Mann-Whitney test) using 3 motifs. Importantly, these models outperformed ones based solely on previously known cis regulatory motifs for mesoderm and FCMs (AUC of 0.59 and 0.72, respectively; see Supplementary Note 2). No statistically significant classifier was found for FCs.
To further demonstrate the practical utility of our models, we tested whether they could predict the activity of cCRMs whose activity had not been measured by eFS. We tested 39 classifier predictions by traditional reporter assays. Six out of 10 cCRMs predicted to be active enhancers in mesoderm drove co-expression of GFP with CD2 (Fig. 5c; Supplementary Fig. 9); 19 out of 29 cCRMs predicted to be inactive drove no expression in CD2+ cells, while 9 of the 10 remaining predicted negative cCRMs drove limited co-expression at stages 11–12 (Supplementary Fig. 9). Consistent with many of the twi:CD2+ eFS-positive enhancers in the training set exhibiting ‘widespread co-expression’ with CD2 and fewer exhibiting ‘limited co-expression’, our classifier appears to perform better in predicting the activity of cCRMs with ‘widespread co-expression’.
DISCUSSION
Our results demonstrate the utility of eFS for highly parallel testing of cCRMs for tissue-specific enhancer activity. No single data type (sequence-specific TF binding, histone modifications, or DHS) was most enriched across all three tissues (Supplementary Fig. 6). Moreover, none of the different classes of genomic features that we used to prioritize cCRMs for testing by eFS (i.e., ChIP-CRMs, CBP-bound regions, DHS) was significantly enriched (p < 0.1) among active cCRMs considering each of the three mesodermal cell populations or their nonredundant union (Supplementary Table 8). It is perhaps not surprising that these regions were not enriched in either the Mef2-I-ED5:CD2+GFP+ or duf:CD2+GFP+ data, since FCMs and FCs are relatively rare cell types and also since many of the putative regulatory regions might drive expression in other cell types as the adjacent genes are often expressed in additional cell types or time points.
Among enhancers found in whole mesoderm, we observed the greatest enrichment for regions bound by Tin at 2–4 hours (i.e., stages 5–7), suggesting that Tin might be a pioneer factor41 that pre-marks mesodermal enhancers active later in development. Indeed, these same Tin-bound enhancers show enhanced Tin binding at 4–6 hrs (i.e., stages 8–9) (data not shown), and are consistent with tin being essential for specification of ventral FCs42 and also with tin activity and putative Tin binding sites being required for the activity in ventral muscle progenitors of an enhancer that does not become expressed until after Tin protein expression has become restricted to the dorsal mesoderm43. Our observed enrichment of Mef2, Twi, and Tin occupancy at 4–6 hrs or 6–8 hrs (i.e., stages 10–11) among enhancers identified from Mef2-I-ED5:CD2+ cells supports the role of Mef2, Twi, and Tin in regulating FCM genes coordinately with Lameduck (Lmd)35.
Future studies will be needed to determine the regulatory functions of the putative mesodermal TFs suggested by the motif analysis results for eFS-identified enhancers in whole mesoderm and FCMs. The enrichment of binding sites for PcG and trxG recruitment factors, and combinations thereof with ubiquitously expressed and mesoderm-specific TFs, suggests that regulatory competence of enhancers requires binding sites of chromatin factors together with those of tissue-specific TFs.
Our classifier analysis results indicate that cis regulation in FCMs is specified by a smaller set of TFs than those used in regulation of a broader class of mesodermal genes expressed in a wider range of cell types, each of which might utilize different cis regulatory codes6,44 (Supplementary Fig. 8b,c). Likewise, the lack of a statistically significant classifier for FCs is likely due to heterogeneity of FCs and their associated enhancers33,44; eFS using CD2 driver lines specific to subsets or even unique FCs should aid in the elucidation of FC-specific cis regulatory codes. Our results on enrichment of various histone modifications (see Supplementary Note 3) are consistent with the model that there exist different classes of active enhancers that show enrichment for different sets of histone modifications26.
