Abstract
Previous binding studies of antibodies that recognized a partially or fully hidden epitope suggest that insect cell-derived dengue virus undergoes structural changes at an elevated temperature. This was confirmed by our cryo-electron microscopy images of dengue virus incubated at 37°C, where viruses change their surface from smooth to rough. Here we present the cryo-electron microscopy structures of dengue virus at 37°C. Image analysis showed four classes of particles. The three-dimensional (3D) map of one of these classes, representing half of the imaged virus population, shows that the E protein shell has expanded and there is a hole at the 3-fold vertices. Fitting E protein structures into the map suggests that all of the interdimeric and some intradimeric E protein interactions are weakened. The accessibility of some previously found cryptic epitopes on this class of particles is discussed.
INTRODUCTION
Dengue virus (DENV) is a mosquito-borne pathogen and the causative agent of dengue fever, dengue hemorrhagic fever (DHF), and the life-threatening dengue shock syndrome (DSS). Currently, DENV infects about 50 to 100 million people per year, resulting in 250,000 to 500,000 cases of DHF or DSS, making it a major health, social, and economic problem (1). This virus belongs to the family Flaviviridae, which also includes other major human pathogens, such as yellow fever virus, West Nile virus (WNV), tick-borne encephalitis virus, Japanese encephalitis virus, etc. The four DENV serotypes share high genetic homology, varying in amino acid sequence by about 25 to 40%. Genotypes within a serotype are even more conserved, containing about only 3% variation in amino acid sequence (2, 3).
Envelope (E) protein is the major antigenic structure on the surface of DENV (4). Crystal structures of the ectodomain of the E protein showed that it consists of three domains (I, II, and III) and that E proteins likely exist as dimers in solution (5–7). Domain III participates in receptor binding, while domain II facilitates virus fusion via the interaction of its fusion loop at the tip of the domain with the endosomal membrane during virus entry into the cell (4). Cryo-electron microscopy (cryo-EM) structures (8, 9) of the mature DENV showed that the surface of the virus is made from 180 copies of E and 180 copies of membrane (M) proteins that are arranged in an icosahedral manner. There are three individual E proteins in each of the 60 asymmetric units (molecules A, B and C; also shown in Fig. 5B) (8, 9). Each of these E proteins is located close to one of the 2-, 3-, or 5-fold vertices, thereby having a different local chemical environment. The E proteins are organized as 90 head-to-tail homodimers. Three of these homodimers lie parallel to each other, forming a raft, and together with the other 29 rafts, they form a herringbone pattern on the surface of the virus.
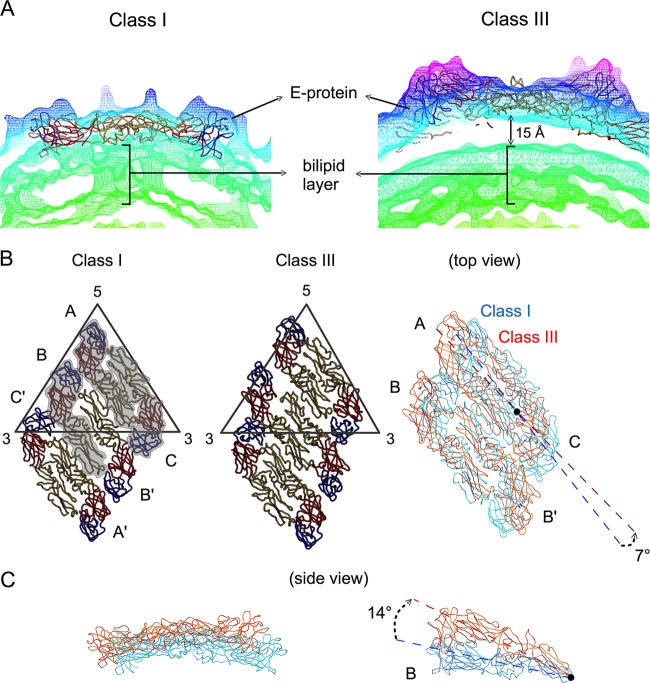
Fig 5.
Structural differences between class I and class III particles. (A) The densities of the cryo-EM maps of classes I and III are shown in mesh and colored according to their radii as shown in Fig. 3. The 15-Å gap between lipid bilayer and E proteins layer in class III particles is indicated. (B) Class I (left) and III (center) E protein rafts, each consisting of two asymmetric units, are shown. The three individual E proteins in the asymmetric unit are shaded and labeled A, B, and C and the same E proteins in the neighboring asymmetric unit are labeled A′, B′, and C′ (left). In the right panel, the two structures are superimposed, colored in cyan and orange for classes I and III, respectively. The E protein dimer (molecules A and C) near the 5-fold vertex in the class III structure rotates about 7° compared to the class I structure. The icosahedral 2-fold E protein dimers (molecules B and B′) in the class I structure have moved apart from each other in the class III structure. (C) Side view of the superimposed class I and III structures of the dimer consisting of molecules A and C (left) and molecule B (right). Molecule B of the class III structure rotates 14° clockwise from the class I structure using the fusion loop (black dot) as a pivot point.
