Abstract
The CCN family of proteins includes six members presently known as CCN1, CCN2, CCN3, CCN4, CCN5 and CCN6. These proteins were originally designated CYR61, CTGF, NOV, and WISP-1, WISP-2, WISP-3. Although these proteins share a significant amount of structural features and a partial identity with other large families of regulatory proteins, they exhibit different biological functions. A critical examination of the progress made over the past two decades, since the first CCN proteins were discovered brings me to the conclusion that most of our present knowledge regarding the functions of these proteins was predicted very early after their discovery. In an effort to point out some of the gaps that prevent us to reach a comprehensive view of the functional interactions between CCN proteins, it is necessary to reconsider carefully data that was already published and put aside, either because the scientific community was not ready to accept them, or because they were not fitting with the « consensus » when they were published. This review article points to avenues that were not attracting the attention that they deserved. However, it is quite obvious that the six members of this unique family of tetra-modular proteins must act in concert, either simultaneously or sequentially, on the same sites or at different times in the life of living organisms. A better understanding of the spatio-temporal regulation of CCN proteins expression requires considering the family as such, not as a set of single proteins related only by their name. As proposed in this review, there is enough convincing pieces of evidence, at the present time, in favor of these proteins playing a role in the coordination of multiple signaling pathways, and constituting a Centralized Communication Network. Deciphering the hierarchy of regulatory circuits involved in this complex system is an important challenge for the near future. In this article, I would like to briefly review the concept of a CCN family of proteins and critically examine the progress made over the past 10 years in the understanding of their biological functions and involvement in both normal and pathological processes.
Keywords: CCN, CCN proteins, CTGF, CYR61, NOV, WISP, Inflammation, Transformation, Cell proliferation, Cell signaling, Cell communication, Centralized communication network, Development, Differentiation
Brief historical and structural context
It is always useful to recall the facts that set the ground rules for current ideas in fields where new comers are not always aware of the succession of events that underlie the prevailing concepts. The history of Science contains many examples of major leaders who have seen their contribution vanish over time.1
The CCN story began in the early 90’s with the discovery in mouse, human and chicken of three proteins that were designated CYR61 (for « Cystein Rich »), CTGF (for Connective Tissue Growth Factor) and NOV (for Nephroblastoma OVerexpressed) (O’Brien et al. 1990; Bradham et al. 1991; Joliot et al. 1992) .
The primary sequence of the genes encoding these proteins revealed that they were very closely related and that they encoded proteins that share partial identity with other major classes of regulatory proteins.
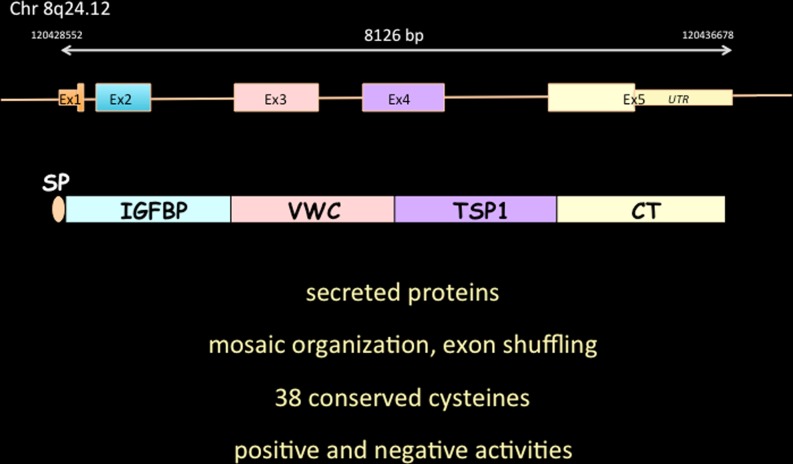
From the predicted amino acid sequences of the proteins, it was also concluded that each of these three proteins was a mosaic assembly of three structural domains that shared partial identity with the N terminus of IGF binding proteins, the type C repeat of the Von Willebrand factor, the type 1 repeat of thrombospondin and a fourth domain, that contained a set of eight cysteines that were shown to form the so-called cystine knot that is present in a whole range of regulatory proteins including growth factors and other secreted proteins. The presence of a sequence encoding a typical signal peptide at the N-terminus of these proteins, strongly suggested that they were secreted, a prediction that was indeed later experimentally confirmed (Fig. 1).
Fig. 1.
The prototypic CCN3 protein. The schematic organization of CCN3 exons on the human genome is represented with the corresponding structural domains contained in the full length version of CCN3. SP is for signal peptide. Salient features of CCN proteins are also indicated. See text for details
Based on these structural features unique to these three proteins, P. Bork (Bork 1993) considered that they should constitute a new family of proteins which he called the « CCN family of proteins ».
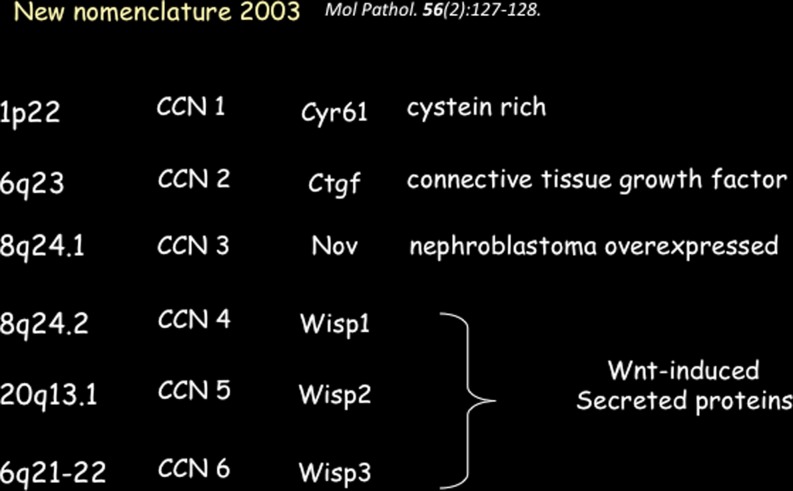
A few years later, with the birth of the International CCN Society, we proposed that the proteins belonging to this family be designated with the CCN acronym followed by a number indicating the chronological order in which they were discovered (Brigstock et al. 2003) (Fig. 2).
Fig. 2.
The CCN family of porteins in human
Both CCN1 (CYR 61) and CCN2 (CTGF) were induced by cell proliferation (Lau & Nathans, 1985, 1987; Simmons et al., 1989, Brunner et al., 1991, Ryseck et al., 1991), whereas CCN3 (NOV) was inhibited when cells were induced to proliferate. In other words, the genes encoding CCN1 and CCN2 could be classified as Immediate Early Genes, whereas CCN3 was not (Joliot et al. 1992; Scholz et al. 1996) .
The fact that NOV was not subject to the same regulatory circuits constituted the very first evidence for CCN3 having distinct biological properties.
The second major difference between CCN3 and the CCN1/CCN2 pair stemmed from the capacity of CCN3 to inhibit cell proliferation and induce cellular transformation when it was truncated at its aminoterminus (Joliot et al. 1992).