Here, we applied the eFS technology to the discovery of muscle enhancers. However, eFS can be used to test cCRMs in any cell type that has at least one known enhancer, by constructing CD2 driver lines using enhancers active in those cell types. Importantly, eFS can be used to screen cCRMs without any prior functional evidence (e.g., ChIP data). Moreover, eFS can be adapted for use in other organisms; the phiC31 integrase system has been employed successfully in other species, including zebrafish45, human and mouse cells46, and mice47. In addition, eFS could be implemented using a different site-specific recombinase or other transformation method. Broader application of eFS should greatly expedite and expand the repertoire of well-defined enhancers and facilitate the development of a more comprehensive picture of their landscape and organization of CRMs across genomes.
ONLINE METHODS
PCR amplification of candidate CRMs (cCRMs)
The composition of our cCRM library is detailed in Supplementary Note 2. All cCRMs were chosen to be 900–1,100 bp long to avoid potential PCR bias. A two-step PCR amplification was used to include Gateway attB sites, and specific forward and reverse sequencing primers with Phusion enzyme (New England Biolabs) using D. melanogaster OreR genomic DNA as template, followed by amplification with common PCR primers (SEQ1, SEQ2), as described in Supplementary Note 2.
Design of reporter vector pEFS-Dest
We created the vector for enhancer-FACS-Seq, pEFS-Dest (Supplementary Note 1), by blunt-end cloning the 1.8 kb HindIII-SpeI fragment of pPelican48 (containing a nuclear-localized GFP reporter construct with a gypsy insulator element upstream of the MCS and minimal promoter) into pWattB, then replacing the Multiple Cloning Site with a cassette providing attR1 and attR2 sites for Gateway cloning. pWattB was made by inserting (1) the ϕC31 attB site from Streptomyces lividans22 and (2) the mini-white gene into the small cloning vector pSP73 (Promega). The reporter cassette comprises the Hsp70 minimal promoter driving expression of a nuclear localization signal-tagged EGFP gene with an SV40 polyadenylation sequence48.
Purification, normalization and cloning of cCRM library into eFS reporter vector
Aliquots of all PCR reactions were run on agarose gels with High DNA Mass Ladder (Invitrogen) and quantified using Quantity One software (BioRad). Equal masses of each 900–1,100 bp band were then pooled, precipitated, and gel-purified, then cloned as a pool using Gateway BP Clonase II (Invitrogen) into pDONR221 (Invitrogen). Cloning reactions were transformed into E. coli Top10 cells (Invitrogen) and plated on LB+kanamycin agar. A plasmid pool was purified from the resulting colonies, from which the combined inserts were cloned using Gateway LR Clonase II (Invitrogen) into pEFS-Dest. Transformed cells were plated on LB+ampicillin agar, yielding colonies from which the final library plasmid pool was prepared for embryo injection.
Generation of CD2 vector pETWCD2
A minimal promoter was fused to rat CD2 and subsequently cloned into P-element transformation vectors by PCR-amplifying the TATA box from pUAST-NTAP, and CD2 from twi-CD214. These PCR products served as templates for an assembly PCR reaction, the product of which was subcloned into pCR (Invitrogen), sequence-verified, digested with NheI and cloned into XbaI-digested pETWN49, resulting in our CD2 vector pETWCD2. Primer sequences are provided in Supplementary Note 2.
Fly embryo injections and husbandry
The pooled plasmid cCRM library was injected posteriorly into syncytial embryos carrying the nos-ϕC31\int.NLS transgene50 on the X chromosome and the attP40 insertion51 on the 2nd chromosome. Surviving males were crossed to excess y w virgin females. Transformant male progeny were selected by eye color. We collected several thousand transformant males and, separately, several thousand virgin females from each tissue-specific CD2 line of interest (see main text). These flies were combined in population cages ~36 hours before the beginning of embryo collections. Population cages were collected from twice "pre-lays" to minimize the presence of older embryos due to retention of fertilized eggs by females, then two collections of 2 hours (for twi:CD2 sorting) or 2.5 hours (for Mef2-I-ED5:CD2 and duf:CD2 sorting) were performed. These plates were aged 10–11 hours at 18°C, after which embryos were collected and dechorionated, and single cell suspensions were prepared for FACS.