The previously published cryo-EM structures were done with DENV grown in mosquito cell lines incubated at 28 to 30°C for several days and then kept at 4°C during virus purification prior to freezing on cryo-EM grids. This resulting structure is contradictory to those in some antibody binding studies. Crystal structures of antibodies 1A1D-2, E53, and E111 in complex with either whole ectodomain or just domain III of the E protein showed that the epitopes of these antibodies would be either partially or completely hidden in the cryo-EM mature virus structure (10–12), making them “cryptic” epitopes. However, these antibodies were found to be neutralizing when the virus was incubated at 37°C. The authors thus suggest that the virus may undergo some structural changes when the temperature is increased, making the cryptic epitopes accessible.
Here we report cryo-EM structures of the DENV2 New Guinea C (NGC) strain incubated at 37°C. Cryo-EM images of the sample showed that the virus appearance has changed from a smooth particle to a rough one. Classification of these images showed the presence of four classes of particles. Interpretation of one of these maps showed that the E protein inter- and intradimer interactions loosen. This may expose the previously hidden cryptic epitopes, allowing antibodies to bind.
MATERIALS AND METHODS
Virus sample preparation for cryo-EM studies.
The methods for virus production and purification are similar to those described elsewhere (8). Briefly, the DENV2 NGC strain was grown in C6/36 mosquito cells. The cells were infected at a multiplicity of infection of 0.1 at 28°C under 5% CO2. After 2 h of infection, the inoculant was replaced with fresh RPMI medium supplemented with 2% fetal bovine serum (FBS) and was further incubated for 4 days. The virus-containing supernatant was clarified from cell debris by centrifugation, and virus particles were collected by precipitation with 8% polyethylene glycol 8000 (PEG 8000) in NTE buffer (10 mM Tris-HCl [pH 8.0], 120 mM NaCl, 1 mM EDTA). Virus particles were resuspended and then purified by centrifugation using a 30% sucrose cushion and then a 10 to 30% potassium tartrate gradient. The virus band was collected and concentrated, and buffer was exchanged into NTE buffer using an Amicon Ultra-4 centrifugal concentrator (Millipore) with a 100-kDa molecular-mass cutoff filter. The virus preparation contained little or no prM, indicating low levels of contamination by immature virus, as determined by Coomassie blue-stained SDS-PAGE. To estimate the envelope protein concentration, the corresponding band was compared with bovine serum albumin (BSA) at different concentrations.
Cryo-EM image collection.
The virus sample was grown in C6/36 cells at 28°C and kept at 4°C throughout the purification steps. The sample was incubated for 30 min at 37°C, followed by another 2 h of incubation at 4°C prior to freezing. The step involving 30 min of incubation at 37°C was omitted for the control. A virus sample of 2.5 µl was applied onto ultrathin carbon-coated lacey carbon grids (Ted Pella), and blotted with filter paper for ∼2.5 s before being plunged into liquid ethane using the FEI Vitrobot Mark IV. The frozen grid was stored at liquid nitrogen temperature. The virus particles were imaged using a 300-kV FEI Titan Krios electron microscope at a nominal magnification of 47,000 with an electron dose of 18.5 e/Å2. The images were captured on a charge-coupled device (CCD) camera (giving a pixel size of 1.9 Å) at an underfocus range of 1.5 to 3 μm. Images showing drift and strong astigmatism were discarded, and 302 images were selected for further processing.
Cryo-EM reconstruction.
The sample incubated at 37°C had mostly particles with a rough surface, and there were only a few smooth particles (Fig. 1). Reconstruction of the particles was initiated by picking the rough-looking unbroken particles using the e2boxer tool in the EMAN2 software package (13). In total, 15,111 particles were selected. Contrast transfer function (CTF) parameters were determined using EMAN2. The orientation search was done by using the multipath simulated annealing (MPSA) procedure (14). The selected virus particles were initially treated as a homogenous population, and thus, only one search model was used: a three-dimensional (3D) map of the mature DENV (15). Following the orientation search, a 3D reconstruction was done with make3d from EMAN (16) with the result used as the reference model for the next cycle. Default parameters were used in the orientation search using the MPSA program, with the exception of the numbers of reference projections and the data resolution range in the Fourier space. Three reference projections and a data resolution range of 53 to 160 Å were used for the first 3 cycles; once the model was improving, we increased the number of projection references as well as the resolution range.
Fig 1.
