Abstract
The kinetochore is the macromolecular protein complex that mediates chromosome segregation. The Dsn1 component is crucial for kinetochore assembly and is phosphorylated by the Aurora B kinase. We found that Aurora B phosphorylation of Dsn1 promotes the interaction between outer and inner kinetochore proteins in budding yeast.
Keywords: Ipl1/Aurora B protein kinase, Dsn1, chromosome segregation, kinetochore
ACCURATE chromosome segregation is essential to avoid aneuploidy and maintain genomic stability (Holland and Cleveland 2012). Segregation is directed by the kinetochore, the macromolecular complex that interacts with spindle microtubules during cell division (Westermann et al. 2007; Cheeseman and Desai 2008; Bloom and Joglekar 2010). The kinetochore is composed of multiple copies of distinct subcomplexes that assemble into a macromolecular structure estimated to be >5 MDa (Joglekar et al. 2006; Lawrimore et al. 2011; Gonen et al. 2012). The inner kinetochore contains proteins that closely associate with centromeric DNA, whereas the outer kinetochore contains the proteins that bind to microtubules. At the base of the kinetochore, the CENP-A centromeric histone H3 variant forms a specialized chromatin environment essential for recruiting other kinetochore proteins such as the CCAN (constitutive centromere-associated network) (Buscaino et al. 2010; Perpelescu and Fukagawa 2011). These inner kinetochore components are essential for the assembly of the outer kinetochore, which includes the KMN (KNL1, Mis12, Ndc80 complexes) network that exhibits robust microtubule-binding activity (Cheeseman et al. 2006; Okada et al. 2006; Powers et al. 2009; Tanaka et al. 2009; Akiyoshi et al. 2010; Carroll et al. 2010; Przewloka et al. 2011; Screpanti et al. 2011). Although the core kinetochore components have been identified, the requirements for their assembly onto the centromere to form the macromolecular kinetochore structure are still unknown (Gascoigne and Cheeseman 2011).
Dsn1, a subunit of the Mis12 complex, is essential for kinetochore assembly (Kline et al. 2006), and may be a keystone for outer kinetochore assembly (Cheeseman and Desai 2008; Davies and Kaplan 2010; Maskell et al. 2010; Petrovic et al. 2010; Malvezzi et al. 2013). The Ipl1/Aurora B kinase is known to destabilize erroneous kinetochore-microtubule attachments (for reviews, see Ruchaud et al. 2007; Lampson and Cheeseman 2011). Interestingly, two studies suggested that Aurora B also promotes outer kinetochore assembly by phosphorylating Dsn1 (Emanuele et al. 2008; Yang et al. 2008). However, more recent work challenged these findings and instead concluded that Dsn1 phosphorylation contributes to destabilizing erroneous microtubule attachments (Welburn et al. 2010). Therefore, the role of Dsn1 phosphorylation by Aurora B remains unclear.
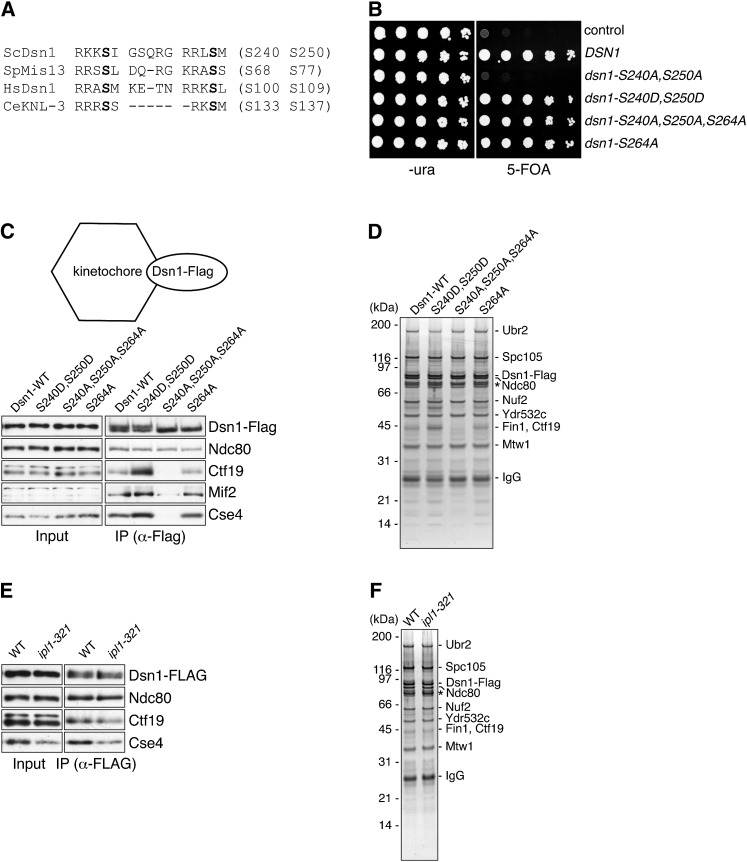