Fluorescence activated cell sorting (FACS)
We modified a previous protocol for isolation of single cells for FACS from live Drosophila embryos at stage 1152 by incorporating a step in which dissociated cells are resuspended in Drosophila cell culture medium and incubated on ice with Alexa647-conjugated anti(rat CD2) antibody (AbD-Serotec, cat. #MCA154A647), as described in Supplementary Note 2. After collection of cells by centrifugation, samples were filtered with Nytex mesh and supplemented with DAPI. Cells were washed, and then analyzed and separated by FACS (see Supplementary Note 2).
cCRM insert amplifications from collected cells
Crude cell extracts were pooled according to sample where necessary to achieve sufficient numbers for accurate quantification of insert abundance (see Supplementary Fig. S2 and Supplementary Note 2), then split five-fold before nested PCR amplification to recover library inserts from genomic DNA (Supplementary Fig. 2). PCRs were performed using KAPA Hi-Fi HotStart ReadyMix (Kapa Biosystems), as described in Supplementary Note 2. PCR products were agarose gel-purified and quantified by NanoDrop and used for Illumina library preparation.
Illumina sequencing
Illumina sequencing libraries were prepared using minor modifications of standard protocols53 and the Multiplexing Sample Preparation Oligonucleotide Kit (Illumina). Pooled PCR product was sonicated by Covaris S2 as described53, and then end-repaired with the End-IT DNA End-Repair Kit (EpiCentre Biotechnologies) and A-tailed with Klenow exo- (New England Biolabs). Standard adapters (Index PE Adapter Oligo Mix) were ligated using Quick T4 DNA Ligase (New England Biolabs). Ligation products were size-selected from agarose gels, and quantified and checked for concentration and size distribution by Agilent 2200 TapeStation. Enrichment PCRs were performed using Phusion thermostable polymerase (New England Biolabs), as described in Supplementary Note 2. Purified enrichment PCR products were assessed by Agilent 2200 TapeStation and submitted to the Partners Center for Personalized Genetic Medicine for concentration measurement by PicoGreen fluorescence and qPCR, followed by equimolor index pooling and sequencing (50 base single-end read) on the Illumina HiSeq 2000.
Mapping Illumina sequencing reads
We used segemehl (version 0.0.9.4, current version is 0.1.3)23 with parameter settings -M 100 -E 5 -D 2 -A 80 to map Illumina sequencing reads to the D. melanogaster genome. For cCRM detection, we required: (1) ≥1 read from each of the 5’ and 3’ ends; (2) ≥5 positions covered by center reads (i.e., without the SEQ1 or SEQ2 primers); and (3) ≥10 total reads. Where overlapping cCRM windows contribute indistinguishable reads to the same genomic regions, we used the unambiguous end reads as weights for dividing the reads that map to overlapping cCRM windows. Analysis of random sets of genomic windows matched for length, sequence context (e.g., intronic, intergenic) and GC content to our foreground windows indicated that the FDR for cCRM detection was less than 5×10−5.
Statistical analysis of enhancer-FACS-Seq (eFS) data
We collected the number of reads mapped to each cCRM for each replicate population and control ‘input’ population, and filtered out cCRMs not detected in any input sample replicate. Enrichment and statistical significance were calculated using DESeq24 with standard parameters and size factor estimation, as described in Supplementary Note 2.
Statistical analysis of genomic features
For a given type of genomic feature, we calculated the scores for each cCRM as the weighted average of the score (e.g., ChIP signal intensity) for feature intervals that overlap the peak as reported in the published Browser Extensive Data (BED) or Wiggle Track Format (WIG) file associated with that experiment. We defined the weights by the amount of overlap (bp) between the cCRM and the feature’s genomic coordinates. All comparisons of enrichment (or depletion) of various genomic marks were performed by calculating enrichment in the eFS-positive enhancers (DESeq P_adj < 0.1) as compared to an equally sized set of inactive cCRMs (DESeq P_adj > 0.8) chosen from the bottom of the ranked list (ranked by decreasing fold-enrichment value). The statistical significance of any such enrichment (or depletion) was determined as P < 0.05 by Wilcoxon-Mann-Whitney U-test.
