Abstract
Protein N-myristoylation is a 14-carbon fatty-acid modification that is conserved across eukaryotic species and occurs on nearly 1% of the cellular proteome1,2. The ability of the myristoyl group to facilitate dynamic protein–protein and protein–membrane interactions (known as the myristoyl switch) makes it an essential feature of many signal transduction systems3. Thus pathogenic strategies that facilitate protein demyristoylation would markedly alter the signalling landscape of infected host cells. Here we describe an irreversible mechanism of protein demyristoylation catalysed by invasion plasmid antigen J (IpaJ), a previously uncharacterized Shigella flexneri type III effector protein with cysteine protease activity. A yeast genetic screen for IpaJ substrates identified ADP-ribosylation factor (ARF)1p and ARF2p, small molecular mass GTPases that regulate cargo transport through the Golgi apparatus4. Mass spectrometry showed that IpaJ cleaved the peptide bond between N-myristoylated glycine-2 and asparagine-3 of human ARF1, thereby providing a new mechanism for host secretory inhibition by a bacterial pathogen5,6. We further demonstrate that IpaJ cleaves an array of N-myristoylated proteins involved in cellular growth, signal transduction, autophagasome maturation and organelle function. Taken together, these findings show a previously unrecognized pathogenic mechanism for the site-specific elimination of N-myristoyl protein modification.
While seeking to understand how bacterial pathogens regulate the host secretory pathway7, we found that S. flexneri potently inhibited cargo transport through the Golgi apparatus (Supplementary Fig. 1). Further investigation showed that the Golgi was severely fragmented after Shigella infection (Fig. 1a). By contrast, neither Listeria monocytogenes nor Salmonella typhimurium disrupted the Golgi, suggesting that Shigella has evolved a specific, yet poorly understood, mechanism to regulate host organelle function.
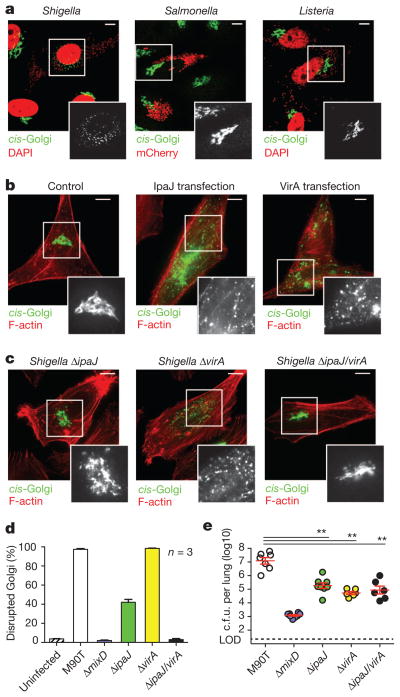
Figure 1. Shigella IpaJ and VirA disrupt Golgi morphology.
a, Fluorescence microscopy HeLa cells infected with Shigella and Listeria visualized with 4′,6-diamidino-2-phenylindole (DAPI) (pseudo-coloured red), and Salmonella carrying mCherry-expressing vector (red). The cis-Golgi (green) was detected by α-GM130 antibodies. Scale bar, 10 μm. b, Fluorescence microscopy of HeLa cells transfected with either IpaJ or VirA. The cis-Golgi (GM130, green) and F-actin (red) are shown. Scale bar, 10 μm. c, Fluorescence microscopy of HeLa cells infected with indicated Shigella mutant strains. Scale bar, 10 μm. d, Percentage of HeLa cells with disrupted Golgi morphology in 100 cells infected with either wild-type Shigella (M90T) or the indicated mutants. Error bars, means ± s.e.m. calculated from three independent experiments. e, Number of recoverable c.f.u. per mouse lung 24 h after intranasal inoculation with 1 ×106 of wild-type M90T or each mutant Shigella strain as indicated. Limit of detection (LOD) for this assay is indicated by the dotted line. Error bars, geometrical means ± s.e.m. **P < 0.001.
Shigella infection requires translocation of more than 20 bacterial ‘effector’ proteins into host cells through the Mxi-SPA Type III Secretion System (T3SS)8. Deletion of mxiD, a component of the needle complex required for T3SS, eliminated Golgi destruction by Shigella infection (Fig. 1d). To identify the specific effector protein required for this activity, Golgi morphology was assessed after transient transfection of 20 effector genes (Supplementary Fig. 2a, b). Both IpaJ and VirA induced profound Golgi fragmentation (Fig. 1b) and inhibited hormone trafficking through the secretory pathway (Supplementary Fig. 2c). However, the other T3SS effector proteins had no discernible effect on Golgi structure or function (Supplementary Fig. 2a).
Comparison of cells infected with Shigella ΔipaJ and ΔvirA gene deletion strains showed that these mutants induced abnormal Golgi morphologies (Fig. 1c) and variable degrees of Golgi disruption (Fig. 1d), yet neither mutant alone fully attenuated Golgi destruction. By contrast, the Golgi remained intact and functional in cells infected with ΔipaJ/virA double mutant strain (Fig. 1c, d). The Shigella ΔipaJ/virA strain showed normal host cell invasion, intracellular replication and actin-based motility, suggesting Golgi disruption was specifically caused by these effector genes (Supplementary Fig. 3). Next, in vivo bacterial replication was evaluated using an established mouse model of mucosal infection9. The number of recoverable bacteria was sharply reduced (by 100-fold, P < 0.001) by deleting either ipaJ or virA genes compared with wild-type Shigella (Fig. 1e). As expected, bacterial replication was further attenuated in the Shigella ΔmixD mutant lacking all T3SS function (Fig. 1e). We also found reductions in host inflammatory cytokines in mice infected with each Shigella mutant strain (data not shown). Taken together, these data demonstrate that two Shigella effector proteins, IpaJ and VirA, each harbour Golgi inhibitory activity, and are essential and play non-redundant roles for optimal in vivo virulence.
Although VirA was recently shown to inactivate Rab1 GTPase signalling pathways5, extra mechanisms are necessary for Golgi destruction during Shigella infection6. We therefore focused on IpaJ because its molecular mechanism of action is poorly understood10,11. BLAST database searches identified IpaJ homologues in numerous bacterial species, but this sequence-based alignment did not offer any clues to its function (Supplementary Fig. 4). In contrast, structural-based bioinformatics showed that IpaJ is distantly related to the cysteine peptidase C39-like family of domains of unknown function (annotated DUF3335) (Fig. 2a). C39-family members cleave leader peptides from bacteriocins with anti-microbial and quorum-sensing activities12. Although IpaJ is unlikely to function in these capacities, it harbours the catalytic cysteine (C), histidine (H) and aspartate (D) residues required for peptide bond hydrolysis (Fig. 2a). As predicted by this alignment, alanine substitutions at C64, H206 or D218 abolished the ability of IpaJ to disrupt host Golgi morphology in transfection studies (Fig. 2b). In addition, complementation of Shigella ΔipaJ/virA deletion strain with plasmid expression of IpaJ induced Golgi disruption phenotype whereas IpaJ C64A, H206A and D218A had no effect (Supplementary Fig. 5).