To elucidate the function of Aurora B-dependent phosphorylation of the budding yeast Dsn1 protein, we mutated the two serine residues in the conserved Aurora B consensus sites (S240 and S250, Figure 1A) to either alanine, which prevents phosphorylation, or aspartic acid, which may mimic phosphorylation. When expressed from the endogenous DSN1 promoter, the single site mutants (dsn1-S240A and dsn1-S250A) supported growth in the absence of wild-type (WT) DSN1 (Westermann et al. 2003 and data not shown). However, the dsn1-S240A, S250A double mutant failed to complement dsn1Δ, suggesting that phosphorylation of at least one of these sites is essential in budding yeast (Akiyoshi et al. 2013 and Figure 1B). In contrast, the phosphomimic mutant (dsn1-S240D, S250D) is viable. Dsn1 is also phosphorylated at three Cdk1 consensus sites (T12, S69, and S264), among which only S264 is conserved within the Saccharomyces lineage (Akiyoshi and Biggins 2010). While studying Dsn1 phosphorylation, we found that the dsn1-S240A, S250A, S264A triple mutant supports growth in the absence of wild-type DSN1 (Figure 1B). We previously found that the Dsn1-S240A, S250A mutant protein is targeted for degradation and the addition of the S264A mutation restores protein levels, suggesting that Cdk1 phosphorylation is involved in regulating Dsn1 protein levels (Akiyoshi et al. 2013 and data not shown).
Figure 1.
The Dsn1-S240A, S250A mutant that lacks Ipl1 phosphorylation sites is inviable and has defective interactions with inner kinetochore proteins. (A) Dsn1 contains two Ipl1/Aurora B phosphorylation motifs (R/K-R/K-X-pS/T-V/I/L/X). Alignment shows the conservation of Ser residues (shown in boldface type). (B) Dsn1-S240A, S250A cells are inviable and rescued by the additional mutation of S264A. Serial dilutions (fivefold) of dsn1Δ cells containing DSN1 on a URA3, CEN vector and the indicated integrated point mutants (SBY2318, SBY5948, SBY5949, SBY5950, SBY5952, and SBY6072) were plated on −ura and 5-FOA plates. Cells that need to maintain the URA3, DSN1 vector for viability are sensitive to 5-FOA. (C and D) Dsn1-S240A, S250A, S264A shows a weakened interaction with the inner-kinetochore components Cse4/CENP-A, Mif2/CENP-C, and Ctf19. Dsn1-Flag proteins containing the indicated mutations (SBY7902, SBY8037, SBY7904, and SBY8123) were immunoprecipitated with anti-Flag antibodies and analyzed by SDS–PAGE followed by immunoblotting against representative kinetochore proteins (C) or silver staining (D) as previously described (Akiyoshi et al. 2010). Note that Mif2/CENP-C is not detectable in lysate and the asterisk (*) indicates proteins that nonspecifically co-purify with all Flag-tagged proteins in D. (E and F). Dsn1-Flag was immunoprecipitated from lysates prepared from WT (SBY7441) or ipl1-321 (SBY8120) strains shifted to 37° for 3 hr and analyzed by SDS–PAGE followed by immunoblotting against representative kinetochore proteins (E) or silver staining (F). Strains are listed in supporting information, Table S1.
