Abstract
Mutations in PIK3CA, the gene encoding the p110α catalytic subunit of phosphatidylinositol-3 kinase (PI3K), have been shown to transform mammary epithelial cells (MECs). Studies suggest this transforming activity requires binding of mutant p110α via p85 to phosphorylated YXXM motifs in activated receptor tyrosine kinases (RTKs) or adaptors. Using transgenic mice, we examined if ErbB3, a potent activator of PI3K, is required for mutant PIK3CA-mediated transformation of MECs. Conditional loss of ErbB3 in mammary epithelium resulted in a delay of PIK3CAH1047R-dependent mammary gland hyperplasia, but tumor latency, gene expression and PI3K signaling were unaffected. In ErbB3-deficient tumors, mutant PI3K remained associated with several tyrosyl phosphoproteins, potentially explaining the dispensability of ErbB3 for tumorigenicity and PI3K activity. Similarly, inhibition of ErbB RTKs with lapatinib did not affect PI3K signaling in PIK3CAH1047R-expressing tumors. However, the p110α-specific inhibitor BYL719, in combination with lapatinib impaired mammary tumor growth and PI3K signaling more potently than BYL719 alone. Further, co-inhibition of p110α and ErbB3 potently suppressed proliferation and PI3K signaling in human breast cancer cells harboring PIK3CAH1047R. These data suggest that PIK3CAH1047R-driven tumor growth and PI3K signaling can occur independently of ErbB RTKs. However, simultaneous blockade of p110α and ErbB RTKs results in superior inhibition of PI3K and mammary tumor growth, suggesting a rational therapeutic combination against breast cancers harboring PIK3CA activating mutations.
Keywords: ErbB3, PIK3CA, PI3K, Breast Cancer, transgenic mice
INTRODUCTION
Phosphatidylinositol-3-kinase (PI3K) is the most frequently mutated signaling pathway in cancer, affecting tumor cell survival, proliferation, migration and metabolism (1, 2). PI3K is a lipid kinase composed of a p85 regulatory subunit dimerized with a p110 catalytic subunit (1, 3). The N-terminal SH2 domain of p85 binds to phosphorylated tyrosines in receptors or adaptors; this binding relieves the p85-mediated inhibition of p110 which, as a result, becomes activated and catalyzes the conversion of phosphatidylinositol-4,5-bisphosphate (PIP2) to phosphatidylinositol-3,4,5-trisphosphate (PIP3), a second messenger that recruits signal transducers (Akt, PDK1, SGK, etc.) to the plasma membrane, where they become activated (1).
The PI3K pathway is aberrantly activated by gain-of-function mutations in p110α (PIK3CA) and p85α (PIK3R1), amplification of wild type PIK3CA, p110β (PIK3CB) and PDK1, loss/inactivation of the PIP3 phosphatases PTEN and INPP4B, mutation and/or amplification of AKT1-3, and amplification of RTKs (4, 5). Three “hotspot” mutations have been identified in PIK3CA: E542K, E545K, and H1047R, accounting for ~80% of PIK3CA mutations (2, 6, 7). PIK3CA mutations occur in approximately 40% of breast cancers, mainly in tumors with luminal and HER2-enriched gene expression (8), where they have been associated with a more virulent phenotype and resistance to anti-estrogen and anti-HER2 therapy.
HER2/ErbB2 gene amplification occurs in 25% of breast cancers where it associates with poor patient outcome. The main dimerization partner of HER2 is HER3/ErbB3. Six p85 binding motifs in ErbB3 activate PI3K (9, 10). ErbB3 is essential for PI3K activation and survival of HER2-overexpressing breast cancer cells (9, 11, 12). Therapies that inhibit PI3K induce ErbB3 expression and reactivation via feedback mechanisms which partially maintain PI3K and counteract drug action. As such, the efficacy of HER2 and PI3K inhibitors is improved by co-inhibition of ErbB3 (11, 13, 14).
In addition to ErbB2/ErbB3, other RTKs activate PI3K via insulin receptor substrate (IRS) and Gab family molecules. These adaptors lack enzymatic activity, but when tyrosine phosphorylated by RTKs, they recruit p85 (PI3K) and other signaling molecules (15, 16). Activated RTKs, including insulin and IGF receptors, VEGFR, EGFR and ALK recruit IRS adaptors (16, 17). IRS-1 has nine p85 binding motifs (18) and, like ErbB3, strongly activates PI3K. Similarly, Gab1 and Gab2 contain three p85 binding motifs and are tyrosine phosphorylated by ErbB2, MET, Abl, FGFR, EGFR and Src kinases (15). The p85 subunit was discovered by its association with PDGFR, a potent activator of PI3K (19). Biochemical analyses have shown that both PIK3CAE545K and PIK3CAH1047R exhibit ~two-fold higher catalytic activity than wild type PI3K. The association of PIK3CAH1047R with PDGFR or IRS-1 phosphopeptides further increases the catalytic activity of the mutant enzyme (20). Additionally, PIK3CAH1047R association with p85 is required for transformation induced by mutant PI3K (6, 21). These data suggest a role for upstream RTKs in the signaling output of mutant PI3K, leading us to hypothesize that mutant PIK3CA requires upstream adaptors, such as ErbB3, to induce epithelial transformation and tumor progression.
We show herein that mammary gland hyperplasia induced by temporally-regulated expression of mutant PIK3CA was delayed in mice lacking ErbB3 in the mammary epithelium. In contrast, tumor formation and PI3K activity were unaffected by ErbB3 ablation. In tumors expressing ErbB3, mutant PI3K associated with several tyrosine-phosphorylated proteins, including ErbB3. In tumors lacking ErbB3, PI3K still associated with other upstream adaptors and RTKs. Inhibition of RTKs or adaptors known to activate PI3K did not block cell growth or PI3K activity in mammary tumors or PIK3CA-mutant human breast cancer cells. However, simultaneous inhibition of upstream RTKs and mutant p110α more potently inhibited tumor growth and PI3K signaling than inhibition of p110α alone. These data suggest that mutant PIK3CA still relies upon upstream activators and combined inhibition of PI3K and these activators is a rational treatment strategy against tumors harboring PIK3CA activating mutations.
MATERIALS AND METHODS
Cell Culture
MDA-MB-453 and T47D cells were from ATCC. MDA-MB-453 were authenticated in March 2013 by short tandem repeat (STR) DNA analysis (DDC Medical); authentication of T47D cells (March 2011) has been described (22). CAL-148 and BT20 cells were provided and authenticated as described (23). All cells were cultured in DMEM with 10% FBS (Life Technologies). EZN-3920 and EZN-4455 are locked nucleic acid antisense molecules provided by Enzon Pharmaceuticals (11, 24); they were resuspended in sterile PBS to a stock concentration of 5 mM and applied to cells at 5 μM in the absence of transfection reagent. EZN-3920 targets ErbB3 and EZN-4455 is a scrambled control antisense. BYL719 (25) and LJM716 (26), provided by Novartis, were resuspended to a stock concentration of 1 mM in DMSO or 10 mg/ml in sterile PBS, respectively. siRNA (Qiagen) were transfected at a final concentration of 50 nM total siRNA using Lipofectamine RNAiMAX (Life Technologies) following the manufacturer’s protocol (see Supplementary Methods). The duration and concentration of each drug or siRNA treatment is described with each figure. Media and inhibitors were replenished every three days. Colony growth was assessed by plating cells and staining with crystal violet as detailed in Supplementary Methods.