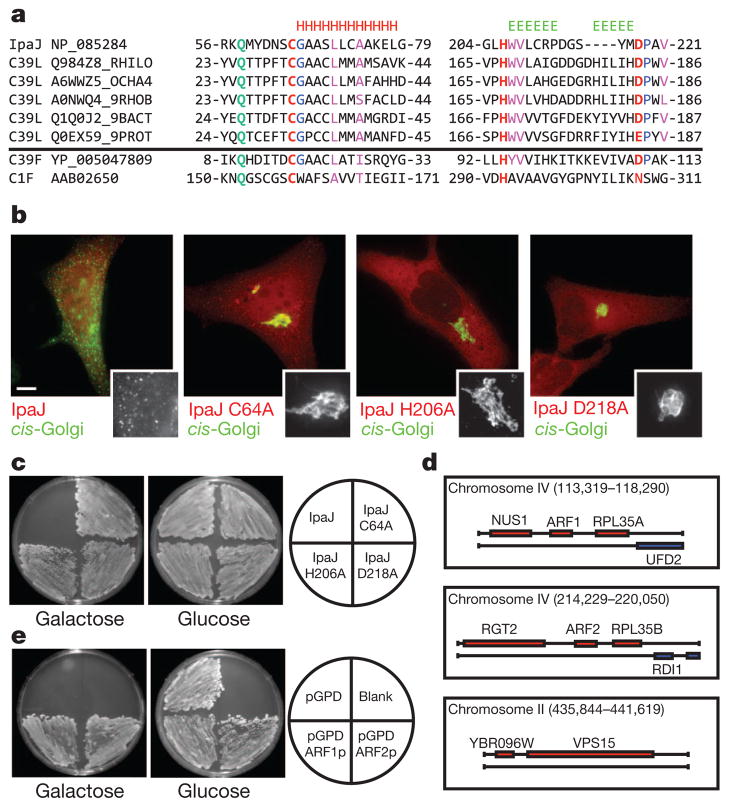
Figure 2. IpaJ belongs to the C39-like family of cysteine proteases and targets ARF-family GTPases.
a, Sequence–structure alignment of S. flexneri IpaJ with members of C39-peptidase-like family identified by HHPred. IpaJ possesses invariant catalytic triad residues Cys 64, His 206 and Asp 218 (red). The conserved Gln 58, which helps form the oxyanion hole found in many proteases, is shown in green. Invariant residues and core hydrophobic residues found in the C39-family are shown in blue and magenta, respectively. Characterized C39-family member (GCN5-acyetyl transferase) and a C1-family member (Papain) are shown. b, Fluorescence microscopy of HeLa cells transfected with the indicated IpaJ mutants. The cis-Golgi (GM130, green) and F-actin (red) are shown. Scale bar, 10 μm. c, IpaJ or its catalytic mutants were expressed from a galactose-inducible promoter (pGal413 vector) and assayed for a growth arrest phenotype on galactose or glucose (control) carbon source. d, Illustration of the yeast genomic clones isolated from the IpaJ suppressor screen. ARF1p, ARF2p and VPS15p are highlighted. e, Yeast strain harbouring a galactose-inducible IpaJ gene were transformed with a multi-copy vector containing the indicated genes. Yeast were assayed for a growth arrest phenotype on galactose or glucose (control) carbon source.
Previous studies have shown that IpaJ induces growth arrest phenotype in yeast13. A similar growth inhibitory activity was found with IpaJ, whereas yeast grew normally in the presence of IpaJ C64A, H206A or D218A catalytic mutants (Fig. 2c). Using a multicopy suppressor screen, we identified three distinct genomic loci that, when introduced on high-copy plasmids, suppressed IpaJ activity in yeast. Two loci encoded ARF1p and ARF2p GTPases, and the third loci encoded VPS15p, a phosphatidylinositol kinase required for yeast vacuole fusion14 (Fig. 2d). Although these data suggest that IpaJ may regulate several host signalling pathways, we focused our initial efforts on ARF GTPases because these enzymes are master regulators of cargo trafficking through the Golgi apparatus4. Overexpression of either ARF1p or ARF2p rescued the yeast growth arrest phenotype, thereby defining ARF GTPases as potential substrates of IpaJ (Fig. 2e).
ARF1 functionally couples guanine-nucleotide exchange (GDP for GTP) and membrane binding (through N-myristoylation) to the recruitment of COPI coat-complex to Golgi membranes as a key step in vesicle fission4. Surprisingly, IpaJ had no effect on the guanine-nucleotide cycle or GTPase-dependent interactions of ARF1 in vitro (Supplementary Fig. 6). We therefore searched for IpaJ-induced post-translational modifications on ARF1 by ‘top-down’ mass spectrometry15,16. ARF1 bearing a carboxy (C)-terminal Strep affinity tag (ARF1-strep) was expressed in HEK293T cells and purified by Strep-Tactin chromatography. We observed an intact monoisotopic molecular mass of 26,895.95 Da and subsequent tandem mass spectrometry (MS/MS) resulted in 30 fragment ions that, together, provided an unambiguous assignment of N-myristoylated ARF1-strep protein (Fig. 3a and Supplementary Fig. 7a). Cellular expression of IpaJ resulted in a 266.35 Da decrease in molecular mass of ARF1-strep, which corresponds closely to the mass of the N-myristoyl group and the amino (N)-terminal glycine (267.22 Da) (Fig. 3b). Indeed, the MS/MS data support the idea that IpaJ cleaved the amide bond between glycine-2 and asparagine-3, liberating the N-myristoylated glycine from the ARF1 GTPase domain (Fig. 3d and Supplementary Fig. 7b). ARF1 cleavage was not observed in IpaJ C64A-treated samples, consistent with the function of IpaJ as cysteine protease (Fig. 3c and Supplementary Fig. 7c).