Given the controversial role of Aurora B-dependent phosphorylation in regulating Dsn1 (Emanuele et al. 2008; Yang et al. 2008; Welburn et al. 2010), we analyzed kinetochore assembly in the viable Dsn1 phosphomutants and the ipl1-321 temperature-sensitive mutants using two complementary assays that we established (Akiyoshi et al. 2009, 2010). In one technique, kinetochore particles are isolated via the purification of Dsn1, which yields kinetochore particles that contain both inner and outer kinetochore proteins (Akiyoshi et al. 2010). However, centromeric DNA and the centromere-binding complex CBF3 do not co-purify, indicating that these particles are not bound to endogenous centromeres after the purification. When we purified Dsn1-Flag from strains containing either wild-type Dsn1 or the Dsn1 phosphomutants that can support viability, there was an equivalent association of the outer KMN network components in all strains (Figure 1, C and D). However, the amounts of co-purifying inner kinetochore components (Ctf19, Mif2/CENP-C, and Cse4/CENP-A) were greatly reduced in the Dsn1-S240A, S250A, S264A mutant strain (Figure 1, C and D). In contrast, the level of inner kinetochore proteins that co-purified with the phosphomimic Dsn1-S240D, S250D mutant was increased (Figure 1C). Together, these data suggest that phosphorylation of Dsn1 promotes its association with inner kinetochore proteins. Importantly, there were no detectable defects in the single Dsn1-S264A strain, suggesting that the effects we observe in the Dsn1-S240A, S250A, S264A strain are largely due to a lack of Aurora B phosphorylation. Consistent with this, we also found that lower amounts of the inner kinetochore proteins Ctf19 and Cse4 co-purified with Dsn1 from ipl1-321 mutant cells (Figure 1E). The levels of the outer kinetochore components were normal based on immunoblotting of Dsn1 and Ndc80 as well as silver-stained PAGE stoichiometry estimates (Figure 1, E and F).
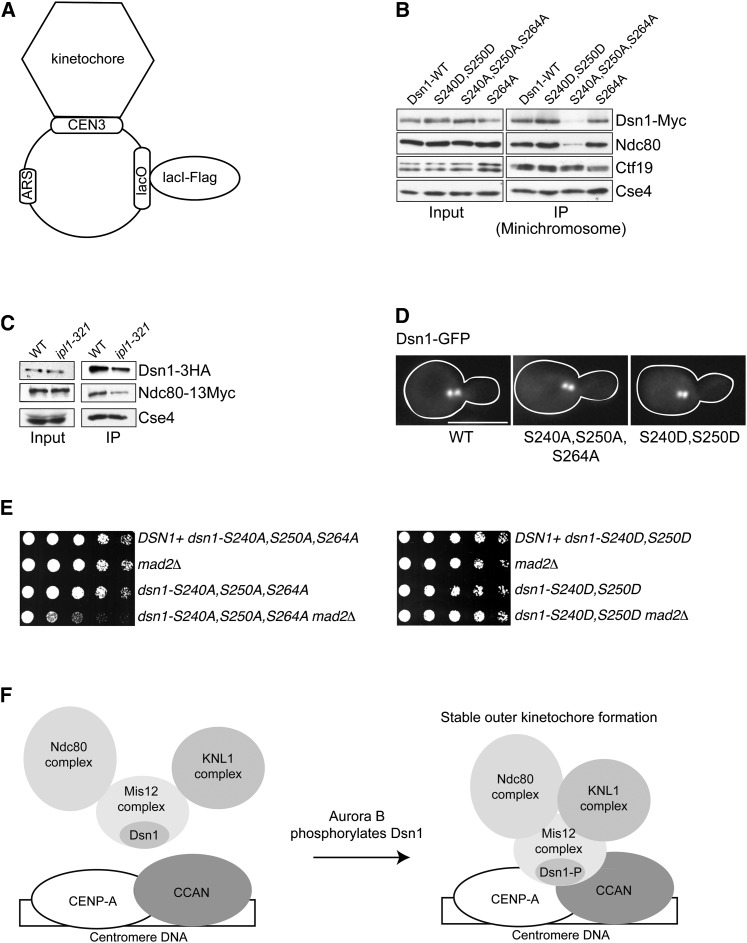
In a complementary approach, we utilized a centromeric minichromosome purification method where only centromere-bound kinetochore proteins are co-purified (Figure 2A and Akiyoshi et al. 2009). We purified minichromosomes from cells expressing Myc-tagged Dsn1 mutants and examined the amount of kinetochore proteins that remain associated with centromeric DNA by immunoblotting (Figure 2B). Although normal amounts of inner kinetochore proteins were associated with the minichromosome in all strains, the levels of co-purifying outer kinetochore proteins (Dsn1 and Ndc80) were reduced in the dsn1-S240A, S250A, S264A mutant compared to controls (Figure 2B). The single dsn1-S264A mutation did not exhibit any obvious defect in kinetochore assembly (Figures 1C and 2B), suggesting that