DNA sequence motif over-representation analysis
We compiled a dictionary of 567 publicly available Drosophila TF binding site motifs6,32–35. Motifs were trimmed, redundant motifs were removed, and motif exemplars were chosen, as described in Supplementary Note 2. To identify over-represented motifs in the twi:CD2+GFP+, Mef2-I-ED5:CD2+GFP+, and duf:CD2+GFP+ foreground (FG) sequence sets, we used PhylCRM and Lever19. Lever calculates the over-representation of individual motifs or combinations thereof, according to their density and evolutionary conservation, as quantified by the PhylCRM scoring scheme19, in each FG sequence set as compared to a random set of background (BG) sequences. BG sets were chosen to be about 20 times the size of the FG sets, and matched for length, GC content, and repeat content. All settings were as previously described54, except that repeats were not masked and length correction was not used since all sequences were roughly the same length. Any motif that did not have occurrences in at least one-quarter of the FG sequences was removed from further consideration. We then used Lever to inspect the FG sets for over-representation of all single and pairwise combinations of the resulting 86-exemplar motif dictionary. Motif PWMs are provided in Supplementary Table 7.
Classifier analysis
For each cCRM, we generated a feature vector of scores that quantify the presence of motif matches for each PWM in the motif exemplar dictionary. The score for a particular PWM and a particular cCRM was defined as the sum of the log-odds ratios of PWM matches in the cCRM sequence that exceeded a permissive match threshold (log-odds ratio > 3.0). For classification we used the Gaussian Naïve Bayes implementation in the scikit-learn package55 for Python (see Supplementary Note 2). As for the motif over-representation analysis, positive cCRMs are those with DESeq P_adj < 0.1; here, negative cCRMs are those from an equally sized set chosen from the bottom of the cCRM list ranked by eFS P_adj. To evaluate classification accuracy, we split the labeled cCRM feature vectors into training and test sets using stratified 10-fold cross-validation. Feature selection was performed independently for each of the folds: in each, the k motifs with the highest individual AUC values in the training set were selected. The classifier was then trained using features corresponding only to those k motifs. We evaluated performance across multiple values of k and selected the value that maximized performance accuracy in cross-validation tests.
Traditional reporter assays
Homozygous or balanced heterozygous transformant males were crossed to homozygous twi:CD2 females in small population cages, and broad collections (~2–17 hours after egg deposition) of embryos were fixed and stained for immunofluorescence by standard protocols49 (see Supplementary Note 2). Stained embryos were imaged with a Zeiss Imager Z1 with Apotome in optical sectioning mode. Co-expression of GFP with CD2 (Supplementary Table 5) was evaluated in individual optical sections with the annotator being blind to the predicted activity of the cCRMs. Co-expression is observed as GFP and CD2 being present in the same cells, since the GFP in these embryos is nuclear, while CD2 is expressed on the cell surface. For validations of CD2− eFS-positive cCRMs as being active enhancers, we assayed for activity anywhere in the embryo at this developmental stage.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported in part by a National Science Foundation Graduate Research Fellowship to L.A.B. and by grant R01 HG005287 from the U.S. National Institutes of Health to M.L.B. We thank G. Losyev and C. Durkin for technical assistance, K.G. Guruharsha and K. VijayRaghavan (Tata Institute of Fundamental Research) for sharing coordinates of the duf enhancer prior to its publication, R. McCord, M. Markstein, and O. Iartchouk for helpful discussion, and R. Gordân, M. Markstein, and T. Siggers for critical reading of the manuscript.
Footnotes
Accession codes. All analyzed sequence data has been deposited in NCBI GEO under Series ID #GSE41503.
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
M.L.B. designed the study, S.S.G., P.W.E., A.M.M, and M.L.B. developed the eFS technology, S.S.G. and A.V. sorted flies, S.S.G., L.A.B., M.P., and A.A. performed computational data analysis, S.S.G., P.W.E., A.V., Y.K., and X.Z. performed PCRs, B.W.B., X.Z., A.S. and C.E.G. generated CD2 fly lines, S.S.G., A.V., A.P., and A.I. performed validation assays, S.S.G., L.B, M.P., A.A., B.W.B., and M.L.B. wrote Supplementary Note 2, S.S.G., L.B., M.P., A.A., and M.L.B. prepared figures and tables, S.S.G. and M.L.B. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare they have no competing financial interests.
