Abstract
The field of antibiotic drug discovery and the monitoring of new antibiotic resistance elements have yet to fully exploit the power of the genome revolution. Despite the fact that the first genomes sequenced of free living organisms were those of bacteria, there have been few specialized bioinformatic tools developed to mine the growing amount of genomic data associated with pathogens. In particular, there are few tools to study the genetics and genomics of antibiotic resistance and how it impacts bacterial populations, ecology, and the clinic. We have initiated development of such tools in the form of the Comprehensive Antibiotic Research Database (CARD; http://arpcard.mcmaster.ca). The CARD integrates disparate molecular and sequence data, provides a unique organizing principle in the form of the Antibiotic Resistance Ontology (ARO), and can quickly identify putative antibiotic resistance genes in new unannotated genome sequences. This unique platform provides an informatic tool that bridges antibiotic resistance concerns in health care, agriculture, and the environment.
INTRODUCTION
Antibiotic resistance is an increasing crisis as both the range of microbial antibiotic resistance in clinical settings expands and the pipeline for development of new antibiotics contracts (1). This problem is compounded by the global genomic scope of the antibiotic resistome, such that antibiotic resistance spans a continuum from genes in pathogens found in the clinic to those of benign environmental microbes along with their proto-resistance gene progenitors (2, 3). The recent emergence of New Delhi metallo-ß-lactamase (NDM-1) in Gram-negative organisms (4), which can hydrolyze all β-lactams with the exception of monobactams, illustrates the capacity of new antibiotic resistance genes to emerge rapidly from as-yet-undetermined reservoirs. Surveys of genes originating from both clinical and environmental sources (microbes and metagenomes) will provide increasing insight into these reservoirs and offer predictive capacity for the emergence and epidemiology of antibiotic resistance.
The increasing opportunity to prepare a broader and comprehensive antibiotic resistance gene census is facilitated by the power and falling costs of next-generation DNA sequencing. For example, whole-genome sequencing (WGS) is being increasingly used to examine new antibiotic-resistant isolates discovered in clinical settings (5). Additionally, culture-independent metagenomic surveys are adding tremendously to the pool of known genes and their distribution outside clinical settings (6, 7). These approaches have the advantage of providing a rapid survey of the antibiotic resistome of new strains, the discovery of newly emergent antibiotic resistance genes, the epidemiology of antibiotic resistance genes, and the horizontal gene transfer (HGT) of known antibiotic resistance genes through plasmids and transposable elements. However, despite the existence of tools for general annotation of prokaryotic genomes (see, e.g., reference 8), prediction of an antibiotic resistance phenotype from a genome sequence is not straightforward and, to date, computational tools for comprehensive prediction of antibiotic resistance genes within genomes have been lacking.
The proliferation of genetic and biochemical information on antibiotic resistance is resulting in a massive increase in molecular information that will facilitate our understanding of the evaluation, spread, and mechanism of antibiotic resistance. However, mining of this information is greatly hampered by the lack of a database that can unify information in a fashion that enables the gathering of over 5 decades of literature and data and includes up-to-date entry of new antibiotic resistance elements and curation of known and new genes. Such databases are increasingly common in other areas of biology and medicine, for example, InnateDB for innate immunity interactions and pathways (http://innatedb.ca/) (9). There have been efforts to establish such knowledge resources in the area of antibiotic resistance: for example, the Lahey clinic database on Ser β-lactamases (www.lahey.org/studies/), the Repository of Antibiotic Resistance Cassettes (http://www2.chi.unsw.edu.au:8080/rac/) (10) that lists a number of antibiotic resistance elements and provides some automatic annotation, the Resistance Map (http://www.cddep.org/resistancemap/) that offers an interactive format to view antibiotic resistance surveillance data, and the Antibiotic Resistance Genes Database (http://ardb.cbcb.umd.edu/) (11). These databases have proven too narrow in scope, are not regularly updated, or offer limited resources and data to integrate molecular information from genes and their products, antibiotics, and the associated literature.
Here we describe our effort to provide a unifying resource for the antibiotic resistance community: the Comprehensive Antibiotic Resistance Database (CARD; http://arpcard.mcmaster.ca). The CARD seeks to include data describing antibiotics and their targets along with antibiotic resistance genes, associated proteins, and antibiotic resistance literature. At the core of the CARD is a highly developed Antibiotic Resistance Ontology (ARO) for the classification of antibiotic resistance gene data. Ontologies, also known as controlled vocabularies, form the foundation of genomic bioinformatics; they provide consistent vocabularies for genes and their products that link them to their activities and enable robust investigation of molecular data (12). Furthermore, the CARD includes bioinformatic tools that enable the identification of antibiotic resistance genes from whole- or partial-genome sequence data, including unannotated raw sequence assembly contigs. The result is a robustly curated database in a user-friendly format that assembles over 1,600 known antibiotic resistance genes, enabling sophisticated analysis and query of antibiotic resistance in a fashion that will serve the broader biomedical research community.