Mice
To generate the iPIK3.iCre.ErbB3FL/FL model on a congenic FVB background, the following FVB mouse strains were interbred: MMTV-rtTA (27), Tet-Op-HA-PIK3CAH1047R-IRES-Luc (28), Tet-Op-Cre (29) and ErbB3FL/FL (30) as described in Supplementary Methods. Details of tumor transplantation are also provided in Supplementary Methods. Briefly, harvested tumors were homogenized in serum-free media with gentleMACS C Tubes (Miltenyi Biotec) and resuspended in 7 ml matrigel diluted with 50% PBS. Tumor homogenates (100 μl) were injected into both inguinal (#4) mammary fat pads of ~4 week old female athymic mice (Harlan laboratory) using a 25-gauge needle. When tumors reached ≥125 mm3, mice were randomized to four treatment groups as indicated in figure legends. Lapatinib di-p-toluenesulfonate salt and imatinib methanesulfonate salt were purchased from LC Laboratories.
Lapatinib, imatinib and BYL719 were resuspended in 0.5% methyl cellulose, 0.1% Tween-80 for treatment by orogastric gavage. Lapatinib and imatinib were administered twice daily at 100 mg/kg/dose and BYL719 once daily at 30 mg/kg/dose. In the first study, all mice were sacrificed when tumors in vehicle treated mice exceeded 1.5 cm3. In the second study, animals with tumors greater than 1.0 cm3 were sacrificed and all remaining animals were sacrificed on day 21. Mice were always sacrificed one hour after drug treatment.
Protein and Histological Analyses
Cell line and tumor protein lysates were prepared as described in Supplementary Methods. Immunoprecipitation was performed with a p85 (Millipore) or an HA antibody (Cell Signaling) using a ratio of 1 μg antibody: 250 μg lysate: 5 μl Dynal protein G beads (Life Technologies) with end-over-end rotation at 4°C for 4 h. For immunoblot analysis, equal amounts of protein/lane were subjected to SDS-PAGE, transferred to nitrocellulose membranes and analyzed with antibodies as described in Supplementary Methods. Phospho-RTK arrays were purchased from R&D systems and incubated with 225 μg cell lysates following manufacturer’s directions. Details of guinea pig anti-cytokeratin 8 (RDI-Fitzgerald) and rabbit anti-cytokeratin 5 (Covance) immunofluorescent staining of tissue sections and hematoxylin staining of whole mount mammary glands are provided in Supplementary Methods.
Gene Expression Analyses
Tumor RNA was harvested from ErbB3FL/+ and ErbB3FL/FL iPIK3.iCre tumors and analyzed by cDNA microarray as described previously (31, 32). Briefly, RNA isolated by Qiagen RNeasy mini kit was hybridized to Agilent custom 4X180K microarrays as previously described (31); the signal from iPIK3.Cre RNA was normalized to the Herschkowitz et al 2011 murine dataset (32). The mutant PIK3CA gene signature (33) was calculated for every genotype in the Herschkowitz et al 2011 mouse tumor dataset (32) and the iPIK3.iCre tumors. The average gene signature scores for each tumor class were plotted as boxplots to compare mutant PIK3CA-induced gene expression across previously defined classes (32).
Statistical Analyses
Significant differences (P <0.01) were determined by ANOVA and Bonferroni post-hoc tests (multiple testing-corrected) or Student’s T-test using Graphpad Prism software.
RESULTS
ErbB3 inhibition sensitizes mutant PIK3CA breast cancer cells to a p110α inhibitor
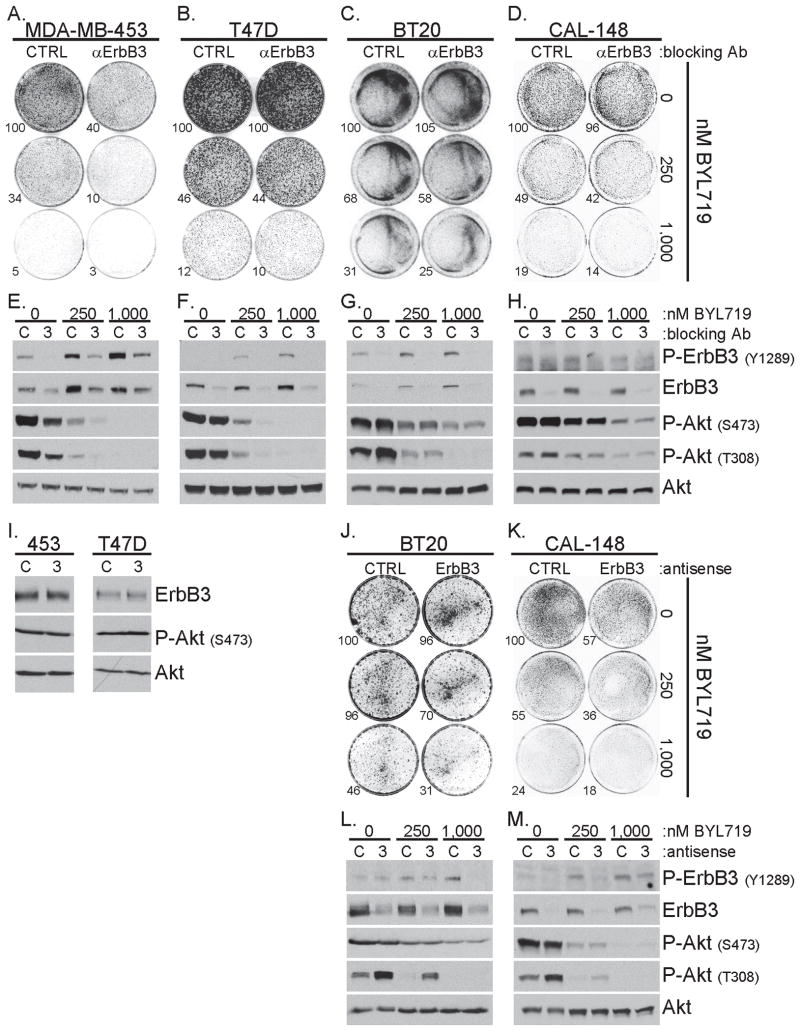
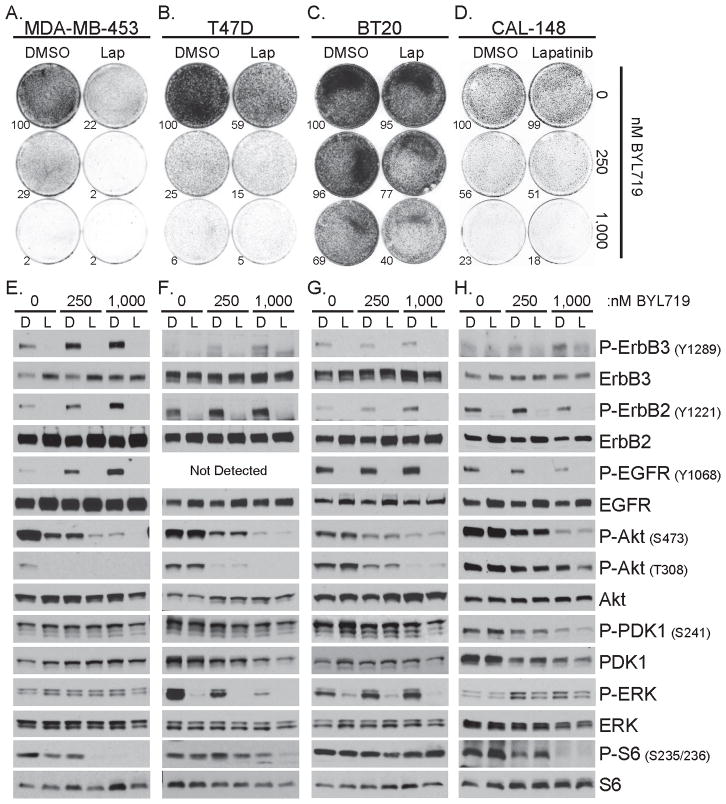
ErbB3 silencing impairs the proliferative advantage conferred by mutant PIK3CA in HER2-amplified breast cancer cells (34). However, the impact of ErbB3 in PIK3CA-mutant breast cells lacking HER2 amplification is less clear. We treated, MDA-MB-453, T47D, BT20 and CAL-148 breast cancer cells, all harboring PIK3CAH1047R, with the ErbB3 neutralizing antibody, LJM716 (26), and BYL719, a p110α-specific inhibitor with an IC50 against wild type and mutant p110α of ≤5 nM (25). BYL719 decreased the proliferation (Fig. 1A–D) and Akt phosphorylation (Fig. 1E–H) of all four cell lines in a dose-dependent fashion, suggesting p110αH1047R is a driver of proliferation and PI3K activity in these cells. LJM716 reduced basal and BYL719-induced total and Y1289-P-ErbB3 (Fig. 1E–H) and enhanced BYL719-mediated growth inhibition in each cell line (Fig. 1A–D), albeit weakly in T47D and CAL-148 cells. We also utilized EZN-3920, a locked nucleic acid (LNA) ErbB3-targeted antisense or a scrambled control LNA antisense, EZN-4455 (11, 24), in combination with BYL719. EZN-3920 did not reduce ErbB3 expression in MDA-MB-453 or T47D cells (Fig. 1I). EZN-3920 reduced ErbB3 levels and BYL719-induced Y1289-P-ErbB3 in both BT20 and CAL-148 cells (Fig. 1L–M). The combination of BYL719+EZN-3920 exhibited a better anti-proliferative effect than either agent alone (Fig. 1J–K). Additionally, increased T308-P-Akt induced by EZN-3920 was abrogated by BYL719 (Fig. 1L–M). These data suggest that co-inhibition of ErbB3 and p110α impairs the proliferative advantage conferred by mutant PIK3CA in breast cancer cells without HER2 gene amplification.