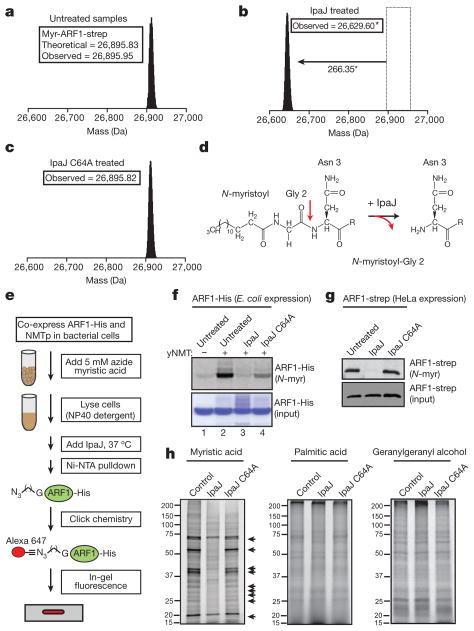
Figure 3. IpaJ cleaves the N-myristoylated glycine of lipidated substrates.
a–c, Mass spectra of purified ARF1-strep in untreated (a), IpaJ-treated (b) or IpaJ C64A-treated (c) cells, with the observed monoisotopic mass reported in daltons. *Observed 1 Da shift in molecular mass is accounted for in MS/MS data (Supplementary Fig. 7). d, Structure of N-myristoylated ARF1 N terminus in untreated (left) and IpaJ-treated (right) samples. R group following Asn 3 denotes ARF1-strep protein residues 4–240. e, Reconstitution of ARF1-His N-myristoylation (N-myr) by NMTp in bacterial cells and IpaJ cleavage reaction in vitro (see Supplementary Information and Methods). f, In-gel fluorescence assay (top panel) visualizing Alexa Fluor 647-labelled myristoylated ARF1-His isolated from bacteria either not expressing (lane 1) or expressing (lane 2) NMTp. Bacterial cell lysates treated with recombinant IpaJ or IpaJ C64A as indicated. The expression levels of ARF1-His were determined by Coomassie blue stain. g, In-gel fluorescence assay (top panel) visualizing Alexa Fluor 647-labelled myristoylated ARF1-strep purified from HeLa cell lysates left untreated or incubated with IpaJ or its catalytic mutant as indicated. The expressed amounts of ARF1-strep are shown (bottom panel). h, In-gel fluorescence assay visualizing protein extracts isolated from HeLa cells incubated with azide myristic acid, azide palmitic acid or geranylgeranyl alcohol azide and subsequently labelled with Alexa Fluor 647 alkyne by click chemistry. Arrows indicate proteins that are proteolytically demyristoylated by IpaJ.
Next, we devised a protocol to reconstitute the ARF1 cleavage reaction in vitro (Fig. 3e). Because Escherichia coli does not express a system for N-myristoylation, ARF1-His was co-expressed with yeast N-myristoyl transferase (NMTp) in the presence of exogenous azide-conjugated myristic acid. The lipid state of purified ARF1-His was visualized by fluorescently labelling the azido-myristoyl group by click chemistry17 (Fig. 3f, lanes 1 and 2). Importantly, IpaJ cleaved N-myristoylated ARF1 in vitro whereas this reaction was less efficient with IpaJ C64A (Fig. 3f, lane 3 and 4). Similar results were also observed when IpaJ was incubated with ARF1-strep isolated from mammalian cellular lysates following a similar procedure (Fig. 3g).
Having established that IpaJ cleaves ARF1, we revisited our studies in yeast showing that IpaJ may target several host substrates including VPS15p, a lipidated kinase14. Although ARF1 and VPS15p are both N-myristoylated on their glycine-2 residue, they do not share any sequence or functional similarity. These observations suggested that IpaJ might cleave a broader range of substrates than initially suspected. To test this hypothesis, we labelled numerous of N-myristoylated proteins found in HEK293T cells and found that a large proportion of these proteins were demyristoylated in IpaJ-treated samples (Fig. 3h, arrows). The spectrum of N-myristoylated proteins was unaltered by IpaJ C64A mutant, thereby verifying the proteolytic-basis of this reaction. IpaJ had no effect on either palmitoylation or geranylgeranylation, indicating that the protease specifically cleaves proteins modified by the myristoyl group (Fig. 3h).
N-myristoylated proteins share a conserved glycine but their sequences are otherwise highly variable (Fig. 4a). To determine how IpaJ recognizes diverse substrates, we first generated chimaeric enhanced green fluorescent protein (EGFP) fusion proteins that expressed 10 amino-acid leader peptides derived from c-Src, MARCKs, hVPS15 and GRASP65, N-myristoylated proteins involved in cellular growth, signal transduction, autophagasome maturation and organelle function, respectively (Fig. 4a)18–21. IpaJ cleaved each of these lipid-modified peptides in a reaction requiring its cysteine protease activity (Fig. 4b). The ability of IpaJ to cleave unrelated peptides suggested that the protease might recognize the lipidated glycine-2 common to all of its substrates. We found that soluble myristic acid was unable to inhibit ARF1 proteolysis, suggesting that IpaJ does not simply recognize long hydrocarbon chains (Supplementary Fig. 8). However, protein lipidation was essential for proteolysis because IpaJ was unable to cleave non-myristoylated ARF1 in vitro (Supplementary Fig. 9). When taken together, these data indicate that IpaJ specifically recognizes the conserved glycine-2 of substrates in context of its amide linkage to N-myristic acid.
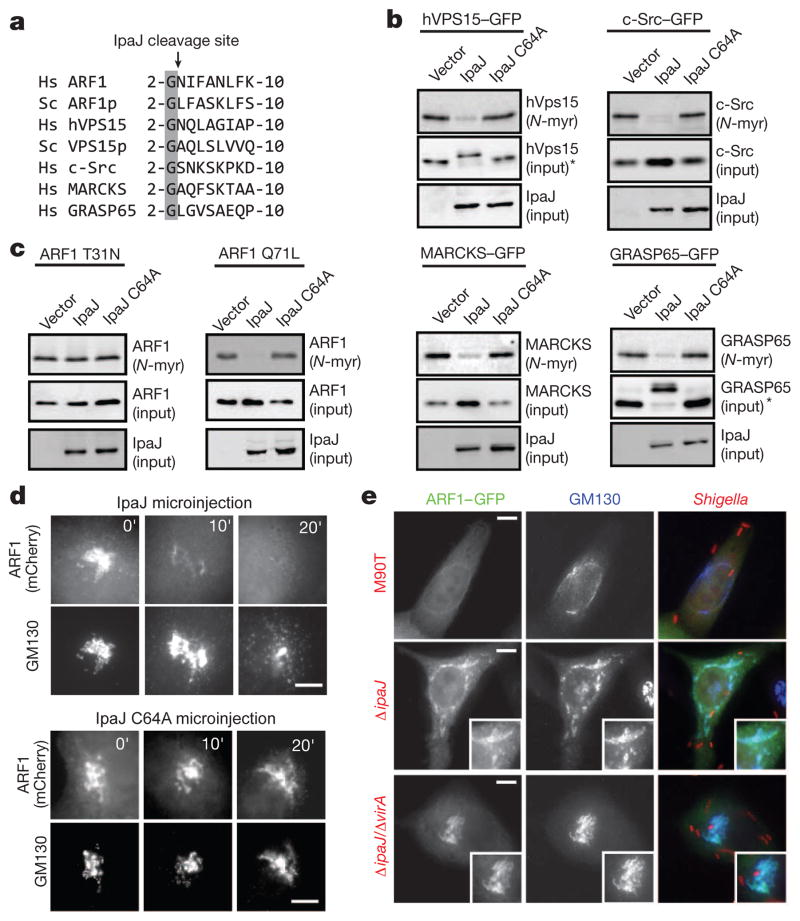
Figure 4. Conformational-dependent cleavage of lipidated substrates by IpaJ.