MATERIALS AND METHODS
The CARD runs on a HP Proliant BL460C G6 Blade Special server using Ubuntu linux 10.04 (64 bit). The CARD was developed using the Generic Model Organism Database (GMOD) Chado database schema (version 1.1) (13) running on PostgreSQL (version 8.4.4) as the underlying organizing principle for storing sequence data, ontologies, citations, and other data. GMOD is a collection of open source software tools for development of genomic database tools and underlies several widely used model organism databases (yeast, Drosophila, Caenorhabditis elegans, etc.) (13). In particular, GMOD's Chado relational database schema provides a mature, modular, flexible, and extensible schema for storage of molecular and related data. The Web front end for the CARD is provided using a combination of Apache 2.2.17, PHP 5.3.2, Drupal 6.17, and custom Drupal modules and themes developed specifically for the CARD. Visualization of molecular sequence data (genes, plasmids, genomes) is provided by GMOD's GBrowse tool (version 2.1) (14) configured to use the Bio:DB:Das:Chado adaptor to have GBrowse read data directly from Chado. The CARD's BLAST tools rely upon NCBI's open source BLAST software (15).
Molecular sequences are imported into the CARD from GenBank using custom software developed specifically for the CARD, with retention of all annotations, NCBI accession numbers, taxonomy identification (ID) numbers of the host pathogen, and associated PubMed publications. By rule, only sequences available in GenBank and associated with a peer-reviewed publication(s) are included in the CARD. Import follows a two-step process in which sequences are first acquired from GenBank in GFF3 format (www.sequenceontology.org) and then loaded into the CARD's Chado database. All GFF3 files are curated prior to loading into Chado to ensure accuracy and kept in a file repository for future reference. To record distributions of genes among pathogens, the CARD additionally uses a pruned form of NCBI's taxonomy system (16) in the form of a custom NCBI Taxonomy ontology as described below. The CARD's GFF3 loader script tags all new sequences with terms from the CARD's NCBI Taxonomy ontology to provide organismal context and similarly loads any associated PubMed publications into the CARD Publication module. Tagging of imported genes with Antibiotic Resistance Ontology (ARO) terms is performed by annotation text mining of regular expressions (RegEx). Additional antibiotic resistance annotation of new sequences is often developed with the aid of the Resistance Gene Identifier (RGI). Details on the ARO, text mining RegEx, and RGI are provided below.
The Antibiotic Resistance Ontology (ARO) was developed via ontology jamborees, review of the antibiotic resistance literature, and extensive curation using custom command line and Web interface tools developed specifically for the CARD. External ontologies such as the Gene Ontology (17) and Sequence Ontology (18) were loaded into the CARD using available GMOD tools. The CARD's NCBI Taxonomy ontology was additionally developed using custom command line and Web interface tools developed specifically for the CARD but in a manner such that ontology accession numbers directly mirror NCBI's Taxonomy ID numbers (16) (e.g., NCBITaxon:470 mirrors Taxonomy ID 470 for Acinetobacter baumannii). The CARD uses only the subset of the available NCBI Taxonomy that is relevant to antibiotic-resistant bacteria, with a simplified taxonomic organization.
Individual Antibiotic Resistance Ontology (ARO) terms in the CARD have been associated with specific computational tools and models via Chado's cvtermprop table. The majority of ARO terms have text mining of regular expressions (RegEx) to correctly assign ARO terms to molecular sequences imported into the CARD based on the text annotations provided in the GenBank records. These RegEx entries are actively curated. The resulting tagged sequences, particularly those of polypeptides, form the underlying BLAST database used by the Resistance Gene Identifier (RGI) to identify antibiotic resistance genes in raw sequence data as outlined below. For antibiotic resistance involving specific mutations (i.e., single nucleotide polymorphisms [SNPs]), ARO terms are additionally associated with hidden Markov models (HMMs), alignment reference sequences, and position-specific SNPs for positional alignment of sequences and detection of position-specific SNPs. HMMs are constructed by the CARD curators and utilized by the CARD software using the HMMer software (19).