Figure 1. ErbB3 inhibition sensitizes mutant PIK3CA breast cancer cells to a p110α inhibitor.
A–D) The indicated cell lines were treated ±10 μg/ml LJM716, ErbB3 blocking antibody (αErbB3), and 0, 250 or 1,000 nM BYL719 for 15 days (MDA-MB-453 and T47D), 9 days (BT20) or 12 days (CAL-148). Colonies were stained with crystal violet and quantitated using Image J software. Values shown are percent growth relative to control treated cells
E–H) Cells were treated as described in panel A–D; after 4 days of treatment cell lysates were prepared and subjected to immunoblot analysis with the indicated antibodies
I) MDA-MB-453 or T47D cells were treated 7 days with 5 μM LNA antisense molecules without transfection reagent. EZN-4455, control LNA antisense (C); or EZN-3920, ErbB3-targeted LNA antisense (3), were used. Lysates were prepared and subjected to immunoblot analysis with the indicated antibodies
J–K) BT20 cells were treated 15 days or CAL-148 cells were treated 12 days with 5 μM control LNA antisense (CTRL) or ErbB3-targeted LNA antisense (ErbB3) without transfection reagent and 0, 250 or 1,000 nM BYL719. Growth was assessed as described in panels A–D
L–M) BT20 or CAL-148 cells were treated as described in panels J–K; after 7 days of treatment cell lysates were prepared and subjected to immunoblot analysis with the indicated antibodies
Loss of ErbB3 delays mammary gland hyperplasia induced by mutant PIK3CA, but does not delay tumor formation
To evaluate whether ErbB3 is required for PIK3CAH1047R-induced mammary epithelial cell (MEC) transformation in vivo, we generated transgenic mice in which doxycycline (DOX)-induced PIK3CAH1047R drives mammary tumor formation in the presence or absence of ErbB3. These mice expressed three transgenes [MMTV-rtTA (27), Tet-Op-HA-PIK3CAH1047R-IRES-Luc (28) and Tet-Op-Cre (29)] and harbored homozygous or heterozygous floxed ERBB3 alleles (11, 30, 35) (Fig. S1). In this model, referred to as iPIK3.iCre, DOX treatment simultaneously induced the expression of hemagglutinin (HA)-tagged PIK3CAH1047R and Cre recombinase in the mammary epithelium, resulting in Cre-mediated recombination of floxed ERBB3 alleles in MECs expressing PIK3CAH1047R. The internal ribosomal entry site (IRES)-Luciferase allows bioluminescent detection of cells/tissues expressing PIK3CAH1047R.
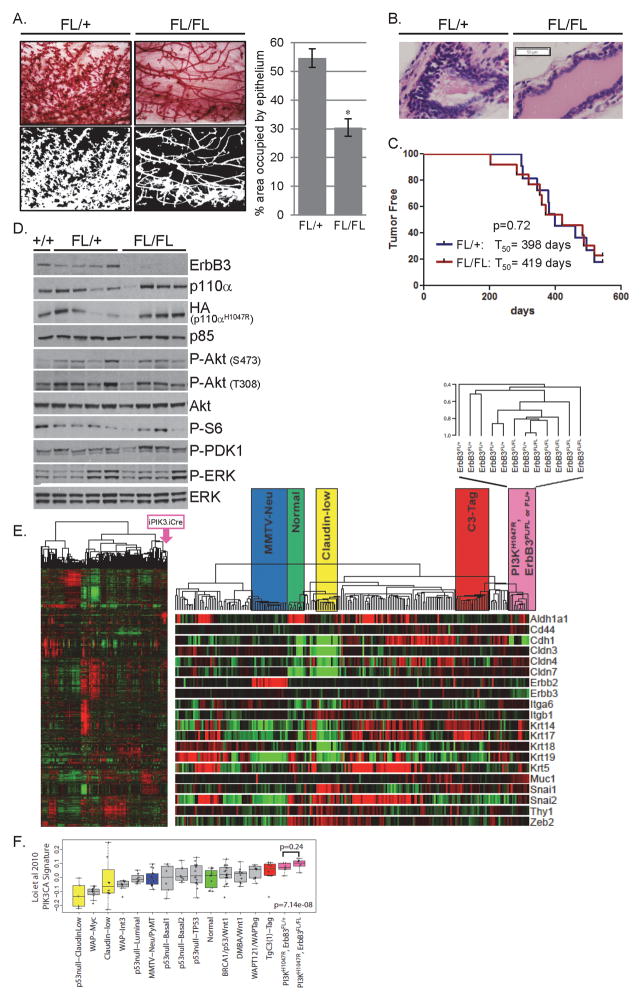
The mammary ductal epithelium in six-week old ErbB3FL/+(control) animals extended distally past the central lymph node whereas the epithelium of ErbB3FL/FL mice was maintained proximal to the lymph node (Fig. S2A) (36). At nine weeks, the ErbB3FL/FL ductal epithelium extended past the lymph node, but remained shorter than that of ErbB3FL/+ animals (Fig. S2B). Mammary ductal hyperplasia (induced by mutant PIK3CA) was not apparent in either group at six or nine weeks of age (Fig. S2A,B). By twelve weeks, ErbB3FL/+ glands exhibited thickened, irregular ductal epithelium indicative of hyperplasia. However, ErbB3FL/FL iPIK3.iCre glands contained smooth, normal-appearing ducts (Figs. 2A and S2C). Quantitation of epithelial content in whole mounts showed that ErbB3FL/+ glands contained 80% more epithelium than ErbB3FL/FL glands (Fig. 2A). Histological examination of mammary gland sections from 12-week old animals showed that the ErbB3FL/+ iPIK3.iCre glands contained multiple cell layers of ductal epithelium, while the ErbB3FL/FL iPIK3.iCre epithelium was arranged in a single, smooth cell layer (Fig. 2B).