The inhibition of cell proliferation by CCN3 contrasted with the stimulatory effects of CCN1 and CCN2, therefore suggesting that, although these three proteins might belong to the same group, they were representing both positive and negative regulators of cell proliferation. These observations also argued against the possibility that the three CCN proteins had redundant functions (Perbal 2001).
There are many other examples of protein families containing both positive and negative effectors and it makes a lot of sense for cells to make use of both types of signals to smoothly regulate their biological behavior.
At this stage in the understanding, we could already make the following prédictions : 1) as CCN proteins contain a consensus signal peptide, they are likely to be secreted, 2) based on the presence of the IGFBP-like module at their N terminus CCN proteins could possibly interact with IGF signaling, 3) CCN proteins probably exist as large multimeric complexes that regulate cell adhesion and proliferation, via their modules 2 and 3, and 4) the cystine knot contained in their C-terminal module is responsible for the formation of homo and heterodimers.
The presence of a hinge région between the second and third modules of the CCN proteins suggested possible translational processing by proteolytic digestion (Perbal et al. 1999, 2003), as also discussed by D. Brigstock, L. Lau and myself, at the first International Workshop on the family of Genes in Saint-Malo (Ayer-Lelievre et al. 2001)
Interestingly, the amino-truncated form of CCN3 that was expressed in one chicken nephroblastoma cell line, as the result of a MAV1-proviral insertion in the nov gene (Perbal 1994, 1995) was shown to induce morphological transformation of chicken fibroblasts when expressed under the control of a RSV-derived promoter (Joliot et al. 1992). On the contrary, the full length protein expressed in the same conditions induced cell growth arrest.
These observations suggested that the inhibitory effect of CCN3 on cell proliferation required the expression of a full length protein. Results that we obtained later on (see below) challenged that conclusion.
A few years later, in 1998, a new member of the CCN family was discovered (Zhang et al. 1998) and designated r-Cop1. In their manuscript, the authors claim that i) « rCOP-1 represents a new class of CCN proteins that have functions opposing those of previously indentified members » ii) that the studies performed on Nov « established the importance of the CCN family in positive cell growth regulation and their involvement in carcinogenesis »; and iii) that « ..from the pattern of its gene expression, Nov may be a negative cell growth regulator like rCop-1 » .
It seems that the authors did not fully appreciate that the induction of CEF cell growth arrest induced by the full length NOV protein was a critical biological feature that we had considered being sufficient enough to distinguish NOV from other CCN proteins, as it was the first member of the family showing antiproliferative activity.
The identification of rCOP-1 not only indicated that the CCN family of proteins contained at least another member with inhibitory effects on cell growth, but it raised a series of very interesting questions that are still open today (see below).
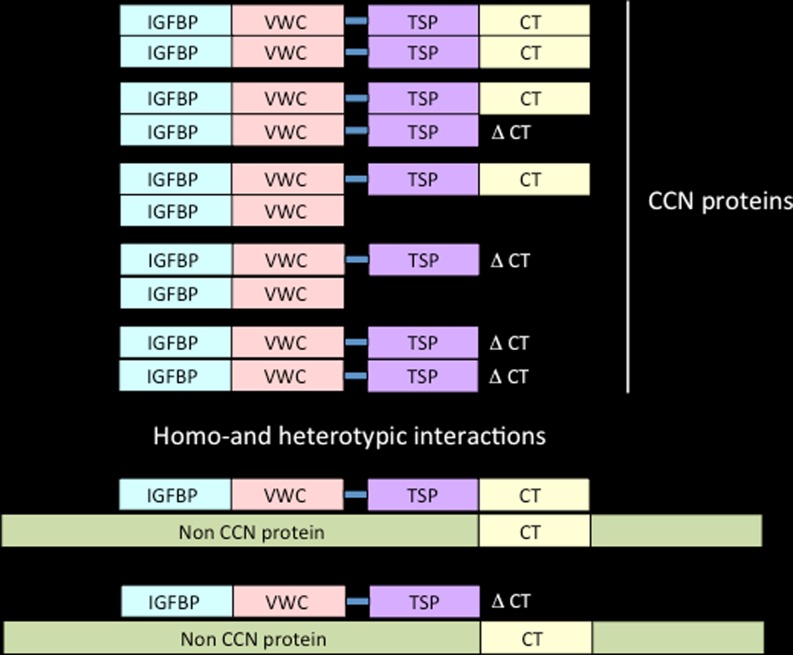
Since the rCOP-1 protein lacks the C-terminal module of the canonical CCN family members, one can expect it to act as a « dominant negative » effector in the homo and heterotropic multimerization of CCN proteins, either with other members of the CCN family or with proteins which do not belong to the CCN family but physically interact with CCN family members (Fig. 3).
Fig. 3.
Model for homotypic and heterotypic interactions. CCN proteins can make use of their CT and VWC domains to interact with other proteins harboring the same type of domains. Proteins lacking one or more of these domains may act as dominant negative. The importance of these potential interactions should be considered in the context of previously reported CCN isoforms lacking domains (Perbal 2009). Non-CCN proteins with CT-like structure can also interact with CCN proteins. This type of interaction is proposed to account for the transport of CCN3 to the nucleus
Soon after the discovery of rCOP-1, two new members of the CCN family (WISP-1 and WISP-2) were identified as proteins encoded by genes whose expression was up-regulated in Wnt-1 transformed cells (Pennica et al. 1998). A search for related proteins in expressed sequence tag (EST) database also identified a homologous protein that was called WISP-3.
In their seminal article the authors proposed that WISP-1, WISP-2, and WISP-3 defined another functional subfamily in the CCN family of proteins. Interestingly, WISP-2 turned out to be the human ortholog of rCOP-1 and confirmed the previously established antiproliferative activity of this protein.
In light of these observations the picture emerged of a « functionally bipartite » CCN family of proteins, comprising members that are involved either in the stimulation or in the inhibition of cell proliferation (Perbal 2001).
From these structural considerations, also stemmed the concept of « combinatorial events » governing the wide variety of biological functions attributed to the CCN proteins (Perbal 2001).
The basics are the following :
-
i)
the expression profile of CCN proteins is tightly regulated both at temporal and spatial levels,
-
ii)
the four CCN modules interact with a large number of diverse ligands and partners.
-
iii)
the interactions, and their biological consequences, are possible, and these occur only if the components of the system are present at the same place at the same time.
Of course, what is true for one pair also stands for tripartite or tetrapartite combinations, involving one or more assembled or free proteins.
The biological functions of the CCN proteins are predicted to be highly dependent upon the microenvironment in which they exist at any specific time.
Furthermore, the nature of the interactions that are permitted in a defined micro-environment, can also govern the ability of CCN proteins to physically interact with other members of the CCN family or other proteins outside of the family, as discussed above.
Variations around this theme would provide a solid basis for reconciling many « apparently opposite, or adverse » results.