the observed defects of dsn1-S240A, S250A, S264A are largely attributable to dsn1-S240A, S250A. Consistent with this, the levels of Cse4 that co-purify with minichromosomes from an ipl1-321 temperature-sensitive mutant are normal, but there are reduced levels of the Ndc80 and Dsn1 kinetochore components (Figure 2C). Our data suggest that the amount of outer kinetochore proteins that assemble onto a centromere in cells defective for Aurora B-mediated phosphorylation of Dsn1 might be low. Alternatively, the association between Dsn1 and inner kinetochore components may be weakened in the absence of Aurora B phosphorylation, leading to loss of their interaction during purification. Because the phosphodeficient Dsn1 mutant properly localizes to kinetochores in vivo (Figure 2D) and the mutant supports viability (Figure 1B), we favor the latter possibility. To determine whether there may be subtle structural defects in vivo, we tested whether the Dsn1 phosphomutants exhibit genetic interactions with mutants in the spindle checkpoint. The dsn1-S240A, S250A, S264A mutant strain exhibited genetic interactions with a deletion in the MAD2 spindle checkpoint gene, but the dsn1-S240D, S250D did not (Figure 2E). These data support the model that phosphorylation of Dsn1 by Aurora B promotes outer kinetochore assembly by increasing its affinity toward inner kinetochores (Figure 2F).
Figure 2.
Outer kinetochore proteins have a weakened association with inner kinetochores in dsn1-S240A, S250A, S264A cells. (A) Centromeric minichromosomes containing kinetochores were purified from strains via a LacI-Flag protein that recognizes lacO repeats in the minichromosomes as previously described (Akiyoshi et al. 2009). (B) Minichromosomes were purified from strains containing the indicated Dsn1 mutants (SBY8338, SBY8339, SBY8340, and SBY8341) and analyzed via immunoblot with the indicated antibodies. (C) Minichromosomes were purified from WT and ipl1-321 strains (SBY7824 and SBY7823) that had been shifted to 37° for 3 hr and analyzed via immunoblot with the indicated antibodies. (D) All cells expressing GFP-tagged Dsn1 (SBY7774), Dsn1-S240A, S250A, S264A (SBY7776), and Dsn1-S240D, S250D (SBY7775) mutants show a normal bilobed kinetochore distribution in vivo. Bar, 5 µm. (E) Dsn1-S240A, S250A, S264A cells, but not dsn1-S240D, S250D cells, exhibit a genetic interaction with the spindle checkpoint mutant mad2Δ. Serial dilutions (fivefold) of dsn1-S240A, S250A, S264A (left; SBY5952, SBY11599, SBY5956, and SBY11601, respectively) or dsn1-S240D, S250D cells (right; SBY5950, SBY11599, SBY5954, and SBY11600, respectively) with or without mad2Δ were plated on YPD at 30°. (F) Model for the role of Dsn1 phosphorylation by the Aurora B kinase. Strains are listed in Table S1.