Figure 2. Loss of ErbB3 delays mammary gland hyperplasia induced by mutant PIK3CA, but does not delay tumor formation or alter PI3K signaling or gene expression.
A) Whole mount inguinal mammary glands from 12 week old ErbB3FL/+ and ErbB3FL/FL iPIK3.iCre mice were stained and photomicrographed at 40x power (top) followed by conversion to binary pictures (bottom). The average percentage of area occupied by epithelium ±SEM was quantitated from four representative binary photomicrographs (n = 3 per genotype; right). * P < 0.01
B) Representative H&E stained sections of mammary glands harvested from 12 week old ErbB3FL/+ and ErbB3FL/FL iPIK3.iCre mice photomicrographed at 400x power.
C) Kaplan-Meier analysis of tumor-free survival of 11 ErbB3FL/+ and 13 ErbB3FL/FL iPIK3.iCre mice. Average tumor latency = T50. P value calculated using log-rank test.
D) Lysates were prepared from tumors harvested from one ErbB3+/+, four ErbB3FL/+ and four ErbB3FL/FL iPIK3.iCre mice and were subjected to immunoblot analysis with the indicated antibodies.
E) Left: Unsupervised hierarchical cluster of six ErbB3FL/+ and six ErbB3FL/FL iPIK3.iCre tumors with thirteen previously characterized breast cancer GEMMs (32) using all probes with at least an absolute log2 expression value greater than two on at least three arrays (2203 genes). Right: Enlargement of the array dendrogram with common murine groups that represent human phenotypes highlighted for reference: MMTV-Neu (luminal), Normal breast, Claudin-low, and C3-Tag (basal-like). Beneath the dendrogram are twenty classic genes that segregate the intrinsic subtypes. The iPIK3.iCre dendrogram is enlarged to discern individual ErbB3FL/+ and ErbB3FL/FL tumors.
F) Genes with a positive fold change in the mutant PIK3CA gene signature of Loi et al (33) were averaged for each tumor in the combined murine dataset. These values were plotted by median expression for the defined murine classes (32) and the ErbB3FL/+ and ErbB3FL/FL iPIK3.iCre tumors.
Mice were monitored for expression of PIK3CA-IRES-Luciferase by IVIS bioluminescence imaging (Fig. S3). Tumor formation was monitored by mammary gland palpation through 600 days of age. Mice lacking the iPIK3 transgene or not induced with DOX failed to develop tumors or exhibit luciferase activity at any point. The mean mammary tumor latency was 398 days in ErbB3FL/+ mice as compared to 419 days in ErbB3FL/FL mice (p=0.72; Fig. 2C). Loss of ErbB3 did not affect the average number of tumors per animal: 1.75 tumors per ErbB3FL/+ mouse and 1.55 tumors per ErbB3FL/FL mouse (p=0.71). These data suggest that loss of ErbB3 delays early mammary hyperplasia but not cancer formation in PIK3CAH1047R-expressing mice.
Loss of ErbB3 does not alter PI3K signaling, histology or gene expression in tumors induced by mutant PIK3CA
Immunoblot analysis revealed loss of ErbB3 expression in ErbB3FL/FL tumors. All tumors expressed p110α and p85 subunits of PI3K as well as the HA-tagged p110αH1047R transgene. Levels of phosphorylated Akt, S6 and PDK1 were similar in ErbB3FL/+ and ErbB3FL/FL tumors, suggesting loss of ErbB3 did not attenuate PI3K signaling in PIK3CAH1047R-driven mammary tumors (Fig. 2D).
Global gene expression patterns in ErbB3-deficient and ErbB3-competent tumors were assessed using cDNA microarrays. Unsupervised, hierarchical cluster analysis of iPIK3 tumors and 13 other genetically engineered mouse models (GEMM) of breast cancer (31, 32) demonstrated ErbB3FL/+ tumors did not segregate from ErbB3FL/FL tumors (Fig. 2E). Two-class significance analysis of microarray (SAM) analysis did not identify any differentially expressed genes in ErbB3FL/+ and ErbB3FL/FL tumors. A previously reported mutant PIK3CA gene expression signature (33) demonstrated that the PIK3CA signature was higher in the iPIK3.iCre models compared to the other models (Fig. 2F), consistent with activation of the PI3K pathway. The mutant PIK3CA signature score was similar in ErbB3FL/+ and ErbB3FL/FL tumors, suggesting loss of ErbB3 did not reduce PI3K signaling output.
Histological examination of iPIK3 tumors revealed heterogeneous histopathologies (Fig. S4A). The most frequent histologies were adenocarcinoma and adenocarcinoma with squamous metaplasia. Squamous metaplasia occurred more frequently in ErbB3FL/FL samples while metaplastic carcinomas and adenomyoepithelioma-like lesions were unique to ErbB3FL/+ tumors. ErbB3FL/+ and ErbB3FL/FL tumors often displayed cribriform architecture, but tumors with papillary architecture occurred more frequently in ErbB3FL/FL mice and tumors with solid architecture were only seen in ErbB3FL/+ mice (Fig. S4B). PIK3CA mutant mammary tumors have been reported to co-express both luminal-type (i.e., CK8) and basal-type cytokeratins (i.e., CK5) (37, 38). This was seen in ErbB3FL/+ and ErbB3FL/FL cancers (Fig. S4C). Thus, loss of ErbB3 did not enrich for either the luminal or basal population. These data are consistent with two recent reports (28, 38).
Mutant p110α binds to ErbB3 and other tyrosine-phosphorylated proteins
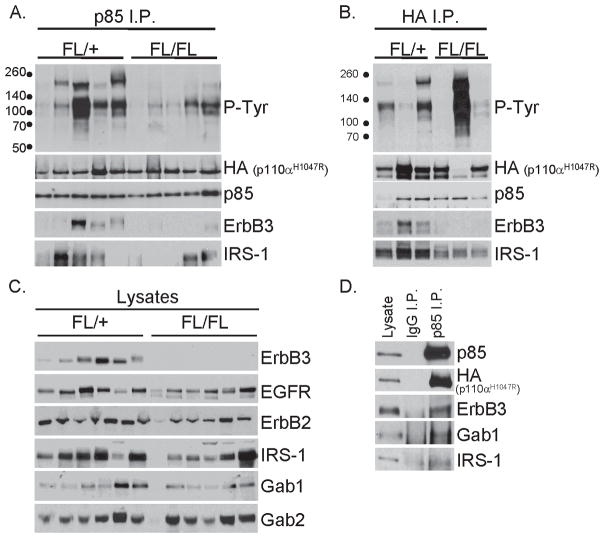
Since ErbB3 loss did not attenuate iPIK3.iCre tumorigenesis or PI3K signaling, we sought to determine if PIK3CAH1047R engaged other tyrosine-phosphorylated adaptors or receptors. We detected HA-tagged p110αH1047R in p85 immunoprecipitates from ErbB3FL/+ and ErbB3FL/FL tumor lysates (Fig. 3A, 2nd and 3rd rows). ErbB3 also co-precipitated with p85 in ErbB3FL/+, but not ErbB3FL/FL tumors. Several tyrosine-phosphorylated proteins co-precipitated with p85 in ErbB3-deficient tumors (Fig. 3A). Immunoprecipitation of mutant p110α using an anti-HA antibody pulled down p85 and several tyrosine-phosphorylated proteins in ErbB3FL/+ and ErbB3FL/FL tumor lysates, including ErbB3 in the ErbB3FL/+ tumors (Fig. 3B). p85 or HA frequently co-precipitated IRS-1, a known PI3K scaffold (Fig. 3A–B). ErbB3FL/FL tumors lacked detectable ErbB3, but expressed EGFR, ErbB2, IRS-1 and Gab1/2 at levels similar to ErbB3FL/+ tumors (Fig. 3C). Association of p85 with ErbB3, Gab1 and IRS-1 in iPIK3 tumor lysates (Fig. 3D) suggests that multiple adaptors can simultaneously engage mutant p110α, perhaps rendering ErbB3 dispensable for activity and plasma membrane localization of PIK3CAH1047R.