Biological properties of CCN proteins
A number of sound reviews have addressed key aspects of the theme since 1999, and have blossomed over the past decade (Brigstock 1999, 2003; Lau and lam 1999; Perbal 2001, 2003, 2004, 2006a; Blom et al. 2002; Planque and Perbal 2003; Bleau et al. 2005; Chaqour and Goppelt-Struebe 2006; Leask and Abraham 2006; Kubota and Takigawa 2007; Kleer et al. 2007; Holbourn et al. 2008; Shi-Wen et al. 2008; Hall-Glenn and Lyons 2011, Jun and Lau 2011; Lau 2011; Arnott et al. 2011; Ouellet and Siegel 2012; Mason 2013). Those who are interested in the biological functions of CCN proteins should go back to these articles which contain thoughtfull considerations, models, and sometimes provocative ideas. Many views discussed in these articles still represent challenges that should be addressed in the light of the widening interest of the scientific community for the field.
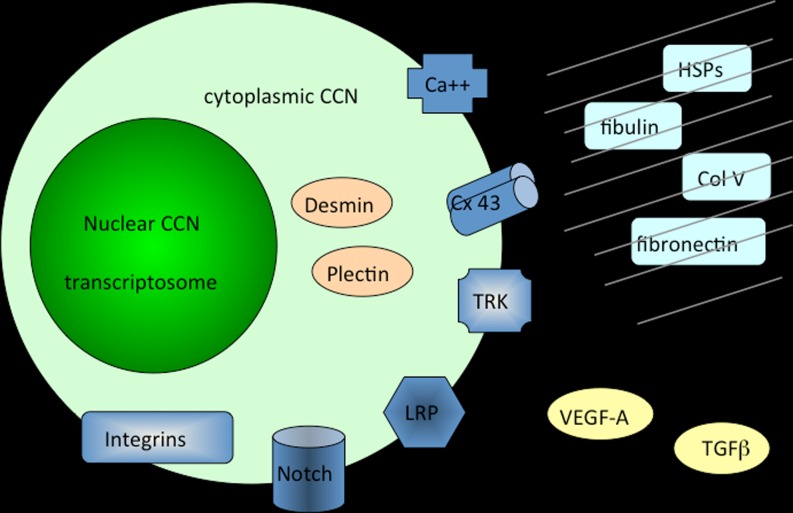
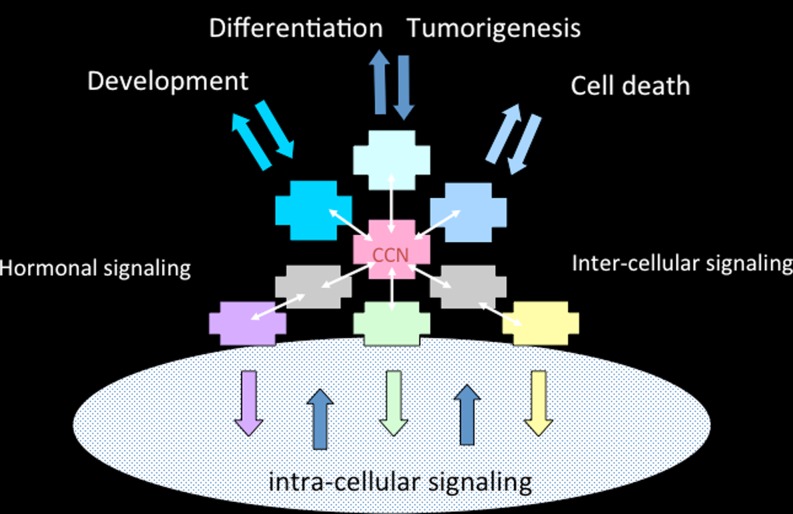
Considering the tetramodular structure of the CCN proteins, it was not much of a surprise that they were found to participate in many essential biological functions including cell communication, control of proliferation, adhesion and migration, regulation of growth, development and differentiation, wound healing, regeneration, and cell death (Yeger and Perbal 2007) (Fig. 4).
Fig. 4.
Schematic drawing of CCN proteins partners. Interactions of CCN proteins with other ligands and regulators are shown here to occur in the extracellular matrix, at the cell membrane and inside the cytoplasm and the nucleus of cells
In order to better understand the antiproliferative activity of CCN3 that we had established in avian primary chicken embryo fibroblasts we first studied the effects on CCN3 on the progression of cell cycle.
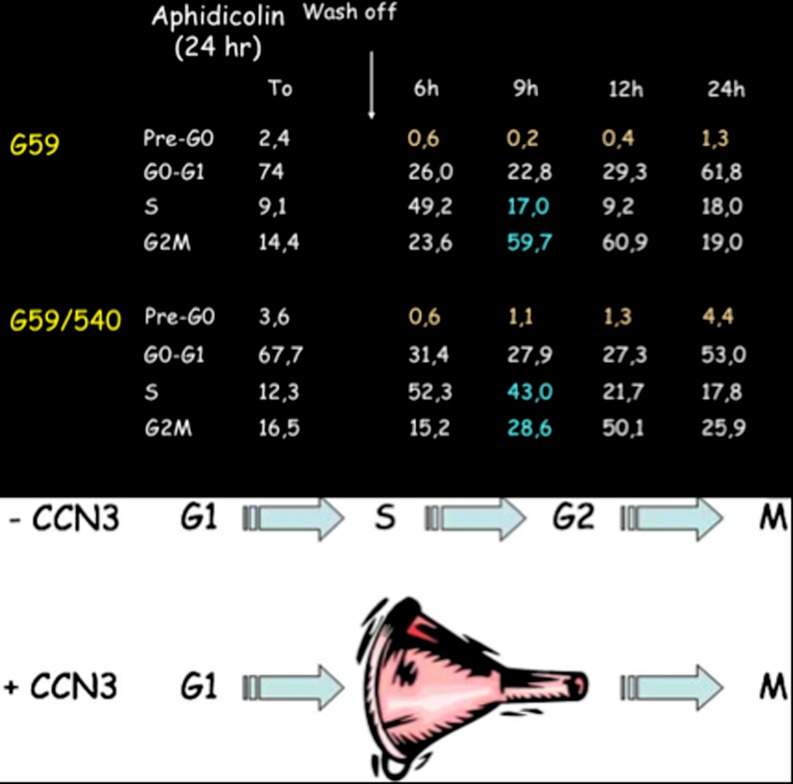
Our initial measurements did not permit us to establish whether CCN3 expression modified the ratio of cell subpopulations at various phases of the cell cycle. When synchronized cell cultures were used, we could establish that the expression of CCN3 interfered with the S/G2 transition of the cell cycle, thereby inducing an artificial accumulation of cells at the S phase (Bleau et al. 2007). These results not only confirmed the antiproliferative activity of CCN3 but they permitted reconciling our present observation with a set of results published by another group who claimed that CCN3 stimulated cell growth, based on Brdu incorporation (Perbal 2008) (Fig. 5).
Fig. 5.
Inhibition of cell proliferation by CCN3 results from a break at the S/G2 transition in the cell cycle. Synchronized cells that express CCN3 accumulate at the S phase. As a result the cells go less efficiently through the cycle. Data are from Bleau et al. 2007 (see text)
In spite of our demonstration that the increase in Brdu incorporation was a direct result of the CCN3 inhibitory effect on cell growth, it is still common to find in the literature, groups who claim that CCN3 shows growth stimulatory properties (based on the Brdu paper !).