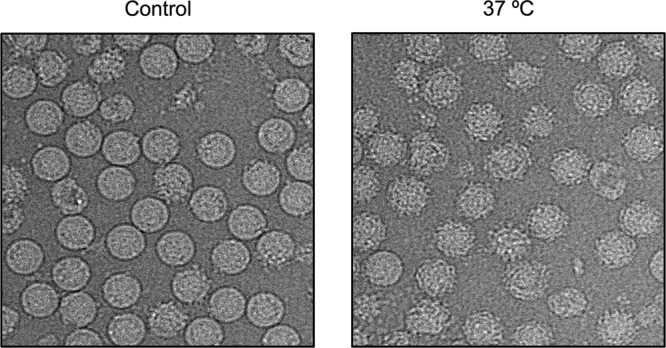
The DENV2 NGC strain undergoes structural changes after incubation at 37°C. The control (left) consists of DENV2 grown in a mosquito cell line at 28°C and then kept at 4°C during purification. The virus particles after incubation at 37°C change their morphology from a smooth surface (left) to a rough surface (right).
The 3D reconstruction of the rough-looking virus particles, by using the known mature dengue virus structure as the initial model (15), resulted in a map showing a smooth surface at a resolution of about 30 Å (result not shown). This map showed that the particles expanded from ∼240 Å to ∼ 280 Å with a small protrusion corresponding to the position of the dimer near the icosahedral 2-fold vertex. Since it was a very-low-resolution map, we had to check for the correctness of this reconstruction. Two separate tests were done. In the first test, we did the reconstruction using another starting model (i.e., a poor model obtained from the program starticos in the EMAN suite). A similar cryo-EM map was obtained, indicating the reliability of the previous result. In the second test, the data set was split into two halves and reconstructions were done separately using the previously published mature DENV map for the initial models. The reconstruction of the first half gave a similar map as before. However, the other half resulted in a map that was very similar to the mature virus model. This result suggested that the particle population is not homogeneous and contained some unexpanded mature virus and, possibly, some other particle classes.
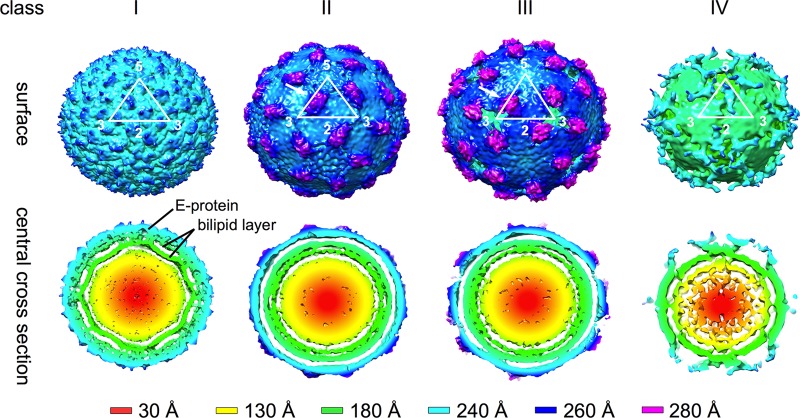
To improve the reconstruction, we decided to sort particles into classes and then use the 3D maps generated from these classes as initial models for reconstructions. Using the K-mean classification method implemented in EMAN2, the particle images were sorted by performing iterative reference-free classification to get a set of representative class averages. There were three visibly different classes of particles: particles with rough edges (called class II), particles with relatively smooth edges (class III), and particles with apparently smaller diameters (Fig. 2) (class IV). The initial models were generated for each class using e2initialmodel from EMAN2. Two separate 3D reconstructions were done. For the first reconstruction, 1,584 particle images corresponding to class IV were extracted from the initial particle set and the reconstruction was done by using the initial model created from this particle class. The particles from classes II and III were combined (total of 13,527 particles), and an orientation search was done using four starting models simultaneously. The four models were the published mature dengue virus structure (named class I) (15), two initial models generated from the class II and III particles, and immature DENV models (7). The projections were compared to all four models and grouped into the model class that had the highest correlation. Even though the control sample (virus not exposed to a temperature of 37°C) indicated only low levels of immature virus in the virus preparation, we included the immature DENV model in the multimodel orientation search to eliminate possible interference by the incidentally picked immature particles. In the final cycles of reconstruction of class IV, 1,327 particles were selected for the reconstruction, resulting in an ∼23-Å resolution map (as measured using Fourier shell correlation of independent half-data sets, with a cutoff at 0.5). The final resolutions of the class I, II, and III maps are 13, 14, and 14 Å, respectively. The particle numbers selected for the final reconstructions are 2,366, 5,085, and 5,861 particles, respectively. Approximately 200 of the 13,527 particles were immature dengue virus particles.
Fig 2.
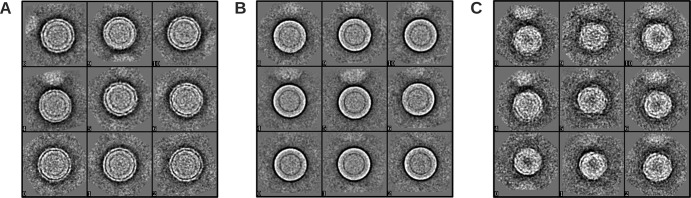
Three different classes of DENV2 NGC particles obtained with EMAN2. (A) Class II particles that have a rough surface. (B) Class III particles that have a relatively smooth surface. (C) Class IV particles that are smaller than the other classes.