The evolutionary conserved Ipl1/Aurora kinase performs various functions throughout mitosis by phosphorylating multiple substrates. It is therefore critical to understand the molecular basis of its action. Although Dsn1 is an Aurora B substrate, the functional relevance of its phosphorylation remains controversial (Westermann et al. 2003; Emanuele et al. 2008; Yang et al. 2008; Welburn et al. 2010). Using the genetically tractable budding yeast, our analyses showed that the phosphodeficient Dsn1 mutant has a reduced affinity for inner kinetochore proteins. These results are consistent with previous findings suggesting that Dsn1 phosphorylation by Aurora B is important for outer kinetochore assembly (Emanuele et al. 2008; Yang et al. 2008). We previously found that the Dsn1-S240A, S250A mutant is targeted for degradation by the Mub1/Ubr2 ubiquitin ligase complex (Akiyoshi et al. 2013), suggesting that defects in kinetochore assembly may be monitored by a quality control system. However, in contrast to these findings, kinetochore assembly appears intact in the budding yeast ipl1-321 temperature-sensitive mutant (Pinsky et al. 2006) as well as in Aurora B depleted/inactivated mammalian cells (e.g., Ditchfield et al. 2003; Hauf et al. 2003). To reconcile these differences, we propose that a minimal level of Aurora B activity is sufficient to phosphorylate Dsn1 and promote kinetochore assembly, whereas higher activity is required to fulfill its other functions (e.g., error correction, spindle assembly, and cytokinesis) (Norden et al. 2006; Xu et al. 2009). Consistent with this idea, we previously showed that spindle assembly requires higher levels of Ipl1 kinase activity than other Aurora-dependent functions (Kotwaliwale et al. 2007). In addition, histone H3 Ser10, a well-established Aurora B substrate, often remains phosphorylated upon Aurora B inactivation (e.g., Kiyomitsu et al. 2010). We also note that budding yeast shows differential requirements in the level of Cdk1 needed to complete various cell cycle events (Bishop et al. 2000). In the future, it will therefore be important to determine whether the difference is due to altered levels of kinase activity, as well as to understand the underlying basis of the requirement for differential kinase activity. It will also be critical to understand how Dsn1 phosphorylation promotes the interaction between the inner and outer kinetochore to fully understand how kinetochores are assembled and maintained, thus ensuring genomic stability.
Supplementary Material
Acknowledgements
We thank Arshad Desai for reagents and Nitobe London for critical comments on the manuscript. This work was supported by National Institutes of Health grants (GM078079 and GM064386) to S.B.
Footnotes
Communicating editor: M. P. Colaiácovo
Literature Cited
- Akiyoshi B., Biggins S., 2010. Cdc14-dependent dephosphorylation of a kinetochore protein prior to anaphase in Saccharomyces cerevisiae. Genetics 186: 1487–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyoshi B., Nelson C. R., Ranish J. A., Biggins S., 2009. Quantitative proteomic analysis of purified yeast kinetochores identifies a PP1 regulatory subunit. Genes Dev. 23: 2887–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyoshi B., Sarangapani K. K., Powers A. F., Nelson C. R., Reichow S. L., et al. , 2010. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature 468: 576–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyoshi B., Nelson C. R., Duggan N., Ceto S., Ranish J. A., et al. , 2013. The Mub1/Ubr2 ubiquitin ligase complex regulates the conserved Dsn1 kinetochore protein. PLoS Genet. 9: e1003216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop A. C., Ubersax J. A., Petsch D. T., Matheos D. P., Gray N. S., et al. , 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407: 395–401 [DOI] [PubMed] [Google Scholar]
- Bloom K., Joglekar A., 2010. Towards building a chromosome segregation machine. Nature 463: 446–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaino A., Allshire R., Pidoux A., 2010. Building centromeres: home sweet home or a nomadic existence? Curr. Opin. Genet. Dev. 20: 118–126 [DOI] [PubMed] [Google Scholar]
- Carroll C. W., Milks K. J., Straight A. F., 2010. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 189: 1143–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I. M., Desai A., 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9: 33–46 [DOI] [PubMed] [Google Scholar]
- Cheeseman I. M., Chappie J. S., Wilson-Kubalek E. M., Desai A., 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127: 983–997 [DOI] [PubMed] [Google Scholar]