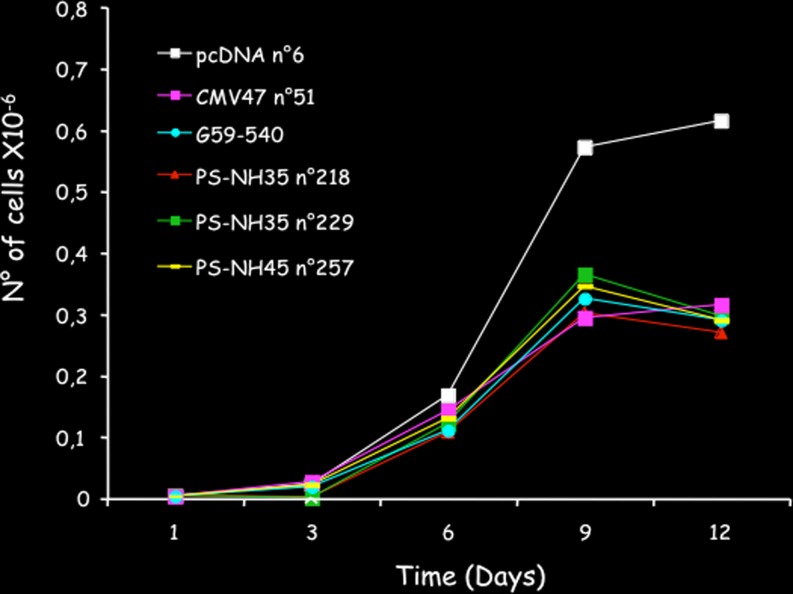
The antiproliferative activity of CCN3 has been confirmed in many different cell types, both by our group and others (Gupta et al. 2001; Sakamoto et al. 2002; McCallum et al. 2006; Bleau et al. 2007; Benini et al. 2005; Fukunaga-Kalabis et al. 2006; Planque et al. 2006; van Roeyen et al. 2008; Vallacchi et al. 2008; Shimoyama et al. 2010; Lin et al. 2010). Experiments performed in our laboratory also established that the inhibitory effects on cell growth do not require the two first domains of CCN3 (Planque et al. 2006) (Fig. 6).
Fig. 6.
The C-terminal half of CCN3 is sufficient to promote antiproliferative activity. The drawing show the growth curves of stably transfected cell lines constructs expressing either the full length CCN3 protein CMV47 and G59-540, the three first domains of CCN3 (NH35), or the C-terminal half of CCN3 (NH45). All constructs induce a dramatic inhibition of cell proliferation. The effects obtained with the NH45 transfected cells indicated that the inhibitory potential is contained within the last two domains of CCN3. Data are fom Bleau et al. 2007 (see text)
Since CCN5 which lacks the CT domain also shows a growth inhibitory effect it is tempting to assign the growth inhibition property to the TSP1 domain of CCN proteins but this needs to be confirmed experimentally.
Along the same line, we could establish that CCN3 also shows an anti-tumorigenic activity, in choriocarcinomas, glioblastomas and Ewing’s tumors (Gellhaus et al. 2004; Gupta et al. 2001; Benini et al. 2005) . In the case of Ewing’s and osteosarcomas, we could also establish that CCN3 is a marker of poor prognosis. Out of 45 patients with primary Ewing’s tumors, those who did not express CCN3 in the primary tumors did not develop metastases whereas, 50 % of the patients with primary tumors positive for CCN3 developed metastastic tumors (Manara et al. 2002).
Similar observations were made in our studies on osteosarcomas where we could establish that expression of CCN3 significantly reduced survival (Perbal et al. 2008).
Pathological conditions are often seen as useful to decipher the role(s) of proteins whose functions may be altered by the environment.
However, one should keep in mind that the protein functions that are uncovered in the context of a given pathological condition might not be comparable to the functions of the same protein in other pathological conditions or in normal conditions.
Thresholds are critical in many instances and the balance between negative and positive signals that is maintained in a normal context might be distrupted in a completely aberrant way in the pathological context.
An illustration of this complexity was shown in a set of two collaborative approaches where the results obtained with skin reconstructs were integrated, and analyzed in the light of the situation encountered in melanomas (Fukunaga-Kalabis et al. 2006, 2008; Vallacchi et al. 2008).
First, the use of skin reconstructs established that CCN3 was essential for the 3D localisation of melanocytes at the basal membrane of normal skin, suggesting that a dysruption of the normal crosstalk between melanocytes and keratinocytes would result in a dermal invasion by melanocytes comparable to the situation that occurs in melanomas. Indeed, CCN3 was found to be inversely correlated with melanoma invasion, as measured in a set of human melanocytic nevi and melanoma samples.
However, in another set of samples, the metastatic potential of melanomas was directly related to the overexpression of CCN3 and the survival rate was much reduced in patients expressing the nuclear truncated CCN3 species previously identified in other agressive tumors.
These observations highlighted the difficulties in simply relating the expression of a marker such as CCN3 to the complexe biology of tumors, even though the levels of CCN3 expression could be used as an indicator for the staging of tumors and in some cases for a molecularly based therapeutics.
CCN proteins: matricellular proteins only?
Most recently published articles quote in their introductory comments, that CCN proteins are secreted and play their biological functions as essential components of the extracellular matrix. Indeed, for the past decade, the « scientifically-correct » way of thinking in the CCN community, was focused on the these proteins being « matricellular proteins ».
Although there is no doubt that some of the CCN proteins are associated to the extracellular matrix (ECM), the situation may not be that simple.
First of all, published data regarding subcellular localization of the CCN proteins clearly indicate that the distribution of the CCN proteins is not similar for all members of the family. For example, CCN2 is barely detected in the cell culture medium (Ball et al. 1998) and is mainly detected at the cell membrane, whereas newly synthesized CCN1 is not heavily detected in the culture medium, as it associates quickly with the ECM where it appears to be stabilized (half-life of greater than 24 h as compared to 30 min for intracellular and cell surface associated CCN1 (Yang and Lau 1991). We have established that CCN3 is efficiently secreted and detected as a long half-life protein in conditioned medium from CCN3 positive cells. It is also detected at the cell membrane and in the ECM (Joliot et al. 1992, Kyurkchiev et al. 2004).
The subcellular localization of rCop-1 is also informative, as it was neither detected in the conditioned medium of infected cells nor in the ECM, and it appeared to be retained in the cytoplasm of cells (Zhang et al. 1998). These data should be taken in careful consideration as it may inform us not only about the relative importance of each structural module in the secretion process, but also about the fate of the CCN proteins when they are not secreted .
Considering the major influence of the complex environment that constitutes the ECM, and the potential physical and functional interactions of CCN proteins with other matricellular proteins, is it wise and meaningful to study CCN protein function(s) by using a soluble semi-purified protein preparation or to use a test outside of a reconstructed matrix environment when one addresses the matricellular functions of CCN proteins ?
The studies that we had performed to better understand the transforming properties of CCN3 in avian nephroblastomas led us to uncover an unexpected aspect of the biological functions for CCN proteins.