a, Multiple-sequence alignment of N-myristoylated glycine and downstream sequences from the indicated proteins (Hs, Homo sapiens; Sc, S. cerevisiae). b, In-gel fluorescence assay showing myristoylated peptides of the indicated proteins in cells expressing IpaJ or catalytic mutant. The peptide–eGFP and IpaJ inputs are indicated. *Demyristoylation of GRASP65 and hVPS15 resulted in slower mobility of the resulting peptide, potentially owing to reduced mobility in SDS–polyacrylamide gel electrophoresis caused by proteolytic reaction. c, In-gel fluorescence assay showing myristoylated ARF1 T31N mutant (a GDP-locked mutant) or ARF1 Q71L (a GTP-locked mutant) after in cells co-expressing IpaJ or catalytic mutants as indicated. Equal loading of ARF1 (middle panel) and IpaJ (lower panel) was determined by western blot analysis. d, Fluorescence microscopy showing ARF1–mCherry and the Golgi apparatus (GM130) after cellular microinjection of recombinant IpaJ or IpaJ C64A, over the indicated period. Scale bar, 10 μm. e, Fluorescence microscopy showing ARF1–eGFP (green) and Golgi membranes (blue, GM130) after infection with Shigella M90T and the indicated mutants (red; mCherry-expressing bacteria). Scale bar, 10μm.
N-myristoylation is unique among lipid modifications in that it undergoes a dynamic conformational switch in response to upstream signalling events (termed the ‘myristoyl switch’)3. For example, the myristoyl group is sequestered in GDP-inactive ARF1 and liberated for membrane binding and signalling by GTP exchange (Supplementary Fig. 10)4,22–24. We found that IpaJ was unable to cleave ARF1 T31N (a GDP-bound mutant form), but efficiently cleaved ARF1 Q71L (a mutant locked it the GTP-bound active state) in cells (Fig. 4c). These findings suggest IpaJ recognizes substrates as they cycle between activity-dependent conformational states. Consistent with this interpretation, microinjection of purified IpaJ induced the release of activated ARF1 from Golgi membranes just before organelle fragmentation (Fig. 4d). ARF1 was also displaced from Golgi membranes by wild-type Shigella infection but not by ΔipaJ single mutant (note that the Golgi is fragmented by VirA in this infection) or ΔipaJ/ΔvirA double mutant strains (Fig. 4e). Extra cell-based experiments support the notion that GTP-active ARF1 is a physiological target of IpaJ during Shigella infection (Supplementary Figs 11–13), thereby defining a new mechanism for inhibition of cytokine secretion and immune detection by a bacterial pathogen.
Here we have determined that IpaJ is a cysteine protease that cleaves the N-myristoylated glycine from ARF1 and potentially numerous host cellular substrates. This discovery adds significantly to the small but increasing number of bacterial proteases that cleave lipidated proteins as a mechanism of microbial pathogenesis25,26. Importantly, the ability of IpaJ to eliminate the N-myristoyl modification of unrelated proteins may allow Shigella to exploit signalling pathways in a variety of host cellular contexts27. Although extra studies will be needed to unravel the hierarchy of these events, our discoveries provide a unique framework from which to explore the functional consequences of protein N-myristoylation in human health and disease.
METHODS
Plasmids and cloning
Bacterial effector genes were amplified by PCR from S. flexneri M90T and cloned into pEntr/D, generating a Gateway™ compatible entry clone (Invitrogen) according to the manufacturer’s recommendations. Twenty S. flexneri effectors were cloned into pEntr/D vector: IpaA, AAK18443.1; IpaB, AAK18446; IpaH1.4, AAK18594.1; IpaH2.5, AAK18367; IpaH4.5, AAK18395; IpaH7.8, AAK18394.1; IpaH9.8, AAK18544; IpaJ, AAK18440; IpgD, AAK18452; IcsB, AAK18450; OspB, AAL72323.1; OspC1, AAL72322.1; OspC2, AAW64906; OspD1, AAW64782; OspE1, AAW64916; OspE2, AAW64805; OspF, AAW64770; OspG, NP_085391; VirA, AAK18501. For mammalian expression, the effectors were then recombined into modified pcDNA3.1 vector carrying N-terminal eGFP or mCherry in frame with the Gateway cassette (Gateway™ vector conversion system, Invitrogen). For bacterial expression, a 50-amino-acid N-terminal deletion of IpaJ (IpaJΔ50; residues 51–259) was cloned in frame into pGEX-4T1 (GST-tag) (Amersham), yielding intact catalytic domain of IpaJ C-terminally fused to GST-tag. N-terminal deletion of ARF1 (ARF1Δ17; residues 18–181) was subcloned by PCR into pGEX-4T1 vector. Human GGA1 (GAT domain, residues 76–215) was amplified and cloned by PCR in frame into pET28b-MBP-His vector. For complementation of S. flexneri knockout strains, ipaJ was amplified and cloned by PCR into pBad/Myc-His vector to be expressed under arabinose-inducible promoter. mCherry was expressed from the rpsD promoter in pDP151 vector in Shigella strains. For galactose-induced yeast expression, ipaJ was recombined from pEntr/ D into Gateway™-compatible pYes-Dest52 vector (Invitrogen) for expression under galactose-inducible promoter. Yeast arf1 and arf2 (SGD: S000002351, SGD: S000002296) were amplified by PCR from Yep13 genomic clones and cloned into p415 vector for yeast expression from a constitutively active promoter (GAPDH). Human full-length arf1 was cloned in frame into modified pcDNA3.1 vector carrying C-terminal mCherry or eGFP. Human GRASP65 was cloned in frame into peGFP-N2 vector. For expression of Strep-tagged full-length human ARF1 in mammalian cells, the arf1 complementary DNA (cDNA) (Missouri S&T cDNA Resource Center; ARF0100000) was first cloned into pEntr/D entry vector. The gene was then recombined into modified pcDNA4T/O vector carrying Gateway™ cassette and two repeats of C-terminal Strep-tag (Trp-Ser-His-Pro-Gln-Phe-Glu-Lys, IBA GmbH). For expression of leader peptides bearing myristoylation signal sequences derived from c-Src, MARCKs, hVPS15 and GRASP65, cDNA sequences encoding 10 N-terminal amino acids (see Fig. 4a) were cloned into modified pcDNA4T/O vector carrying C-terminal GFP and two repeats of Strep-tag. All site-directed mutations were generated using the QuickChange Site-Directed Mutagenesis Kit (Stratagene). More primer and plasmid information is available upon request.