- Davies A. E., Kaplan K. B., 2010. Hsp90-Sgt1 and Skp1 target human Mis12 complexes to ensure efficient formation of kinetochore-microtubule binding sites. J. Cell Biol. 189: 261–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditchfield C., Johnson V. L., Tighe A., Ellston R., Haworth C., et al. , 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161: 267–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele M. J., Lan W., Jwa M., Miller S. A., Chan C. S., et al. , 2008. Aurora B kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. J. Cell Biol. 181: 241–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne K. E., Cheeseman I. M., 2011. Kinetochore assembly: if you build it, they will come. Curr. Opin. Cell Biol. 23: 102–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonen S., Akiyoshi B., Iadanza M. G., Shi D., Duggan N., et al. , 2012. The structure of purified kinetochores reveals multiple microtubule-attachment sites. Nat. Struct. Mol. Biol. 19: 925–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S., Cole R. W., LaTerra S., Zimmer C., Schnapp G., et al. , 2003. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161: 281–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A. J., Cleveland D. W., 2012. Losing balance: the origin and impact of aneuploidy in cancer. EMBO Rep. 13: 501–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar A. P., Bouck D. C., Molk J. N., Bloom K. S., Salmon E. D., 2006. Molecular architecture of a kinetochore-microtubule attachment site. Nat. Cell Biol. 8: 581–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T., Iwasaki O., Obuse C., Yanagida M., 2010. Inner centromere formation requires hMis14, a trident kinetochore protein that specifically recruits HP1 to human chromosomes. J. Cell Biol. 188: 791–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline S. L., Cheeseman I. M., Hori T., Fukagawa T., Desai A., 2006. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J. Cell Biol. 173: 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotwaliwale C. V., Frei S. B., Stern B. M., Biggins S., 2007. A pathway containing the Ipl1/aurora protein kinase and the spindle midzone protein Ase1 regulates yeast spindle assembly. Dev. Cell 13: 433–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson M. A., Cheeseman I. M., 2011. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 21: 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrimore J., Bloom K. S., Salmon E. D., 2011. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J. Cell Biol. 195: 573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvezzi F., Litos G., Schleiffer A., Heuck A., Mechtler K., et al. , 2013. A structural basis for kinetochore recruitment of the Ndc80 complex via two distinct centromere receptors. EMBO J. 32: 409–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskell D. P., Hu X. W., Singleton M. R., 2010. Molecular architecture and assembly of the yeast kinetochore MIND complex. J. Cell Biol. 190: 823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norden C., Mendoza M., Dobbelaere J., Kotwaliwale C. V., Biggins S., et al. , 2006. The NoCut pathway links completion of cytokinesis to spindle midzone function to prevent chromosome breakage. Cell 125: 85–98 [DOI] [PubMed] [Google Scholar]
- Okada M., Cheeseman I. M., Hori T., Okawa K., McLeod I. X., et al. , 2006. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8: 446–457 [DOI] [PubMed] [Google Scholar]
- Perpelescu M., Fukagawa T., 2011. The ABCs of CENPs. Chromosoma 120: 425–446 [DOI] [PubMed] [Google Scholar]
- Petrovic A., Pasqualato S., Dube P., Krenn V., Santaguida S., et al. , 2010. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J. Cell Biol. 190: 835–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky B. A., Kung C., Shokat K. M., Biggins S., 2006. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat. Cell Biol. 8: 78–83 [DOI] [PubMed] [Google Scholar]
- Powers A. F., Franck A. D., Gestaut D. R., Cooper J., Gracyzk B., et al. , 2009. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell 136: 865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przewloka M. R., Venkei Z., Bolanos-Garcia V. M., Debski J., Dadlez M., et al. , 2011. CENP-C is a structural platform for kinetochore assembly. Curr. Biol. 21: 399–405 [DOI] [PubMed] [Google Scholar]
- Ruchaud S., Carmena M., Earnshaw W. C., 2007. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 8: 798–812 [DOI] [PubMed] [Google Scholar]
- Screpanti E., De Antoni A., Alushin G. M., Petrovic A., Melis T., et al. , 2011. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr. Biol. 21: 391–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Chang H. L., Kagami A., Watanabe Y., 2009. CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev. Cell 17: 334–343 [DOI] [PubMed] [Google Scholar]
- Welburn J. P., Vleugel M., Liu D., Yates J. R., 3rd, Lampson M. A., et al. , 2010. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell 38: 383–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S., Cheeseman I. M., Anderson S., Yates J. R., 3rd, Drubin D. G., et al. , 2003. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J. Cell Biol. 163: 215–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S., Drubin D. G., Barnes G., 2007. Structures and functions of yeast kinetochore complexes. Annu. Rev. Biochem. 76: 563–591 [DOI] [PubMed] [Google Scholar]
- Xu Z., Ogawa H., Vagnarelli P., Bergmann J. H., Hudson D. F., et al. , 2009. INCENP-aurora B interactions modulate kinase activity and chromosome passenger complex localization. J. Cell Biol. 187: 637–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Wu F., Ward T., Yan F., Wu Q., et al. , 2008. Phosphorylation of HsMis13 by Aurora B kinase is essential for assembly of functional kinetochore. J. Biol. Chem. 283: 26726–26736 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.