To validate these structures, we used a different method of classification. Using the entire data set and mature virus as a model for reconstruction by MPSA, a structure with a larger diameter than that in the mature virus model was generated. (This was later used as the first model.) However, not all of the particles were selected from this reconstruction, indicating that there are other classes of particles. We then generated a random model by simply averaging the remaining projections. (This was later used as the second model.) We then started another reconstruction using the first two models and the original mature virus maps as initial models running simultaneously. Maps generated from these three models were similar to the class I, II, and III maps obtained when classifications were done by K-mean classification.
Model fitting.
The cryo-EM structure of the DENV2 E protein (Protein Data Bank [PDB] accession code 1P58) (15) was used for fitting into the cryo-EM maps. The structure fits well into the class I map since it is similar to the previously published cryo-EM DENV2 map. For the class III map, the handedness of the map was determined by superimposing it with the envelope protein structure mentioned above. The E proteins were manually fitted into the map density using the program O (17). Briefly, the three E proteins in the icosahedral asymmetric unit had to be moved to a higher radius in order to fit into the density. The dimer (molecules A and C) that lies closer to 5-fold axis required translation and rotation adjustments, whereas the 2-fold E protein dimer (molecules B and B′) required some rotational adjustments. The final fitting results showed no clashes between the E proteins, and all densities were interpreted. The Fit in Map tool implemented in the program Chimera (18) was then used to optimize the fit. Although the class II map is estimated to have a 14-Å resolution, visual inspection of the map showed that it did not have enough visible features for the interpretation of the map. One possibility is there is a heterogenous population of virus in this class, resulting in smeared densities. Thus, the class II map was not interpreted. Similarly, no model fitting was done for the class IV map as the map only shows the lipid bilayer with only small patches of density left for the E proteins.
Comparison of the infectivities of DENV preincubated at 28 and 37°C.
Briefly, BHK cells were cultured in RPMI supplemented with 10% FBS at 37°C in 5% CO2. When cells reached approximately 80% confluence, virus infection was carried out. Virus was serially diluted 10-fold from 1:102 to 1:108 using RPMI supplemented with 2% FBS. For the 28°C experiment, the virus was preincubated at 28°C for 30 min or 1 h and then 100 μl of virus was layered onto the BHK cells, which were preequilibrated at 28°C for 2 h. For the 37°C experiment, the virus and cells were treated in a similar manner to the 28°C experiment, except the temperature was set at 37°C. The cells were then further incubated at their respective temperatures for 2 h. The infected cells were overlaid with RPMI supplemented with 2% FBS and 1% Aquacide. The plates were further incubated at 37°C in 5% CO2 for 4 days. The cells were then stained by first discarding the supernatant and then adding 1 ml of 0.5% (wt/vol) crystal violet–25% formaldehyde to the cell monolayer and further incubated for 1 h. The cell monolayers were then washed with water, and plaques were counted by visual inspection. The experiments were repeated twice.
Protein structure accession number.
The cryo-EM map reconstructed from class III particles was deposited in the Electron Microscopy Database under accession no. EMD-2296. The modeled protein structure was deposited in the PDB under accession no. 3ZKO.
RESULTS
Virus morphology changed when the virus was incubated at 37°C.
The cryo-EM images of the control sample of the DENV2 NGC strain that was not exposed to 37°C (Fig. 1, left) showed about ∼80% of the particles had a smooth surface and round shape indicative of the mature virion. The rest of the virus population consisted of either broken nonrounded particles or the spiky immature virus. When incubated at 37°C, the majority of the virus particles showed rough surface (Fig. 1, right). The same observation was also found with DENV-2 WRAIR strain S16803 (data not shown).
DENV incubated at 28 and 37°C showed similar infectivities in BHK cells.
When tissue culture supernatant containing virus was incubated at 28 or 37°C for 30 min and then layered onto BHK cells for another 2 h at their respective temperatures, the number of plaques obtained were 3.6 ×106 PFU/ml and 3.1 × 106 PFU/ml, respectively. Increasing the incubation time of the virus to 1 h prior to infection showed plaque counts of 2.4 × 106 PFU/ml and 2.6 × 106 PFU/ml for 28 and 37°C, respectively. This suggests that increasing the temperature to 37°C does not alter the infectivity of the virus.
Reconstruction of four different classes of DENV particles observed at 37°C.
The cryo-EM maps of classes I, II, and III are shown in Fig. 3. The class I map is similar to the previously published mature DENV map (7–9, 15). Briefly, the particle has a radius of ∼240 Å, and the surface is relatively smooth with small bumps at positions corresponding to all E protein domain IIIs. The central cross-section shows the well-resolved lipid bilayer inside the envelope protein shell. The class II particle is slightly larger than the class I virion and has protrusions between the 5- and 3-fold vertices. The E protein layer is located at a higher radius of ∼250 Å compared to class I particles, indicating an expansion in the outer protein shell. Class III particles are the largest among the three classes, with a radius of ∼260 Å. The class III particle shows more pronounced protrusions between 5- and 3-fold vertices that reach out to 280 Å from the center. There is a hole at all of the 3-fold vertices (Fig. 4, right).