Bacterial infection of cultured cells
For Shigella infections, HeLa cells or HeLa cells stably expressing a GFP tag attached to the N-terminal Golgi retention signal of N-acetylglucosaminyltransferase I31 (NAGFP HeLa) were seeded onto cover-slips in six-well dishes the day before infection. Sodium butyrate at 5 μM was added to NAGFP cells after seeding to stimulate production of Golgi-localized GFP marker31. Shigella strains were inoculated from frozen stocks and grown overnight at 30 °C in brain–heart infusion media (BHI) (Difco™, BD Biosciences). Bacteria were then back-diluted 1:50 and incubated at 37 °C until reaching D600 ~ 0.5–0.6. Bacteria were then washed in 1× phosphate buffer saline (PBS) and incubated at 37 °C for 15 min in 0.003% Congo red. Twenty microlitres of bacterial suspension were then added to each well of a six-well dish of semi-confluent HeLa or NAGFP cells and centrifuged for 10 min at room temperature (1000g) to facilitate bacterial adherence. The plates were then incubated for 90 min at 37 °C, 5% CO2. The media was removed and the wells were washed three times with gentamicin (150 μg ml−1) followed by three washes with sterile PBS. Fresh antibiotic-free DMEM was added to each well after that and cells were incubated for an extra 2–4 h (37 °C, 5% CO2). After incubation, the slides were fixed in 3.7% formaldehyde and prepared for microscopy. Golgi and F-actin were visualized with anti-GM130 (BD Transduction Laboratories) and rhodamine-phalloidin (Molecular Probes) respectively. For infections with S. flexneri knockout strains complimented with IpaJ (wild type and C/H/D mutants), overnight bacterial cultures were diluted 1:50 in BHI containing 2% arabinose and incubated until reaching D600 ~ 0.5–0.6 as described above. Infections were performed in low-glucose media containing 2% arabinose to allow expression of IpaJ from arabinose-inducible promoter. For infections with S. flexneri strains expressing mCherry, ampicillin was added both to BHI and DMEM culture media.
A protocol for Salmonella infection of HeLa cells was adapted from previous studies32. S. typhimurium was grown overnight at 37 °C in a glass flask with shaking then subcultured (1:30) and grown for 3 h. One millilitre was pelleted, suspended in 1× PBS and added dropwise at various concentrations to semi-confluent HeLa cells in low-glucose DMEM+10% FBS. Cells were incubated (37 °C, 5% CO2) for 10 min, washed three times with PBS and incubated for an extra 15 min (37 °C, 5% CO2) in fresh low-glucose DMEM + 10% FBS. Cells were washed again with PBS and incubated in DMEM + 10% FBS + 100 mg ml−1 gentamicin before fixing with 3.7% formaldehyde at various time points. Cellular phenotypes were visualized as described.
L. monocytogenes was grown and prepared for HeLa infection as described33. Various concentrations of bacteria were added to semi-confluent HeLa cells in DMEM + 10% FBS from an overnight culture of Listeria grown in BHI media. Cells were centrifuged (250g, room temperature, 5 min) and incubated (37 °C, 5% CO2) for 10 min, then washed three times with 1× PBS. Fresh media containing 10 mg ml−1 gentamicin were added and cells incubated for an extra 4–6 h before fixation and visualization.
Gene disruption in Shigella
The virA, ipaJ and mxiD genes were individually disrupted using the λ red recombinase-mediated recombination system28. Briefly, a kanamycin resistance cassette flanked with 50 base pairs homologous to the gene of interest (virA, ipaJ or mxiD) was amplified from plasmid DNA (pKD3) (primer sequences are available upon request). PCR products were electroporated into S. flexneri strain M90T carrying the red recombinase plasmid pKD46. Transformants were selected by growth on Luria broth agar plates containing kanamycin (50 μg ml−1) and simultaneously cured of pKD46 by growth at 42 °C overnight. The kanamycin resistance gene was eliminated through the introduction of the pCP20 helper plasmid that contained the FLP recombinase. Subsequent curing of pCP20 was done by growing strains at 42 °C for 5 h. Disruption of the virA and ipaJ genes was confirmed through DNA sequencing of the respective genetic loci. To generate the double strain (Shigella ΔvirA/ΔipaJ), the ipaJ locus was disrupted from a Shigella ΔvirA strain following the protocol described above.
Shigella infection studies in mice
Female C57Bl/6 mice were purchased from the National Cancer Institute and used between 6 and 8 weeks of age. For infection, S. flexneri M90T or mutants were grown and back-diluted from the stationary to early log phase growth (D600 ~ 0.1) in BHI media (BD Biosciences) at 37 °C, washed and diluted in sterile saline, and administered dropwise (106 c.f.u. in 30 μl volume) into the external nares of mice anaesthetized with ketamine/xylazine as described9. At the indicated time points after infection, mice were euthanized, and the recoverable bacterial c.f.u. enumerated by plating serial dilutions of the lung homogenate on BHI plates. Statistical significance was determined by a one-way analysis of variance (P < 0.01) Tukey post hoc test. The amounts of the TNF-a and IL-6 in the serum and lung homogenate were enumerated using commercially available enzyme-linked immunosorbent assay (ELISA) reagents (R&D Systems). All experiments were performed under University of Minnesota or Cincinnati Children’s Hospital Institutional Animal Care and Use Committee approved protocols (S.S.W.).
hGH trafficking assay
As previously described7,34, HeLa cells (50% confluence) were transfected with 1 μg of 4× FKBP-hGH (Ariad Pharmaceutical; source of material, D. Bernstein) and either 0.5 μg eGFP–IpaJ, eGFP–VirA or pEGFP control plasmid with Fugene6 (Roche). Sixteen hours later, the medium was replaced with medium containing AP21998 (final concentration 2 mM) or vehicle control. AP21998 was incubated with the cells for 2 h before the supernatant was collected. The supernatant was then diluted 100-fold and compared against an hGH standard curve (12.5–400 pg ml−1) for the quantification of hGH released using an hGH enzyme-linked ELISA (Roche). For no drug controls, 100% ethanol (2 μl) was incubated with the cells for 2 h.
IpaJ bioinformatics
HHpred was used to detect known Pfam domains (profile database search used: pfam version 25.0) with distant structural homology relationships to full-length IpaJ (GenInfo Identifier (GI): 12329066) with default parameter settings35. The DUF3335 (C39-like peptidase family) gave a probability score of 90.0. One hundred proteins in the Pfam database contain this domain. PROMALS3D was used to produce a multiple-sequence alignment with the eight IpaJ and 100 DUF3335 family members to identify invariant catalytic residues and confirm conserved secondary and hydrophobicity patterns29.