First of all, the nucleus of several tumor cell lines stained positive when immunostaining was performed with a CCN3-specific antibody that was raised against the C-terminus of the protein (Perbal 1999, 2004, 2006b). In spite of a whole set of additional controls that were performed in response to reviewers’ comments, the CCN community did not receive this piece of data with enthusiasm. Several other growth factors and receptors had been previously shown to reside in the nucleus of cells but obviously, some of the CCN cell biologists did not like to see « secreted » proteins partitioning between the ECM and the nucleus …
At the same time, IGF binding proteins were also detected in the nucleus. The response of the IGF community was much more positive and a few groups addressed very nicely the problem. It is important to remember, at that stage in the investigations, IGFBPs and CCN proteins were found to share a partial identity at their aminoterminal ends.
Those who are interested in these considerations should read the communication of Professor Rob Baxter in this issue.
A few years after our initial report, a nuclear form of CCN2 was also described (Wahab et al. 2001). From what I undestood, and in spite of the high quality of the data that was presented, the publication of this manuscript was not easy.
Other examples of nuclear CCN1, CCN2, CCN3 and CCN5 proteins were reported (Hirschfeld et al. 2009; Sha and Leask 2011; Wiesman et al. 2010; Rittié et al. 2011), but it seems that nuclear CCN proteins do not trigger much interest in the CCN community in spite of accumulating evidence that attributes to nuclear proteins a role in the control of transcription.
A careful examination of the truncated CCN3 protein that was identified in one of the MAV1-induced avian nephroblastoma, led to the conclusion that the aminotruncation of CCN3 might affect the fate of the protein within the tumor cells. Indeed, we could demonstrate that the proviral insertion within the ccn3 gene removed the sequences encoding the signal peptide, and resulted in a CCN3 protein that was addressed to the nucleus (Planque et al. 2006).
Further experiments established that although it does not contain a typical nuclear localisation signal, the C-terminal module of CCN3 was responsible for the nuclear addressing of CCN3 deprived of its signal peptide (Planque et al. 2006).
One possible interpretation is that the truncated CCN3 protein associates with a carrier that belongs to the group of proteins which contain a cystine knot similar to the one contained in the CT module of CCN3 (see Fig. 3).
In support of this hypothesis it is worth noting that imunogold staining helped to identify CCN3 protein at some nucleus pores, and a careful examination of the images indicated that the gold label was not associated with a vesiculated structure (Thomopoulos et al. 2001). Of course further studies are required to firmly establish these conclusions.
Future directions
This critical overview brings me to address a few questions that are still open and that constitute, in my eyes, very promising fields for future studies.
A single protein and/or complexes ?
Many of us have faced situations where several unexpected protein « bands » are detected on a blot ! Of course there are basic explanations for this result, including the poor quality of the reagents used, a problem that was addressed several times during the CCN workshops by our colleagues. For example, L. Lau raised the need to distribute reference samples of tested reagents and G. Fisher proposed an « ICCNS qualification » that could be given to satisfactory batches of reagents (commercially available or not) once they are tested by reference laboratories.
The use of good reagents also leads to the immunodetection of several other relevant bands in addition to the major canonical one described in the literature.
When this issue was raised at the first CCN Workshop, many participants in the audience admitted that they also observed, larger and shorter bands, that were running at similar apparent molecular weights in different laboratories, using different sources of antibodies and samples.
Although there is no published thorough study of these extra bands, it is generally accepted that they may represent CCN-related proteins. To my knowledge, there is at least one unpublished, as yet, proof of evidence that links a high molecular weight band to the canonical form of a CCN protein.
Ignoring these extra bands is not a satisfactory way of dealing with that important question.
Considering the structural features of the CCN proteins and the presence of sequences known to generate high molecular weight complexes in other proteins, it is possible that complexes resistant to the denaturing agents that are usually used to run gels, are formed between CCN proteins and other components, with the possibility that covalent links might even be involved in some cases.
In my opinion, the existence of CCN isoforms (Perbal 2004, 2009) is an aspect of the CCN biology that has been underestimated for years, and one that will need to be addressed. As post translational modifications of CCN proteins other than glycosylation will be uncovered, it will become obvious that they are key aspects of the regulatory events governing the biology of these proteins.
The implications of post translational modifications in the regulation of protein interactions with receptors and other partners is well documented in other systems. Recognizing their importance in the CCN field will certainly have profound consequences on the understanding of CCN protein biology
Interactivity and cross regulation
Cross regulation between expression of two CCN proteins was first observed by ChangLong Li in my laboratory, who reported that the induction of CCN2 in adrenocortical tumor cells which express high levels of CCN3, resulted in a dramatic down regulation of CCN3 expression.
Based on this observation, a collaborative study performed with the laboratory of B. Riser confirmed and extended that observation, in establishing that the expression of CCN2 and CCN3 was inversely correlated and that expression of CCN3 inhibited CCN2 (Riser et al. 2009).
Interestingly, CCN2 knocked-down cells express high levels of CCN3, and the antagonistic effects of CCN2 (stimulation of cell growth) and CCN3 (inhibition of cell growth) are key factors in bone and cartilage differentiation (Kawaki et al. 2008).
The isolation of the first true CCN3 Knock Out mice (Shimoyama et al. 2010; Perbal 2007) will permit to better decipher the functionnal relationships between the CCN2 and CCN3 proteins.
The inverse functional relationship between these two CCN proteins is quite important in the light of their documented opposite effects on cell proliferation and differentiation in various cellular systems.
Indeed, future therapeutic approaches based on CCN protein targeting should take these observations into account in order to selectively alter the expression of CCN2 or CCN3.
One or more receptors? The concept of a Centralized Communication Network (CCN)
In the same vein, the search for a CCN receptor has suffered from a lack of « open minded » approaches. Evidence accumulated quickly in favor of the CCN proteins interacting not with a single receptor, but with a whole variety of receptors on the cell membrane.
The question now is, not to decide whether these represent true receptors, but rather to establish whether the CCN proteins that were shown to interact with receptor A and B are binding to these two at the same time and in the same environment. The simultaneous interaction of CCN proteins with more than one receptor might allow establishing a physical connection, hence a physical support to the coordination, of several distinct regulatory pathways.
Along with this view I had previously proposed that the tetramodular structure of the CCN proteins allows them to engage with a large range of effectors (Perbal 2001), and thereby to act as scaffolds permitting a coordinated and integrated control of pathways that are activated towards the same end point, such as differentiation, growth, cell movement, etc. (Perbal and Perbal 2007) (Fig. 7).
Fig. 7.