Fig 3.
Cryo-EM maps reconstructed from class I to IV particles present in the DENV2 sample incubated at 37°C. The surfaces of the maps (above) and their central cross-sections (below) are colored according to their radii (shown in the lower panel). The white triangle represents an icosahedral asymmetric unit, and the corresponding 2-, 3-, and 5-fold symmetry vertices are indicated. Maps generated from the class I particles are similar to the previous published DENV-2 maps (7–9, 15). Class II and III maps showed that the particles in these classes have bigger radii, indicating that the virus had expanded. There are protruding densities on the virus surface between the 5- and 3-fold vertices (white arrows). The class IV map showed very poor density on the E protein layer, indicating that the E protein had lost its icosahedral symmetry.
Fig 4.
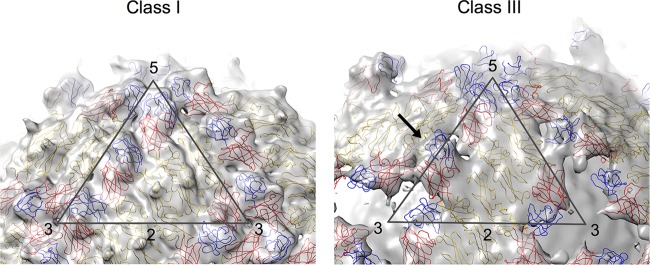
The fit of E proteins into cryo-EM maps of class I and III particles. The maps are colored in light gray, whereas the fitted E protein structures are drawn in a wire representation and colored in red, yellow, and blue for domains I, II, and III, respectively. The icosahedral asymmetric unit of both maps is depicted as a black triangle with the corresponding symmetry axis indicated by numbers. The class I map (left) is similar to the previously published DENV-2 cryo-EM structure (7–9, 15). The class III map (right) showed the presence of a hole at each of the 3-fold vertices. Also, domains I and III of the E protein molecule at the icosahedral 2-fold vertices are observed to be protruding (indicated by a black arrow).
The cryo-EM map reconstructed from class IV particles showed a radius of only ∼180 Å with some irregular densities around the 5-fold vertices (Fig. 3). The cross-section shows that most of the densities corresponding to the E protein shell have disappeared, indicating that the E protein layer has lost its icosahedral symmetry.
Structure fitting to the cryo-EM map.
The organization of E proteins in the class I particle is identical to those of previously reported DENV structures (7–9, 15). This class has the most compact structure on its outer layer compared to the other classes. There are 180 copies of envelope proteins on the virus surface forming the herringbone pattern (Fig. 4, left).
The map generated from the class II particles cannot be interpreted as the surface is very smooth and continuous, with no obvious features or boundaries, except at the protrusion between the 5- and 3-fold vertices (Fig. 3). Attempts to fit the map produced numerous possibilities with no clear optimal fit. In contrast to the class II map, the class III map had more features and showed clearer boundaries guiding the fit of the E protein molecules. The fit was done by manually fitting the E protein dimer as a rigid body into the densities near the 5-fold vertex and assigning the remaining density to the E proteins at the icosahedral 2-fold vertex (Fig. 4, right). The final structure has a correlation coefficient of 0.84 as calculated by the Fit-in-Map tool in Chimera (18). In the final structure, domains I and III of molecules C of the dimers near the 5-fold vertices define the periphery of the hole at the 3-fold vertices (Fig. 4 and 5B), whereas the protrusions between the 5- and 3-fold vertices (Fig. 4) are filled by domains I and III of molecule B (Fig. 4 and 5B).
Conformational changes of DENV at 37°C.
Increasing the temperature to 37°C induced structural rearrangement of the E proteins on the surface of DENV. We observed in the class III particles that the E protein layer moves outward, while the membrane (lipid bilayer) remains at the same radii as the unexpanded particles, thus creating a gap of about 15 Å (Fig. 5A). The 5-fold dimer (molecules A and C) undergoes translation about ∼14 Å along the dimer axis and also rotates about 7° anticlockwise (Fig. 5B). The dimeric interactions of the E proteins at the icosahedral 2-fold vertices loosen (molecules B and B′), and the two E protein molecules move in opposite directions perpendicular to the dimer axis, creating a distance of ∼6 Å between the molecules. Furthermore, each of the E proteins rotates clockwise by about 14° using the fusion loop as a pivot point (Fig. 5C). The conformational changes of the icosahedral 2-fold E proteins break the raft structure.