References
- 1.Bulyk ML. Computational prediction of transcription-factor binding site locations. Genome Biol. 2003;5:201. doi: 10.1186/gb-2003-5-1-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davidson E. Genomic Regulatory Systems: Development and Evolution. Chapter 2. Academic Press; 2001. pp. 25–62. [Google Scholar]
- 3.Pfeiffer BD, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halfon MS, Gallo SM, Bergman CM. REDfly 2.0: an integrated database of cis-regulatory modules and transcription factor binding sites in Drosophila. Nucleic Acids Res. 2008;36:D594–D598. doi: 10.1093/nar/gkm876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandmann T, et al. A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes Dev. 2007;21:436–449. doi: 10.1101/gad.1509007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EE. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature. 2009;462:65–70. doi: 10.1038/nature08531. [DOI] [PubMed] [Google Scholar]
- 7.Jagalur M, Pal C, Learned-Miller E, Zoeller RT, Kulp D. Analyzing in situ gene expression in the mouse brain with image registration, feature extraction and block clustering. BMC Bioinformatics. 2007;8(Suppl 10):S5. doi: 10.1186/1471-2105-8-S10-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gertz J, Siggia ED, Cohen BA. Analysis of combinatorial cis-regulation in synthetic and genomic promoters. Nature. 2009;457:215–218. doi: 10.1038/nature07521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharon E, et al. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nature Biotechnology. 2012;30:521–530. doi: 10.1038/nbt.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nam J, Dong P, Tarpine R, Istrail S, Davidson EH. Functional cis-regulatory genomics for systems biology. Proc. Natl. Acad. Sci. U.S.A. 2010;107:3930–3935. doi: 10.1073/pnas.1000147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melnikov A, et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nature Biotechnology. 2012;30:271–277. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patwardhan RP, et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nature Biotechnology. 2012;30:265–270. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnold CD, et al. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–1077. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- 14.Dunin-Borkowski OM, Brown NH. Mammalian CD2 is an effective heterologous marker of the cell surface in Drosophila. Dev Biol. 1995;168:689–693. doi: 10.1006/dbio.1995.1115. [DOI] [PubMed] [Google Scholar]
- 15.Bate M. The mesoderm and its derivatives. In: Bate M, Martinez-Arias A, editors. The development of Drosophila melanogaster. Plainview, NY: Cold Spring Harbor Laboratory; 1993. pp. 1013–1090. [Google Scholar]
- 16.Roy S, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Contrino S, et al. modMine: flexible access to modENCODE data. Nucleic Acids Research. 2012;40:D1082–D1088. doi: 10.1093/nar/gkr921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas S, et al. Dynamic reprogramming of chromatin accessibility during Drosophila embryo development. Genome Biology. 2011;12:R43. doi: 10.1186/gb-2011-12-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warner J, et al. Systematic identification of mammalian regulatory motifs’ target genes and functions. Nature Methods. 2008;5:347–353. doi: 10.1038/nmeth.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duan H, Skeath JB, Nguyen HT. Drosophila Lame duck, a novel member of the Gli superfamily, acts as a key regulator of myogenesis by controlling fusion-competent myoblast development. Development. 2001;128:4489–4500. doi: 10.1242/dev.128.22.4489. [DOI] [PubMed] [Google Scholar]
- 21.Guruharsha KG, Ruiz-Gomez M, Ranganath HA, Siddharthan R, Vijayraghavan K. The complex spatio-temporal regulation of the Drosophila myoblast attractant gene duf/kirre. PLoS ONE. 2009;4:e6960. doi: 10.1371/journal.pone.0006960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann S, et al. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Computational Biology. 2009;5:e1000502. doi: 10.1371/journal.pcbi.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallo SM, et al. REDfly v3.0: toward a comprehensive database of transcriptional regulatory elements in Drosophila. Nucleic Acids Research. 2011;39:D118–D123. doi: 10.1093/nar/gkq999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonn S, et al. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nature Genetics. 2012;44:148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- 27.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 29.Pekowska A, et al. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 2011;30:4198–4210. doi: 10.1038/emboj.2011.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kharchenko PV, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu LJ, et al. FlyFactorSurvey: a database of Drosophila transcription factor binding specificities determined using the bacterial one-hybrid system. Nucleic Acids Research. 