A model for the CCN Centralized Communication Network. In this model, CCN proteins constitute the control center of a complex array of interactions which are part of several signaling pathways whose action must be tightly coordinated to provoke responses that must be adapted to the variations of the surrounding microenvironment and on another scale, to the variations of the outside medium in which organisms evolve. The coordination of these pathways by CCN proteins is based on functional interactions that have been reported for example with Integrins, Bone Morphogenic Proteins, Notch1, Calcium channels, fibulin 1C, Wnt, Heparan sulfate proteoglycans, decorin, TGFbeta, Tyrosine receptor kinase A, Low-density lipoprotein receptor related protein, and CCN proteins themselves
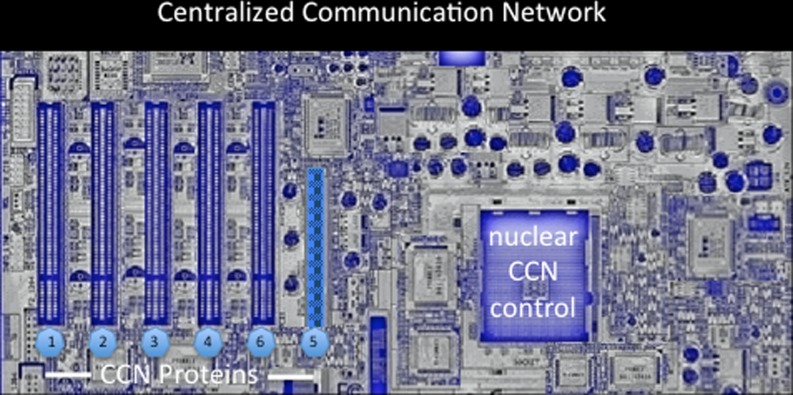
It is more and more obvious now that the interactions of CCN proteins with a mutlitude of effectors involved in the regulation of key signaling pathways, place the CCN proteins at the center of a Centralized Communication Network (Fig. 8).
Fig. 8.
An artistic representation of the Centralized Communication Network (CCN)
Acknowledgments
I am grateful to all the colleagues with whom I could engage very fruitful collaborative projects. The International Workshops on the CCN family of Genes have been the source the inspirational discussions. Again this review provides me the opportunity to deeply thank my wife Annick for her tremendous help in organizing the workshops and my friend and colleague Herman Yeger for his early involvement and support in this venture. The work that was performed in my laboratory was funded by grants from the European Union : PROTHETS (Prognosis and Therapeutic Targets of Ewing Family of Tumors, FP6 Contract 503036), grants from the Ligue Nationale Contre le Cancer, and the French Ministry of Education.
Footnotes
For example, who still remembers the pivotal contribution of JC Chermann in the identification of the HIV as AIDS associated virus see « http://nobelchermann.com/ »
References
- Arnott JA, Lambi AG, Mundy C, Hendesi H, Pixley RA, Owen TA, Safadi FF, Popoff SN. The role of connective tissue growth factor (CTGF/CCN2) in skeletogenesis. Crit Rev Eukaryot Gene Expr. 2011;21(1):43–69. doi: 10.1615/CritRevEukarGeneExpr.v21.i1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayer-Lelievre C, Brigstock D, Lau L, Pennica D, Perbal B, Yeger H. Report and abstracts on the first international workshop on the CCN family of genes. Mol Pathol. 2001;54(2):105–120. doi: 10.1136/mp.54.2.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball DK, Surveyor GA, Diehl JR, Steffen CL, Uzumcu M, Mirando MA, Brigstock DR. Characterization of 16- to 20-kilodalton (kDa) connective tissue growth factors (CTGFs) and demonstration of proteolytic activity for 38-kDa CTGF in pig uterine luminal flushings. Biol Reprod. 1998;59(4):828–835. doi: 10.1095/biolreprod59.4.828. [DOI] [PubMed] [Google Scholar]
- Benini S, Perbal B, Zambelli D, Colombo MP, Manara MC, Serra M, Parenza M, Martinez V, Picci P, Scotlandi K. In Ewing’s sarcoma CCN3(NOV) inhibits proliferation while promoting migration and invasion of the same cell type. Oncogene. 2005;24(27):4349–4361. doi: 10.1038/sj.onc.1208620. [DOI] [PubMed] [Google Scholar]
- Bleau AM, Planque N, Perbal B. CCN proteins and cancer: two to tango. Front Biosci. 2005;10:998–1009. doi: 10.2741/1594. [DOI] [PubMed] [Google Scholar]
- Bleau AM, Planque N, Lazar N, Zambelli D, Ori A, Quan T, Fisher G, Scotlandi K, Perbal B. Antiproliferative activity of CCN3: involvement of the C-terminal module and post-translational regulation. J Cell Biochem. 2007;101(6):1475–1491. doi: 10.1002/jcb.21262. [DOI] [PubMed] [Google Scholar]
- Blom IE, Goldschmeding R, Leask A. Gene regulation of connective tissue growth factor: new targets for antifibrotic therapy? Matrix Biol. 2002;21(6):473–482. doi: 10.1016/S0945-053X(02)00055-0. [DOI] [PubMed] [Google Scholar]
- Bork P. The modular architecture of a new family of growth regulators related to connective tissue growth factor. FEBS Lett. 1993;327:125–130. doi: 10.1016/0014-5793(93)80155-N. [DOI] [PubMed] [Google Scholar]
- Bradham DM, Igarashi A, Potter RL, Grotendorst GR. Connective tissue growth factor: a cysteine-rich mitogen secreted by human vascular endothelial cells is related to the SRC-induced immediate early gene product CEF-10. J Cell Biol. 1991;114:1285–1294. doi: 10.1083/jcb.114.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigstock DR. The connective tissue growth factor/cysteine-rich 61/nephroblastoma overexpressed (CCN) family. Endocr Rev. 1999;20:189–206. doi: 10.1210/er.20.2.189. [DOI] [PubMed] [Google Scholar]
- Brigstock DR. The CCN family: a new stimulus package. J Endocrinol. 2003;178(2):169–175. doi: 10.1677/joe.0.1780169. [DOI] [PubMed] [Google Scholar]
- Brigstock DR, Goldschmeding R, Katsube KI, Lam SCT, Lau LF, Lyons K, Naus C, Perbal B, Riser B, Takigawa M, Yeger H. Proposal for a unified CCN nomenclature. J Clin Pathol Mol Pathol. 2003;56:127–128. doi: 10.1136/mp.56.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaqour B, Goppelt-Struebe M. Mechanical regulation of the Cyr61/CCN1 and CTGF/CCN2 proteins. FEBS J. 2006;273(16):3639–3649. doi: 10.1111/j.1742-4658.2006.05360.x. [DOI] [PubMed] [Google Scholar]
- Fukunaga-Kalabis M, Martinez G, Liu ZJ, Kalabis J, Mrass P, Weninger W, Firth SM, Planque N, Perbal B, Herlyn M. CCN3 controls 3D spatial localization of melanocytes in the human skin through DDR1. J Cell Biol. 2006;175(4):563–569. doi: 10.1083/jcb.200602132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga-Kalabis M, Martinez G, Telson SM, Liu ZJ, Balint K, Juhasz I, Elder DE, Perbal B, Herlyn M. Downregulation of CCN3 expression as a potential mechanism for melanoma progression. Oncogene. 2008;27(18):2552–2560. doi: 10.1038/sj.onc.1210896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellhaus A, Dong X, Propson S, Maass K, Klein-Hitpass L, Kibschull M, Traub O, Willecke K, Perbal B, Lye SJ, Winterhager E. Connexin43 interacts with NOV: a possible mechanism for negative regulation of cell growth in choriocarcinoma cells. J Biol Chem. 2004;279(35):36931–36942. doi: 10.1074/jbc.M404073200. [DOI] [PubMed] [Google Scholar]