DISCUSSION
Structural change in flavivirus at elevated temperature has been indirectly observed through various antibody-mediated neutralization studies, especially in DENV and West Nile viruses (WNV). The epitopes identified by crystal structures of monoclonal antibodies (MAbs) E53 (11), 1A1D-2 (12) and E111 (10) complexed with either whole E protein or domain III, were also confirmed by mutational studies (10, 12, 19–21). The locations of these epitopes in the previously published DENV-2 cryo-EM map (7, 9, 15) suggested that these epitopes would be either partially or completely hidden on the virus surface. This indicates that the antibodies would not be able to bind to the virus; however, the neutralizing properties of the antibodies suggested otherwise (10, 12, 22). Prolonged incubation at 37°C significantly improves the binding and neutralizing activities of these antibodies (10, 12, 22). This points to the possibility of the virus surface undergoing structural changes, thus transiently exposing previously hidden epitopes. This structural change is not unique in dengue serotype 2 NGC strain. DENV2 WRAIR strain S16803 also displayed similar characteristics. In addition, Austin et al. (10) showed that a dengue serotype 1-specific antibody, E111, which binds to a completely hidden epitope, could form a complex with most genotypes, although some of these genotypes required a higher temperature (40°C) and longer exposure times to these antibodies.
Increased temperature stimulates structural rearrangement of the virus surface proteins, and these structural changes do not affect virus infection of mammalian cells, as indicated by the plaque assay. The class III particles are more homogenous in structure compared to the other classes, as indicated by the ability to reconstruct the map to a resolution of 14 Å. About half of the particles in the virus population belong to this class. The cryo-EM map showed structural features that allowed interpretation of the density. In contrast to class III, class II may contain particles with heterogenous structures, thus resulting in a visibly blurrier map. It is probable that other classes of dengue virus particles are present, in addition to classes I to IV, that cannot be solved by cryo-EM single-particle analysis.
The structural changes of DENV stimulated by incubation at higher temperatures have been popularized as “breathing” (12, 22–24). The word “breathing” suggests that the structures of these different classes are reversible. Our work is unable to distinguish if these structures are indeed reversible.
Class II and III particles have an expanded E protein layer. Interpretation of the class III map showed that all interdimeric E protein interactions are weakened. Near the icosahedral 2-fold vertices, the E protein molecules (B and B′) within a dimer also separated. Each E protein consists of an E ectodomain and a stem region that connects the ectodomain to its transmembrane region. The stem contains two α helices, one of which interacts with the viral lipid membrane. The fact that the E protein layer of class II and III particles is now located at a higher radius indicates that the E protein helical stem region that previously was lying on the surface of the lipid layer may now have to be fully extended from the membrane surface. Similar extension of the stem region had been previously reported in the prefusion intermediate structure of WNV complexed with Fab E16 (25).
The organization of the E proteins showing the structural changes on the surface of DENV presented here is different from the virus structure as observed in the cryo-EM Fab 1A1D-2–DENV complex structure (12). Binding of antibody 1A1D-2 requires the structural changes of the virion in order to expose the partially hidden epitopes. Thus, Fab 1A1D-2 was thought to trap the DENV surface in a state of motion. Comparison between the class III structure and the Fab 1A1D-2–DENV complex structure showed vastly different E protein organization, especially near the 5-fold and 3-fold vertices (molecules A and C, respectively) (Fig. 6A). These two E proteins were bound by Fab molecules in the 1A1D-2–DENV complex, suggesting that the conformation changes of these two E proteins were induced by Fab binding. In addition, the fusion loops of these molecules in the Fab 1A1D-2–DENV complex were interacting with its viral membrane. The structure is therefore unlikely to represent part of the natural structural motion as the fusion loop will not be available for infection. For the E protein near the 2-fold vertices (B molecule), the two structures showed that the 1A1D-2 epitope on domain III gradually turned inwards, eventually facing the viral lipid membrane in the 1A1D-2–DENV structure (Fig. 6B). This is consistent with the observation that antibody 1A1D-2 is unable to bind to the B molecule.
Fig 6.
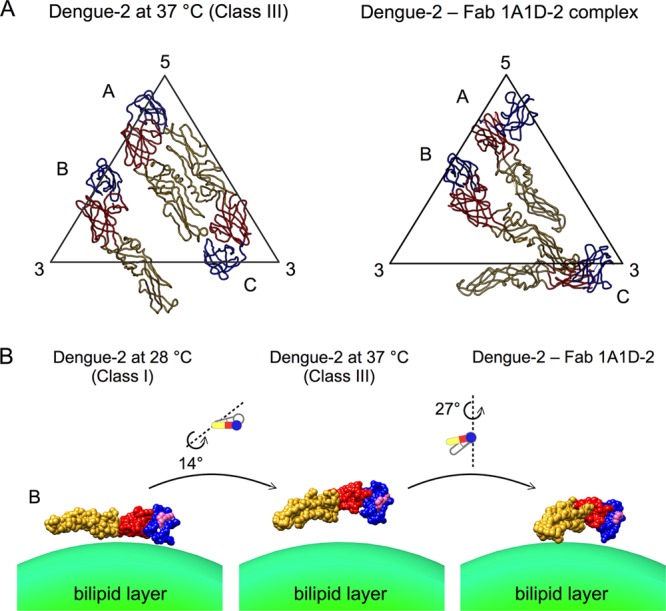
Comparison of the class III structure with the Fab 1A1D-2–DENV complex structure. (A) Organization of the three individual E proteins in an icosahedral asymmetric unit of the class III structure (left) and Fab 1A1D-2–DENV complex structure (right). Domains I, II, and III are colored in red, yellow, and blue, respectively. (B) Differences in the orientations of molecule B in the class I, III, and Fab 1A1D-2–DENV complex structures. In the Fab 1A1D-2–DENV structure, molecule B is not bound by antibody, while the other E protein molecules are bound. The E protein is drawn as a surface representation. The antibody 1A1D-2 epitope is indicated in pink on the E protein surface. Conformational changes from class I to class III to the one observed in Fab 1A1D-2–DENV structures are shown.