Figure 3. Mutant p110α binds to ErbB3 and other tyrosine-phosphorylated proteins.
A) p85 was immunoprecipitated (I.P.) from five ErbB3FL/+ and five ErbB3FL/FL primary tumor lysates. Immune complexes were prepared as indicated in Methods and subjected to immunoblot analysis with the indicated antibodies.
B) HA-tagged p110αH1047R was immunoprecipitated from the lysates of three ErbB3FL/+ and three ErbB3FL/FL tumors. Immune complexes were analyzed as described in panel A.
C) Lysates of six ErbB3FL/+ and six ErbB3FL/FL iPIK3.iCre primary tumors were evaluated by immunoblot analysis with the indicated antibodies.
D) An iPIK3.iCre tumor lysate was subjected to immunoprecipitation with control IgG or p85 antibodies. Whole lysates and antibody pull downs were evaluated by immunoblot analysis with the indicated antibodies
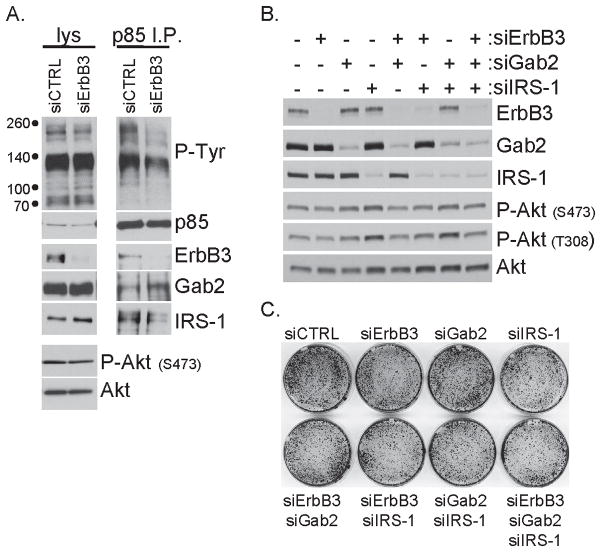
Because the PIK3CAH1047R -mutant human breast cancer cell line T47D requires mutant p110α for growth and Akt phosphorylation (Fig. 1B,F), we assessed the requirement for PI3K-activating adaptors. ErbB3 siRNA reduced co-precipitation of ErbB3 with p85, but Gab2 and IRS-1 continued to associate with p85 following ErbB3 depletion (Fig. 4A). Furthermore, P-Akt levels were sustained following ErbB3 depletion, consistent with maintenance of PI3K signaling. We combined siRNA-mediated knockdown of ErbB3, Gab2 and/or IRS-1. Despite significant reduction of all three PI3K adaptors, Akt remained phosphorylated, suggesting that PI3K activity was not reduced (Fig. 4B). Similarly, proliferation was not dramatically altered by combined depletion of ErbB3, Gab2 and IRS-1 (Fig. 4C). These data suggest that because multiple proteins can engage mutant PI3K, depletion of ErbB3, Gab2 and/or IRS-1 is insufficient to reduce PI3K activity of tumors or cells driven by PIK3CAH1047R.
Figure 4. Loss of ErbB3, Gab2 and/or IRS-1 does not inhibit Akt or growth of T47D breast cancer cells.
A) Lysates (lys) or p85 immunoprecipitates (I.P.) from T47D cells transfected with control siRNA (siCTRL) or HER3 siRNA (siHER3) collected 48 hours after transfection were evaluated by immunoblot analysis with the indicated antibodies.
B) T47D cells were transfected with siRNA targeting ErbB3, Gab2 or IRS-1 as indicated with control siRNA being used such that 50 nM total siRNA was always used. Lysates were prepared 48 hours after transfection and evaluated by immunoblot analysis with the indicated antibodies
C) T47D cells transfected with siRNA as described in panel B were stained with crystal violet 7 days after transfection
ErbB RTK activity is dispensable in PIK3CAH1047R tumors
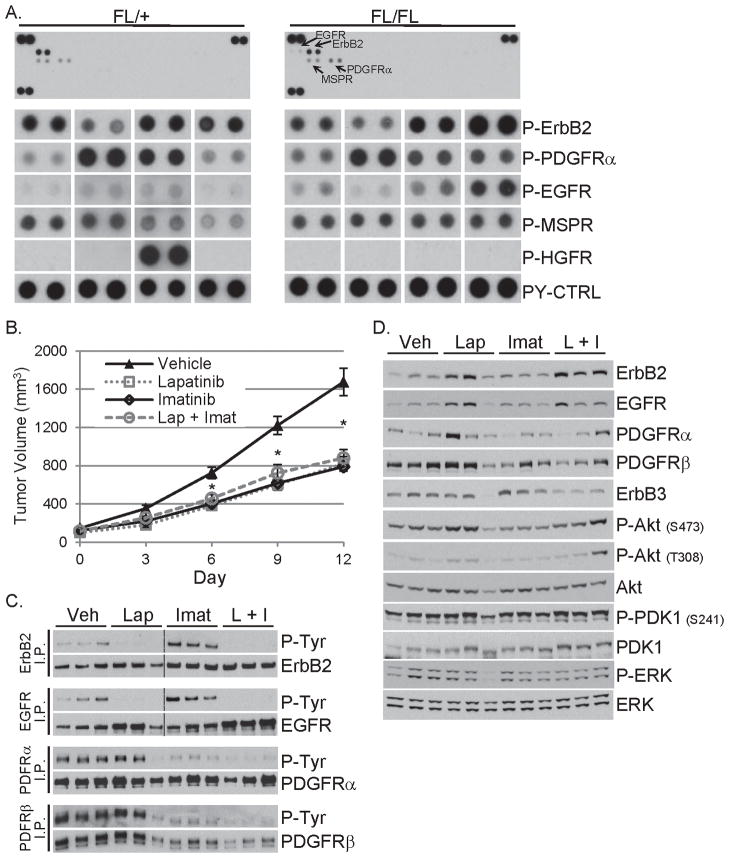
To determine whether RTKs other than ErbB3 are activated in iPIK3.iCre tumors, we performed phospho-RTK arrays. The most prominent tyrosine-phosphorylated RTKs in both ErbB3FL/+ and ErbB3FL/FL tumors were ErbB2, EGFR, PDGFR and macrophage stimulating protein receptor (MSPR/Ron) (Fig. 5A). One of the eight tumors exhibited high P-HGFR, consistent with HGFR (MET) amplification previously reported in this model (28). We speculate the presence of these phosphorylated RTKs, all capable of engaging PI3K, may negate a requirement for ErbB3 in iPIK3 tumors.
Figure 5. ErbB RTK activity is dispensable in PIK3CAH1047R tumors.
A) The lysates of four ErbB3FL/+ and four ErbB3FL/FL primary tumors were applied to phospho-RTK arrays. One representative array of each genotype is presented (top) and enlarged, exposure-matched levels of ErbB2, PDGFRα, EGFR, MSPR and HGFR is presented (lower).
B) Mice bearing ≥125 mm3 orthotopically-transplanted iPIK3.iCre tumors were treated twice daily (100 mg/kg/dose) by orogastric gavage with vehicle, lapatinib, imatinib or lapatinib + imatinib (10 tumors per group). Average tumor volume for each group ±SEM is plotted against time. * P < 0.01 (vehicle vs. treatment)
C) Three tumor lysates per group were subjected to immunoprecipitation (I.P.) with antibodies against ErbB2, EGFR, PDGFRα or PDGFRβ. Antibody pull downs were subjected to immunoblot analysis with a P-Tyr antibody. Membranes were stripped and re-probed to detect the pulled down RTK.