Recombinant protein expression and purification
Recombinant proteins (IpaJΔ50, ARF1ΔN17, GGA176–215) were expressed in BL21–DE3 E. coli strains by induction with 0.4 mM IPTG for 16 h at 18 °C. Pellets were lysed with His buffer (100 mM HEPES, pH 7.5, 300 mM NaCl) or GST buffer (Tris buffer saline (TBS); 50 mM Tris pH 7.5, 150 mM NaCl, 2 mM DTT) supplemented with protease cocktail (Roche). Proteins were purified with nickel agarose (Qiagen) or glutathione Sepharose (Amersham Biosciences) following the manufacturer’s instructions. Eluted proteins were buffer exchanged into TBS using concentration centrifugal columns (Millipore); glycerol was added to 15% and the proteins were then stored at −80 °C.
Cell transfections, microinjections and fluorescence microscopy
HEK293A, HeLa and NAGFP HeLa (see above) cells were transfected using FuGene6 (Roche) and incubated for 16–18 h. For expression of Strep-tagged proteins, HEK293T cells were transfected using calcium phosphate and incubated for 18–24 h. Equal amounts of DNA were used for co-transfection. Cells were then lysed (lysis buffer: 20 mM Tris HCl pH 7.5, 1.5 mM MgCl2, 350 mM NaCl, 0.5% NP-40, 5% glycerol) and sonicated for 5 s. The lysate was then clarified by centrifugation (15,000g, 10 min) and applied to Strep-Tactin Superflow Plus resin (Qiagen). After incubation (90 min, 4 °C) the column was washed three times (washing buffer: 20 mM Tris HCl pH 7.5, 1.5 mM MgCl2, 150 mM NaCl, 0.2% NP-40, 5% glycerol) and Strep-tagged proteins were eluted with 2.5 mM desthiobiotin (100 mM Tris•HCl, 150 mM NaCl, 1 mM EDTA, 2.5 mM desthiobiotin, pH 8.0). Microinjections of IpaJΔ50 were performed using a semiautomatic InjectMan NI2 micromanipulator (Eppendorf). Recombinant proteins were diluted in 1× TBS with Cascade Blue (Invitrogen) fluorescent dyes until the final concentration of the protein of 1 mg ml−1 (21 μM). Cellular concentration of microinjected protein was estimated as 1 μM. For IpaJ C64A and ARF1 co-localization studies, HeLa cells were treated either with Brefeldin A (2.5 μg ml−1) for 20 min or with nocodazole (10 μg ml−1) for 1 h before fixation. Immunofluorescence images in all experiments were acquired with Zeiss AxioVert 200 fluorescence microscope. F-actin was visualized by rhodamine- or Alexa Fluor 350-conjugated phalloidin (Molecular Probes), Golgi was detected by GM130 antibodies (BD Biosciences) and secondary anti-mouse IgG antibodies (Thermo Scientific). NAGFP HeLa cells stimulated for production of GFP marker were additionally stained with anti-GFP antibodies (Clontech) and fluorescein-conjugated anti-rabbit IgG secondary antibodies (Thermo Scientific) to enhance the fluorescent signal.
Galactose-induced yeast growth inhibition and yeast multicopy suppressor screen
Yeast InvscI strain was transformed with pYes-dest52 (Invitrogen) vector carrying ipaJ gene (wild type or C/H/D catalytic mutant) under the GAL1 inducible promoter. Yeast was streaked onto Yc-U agar media containing glucose or galactose and rafinose as carbon source. Yeast was cultured for at 30 °C for 3–5 days and the plates were visually inspected for growth. For the yeast multicopy suppressor screen, the ipaJ gene was stably integrated into Y7092 yeast strain genome (MATα can1Δ::STE2pr-Sp_his5 his3Δ1leu2Δ0ura3Δ0met15Δ0 lyp1Δ) as previously described36. The resulting strain Y7092-IpaJ (MATα can1Δ::STE2pr-Sp_his5 his3Δ1leu2Δ0ura3Δ0met15Δ0 lyp1Δ trp1Δ::GAL1-IpaJ-URA3) carryied a single copy of the ipaJ gene under control of GAL1 galactose-inducible promoter. Y7092-IpaJ cells were then transformed with the S. cerevisiae genomic library in the Yep13 vector (ATCC) and transformants were selected for survival on Yc-UL media containing 2% galactose and 1% raffinose as carbon sources. Positive clones were isolated and library vectors were sequenced. For the complementation assay, PCR-amplified yeast arf1 and arf2 were expressed in yeast under GAPDH promoter (see above).
Top-down mass spectrometry
Strep-tagged ARF1 protein was co-transfected with IpaJ or control vector. ARF1-strep was purified with a Strep-Tactin Superflow Plus column and eluted with 2.5 mM desthiobiotin buffer (see above). Purified ARF1-strep was used for LC-MS/MS analysis.
LC-MS/MS was generally as previously described30. Optima-grade solvents and acids (Thermo Scientific) were used. Reverse-phase liquid chromatography (RPLC) capillary columns were packed in-house to a length of 15 cm with 5 μm diameter C18 Poroshell-300 resin (Agilent Technologies) in 75 μm inner diameter × 360 μm outer diameter Picofrit columns (New Objective) with 15 μm inner diameter integrated micro-electrospray tips. A capillary column heater (Analytical Sales & Services) was used to maintain the column temperature at 60 °C during analysis. RPLC mobile phase A included 0.025% TFA, 0.3% formic acid and 5% acetonitrile in water. RPLC mobile phase B included 0.025% TFA, 0.3% formic acid and 20% isopropanol in acetonitrile. The elution gradient was 0% B at 0–3 min, 45% B at 3.01 min and 45–60% B from 3.01 to 23 min. Flow was regulated by an 1100-nano-LC (Agilent) at a flow rate of 0.5 μl min−1.
Before analysis, the protein was de-salted using a C4 ZipTip (Millipore) as described previously and suspended in 0.2% formic acid in water. Approximately 1600 fmol of protein was loaded onto the capillary column per injection. Analysis was on an LTQ Orbitrap XLTM (Thermo Scientific) in full MS mode at 60,000 Fourier-transform mass spectrometry resolution, two microscans, maximum ion accumulation time of 500 ms, scan range from 800 to 2000 m/z, source-induced dissociation =25 V, tube lens 100 V, capillary voltage 50 V, capillary temperature 275 °C. MS/MS data were collected with the source-induced dissociation =60 V, tube lens 100 V, capillary voltage 50 V, capillary temperature 375 °C. The [M +6H]6+ charge state of ubiquitin was used to tune the mass spectrometer. The ‘Xtract’ function in the XcaliburTM (Thermo Scientific) data system with a signal:noise of 3 was used to extract intact protein and fragment masses from the raw spectral data. All fragmentation data presented were the summation of scans across the entire protein elution profile. MS/MS data sets were interrogated with Prosight PC 2.0TM (Thermo Scientific). Analysis of the MS/MS data against the known ARF1 sequences without the correct modification state led to few or no b-ion matches. Replicate analysis was performed on each sample and assigned fragment ions were manually validated in raw data.