- Gupta N, Wang H, McLeod TL, Naus CC, Kyurkchiev S, Advani S, Yu J, Perbal B, Weichselbaum RR. Inhibition of glioma cell growth and tumorigenic potential by CCN3 (NOV) Mol Pathol. 2001;54(5):293–299. doi: 10.1136/mp.54.5.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Glenn F, Lyons KM. Roles for CCN2 in normal physiological processes. Cell Mol Life Sci. 2011;68(19):3209–3217. doi: 10.1007/s00018-011-0782-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschfeld M, van Hausen A, Bettendorf H, Jäger M, Stickeler E. Alternative splicing of Cyr61 is regulated by hypoxia and significantly changed in breast cancer. Cancer Res. 2009;69(5):2082–2090. doi: 10.1158/0008-5472.CAN-08-1997. [DOI] [PubMed] [Google Scholar]
- Holbourn KP, Acharya KR, Perbal B. The CCN family of proteins: structure-function relationships. Trends Biochem Sci. 2008;33(10):461–473. doi: 10.1016/j.tibs.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliot V, Martinerie C, Dambrine G, Plassiart G, Brisac M, Crochet J, Perbal B. Proviral rearrangements and overexpression of a new cellular gene (nov) in myeloblastosis-associated virus type 1-induced nephroblastomas. Mol Cell Biol. 1992;12:10–21. doi: 10.1128/mcb.12.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun JI, Lau LF. Taking aim at the extracellular matrix: CCN proteins as emerging therapeutic targets. Nat Rev Drug Discov. 2011;10(12):945–963. doi: 10.1038/nrd3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaki H, Kubota S, Suzuki A, Lazar N, Yamada T, Matsumura T, Ohgawara T, Maeda T, Perbal B, Lyons KM, Takigawa M. Cooperative regulation of chondrocyte differentiation by CCN2 and CCN3 shown by a comprehensive analysis of the CCN family proteins in cartilage. J Bone Miner Res. 2008;23(11):1751–1764. doi: 10.1359/jbmr.080615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleer CG, Zhang Y, Merajver SD. CCN6 (WISP3) as a new regulator of the epithelial phenotype in breast cancer. Cells Tissues Organs. 2007;185(1–3):95–99. doi: 10.1159/000101308. [DOI] [PubMed] [Google Scholar]
- Kubota S, Takigawa M. Role of CCN2/CTGF/Hcs24 in bone growth. Int Rev Cytol. 2007;257:1–41. doi: 10.1016/S0074-7696(07)57001-4. [DOI] [PubMed] [Google Scholar]
- Kyurkchiev S, Yeger H, Bleau AM, Perbal B. Potential cellular conformations of the CCN3(NOV) protein. Cell Commun Signal. 2004;2(1):9. doi: 10.1186/1478-811X-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau LF. CCN1/CYR61: the very model of a modern matricellular protein. Cell Mol Life Sci. 2011;68(19):3149–3163. doi: 10.1007/s00018-011-0778-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau LF, Lam SC. The CCN family of angiogenic regulators: the integrin connection. Exp Cell Res. 1999;248(1):44–57. doi: 10.1006/excr.1999.4456. [DOI] [PubMed] [Google Scholar]
- Leask A, Abraham DJ. All in the CCN family: essential matricellular signaling modulators emerge from the bunker. J Cell Sci. 2006;119(Pt 23):4803–4810. doi: 10.1242/jcs.03270. [DOI] [PubMed] [Google Scholar]
- Lin Z, Natesan V, Shi H, Hamik A, Kawanami D, Hao C, Mahabaleshwar GH, Wang W, Jin ZG, Atkins GB, Firth SM, Rittié L, Perbal B, Jain MK. A novel role of CCN3 in regulating endothelial inflammation. J Cell Commun Signal. 2010;4(3):141–153. doi: 10.1007/s12079-010-0095-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manara MC, Perbal B, Benini S, Strammiello R, Cerisano V, Perdichizzi S, Serra M, Astolfi A, Bertoni F, Alami J, Yeger H, Picci P, Scotlandi K. The expression of ccn3(nov) gene in musculoskeletal tumors. Am J Pathol. 2002;160(3):849–859. doi: 10.1016/S0002-9440(10)64908-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason RM. Fell-Muir lecture: connective tissue growth factor (CCN2)—a pernicious and pleiotropic player in the development of kidney fibrosis. Int J Exp Pathol. 2013;94(1):1–16. doi: 10.1111/j.1365-2613.2012.00845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum L, Price S, Planque N, Perbal B, Pierce A, Whetton AD, Irvine AE. A novel mechanism for BCR-ABL action: stimulated secretion of CCN3 is involved in growth and differentiation regulation. Blood. 2006;108(5):1716–1723. doi: 10.1182/blood-2006-04-016113. [DOI] [PubMed] [Google Scholar]
- O’Brien TP, Yang GP, Sanders L, Lau LF. Expression of cyr61, a growth factor-inducible immediate-early gene. Mol Cell Biol. 1990;10:3569–3577. doi: 10.1128/mcb.10.7.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet V, Siegel PM. CCN3 modulates bone turnover and is a novel regulator of skeletal metastasis. J Cell Commun Signal. 2012;6(2):73–85. doi: 10.1007/s12079-012-0161-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennica D, Swanson TA, Welsh JW, Roy M, Lawrence D, Lee J, Brush J, Taneyhill L, Deuel B, Lew M, Watanabe C, Cohen R, Melhem M, Finley G, Quirke P, Goddard A, Hillan K, Gurney A, Botstein D, Levine A. WISP genes are members of the connective tissue growth factor family that are up-regulated in Wnt-1-transformed cells and aberrantly expressed in human colon tumors. Proc Natl Acad Sci USA. 1998;95:14717–14722. doi: 10.1073/pnas.95.25.14717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbal B. Caractérisation et expression du proto-oncogène nov humain dans les tumeurs de Wilms. Bull Cancer (Paris) 1994;81:957–961. [Google Scholar]
- Perbal B. Contribution of MAV-1-induced nephroblastoma to the study of genes involved in human Wilms’ tumor development. Crit Rev Oncog. 1995;5:589–613. [PubMed] [Google Scholar]
- Perbal B. Nuclear localization of NOV protein: a potential role for nov in the regulation of gene expression. Mol Pathol. 1999;52:84–91. doi: 10.1136/mp.52.2.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbal B. NOV (nephroblastoma overexpressed) and the CCN family of genes: structural and functional issues. Mol Pathol. 2001;54:57–79. doi: 10.1136/mp.54.2.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbal B. The CCN3 (NOV) cell growth regulator: a new tool for molecular medicine. Expert Rev Mol Diagn. 2003;3(5):597–604. doi: 10.1586/14737159.3.5.597. [DOI] [PubMed] [Google Scholar]
- Perbal B. CCN proteins: multifunctional signalling regulators. Lancet. 2004;363(9402):62–64. doi: 10.1016/S0140-6736(03)15172-0. [DOI] [PubMed] [Google Scholar]
- Perbal B. The CCN3 protein and cancer. Adv Exp Med Biol. 2006;587:23–40. doi: 10.1007/978-1-4020-5133-3_3. [DOI] [PubMed] [Google Scholar]