D) The same tumor lysates as in panel C were evaluated by immunoblot analysis with the indicated antibodies.
To determine if inhibition of EGFR, ErbB2 and PDGFR would inhibit growth of PIK3CAH1047R-expressing mammary tumors, we treated iPIK3.iCre tumor-bearing mice with 1) vehicle; 2) EGFR/ErbB2 tyrosine kinase inhibitor (TKI) lapatinib; 3) PDGFR TKI imatinib, and 4) lapatinib + imatinib. Tumor growth was delayed by each single inhibitor and the combination (Fig. 5B). P-ErbB2 and P-EGFR levels were reduced by lapatinib while imatinib inhibited P-PDGFRα and P-PDGFRβ (Fig. 5C). Imatinib-treated tumors exhibited higher levels of P-ErbB2 and P-EGFR, which were abolished in tumors co-treated with lapatinib. Although the combination of imatinib and lapatinib slowed tumor growth and inhibited ErbB/PDGFR phosphorylation, Akt and PDK1 remained phosphorylated in tumors treated with both RTK inhibitors (Fig. 5D). Pull down of p85 from tumor lysates co-precipitated IRS-1 and Gab1 as well as other tyrosine-phosphorylated proteins (Fig. S5A,B). These data suggest that mutant PI3K engages multiple upstream adaptors, potentially explaining how it maintains its activity and effect on tumor growth despite inhibition of EGFR, ErbB2 and PDGFR kinases with a combination of small molecule inhibitors.
Inhibition of PIK3CAH1047R–driven tumor growth by a p110α inhibitor is enhanced by co-inhibition of ErbB2/EGFR
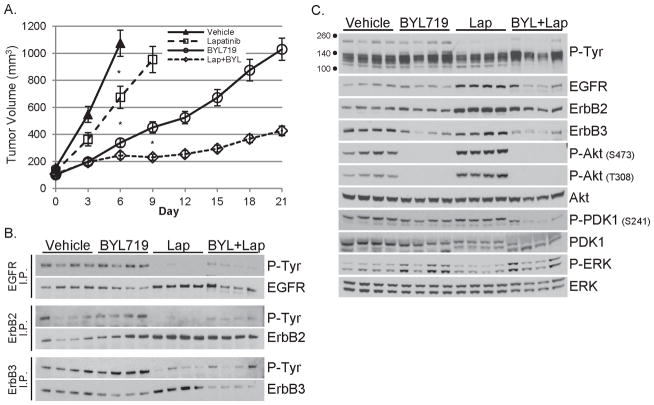
Data shown in Figs. 2C–F, 4 and 5B–D suggest that targeted inhibition of signaling events upstream of PI3K, including ErbB3, EGFR, ErbB2, PDGFR, Gab2 or IRS-1, is insufficient to silence PI3K activity and tumor growth in PIK3CAH1047R-driven cancers. Further, compensatory upregulation of signaling networks upstream of PI3K can limit the therapeutic response to PI3K inhibitors (13, 39). To determine if this occurs in tumors that express PIK3CAH1047R as the pathogenic oncogene, we treated iPIK3.iCre tumors with 1) vehicle; 2) p110α-specific inhibitor BYL719; 3) lapatinib or 4) lapatinib + BYL719. Although lapatinib slowed tumor growth less effectively than BYL719, the combination of lapatinib + BYL719 inhibited tumor growth more potently than either agent alone (Fig. 6A). Lapatinib reduced EGFR, ErbB2 and ErbB3 phosphorylation (Fig. 6B), but did not decrease P-PDK1 or P-Akt (Fig. 6C). BYL719 alone decreased P-Akt, but not P-PDK1. In contrast, BYL719 + lapatinib markedly reduced P-Akt and P-PDK1 levels (Fig. 6C), suggesting a more potent suppression of PI3K when p110α and EGFR/ErbB2/ErbB3 are inhibited simultaneously. BYL719 + lapatinib treatment slowed the weight gain of adolescent, tumor-bearing mice compared to mice treated with single agent BYL719 (P < 0.01), but histopathological analysis revealed no liver damage in any treatment group (Fig. S6).
Figure 6. Growth of PIK3CAH1047R–driven tumors is inhibited by combined blockade of p110α and ErbB2/EGFR.
A) Mice bearing ≥125 mm3 orthotopically-transplanted iPIK3.iCre tumors were treated with vehicle (twice daily), lapatinib (twice daily at 100 mg/kg/dose), BYL719 (once daily at 30 mg/kg/dose) or lapatinib + BYL719 (14-16 tumors per group). Average tumor volume for each group ±SEM is plotted against time. * P < 0.01 reported at earliest significant difference between groups.
B) Tumor lysates were subjected to immunoprecipitation (I.P.) with antibodies against EGFR, ErbB2 or ErbB3 and the resulting immune complexes evaluated by immunoblot analysis with a P-Tyr antibody. Membranes were stripped and re-probed to detect the pulled down RTK.
C) The same tumor lysates as in panel B were evaluated by immunoblot analysis with the indicated antibodies.
To extend these observations, four human breast cancer cell lines with PIK3CAH1047R, but without HER2 gene amplification (MDA-MB-453, T47D, BT20 and CAL-148), were treated with BYL719 and lapatinib. BYL719 reduced proliferation and P-Akt in all four cell lines (Fig. 7). Lapatinib inhibited basal and BYL719-induced phosphorylation of EGFR, ErbB2 and ErbB3 in each cell line (Fig. 7E–H). Lapatinib inhibited the proliferation of two cell lines as a single agent and aided the anti-proliferative action of BYL719 in all four cell lines (Fig. 7A–D). Compared to single agent treatments, the combination of BYL719 + lapatinib resulted in more potent inhibition of S473-P-Akt, T308-P-Akt and P-S6 in MDA-MB-453 cells; S473-P-Akt, P-ERK and P-S6 in T47D cells; P-PDK1 and P-ERK in BT20 cells; and T308-P-Akt, P-PDK1 and P-ERK in CAL-148 cells (Fig. 7E–H). Thus, while signaling downstream of PI3K was differentially affected by BYL719 and lapatinib in the four human cell lines and iPIK3.iCre transgenic mouse tumors, combined inhibition of p110α and ErbB receptors resulted in a more potent antitumor effect in all five PIK3CAH1047R-driven models.
Figure 7. HER2 non-amplified human breast cancer cells with PIK3CAH1047R are inhibited by combination of p110α inhibitor and lapatinib.
A–D) The indicated cell lines were treated with DMSO or 500 nM lapatinib (Lap) and 0, 250 or 1,000 nM BYL719 for 10 days (MDA-MB-453), 14 days (T47D and BT20) or 12 days (CAL-148). Growth was assessed as described in Figure 1.