The CARD includes a number of modifications or additions to the GMOD Chado schema or its use. Foremost of these is the downplaying of the Chado organism table in favor of an ontological approach to storing source pathogen and taxonomic relationships. In the CARD, all bacterial sequences are encoded as “Bacteria” in the organism table but association of sequences with source organism(s) and their overall taxonomy is handled via the feature_cvterm table and the NCBITaxon ontology. The CARD also includes a custom cvterm_crossref table and related materialization tables to provide precomputed feature counts and lists within the context of cross-referencing of ontology subtrees, e.g., “how many aminoglycoside acetyltransferase genes are found in any strain of A. baumannii?” Association of features with their sources (i.e., genes found on a plasmid or genome) is additionally used to reflect physical linkage within cvterm_crossref, e.g., “how many antibiotic resistance genes are plasmid borne?” Ontological cross-referencing is used extensively to provide browsing power to the CARD, and the cvterm_crossref tables serve both to avoid frequent use of complex SQL queries by the CARD (by having results precalculated) and to allow nested complex queries, e.g., “list all plasmid-borne ß-lactamases in any strain of A. baumannii, excluding all TEM ß-lactamases.”
The CARD also deviates from traditional use of Chado due to the prokaryotic nature of antibiotic resistance. In particular, both Chado and its use of the Sequence Ontology (SO) and the GFF3 formats have traditionally been eukaryote centered. The CARD includes development of a prokaryotic gene model, associated GFF3 encoding, and Chado loading protocol based on both custom ideas and those previously posted by GMOD/SO/GFF research community discussion groups. This custom approach remains compliant with the Sequence Ontology Feature Annotation (SOFA) standard (18).
As the CARD is not a static database but instead undergoes constant curation, addition of new molecular data, pathogens, and publications, and continual evolution of the Antibiotic Resistance Ontology (ARO), it necessarily has a number of important maintenance routines. Foremost of these are scripts for updating Chado tables relating to the structure of ontologies (e.g., cvtermpath) and for cross-referencing ontology term tagging (e.g., cvterm_crossref). The CARD also updates text, BLAST, and RGI search reference files on a 24-h basis. Lastly, the Publication module incorporates curator-submitted citations upon their submission and refreshes all citations once a month to capture updated citation information provided by PubMed.
Much of the data, ontological structure, and models in the CARD are utilized by the Resistance Gene Identifier (RGI), a new tool for de novo annotation of gene, complete-genome, or genome assembly sequences for their antibiotic resistome. Gene prediction in the RGI includes only protein-coding genes, as predicted by GeneMark (20). Predicted open reading frames of less than 30 bp are ignored. Antibiotic resistance annotation is based on BLASTP hits to curated protein sequences present in CARD. Any predicted gene with a BLASTP hit to a protein tagged for antibiotic resistance in the ARO is highlighted. The default BLASTP cutoff is an expectation value of e−30, but many types of antibiotic resistance genes use custom cutoffs based on their classification. Where antibiotic resistance is conferred by SNPs, the RGI additionally screens for known resistance SNPs via use of hidden Markov models (HMMs), reference sequences, and position-specific SNP sequences for positional alignment and assessment of query sequences using HMMer (19).
RESULTS
Molecular sequence data in the CARD.
The CARD is based on molecular determinants of antibiotic resistance: the genes and their regulators conferring resistance to antibiotic molecules. The CARD is populated with molecular sequences of over 1,600 antibiotic resistance genes (Table 1). All are the product of active and ongoing curation of sequences available in GenBank associated with peer-reviewed publications and reflect an effort to include all genes and mechanisms involved in antibiotic resistance, providing an exhaustive molecular foundation within the CARD for cataloging and interpreting antibiotic resistance data and for providing new analytical and predictive tools. Addition of new genes occurs regularly and by external users alerting the curation team through the Web portal as new genes are identified.
Table 1.