2011;39:D111–D117. doi: 10.1093/nar/gkq858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Philippakis AA, et al. Expression-guided in silico evaluation of candidate cis regulatory codes for Drosophila muscle founder cells. PLoS Computational Biology. 2006;2 doi: 10.1371/journal.pcbi.0020053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Busser BW, et al. Molecular mechanism underlying the regulatory specificity of a Drosophila homeodomain protein that specifies myoblast identity. Development. 2012;139:1164–1174. doi: 10.1242/dev.077362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Busser BW, et al. Integrative analysis of the zinc finger transcription factor Lame duck in the Drosophila myogenic gene regulatory network. Proc. Natl. Acad. Sci. U.S.A. 2012;109:20768–20773. doi: 10.1073/pnas.1210415109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thisse B, el Messal M, Perrin-Schmitt F. The twist gene: isolation of a Drosophila zygotic gene necessary for the establishment of dorsoventral pattern. Nucleic Acids Research. 1987;15:3439–3453. doi: 10.1093/nar/15.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Furlong EE, Andersen EC, Null B, White KP, Scott MP. Patterns of gene expression during Drosophila mesoderm development. Science. 2001;293:1629–1633. doi: 10.1126/science.1062660. [DOI] [PubMed] [Google Scholar]
- 38.Grimaud C, Negre N, Cavalli G. From genetics to epigenetics: the tale of Polycomb group and trithorax group genes. Chromosome Res. 2006;14:363–375. doi: 10.1007/s10577-006-1069-y. [DOI] [PubMed] [Google Scholar]
- 39.Herrmann C, Van de Sande B, Potier D, Aerts S. i-cisTarget: an integrative genomics method for the prediction of regulatory features and cis-regulatory modules. Nucleic Acids Research. 2012;40:e114. doi: 10.1093/nar/gks543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan Y, Guo L, Shen L, Liu JS. Predicting gene expression from sequence: a reexamination. PLoS Computational Biology. 2007;3:e243. doi: 10.1371/journal.pcbi.0030243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes & Development. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Azpiazu N, Frasch M. tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 1993;7:1325–1340. doi: 10.1101/gad.7.7b.1325. [DOI] [PubMed] [Google Scholar]
- 43.Cripps RM, Zhao B, Olson EN. Transcription of the myogenic regulatory gene Mef2 in cardiac, somatic, and visceral muscle cell lineages is regulated by a Tinman-dependent core enhancer. Dev Biol. 1999;215:420–430. doi: 10.1006/dbio.1999.9446. [DOI] [PubMed] [Google Scholar]
- 44.Busser BW, et al. A machine learning approach for identifying novel cell type-specific transcriptional regulators of myogenesis. PLoS Genetics. 2012;8:e1002531. doi: 10.1371/journal.pgen.1002531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lister JA. Transgene excision in zebrafish using the phiC31 integrase. Genesis. 2010;48:137–143. doi: 10.1002/dvg.20595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thyagarajan B, Olivares EC, Hollis RP, Ginsburg DS, Calos MP. Site-specific genomic integration in mammalian cells mediated by phage phiC31 integrase. Molecular and Cellular Biology. 2001;21:3926–3934. doi: 10.1128/MCB.21.12.3926-3934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hollis RP, et al. Phage integrases for the construction and manipulation of transgenic mammals. Reproductive Biology and Endocrinology. 2003;1:79. doi: 10.1186/1477-7827-1-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barolo S, Carver LA, Posakony JW. GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques. 2000;29:726–728. 730–732. doi: 10.2144/00294bm10. [DOI] [PubMed] [Google Scholar]
- 49.Halfon MS, et al. Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell. 2000;103:63–74. doi: 10.1016/s0092-8674(00)00105-7. [DOI] [PubMed] [Google Scholar]
- 50.Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40:476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Estrada B, et al. An integrated strategy for analyzing the unique developmental programs of different myoblast subtypes. PLoS Genet. 2006;2:e16. doi: 10.1371/journal.pgen.0020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quail MA, Swerdlow H, Turner DJ. Improved protocols for the illumina genome analyzer sequencing system. Current Protocols in Human Genetics. 2009;Chapter 18(Unit 18):2. doi: 10.1002/0471142905.hg1802s62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aboukhalil A, Bulyk ML. LOESS correction for length variation in gene set-based genomic sequence analysis. Bioinformatics. 2012;28:1446–1454. doi: 10.1093/bioinformatics/bts155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pedregosa F, et al. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research. 2011;12:2825–2830. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.