E–H) Cells were treated as described in panels A–D; after 3 days of treatment cell lysates were prepared and subjected to immunoblot analysis with the indicated antibodies. (D=DMSO; L=lapatinib)
DISCUSSION
PI3K is the most frequently mutated signaling pathway in breast cancer. Most common somatic alterations in this pathway are ‘hot spot’ mutations in the helical and catalytic domains of PIK3CA (8). Transgenic mice with conditional expression of PIK3CAH1047R in the mammary gland develop mammary cancers (28). Despite exhibiting a ‘gain-of-function,’ mutant PIK3CA still requires binding via the regulatory subunit p85 to phosphorylated adaptors or receptors. For example, PIK3CAH1047R-mediated transformation of chick embryo fibroblast depends on its interaction with p85 (21) and the catalytic activity of PI3KH1047R is enhanced by coupling to phosphorylated PDGFR or IRS-1 (20). Further, knockdown of ErbB3 or its ligand heregulin inhibits growth and P-Akt levels in cells expressing PIK3CAH1047R (34). Tissue-specific deletion of ErbB3 in the mammary gland results in a delay in ductal extension during puberty, increased apoptosis in terminal end buds (TEBs) and a reduction in P-Akt (36), suggesting ErbB3-activated PI3K has an important role in mammary gland morphogenesis. Finally, Cre-mediated deletion of ErbB3 prolonged mammary tumor latency, reduced lung metastases and reduced P-Akt levels in mammary tumors driven by polyomavirus middle T oncogene (PyV mT) (11). In this mouse model, PyV mT-driven tumor formation is highly dependent upon the association of middle T with p85 (40) and this association is lost upon deletion of ErbB3 or inhibition of ErbB3 phosphorylation with the EGFR/ErbB2 inhibitor lapatinib (11). These data suggest that ErbB3 is a major activator of wild type and mutant PI3K in normal and transformed MECs. Thus, we examined if ErbB3 was required for MEC transformation by mutant PI3K.
We demonstrated herein that loss of ErbB3 delayed mammary gland hyperplasia induced by mutant PIK3CA, but was dispensable for mammary tumorigenesis. This is in contrast to mammary tumors induced by the ErbB2/Neu transgene where loss of ErbB3 blocks mammary tumorigenesis (35). Our data are reminiscent of a recent study by Lahlou et al., in which ErbB2/Neu-mediated hyperplasia, but not MEC transformation, was reduced by an ErbB3 mutant incapable of binding to p85 (PI3K) (41). In tumors expressing the PI3K-uncoupled ErbB3 mutant, ErbB2 and EGFR engaged p85 to maintain PI3K activity. The iPIK3.iCre mice we present herein are phenotypically similar. It is unclear why genetic ablation of ErbB3 inhibits Neu-induced tumor formation (35), while uncoupling PI3K from ErbB3 does not (41), but the results suggest PI3K-independent functions of ErbB3.
Inhibition of EGFR/ErbB2 or PDGFR slowed mutant PIK3CA tumor growth without affecting Akt and PDK1 phosphorylation. This result suggests two not mutually exclusive possibilities. First, many RTKs or adaptors might contribute to PI3K activation in tumors harboring PIK3CA mutations. Second, the heightened activity of the PIK3CAH1047R kinase domain mutant (42, 43) allows for this kinase to signal strongly with fewer/no upstream binding partner(s). However, some upstream binding partners are likely needed because structural data suggest that p85 inhibits the catalytic activity of p110 until the p85/p110 complex binds to consensus phosphotyrosine YXXM motifs in RTKs/adaptors, thus relieving p85-mediated inhibition of p110 (44). E545K mutation in PIK3CA causes a structural change in the PI3K holoenzyme such that p85 no longer inhibits p110α, resulting in increased PI3K activity (45) which cannot be further activated by added tyrosine phosphorylated peptides (20). In contrast, the high kinase activity of H1047R PI3K doubles upon the addition of phospho-PDGFR or phospho-IRS-1 peptides (20). Finally, the transforming action of PIK3CAH1047R, but not PIK3CAE545K, is markedly reduced by loss of p85 binding, suggesting that PIK3CAH1047R requires binding to RTKs/adaptors for full activity (6, 21).
The driving oncogene in iPIK3.iCre tumors is PIK3CAH1047R, making PI3K the most compelling therapeutic target. As a single agent, the p110α-specific inhibitor BYL719 reduced tumor growth and P-Akt in iPIK3.iCre tumors without effecting P-PDK1. Dual EGFR/ErbB2 and PI3K inhibition reduced P-Akt and P-PDK1 and decreased tumor growth better than either agent alone. These data suggest that while individual inhibition of mutant PI3K and RTKs upstream was insufficient to suppress PI3K activity and growth, ErbB inhibition significantly enhanced the effect of BYL719 against PI3K mutant cancers. This was recapitulated in PIK3CAH1047R human breast cancer cells. We speculate, however, that tumor cells not initially eliminated by combined ErbB and PI3K inhibition may allow mutant PI3K to engage other RTKs and adaptors, eventually allowing tumors to evade the antitumor effect of PI3K inhibitors.
Supplementary Material
Acknowledgments
GRANT SUPPORT
This work was supported by R01 grants CA80195 (CLA) and CA143126 (RSC), ACS Clinical Research Professorship Grant CRP-07-234 (CLA), Breast Cancer Specialized Program of Research Excellence (SPORE) P50 CA98131, Vanderbilt-Ingram Cancer Center Support Grant P30 CA68485, a Stand Up to Cancer Dream Team Translational Research Grant from the Entertainment Industry Foundation (SU2C-AACR-DT0209) and Susan G. Komen for the Cure grant KG100677 (RSC). CDY is supported by DOD postdoctoral fellowship grant W81XWH-12-1-0026.
References
- 1.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–19. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 2.Bader AG, Kang S, Zhao L, Vogt PK. Oncogenic PI3K deregulates transcription and translation. Nat Rev Cancer. 2005;5:921–9. doi: 10.1038/nrc1753. [DOI] [PubMed] [Google Scholar]
- 3.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parsons R, Simpson L. PTEN and cancer. Methods Mol Biol. 2003;222:147–66. doi: 10.1385/1-59259-328-3:147. [DOI] [PubMed] [Google Scholar]
- 5.Courtney KD, Corcoran RB, Engelman JA. The PI3K pathway as drug target in human cancer. J Clin Oncol. 2010;28:1075–83. doi: 10.1200/JCO.2009.25.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao L, Vogt PK. Hot-spot mutations in p110alpha of phosphatidylinositol 3-kinase (pI3K): differential interactions with the regulatory subunit p85 and with RAS. Cell Cycle. 2010;9:596–600. doi: 10.4161/cc.9.3.10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bader AG, Kang S, Vogt PK. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc Natl Acad Sci U S A. 2006;103:1475–9. doi: 10.1073/pnas.0510857103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.TCGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holbro T, Beerli RR, Maurer F, Koziczak M, Barbas CF, 3rd, Hynes NE. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc Natl Acad Sci U S A. 2003;100:8933–8. doi: 10.1073/pnas.1537685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schulze WX, Deng L, Mann M. Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol Syst Biol. 2005;1:2005 0008. doi: 10.1038/msb4100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cook RS, Garrett JT, Sanchez V, Stanford JC, Young C, Chakrabarty A, et al. ErbB3 ablation impairs PI3K/Akt-dependent mammary tumorigenesis. Cancer Res. 2011;71:3941–51. doi: 10.1158/0008-5472.CAN-10-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Junttila TT, Akita RW, Parsons K, Fields C, Lewis Phillips GD, Friedman LS, et al. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer Cell. 2009;15:429–40. doi: 10.1016/j.ccr.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 13.Chakrabarty A, Sanchez V, Kuba MG, Rinehart C, Arteaga CL. Breast Cancer Special Feature: Feedback upregulation of HER3 (ErbB3) expression and activity attenuates antitumor effect of PI3K inhibitors. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1018001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garrett JT, Olivares MG, Rinehart C, Granja-Ingram ND, Sanchez V, Chakrabarty A, et al. Transcriptional and posttranslational up-regulation of HER3 (ErbB3) compensates for inhibition of the HER2 tyrosine kinase. Proc Natl Acad Sci U S A. 2011;108:5021–6. doi: 10.1073/pnas.1016140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wohrle FU, Daly RJ, Brummer T. Function, regulation and pathological roles of the Gab/DOS docking proteins. Cell Commun Signal. 2009;7:22. doi: 10.1186/1478-811X-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mardilovich K, Pankratz SL, Shaw LM. Expression and function of the insulin receptor substrate proteins in cancer. Cell Commun Signal. 2009;7:14. doi: 10.1186/1478-811X-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myers MG, Jr, Sun XJ, Cheatham B, Jachna BR, Glasheen EM, Backer JM, et al. IRS-1 is a common element in insulin and insulin-like growth factor-I signaling to the phosphatidylinositol 3′-kinase. Endocrinology. 1993;132:1421–30. doi: 10.1210/endo.132.4.8384986. [DOI] [PubMed] [Google Scholar]