List of antibiotic resistance genes curated in the CARD within which citation, molecular sequence, protein structure, mechanism, and ARO classification details are provideda
| Aminocoumarins |
| Aminocoumarin-resistant DNA topoisomerases |
| Aminocoumarin-resistant GyrB, ParE, ParY |
| Aminoglycosides |
| Aminoglycoside acetyltransferases |
| AAC(1), AAC(2′), AAC(3), AAC(6′) |
| Aminoglycoside nucleotidyltransferases |
| ANT(2″), ANT(3″), ANT(4′), ANT(6), ANT(9) |
| Aminoglycoside phosphotransferases |
| APH(2″), APH(3″), APH(3′), APH(4), APH(6), APH(7″), APH(9) |
| 16S rRNA methyltransferases |
| ArmA, RmtA, RmtB, RmtC, Sgm |
| β-Lactams |
| Class A β-lactamases |
| AER, BLA1, CTX-M, KPC, SHV, TEM, etc.b |
| Class B (metallo-)β-lactamases |
| BlaB, CcrA, IMP, NDM, VIM, etc.b |
| Class C β-lactamases |
| ACT, AmpC, CMY, LAT, PDC, etc.b |
| Class D β-lactamases |
| OXA β-lactamaseb |
| mecA (methicillin-resistant PBP2) |
| Mutant porin proteins conferring antibiotic resistance |
| Antibiotic-resistant Omp36, OmpF, PIB (por) |
| Genes modulating β-lactam resistance |
| bla (blaI, blaR1) and mec (mecI, mecR1) operons |
| Chloramphenicol |
| Chloramphenicol acetyltransferase (CAT) |
| Chloramphenicol phosphotransferase |
| Ethambutol |
| Ethambutol-resistant arabinosyltransferase (EmbB) |
| Mupirocin |
| Mupirocin-resistant isoleucyl-tRNA synthetases |
| MupA, MupB |
| Peptide antibiotics |
| Integral membrane protein MprF |
| Phenicol |
| Cfr 23S rRNA methyltransferase |
| Rifampin |
| Rifampin ADP-ribosyltransferase (Arr) |
| Rifampin glycosyltransferase |
| Rifampin monooxygenase |
| Rifampin phosphotransferase |
| Rifampin resistance RNA polymerase-binding proteins |
| DnaA, RbpA |
| Rifampin-resistant beta-subunit of RNA polymerase (RpoB) |
| Streptogramins |
| Cfr 23S rRNA methyltransferase |
| Erm 23S rRNA methyltransferases |
| ErmA, ErmB, Erm(31), etc.d |
| Streptogramin resistance ATP-binding cassette (ABC) efflux pumps |
| Lsa, MsrA, Vga, VgaB |
| Streptogramin Vgb lyase |
| Vat acetyltransferase |
| Fluoroquinolones |
| Fluoroquinolone acetyltransferase |
| Fluoroquinolone-resistant DNA topoisomerases |
| Fluoroquinolone-resistant GyrA, GyrB, ParC |
| Quinolone resistance protein (Qnr) |
| Fosfomycin |
| Fosfomycin phosphotransferases |
| FomA, FomB, FosC |
| Fosfomycin thiol transferases |
| FosA, FosB, FosX |
| Glycopeptides |
| VanA, VanB, VanD, VanR, VanS, etc.c |
| Lincosamides |
| Cfr 23S rRNA methyltransferase |
| Erm 23S rRNA methyltransferases |
| ErmA, ErmB, Erm(31), etc.d |
| Lincosamide nucleotidyltransferase (Lin) |
| Linezolid |
| Cfr 23S rRNA methyltransferase |
| Macrolides |
| Cfr 23S rRNA methyltransferase |
| Erm 23S rRNA methyltransferases |
| ErmA, ErmB, Erm(31), etc.d |
| Macrolide esterases |
| EreA, EreB |
| Macrolide glycosyltransferases |
| GimA, Mgt, Ole |
| Macrolide phosphotransferases (MPH) |
| MPH(2′)-I, MPH(2′)-II |
| Macrolide resistance efflux pumps |
| MefA, MefE, Mel |
| Streptothricin |
| Streptothricin acetyltransferase (sat) |
| Sulfonamides |
| Sulfonamide-resistant dihydropteroate synthases |
| Sul1, Sul2, Sul3, sulfonamide-resistant FolP |
| Tetracyclines |
| Mutant porin PIB (por) with reduced permeability |
| Tetracycline inactivation enzyme TetX |
| Tetracycline resistance major facilitator superfamily (MFS) efflux pumps |
| TetA, TetB, TetC, Tet30, Tet31, etc.e |
| Tetracycline resistance ribosomal protection proteins |
| TetM, TetO, TetQ, Tet32, Tet36, etc.e |
| Efflux pumps conferring antibiotic resistance |
| ABC antibiotic efflux pump |
| MacAB-TolC, MsbA, MsrA,VgaB, etc.f |
| MFS antibiotic efflux pump |
| EmrD, EmrAB-TolC, NorB, GepA, etc.f |
| Multidrug and toxic compound extrusion (MATE) transporter |
| MepA |
| Resistance-nodulation-cell division (RND) efflux pump |
| AdeABC, AcrD, MexAB-OprM, mtrCDE, etc.f |
| Small multidrug resistance (SMR) antibiotic efflux pump |
| EmrE |
| Genes modulating antibiotic efflux |
| adeR, acrR, baeSR, mexR, phoPQ, mtrR, etc.g |
ARO, Antibiotic Resistance Ontology.