- Perbal B. New insight into CCN3 interactions–nuclear CCN3: fact or fantasy? Cell Commun Signal. 2006;4:6. doi: 10.1186/1478-811X-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbal B. CCN3-mutant mice are distinct from CCN3-null mice. J Cell Commun Signal. 2007;1(3–4):229–230. doi: 10.1007/s12079-008-0020-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbal B. CCN3: Doctor Jekyll and Mister Hyde. J Cell Commun Signal. 2008;2(1–2):3–7. doi: 10.1007/s12079-008-0028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbal B. Alternative splicing of CCN mRNAs .... it has been upon us. J Cell Commun Signal. 2009;3(2):153–157. doi: 10.1007/s12079-009-0051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbal A, Perbal B. CCN proteins, microenvironment, communication and signaling: why did we need a new journal? J Cell Commun Signal. 2007;1(1):1–3. doi: 10.1007/s12079-007-0007-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbal B, Martinerie C, Sainson R, Werner M, He B, Roizman B. The C-terminal domain of the regulatory proteinNOVH is sufficient to promote interaction with fibulin 1C: a clue for a role of NOVH in cell-adhesion signaling. Proc Natl Acad Sci U S A. 1999;96:869–874. doi: 10.1073/pnas.96.3.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbal B, Brigstock DR, Lau L. Report of the second international workshop on the CCN family of genes. J Clin Pathol Mol Pathol. 2003;56:80–85. doi: 10.1136/mp.56.2.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbal B, Zuntini M, Zambelli D, Serra M, Sciandra M, Cantiani L, Lucarelli E, Picci P, Scotlandi K. Prognostic value of CCN3 in osteosarcoma. Clin Cancer Res. 2008;14(3):701–709. doi: 10.1158/1078-0432.CCR-07-0806. [DOI] [PubMed] [Google Scholar]
- Planque N, Perbal B. A structural approach to the role of CCN (CYR61/CTGF/NOV) proteins in tumourigenesis. Cancer Cell Int. 2003;3(1):15. doi: 10.1186/1475-2867-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planque N, Long Li C, Saule S, Bleau AM, Perbal B. Nuclear addressing provides a clue for the transforming activity of amino-truncated CCN3 proteins. J Cell Biochem. 2006;99(1):105–116. doi: 10.1002/jcb.20887. [DOI] [PubMed] [Google Scholar]
- Riser BL, Najmabadi F, Perbal B, Peterson DR, Rambow JA, Riser ML, Sukowski E, Yeger H, Riser SC. CCN3 (NOV) is a negative regulator of CCN2 (CTGF) and a novel endogenous inhibitor of the fibrotic pathway in an in vitro model of renal disease. Am J Pathol. 2009;174(5):1725–1734. doi: 10.2353/ajpath.2009.080241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittié L, Perbal B, Castellot JJ, Jr, Orringer JS, Voorhees JJ, Fisher GJ. Spatial-temporal modulation of CCN proteins during wound healing in human skin in vivo. J Cell Commun Signal. 2011;5(1):69–80. doi: 10.1007/s12079-010-0114-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Yamaguchi S, Ando R, Miyawaki A, Kabasawa Y, Takagi M, Li CL, Perbal B, Katsube K. The nephroblastoma overexpressed gene (NOV/ccn3) protein associates with Notch1 extracellular domain and inhibits myoblast differentiation via Notch signaling pathway. J Biol Chem. 2002;277:29399–29405. doi: 10.1074/jbc.M203727200. [DOI] [PubMed] [Google Scholar]
- Scholz G, Martinerie C, Perbal B, Hanafusa H. Transcriptional down regulation of the nov proto-oncogene in fibroblasts transformed by p60v-src. Mol Cell Biol. 1996;16(2):481–486. doi: 10.1128/mcb.16.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha W, Leask A. CCN2 expression and localization in melanoma cells. J Cell Commun Signal. 2011;5(3):219–226. doi: 10.1007/s12079-011-0128-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoyama T, Hiraoka S, Takemoto M, Koshizaka M, Tokuyama H, Tokuyama T, Watanabe A, Fujimoto M, Kawamura H, Sato S, Tsurutani Y, Saito Y, Perbal B, Koseki H, Yokote K. CCN3 inhibits neointimal hyperplasia through modulation of smooth muscle cell growth and migration. Arterioscler Thromb Vasc Biol. 2010;30(4):675–682. doi: 10.1161/ATVBAHA.110.203356. [DOI] [PubMed] [Google Scholar]
- Shi-Wen X, Leask A, Abraham D. Regulation and function of connective tissue growth factor/CCN2 in tissue repair, scarring and fibrosis. Cytokine Growth Factor Rev. 2008;19(2):133–144. doi: 10.1016/j.cytogfr.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Thomopoulos GN, Kyurkchiev S, Perbal B. Immunocytochemical localization of NOVH protein and ultrastructural characteristics of NCI-H295R cells. J Submicrosc Cytol Pathol. 2001;33:251–260. [PubMed] [Google Scholar]
- Vallacchi V, Daniotti M, Ratti F, Di Stasi D, Deho P, De Filippo A, Tragni G, Balsari A, Carbone A, Rivoltini L, Parmiani G, Lazar N, Perbal B, Rodolfo M. CCN3/nephroblastoma overexpressed matricellular protein regulates integrin expression, adhesion, and dissemination in melanoma. Cancer Res. 2008;68(3):715–723. doi: 10.1158/0008-5472.CAN-07-2103. [DOI] [PubMed] [Google Scholar]
- van Roeyen CR, Eitner F, Scholl T, Boor P, Kunter U, Planque N, Gröne HJ, Bleau AM, Perbal B, Ostendorf T, Floege J. CCN3 is a novel endogenous PDGF-regulated inhibitor of glomerular cell proliferation. Kidney Int. 2008;73(1):86–94. doi: 10.1038/sj.ki.5002584. [DOI] [PubMed] [Google Scholar]
- Wahab NA, Brinkman H, Mason RM. Uptake and intracellular transport of the connective tissue growth factor: a potential mode of action. Biochem J. 2001;359(Pt 1):89–97. doi: 10.1042/0264-6021:3590089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesman KC, Wei L, Baughman C, Russo J, Gray MR, Castellot JJ. CCN5, a secreted protein, localizes to the nucleus. J Cell Commun Signal. 2010;4(2):91–98. doi: 10.1007/s12079-010-0087-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang GP, Lau LF. Cyr61, product of a growth factor-inducible immediate early gene, is associated with the extracellular matrix and the cell surface. Cell Growth Differ. 1991;2(7):351–357. [PubMed] [Google Scholar]
- Yeger H, Perbal B. The CCN family of genes: a perspective on CCN biology and therapeutic potential. J Cell Commun Signal. 2007;1(3–4):159–164. doi: 10.1007/s12079-008-0022-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Averboukh L, Zhu W, Zhang H, Jo H, Dempsey PJ, Coffey RJ, Pardee AB, Liang P. Identification of rCop-1, a new member of the CCN protein family, as a negative regulator for cell transformation. Mol Cell Biol. 1998;18:6131–6141. doi: 10.1128/mcb.18.10.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]