Analysis of the 1A1D-2 epitope in the class I map showed that the epitope is partially hidden in the E proteins near the 5- and 3-fold vertices (molecules A and C) in the asymmetric unit. The epitope in molecules A and C remains hidden in the class III structure (Fig. 7). This situation is different for the 2-fold E protein in the class III structure (molecule B), where the epitope is much more exposed. However, epitope orientation is as important as epitope accessibility. This orientation (facing the virus lipid envelope) prevents the antibody from binding. One way that Fab 1A1D-2 could access the partially hidden epitope is via the E proteins near the 3-fold vertices (C molecules). The domain III of this E protein is located on the periphery of the hole (Fig. 4). The open space would allow flexibility of the domain III, thus increasing its accessibility to the antibody. Another possibility is the antibody may bind to the class IV particle, which has looser E proteins on its surface.
Fig 7.
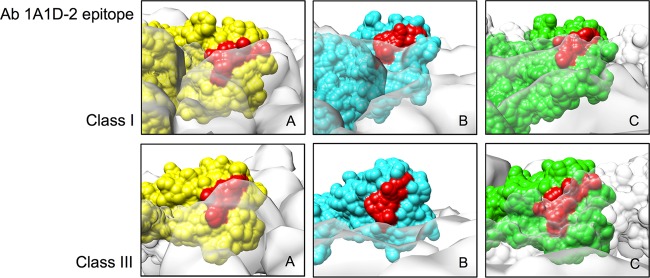
Comparison of the accessibility of the 1A1D-2 epitope on the three individual E proteins in the asymmetric unit of class I and class III structures. Molecules A, B, and C (indicated in Fig. 5B) are colored in yellow, cyan, and green, respectively. The other neighboring E proteins are shown as a transparent white surface. The antibody (Ab) 1A1D-2 epitope is colored in red.
MAb E111 also binds to another cryptic epitope on domain III (Fig. 8) (10). In the class I map, this epitope is completely hidden for all of the E proteins in the asymmetric unit, whereas in the class III map, it is exposed on the E proteins' B and C molecules. These E111 epitopes are located on the protruding densities between the 5- and 3-fold vertices and at the edge of the 3-fold holes and therefore can be readily accessible to antibodies.
Fig 8.
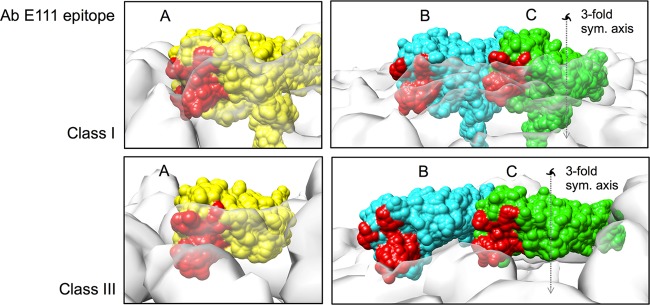
Comparison of the accessibility of the E111 epitope on the three individual E proteins in the asymmetric unit of class I and class III structures. The coloring scheme is the same as in Fig. 7. The 3-fold symmetry axis is indicated as a dashed line with a 3-fold symmetry (sym.) symbol. Holes on the class III surface are located at all 3-fold vertices.
In conclusion, we showed that DENV undergoes structural changes when incubated at an elevated temperature of 37°C, and these changes do not affect infectivity. We also showed the existence of at least four classes of structures in the sample. These findings in general suggest that the virus surface E protein interactions loosen at physiological temperature, thus exposing previously cryptic epitopes for antibody binding. These structures may also be relevant for receptor binding, as DENV would have undergone these structural changes when injected into the human body from the mosquito vector.
ACKNOWLEDGMENTS
The work was supported by Duke-NUS block grant R913-200-039-263, NMRC grant R-913-300-021-213, and NRF fellowship R-913-301-015-281 awarded to S.M.L.