- 18.Backer JM, Myers MG, Jr, Shoelson SE, Chin DJ, Sun XJ, Miralpeix M, et al. Phosphatidylinositol 3′-kinase is activated by association with IRS-1 during insulin stimulation. EMBO J. 1992;11:3469–79. doi: 10.1002/j.1460-2075.1992.tb05426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Escobedo JA, Navankasattusas S, Kavanaugh WM, Milfay D, Fried VA, Williams LT. cDNA cloning of a novel 85 kd protein that has SH2 domains and regulates binding of PI3-kinase to the PDGF beta-receptor. Cell. 1991;65:75–82. doi: 10.1016/0092-8674(91)90409-r. [DOI] [PubMed] [Google Scholar]
- 20.Carson JD, Van Aller G, Lehr R, Sinnamon RH, Kirkpatrick RB, Auger KR, et al. Effects of oncogenic p110alpha subunit mutations on the lipid kinase activity of phosphoinositide 3-kinase. Biochem J. 2008;409:519–24. doi: 10.1042/BJ20070681. [DOI] [PubMed] [Google Scholar]
- 21.Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci U S A. 2008;105:2652–7. doi: 10.1073/pnas.0712169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller TW, Balko JM, Fox EM, Ghazoui Z, Dunbier A, Anderson H, et al. ERalpha-dependent E2F transcription can mediate resistance to estrogen deprivation in human breast cancer. Cancer Discov. 2011;1:338–51. doi: 10.1158/2159-8290.CD-11-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–67. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Qu Z, Kim S, Shi V, Liao B, Kraft P, et al. Down-modulation of cancer targets using locked nucleic acid (LNA)-based antisense oligonucleotides without transfection. Gene Ther. 2011;18:326–33. doi: 10.1038/gt.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang A, Fritsch C, Wilson C, Reddy A, Liu M, Lehar J, et al. Abstract 3749: Single agent activity of PIK3CA inhibitor BYL719 in a broad cancer cell line panel. Proceedings of the 103rd Annual Meeting of the American Association for Cancer Research; 2012. [Google Scholar]
- 26.Garner A, Sheng Q, Bialucha U, Chen D, Chen Y, Das R, et al. Abstract 2733: LJM716: an anti-HER3 antibody that inhibits both HER2 and NRG driven tumor growth by trapping HER3 in the inactive conformation. Proceedings of the 103rd Annual Meeting of the American Association for Cancer Research; 2012. [Google Scholar]
- 27.Gunther EJ, Belka GK, Wertheim GB, Wang J, Hartman JL, Boxer RB, et al. A novel doxycycline-inducible system for the transgenic analysis of mammary gland biology. FASEB J. 2002;16:283–92. doi: 10.1096/fj.01-0551com. [DOI] [PubMed] [Google Scholar]
- 28.Liu P, Cheng H, Santiago S, Raeder M, Zhang F, Isabella A, et al. Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway-dependent and PI3K pathway-independent mechanisms. Nat Med. 2011;17:1116–20. doi: 10.1038/nm.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo ZM, Xu K, Yue Y, Huang B, Deng XY, Zhong NQ, et al. Temporal control of Cre recombinase-mediated in vitro DNA recombination by Tet-on gene expression system. Acta Biochim Biophys Sin (Shanghai) 2005;37:133–8. [PubMed] [Google Scholar]
- 30.Qu S, Rinehart C, Wu HH, Wang SE, Carter B, Xin H, et al. Gene targeting of ErbB3 using a Cre-mediated unidirectional DNA inversion strategy. Genesis. 2006;44:477–86. doi: 10.1002/dvg.20243. [DOI] [PubMed] [Google Scholar]
- 31.Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herschkowitz JI, Zhao W, Zhang M, Usary J, Murrow G, Edwards D, et al. Breast Cancer Special Feature: Comparative oncogenomics identifies breast tumors enriched in functional tumor-initiating cells. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1018862108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loi S, Haibe-Kains B, Majjaj S, Lallemand F, Durbecq V, Larsimont D, et al. PIK3CA mutations associated with gene signature of low mTORC1 signaling and better outcomes in estrogen receptor-positive breast cancer. Proc Natl Acad Sci U S A. 2010;107:10208–13. doi: 10.1073/pnas.0907011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chakrabarty A, Rexer BN, Wang SE, Cook RS, Engelman JA, Arteaga CL. H1047R phosphatidylinositol 3-kinase mutant enhances HER2-mediated transformation by heregulin production and activation of HER3. Oncogene. 2010;29:5193–203. doi: 10.1038/onc.2010.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaught DB, Stanford JC, Young C, Hicks DJ, Wheeler F, Rinehart C, et al. HER3 Is Required for HER2-Induced Preneoplastic Changes to the Breast Epithelium and Tumor Formation. Cancer Res. 2012;72:2672–82. doi: 10.1158/0008-5472.CAN-11-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balko JM, Miller TW, Morrison MM, Hutchinson K, Young C, Rinehart C, et al. The receptor tyrosine kinase ErbB3 maintains the balance between luminal and basal breast epithelium. Proc Natl Acad Sci U S A. 2012;109:221–6. doi: 10.1073/pnas.1115802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tikoo A, Roh V, Montgomery KG, Ivetac I, Waring P, Pelzer R, et al. Physiological Levels of Pik3ca(H1047R) Mutation in the Mouse Mammary Gland Results in Ductal Hyperplasia and Formation of ERalpha-Positive Tumors. PLoS One. 2012;7:e36924. doi: 10.1371/journal.pone.0036924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer DS, Brinkhaus H, Muller U, Muller M, Cardiff RD, Bentires-Alj M. Luminal Expression of PIK3CA Mutant H1047R in the Mammary Gland Induces Heterogeneous Tumors. Cancer Res. 2011 doi: 10.1158/0008-5472.CAN-10-3827. [DOI] [PubMed] [Google Scholar]
- 39.Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Webster MA, Hutchinson JN, Rauh MJ, Muthuswamy SK, Anton M, Tortorice CG, et al. Requirement for both Shc and phosphatidylinositol 3′ kinase signaling pathways in polyomavirus middle T-mediated mammary tumorigenesis. Mol Cell Biol. 1998;18:2344–59. doi: 10.1128/mcb.18.4.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lahlou H, Muller T, Sanguin-Gendreau V, Birchmeier C, Muller WJ. Uncoupling of PI3K from ErbB3 Impairs Mammary Gland Development but Does Not Impact on ErbB2-Induced Mammary Tumorigenesis. Cancer Res. 2012;72:3080–90. doi: 10.1158/0008-5472.CAN-11-3513. [DOI] [PubMed] [Google Scholar]
- 42.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 43.Isakoff SJ, Engelman JA, Irie HY, Luo J, Brachmann SM, Pearline RV, et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65:10992–1000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- 44.Yu J, Zhang Y, McIlroy J, Rordorf-Nikolic T, Orr GA, Backer JM. Regulation of the p85/p110 phosphatidylinositol 3′-kinase: stabilization and inhibition of the p110alpha catalytic subunit by the p85 regulatory subunit. Mol Cell Biol. 1998;18:1379–87. doi: 10.1128/mcb.18.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miled N, Yan Y, Hon WC, Perisic O, Zvelebil M, Inbar Y, et al. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317:239–42. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.