Labelling N-myristoylated proteins by click chemistry and in vitro cleavage reaction
E. coli BL-21 cells expressing ARF1-His with or without yeast NMTp were grown overnight at 37 °C in LB media. Cultures were diluted 1:50 and incubated at 37 °C until reaching D600 ~ 0.5–0.6. Myristic acid and azide myristic acid were added at the concentrations 50 μM and 5 μM respectively, and cells were additionally incubated for 30 min. Protein expression was then induced with 0.4 mM IPTG and cells were further incubated at 37 °C for 3 h. We estimated that N-myristoylated ARF1 is bound in a GDP:GTP ratio of 1:1 as assessed by GGA-binding assays. Expression of IpaJ or IpaJ C64A mutant was performed by the same method without adding myristic acid. ARF1- and IpaJ-expressing cells were lysed by sonication for 1 mn separately (lysis buffer: 20 mM Tris HCl pH 7.5, 1.5 mM MgCl2, 350 mM NaCl, 0.5% NP-40, 5% glycerol) and the lysates were mixed together and incubated for 30 min at 37 °C. ARF1 was then purified using nickel agarose (Qiagen). N-myristoylated proteins were labelled with Alex Fluor 647 Alkyne using Click-iT Reaction Buffer Kit (Invitrogen) according to instructions with modifications. Specifically, click labelling was performed on the proteins still bound to the column. The column was then washed and proteins were eluted with 500 mM imidazole/100 mM HEPES containing 1% SDS. The N-myristoylation status was analysed by in-gel fluorescence and the equal protein load was confirmed by Coomassie stain. For cleavage inhibition, free myristic acid or vehicle control (DMSO) was added to bacterial lysate containing ARF1 before adding IpaJ. After cleavage reaction, ARF1 was then labelled and purified as described above. To test the cleavage of non-myristoylated ARF1, a similar experimental setup was used with the following modifications: E. coli BL-21 cells expressing ARF1-His (in the absence of NMTp or exogenous myristic acid) were purified with Ni-NTA agarose, eluted with 500 mM imidazole/100 mM HEPES and processed for mass spectrometry.
For metabolic labelling in mammalian cells, myristic acid-azide (Invitrogen) was added to the cell cultures 6 h after transfection at final concentration 10 μM and incubation was continued overnight. The next day, Strep-tagged proteins were purified using Strep-Tactin SuperFlow Plus columns (see above). N-myristoylated proteins were labelled with Alex Fluor 647 Alkyne as described above. The column was then washed and proteins were eluted with 2.5 mM desthiobiotin elution buffer. Purified proteins were separated on SDS–polyacrylamide gel electrophoresis gel and transferred onto nitrocellulose paper (Biorad) for probing with STREPtactin-HRP (Biorad). Fluorescence was analysed both in SDS-gel and on nitrocellulose paper (collectively ‘in-gel fluorescence’). Expression of IpaJ–GFP (wild type or catalytic mutants) was confirmed by probing cell lysates with anti-GFP antibodies (Clontech) and secondary HRP-anti-rabbit IgG antibodies (Invitrogen). Expression of GRASP65– and hVPS15–GFP chimaeric proteins was probed in cell lysate, before protein purification. For in vitro cleavage reaction, ARF1-strep transfected cells labelled with myristic acid-azide were lysed and 3 μg of recombinant IpaJ Δ50 (wild type or C64A catalytic mutant) were added into 20 μl of the lysate. After incubation at 37 °C for 30 min, myristoylated ARF1-strep was labelled and analysed as described above. For analysis of cleavage of multiple proteins, HEK293T cells were cultured in the presence of azide-modified moieties (myristic acid, palmitic acid, geranylgeranyl alcohol) according to the Click-iT protocol. Cell lysates were incubated with 3 μg of IpaJ Δ50 (wild type or C64A) for 30 min at 37 °C. Proteins containing azide-modified moieties were then labelled with Alex Fluor 647 Alkyne and purified according to the Click-iT labelling protocol.
Supplementary Material
Acknowledgments
We thank our colleagues at University of Texas Southwestern Medical Center, specifically K. Orth, Y. M. Chook and J. Seeman, for discussions in preparing this manuscript. We are particularly indebted to L. Kinch, N. Grishin, D. Mitchel, X. Guo, D. Trudgian and S. Perelman for their contributions. We acknowledge the services of the University of Texas Southwestern Medical Center proteomics core, supported by a Cancer Prevention and Research Institute of Texas grant RP120613. A.S.S. was supported by the Howard Hughes Medical Institute International Student Research fellowship. B.A.W. was supported by a National Institutes of Health training grant (NIAID; 5T32AI007520) and S.S.W. by the Burroughs Wellcome Fund. This work was supported by grants from the National Institutes of Health (NIAID; RO1AI083359 and NIGMS; R01GM100486), the Welch Foundation (I-1704) and the Burroughs Wellcome Fund to N.M.A.
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions N.B. and N.M.A. conceived the general ideas for this work. N.B. and N.M.A. planned, performed and interpreted experiments. T.G.F. and A.S.S. generated bacterial knockout strains and performed GTPase assays, respectively. J.M.E. and S.S.W. performed mouse infection studies. B.A.W. performed the bioinformatics studies on IpaJ. D.A.P. and S.M.P. performed top-down mass spectrometry. N.M.A. and N.B. wrote the manuscript and all authors provided editorial input.