Footnotes
Published ahead of print 24 April 2013
REFERENCES
- 1. Gubler DJ. 2002. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century Trends Microbiol. 10:100–103 [DOI] [PubMed] [Google Scholar]
- 2. Holmes EC, Twiddy SS. 2003. The origin, emergence and evolutionary genetics of dengue virus. Infect. Genet. Evol. 3:19–28 [DOI] [PubMed] [Google Scholar]
- 3. Rico-Hesse R. 1990. Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology 174:479–493 [DOI] [PubMed] [Google Scholar]
- 4. Roehrig JT. 2003. Antigenic structure of flavivirus proteins. Adv. Virus Res. 59:141–175 [DOI] [PubMed] [Google Scholar]
- 5. Modis Y, Ogata S, Clements D, Harrison SC. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 100:6986–6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Modis Y, Ogata S, Clements D, Harrison SC. 2005. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J. Virol. 79:1223–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang Y, Zhang W, Ogata S, Clements D, Strauss JH, Baker TS, Kuhn RJ, Rossmann MG. 2004. Conformational changes of the flavivirus E glycoprotein. Structure 12:1607–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, Strauss JH. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang X, Ge P, Yu X, Brannan JM, Bi G, Zhang Q, Schein S, Zhou ZH. 2012. Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nat. Struct. Biol. 20:105–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Austin SK, Dowd KA, Shrestha B, Nelson CA, Edeling MA, Johnson S, Pierson TC, Diamond MS, Fremont DH. 2012. Structural basis of differential neutralization of DENV-1 genotypes by an antibody that recognizes a cryptic epitope. PLoS Pathog. 8:e1002930. 10.1371/journal.ppat.1002930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cherrier MV, Kaufmann B, Nybakken GE, Lok SM, Warren JT, Chen BR, Nelson CA, Kostyuchenko VA, Holdaway HA, Chipman PR, Kuhn RJ, Diamond MS, Rossmann MG, Fremont DH. 2009. Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody. EMBO J. 28:3269–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lok SM, Kostyuchenko V, Nybakken GE, Holdaway HA, Battisti AJ, Sukupolvi-Petty S, Sedlak D, Fremont DH, Chipman PR, Roehrig JT, Diamond MS, Kuhn RJ, Rossmann MG. 2008. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat. Struct. Mol. Biol. 15:312–317 [DOI] [PubMed] [Google Scholar]
- 13. Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157:38–46 [DOI] [PubMed] [Google Scholar]
- 14. Liu X, Jiang W, Jakana J, Chiu W. 2007. Averaging tens to hundreds of icosahedral particle images to resolve protein secondary structure elements using a multi-path simulated annealing optimization algorithm. J. Struct. Biol. 160:11–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang W, Chipman PR, Corver J, Johnson PR, Zhang Y, Mukhopadhyay S, Baker TS, Strauss JH, Rossmann MG, Kuhn RJ. 2003. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat. Struct. Biol. 10:907–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ludtke SJ, Baldwin PR, Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82–97 [DOI] [PubMed] [Google Scholar]
- 17. Jones TA, Zou JY, Cowan SW, Kjeldgaard M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110–119 [DOI] [PubMed] [Google Scholar]
- 18. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 19. Oliphant T, Nybakken GE, Engle M, Xu Q, Nelson CA, Sukupolvi-Petty S, Marri A, Lachmi BE, Olshevsky U, Fremont DH, Pierson TC, Diamond MS. 2006. Antibody recognition and neutralization determinants on domains I and II of West Nile virus envelope protein. J. Virol. 80:12149–12159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sukupolvi-Petty S, Austin SK, Purtha WE, Oliphant T, Nybakken GE, Schlesinger JJ, Roehrig JT, Gromowski GD, Barrett AD, Fremont DH, Diamond MS. 2007. Type- and subcomplex-specific neutralizing antibodies against domain III of dengue virus type 2 envelope protein recognize adjacent epitopes. J. Virol. 81:12816–12826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gromowski GD, Roehrig JT, Diamond MS, Lee JC, Pitcher TJ, Barrett AD. 2010. Mutations of an antibody binding energy hot spot on domain III of the dengue 2 envelope glycoprotein exploited for neutralization escape. Virology 407:237–246 [DOI] [PubMed] [Google Scholar]
- 22. Dowd KA, Jost CA, Durbin AP, Whitehead SS, Pierson TC. 2011. A dynamic landscape for antibody binding modulates antibody-mediated neutralization of West Nile virus. PLoS Pathog. 7:e1002111. 10.1371/journal.ppat.1002111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pierson TC, Kuhn RJ. 2012. Capturing a virus while it catches its breath. Structure 20:200–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cockburn JJB, Sanchez MEN, Fretes N, Urvoas A, Staropoli I, Kikuti CM, Coffey LL, Seisdedos FA, Bedouelle H, Rey FA. 2012. Mechanism of dengue virus broad cross-neutralization by a monoclonal antibody. Structure 20:303–314 [DOI] [PubMed] [Google Scholar]
- 25. Kaufmann B, Chipman PR, Holdaway HA, Johnson S, Fremont DH, Kuhn RJ, Diamond MS, Rossmann MG. 2009. Capturing a flavivirus pre-fusion intermediate. PLoS Pathog. 5:e1000672. 10.1371/journal.ppat.1000672 [DOI] [PMC free article] [PubMed] [Google Scholar]