Author Information Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
References
- 1.Farazi TA, Waksman G, Gordon JI. The biology and enzymology of protein N-myristoylation. J Biol Chem. 2001;276:39501–39504. doi: 10.1074/jbc.R100042200. [DOI] [PubMed] [Google Scholar]
- 2.Maurer-Stroh S, et al. MYRbase: analysis of genome-wide glycine myristoylation enlarges the functional spectrum of eukaryotic myristoylated proteins. Genome Biol. 2004;5:R21. doi: 10.1186/gb-2004-5-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McLaughlin S, Aderem A. The myristoyl-electrostatic switch: a modulator of reversible protein-membrane interactions. Trends Biochem Sci. 1995;20:272–276. doi: 10.1016/s0968-0004(00)89042-8. [DOI] [PubMed] [Google Scholar]
- 4.Kahn RA. Toward a model for Arf GTPases as regulators of traffic at the Golgi. FEBS Lett. 2009;583:3872–3879. doi: 10.1016/j.febslet.2009.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong N, et al. Structurally distinct bacterial TBC-like GAPs link Arf GTPase to Rab1 inactivation to counteract host defenses. Cell. 2012;150:1029–1041. doi: 10.1016/j.cell.2012.06.050. [DOI] [PubMed] [Google Scholar]
- 6.Mounier J, et al. Shigella effector IpaB-induced cholesterol relocation disrupts the Golgi complex and recycling network to inhibit host cell secretion. Cell Host Microbe. 2012;12:381–389. doi: 10.1016/j.chom.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 7.Selyunin AS, et al. The assembly of a GTPase-kinase signalling complex by a bacterial catalytic scaffold. Nature. 2011;469:107–111. doi: 10.1038/nature09593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashida H, et al. Shigella are versatile mucosal pathogens that circumvent the host innate immune system. Curr Opin Immunol. 2011;23:448–455. doi: 10.1016/j.coi.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Way SS, Borczuk AC, Dominitz R, Goldberg MB. An essential role for gamma interferon in innate resistance to Shigella flexneri infection. Infect Immun. 1998;66:1342–1348. doi: 10.1128/iai.66.4.1342-1348.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buysse JM, Dunyak DS, Hartman AB, Venkatesan MM. Identification and molecular characterization of a 27 kDa Shigella flexneri invasion plasmid antigen, IpaJ. Microb Pathog. 1997;23:357–369. doi: 10.1006/mpat.1997.0164. [DOI] [PubMed] [Google Scholar]
- 11.Liu M, et al. Sequence analysis and characterization of plasmid pSFD10 from Salmonella choleraesuis. Plasmid. 2002;48:59–63. doi: 10.1016/s0147-619x(02)00008-2. [DOI] [PubMed] [Google Scholar]
- 12.Michiels J, Dirix G, Vanderleyden J, Xi C. Processing and export of peptide pheromones and bacteriocins in Gram-negative bacteria. Trends Microbiol. 2001;9:164–168. doi: 10.1016/s0966-842x(01)01979-5. [DOI] [PubMed] [Google Scholar]
- 13.Slagowski NL, Kramer RW, Morrison MF, LaBaer J, Lesser CF. A functional genomic yeast screen to identify pathogenic bacterial proteins. PLoS Pathog. 2008;4:e9. doi: 10.1371/journal.ppat.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herman PK, Stack JH, DeModena JA, Emr SD. A novel protein kinase homolog essential for protein sorting to the yeast lysosome-like vacuole. Cell. 1991;64:425–437. doi: 10.1016/0092-8674(91)90650-n. [DOI] [PubMed] [Google Scholar]
- 15.Sze SK, Ge Y, Oh H, McLafferty FW. Top-down mass spectrometry of a 29-kDa protein for characterization of any posttranslational modification to within one residue. Proc Natl Acad Sci USA. 2002;99:1774–1779. doi: 10.1073/pnas.251691898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelleher NL. Top-down proteomics. Anal Chem. 2004;76:197A–203A. [PubMed] [Google Scholar]
- 17.Charron G, et al. Robust fluorescent detection of protein fatty-acylation with chemical reporters. J Am Chem Soc. 2009;131:4967–4975. doi: 10.1021/ja810122f. [DOI] [PubMed] [Google Scholar]
- 18.Barr FA, Puype M, Vandekerckhove J, Warren G. GRASP65, a protein involved in the stacking of Golgi cisternae. Cell. 1997;91:253–262. doi: 10.1016/s0092-8674(00)80407-9. [DOI] [PubMed] [Google Scholar]
- 19.Kamps MP, Buss JE, Sefton BM. Mutation of NH2-terminal glycine of p60src prevents both myristoylation and morphological transformation. Proc Natl Acad Sci USA. 1985;82:4625–4628. doi: 10.1073/pnas.82.14.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stumpo DJ, Graff JM, Albert KA, Greengard P, Blackshear PJ. Molecular cloning, characterization, and expression of a cDNA encoding the “80- to 87-kDa” myristoylated alanine-rich C kinase substrate: a major cellular substrate for protein kinase C. Proc Natl Acad Sci USA. 1989;86:4012–4016. doi: 10.1073/pnas.86.11.4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Q, et al. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franco M, Chardin P, Chabre M, Paris S. Myristoylation-facilitated binding of the G protein ARF1GDP to membrane phospholipids is required for its activation by a soluble nucleotide exchange factor. J Biol Chem. 1996;271:1573–1578. doi: 10.1074/jbc.271.3.1573. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Kahn RA, Prestegard JH. Structure and membrane interaction of myristoylated ARF1. Structure. 2009;17:79–87. doi: 10.1016/j.str.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Kahn RA, Prestegard JH. Dynamic structure of membrane-anchored Arf*GTP. Nature Struct Mol Biol. 2010;17:876–881. doi: 10.1038/nsmb.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shao F, Merritt PM, Bao Z, Innes RW, Dixon JE. A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell. 2002;109:575–588. doi: 10.1016/s0092-8674(02)00766-3. [DOI] [PubMed] [Google Scholar]
- 26.Choy A, et al. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science. 2012;338:1072–1076. doi: 10.1126/science.1227026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phalipon A, Sansonetti PJ. Shigellosis: innate mechanisms of inflammatory destruction of the intestinal epithelium, adaptive immune response, and vaccine development. Crit Rev Immunol. 2003;23:371–401. doi: 10.1615/critrevimmunol.v23.i56.20. [DOI] [PubMed] [Google Scholar]
- 28.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roth MJ, et al. Sensitive and reproducible intact mass analysis of complex protein mixtures with superficially porous capillary reversed-phase liquid chromatography mass spectrometry. Anal Chem. 2011;83:9586–9592. doi: 10.1021/ac202339x. [DOI] [PubMed] [Google Scholar]
- 31.Shima DT, Haldar K, Pepperkok R, Watson R, Warren G. Partitioning of the Golgi apparatus during mitosis in living HeLa cells. J Cell Biol. 1997;137:1211–1228. doi: 10.1083/jcb.137.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steele-Mortimer O, Meresse S, Gorvel JP, Toh BH, Finlay BB. Biogenesis of Salmonella typhimurium-containing vacuoles in epithelial cells involves interactions with the early endocytic pathway. Cell Microbiol. 1999;1:33–49. doi: 10.1046/j.1462-5822.1999.00003.x. [DOI] [PubMed] [Google Scholar]
- 33.Shiloh MU, et al. Phenotype of mice and macrophages deficient in both phagocyte oxidase and inducible nitric oxide synthase. Immunity. 1999;10:29–38. doi: 10.1016/s1074-7613(00)80004-7. [DOI] [PubMed] [Google Scholar]
- 34.Rivera VM, et al. Regulation of protein secretion through controlled aggregation in the endoplasmic reticulum. Science. 2000;287:826–830. doi: 10.1126/science.287.5454.826. [DOI] [PubMed] [Google Scholar]
- 35.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alto NM, Dixon JE. Analysis of Rho-GTPase mimicry by a family of bacterial type III effector proteins. Methods Enzymol. 2008;439:131–143. doi: 10.1016/S0076-6879(07)00410-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.