Complete lists of β-lactamase families and individual β-lactamases can be found in the CARD.
Glycopeptide resistance gene clusters include a number of genes encoding proteins with different functions, including sensors, regulators, and enzymes, all of which result in restructuring of the cell wall, providing resistance to glycopeptides. The full list of genes involved can be found in the CARD.
Complete lists of Erm 23S rRNA methyltransferases can be found in the CARD.
Complete lists of tetracycline resistance major facilitator superfamily (MFS) efflux pumps and ribosomal protection proteins can be found in the CARD.
Complete lists of ATP-binding cassette (ABC), MFS, and resistance-nodulation-cell division (RND) antibiotic efflux pumps can be found in the CARD.
Complete lists of efflux regulatory proteins can be found in the CARD, including information on mutations conferring increased rates of antibiotic efflux.
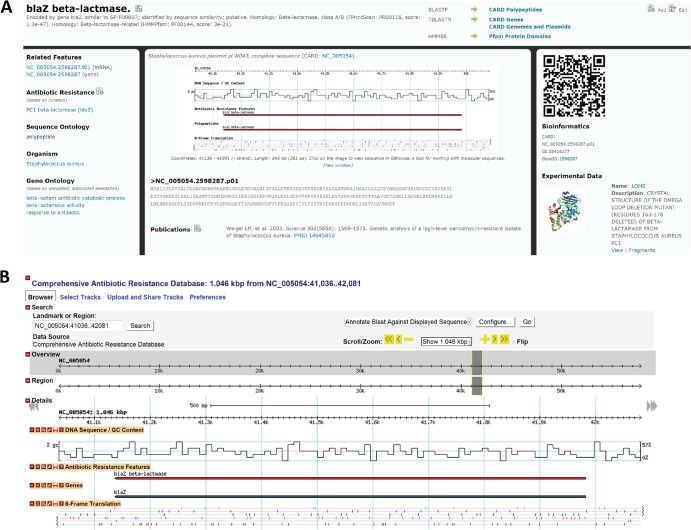
In order to generate a database with optimal modularity, flexibility, and integration with genome and metagenome projects, we chose to use the Generic Model Organism Database (GMOD; www.gmod.org) open source software for construction of the CARD. With minor modifications (see Materials and Methods), all of molecular sequence data in CARD have been loaded into GMOD's Chado schema and can be browsed throughout the CARD and within GMOD's powerful, interactive genome visualization and analysis tool GBrowse (14) (Fig. 1). All molecular sequences within the CARD are classified and organized using the Sequence Ontology (18), which is comprised of a set of ontology terms describing the many levels of organization and relationships between different molecular sequences (e.g., a polypeptide derives_from a gene, which is part_of a genome). This Sequence Ontology-based organization was critical for adapting Chado to a prokaryotic gene model (see Materials and Methods) and for development of advanced search and analysis tools as outlined below.
Fig 1.
Presentation of the Staphylococcus aureus blaZ ß-lactamase gene in the CARD. (A) The gene's Web page in the CARD, providing annotation, accession, source information, ontological classification (SO, ARO, GO), and associated molecular features (mRNA, polypeptide, coding sequence [CDS]). (B) Dynamic browsing and analysis of the blaZ gene using the Web-based genome visualization and analysis tool GBrowse.
The Antibiotic Resistance Ontology.
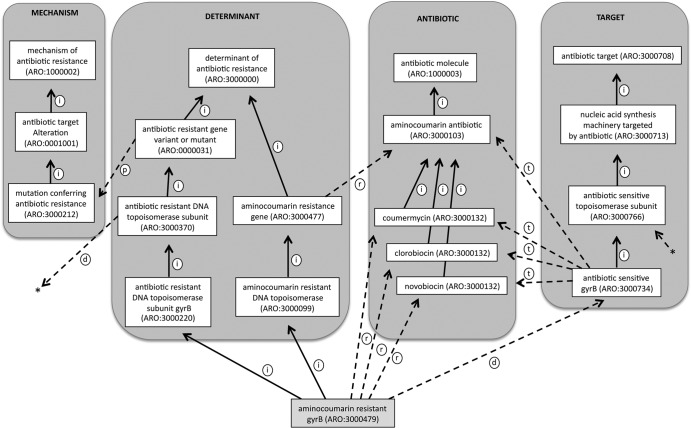
While all molecular sequences within the CARD are classified and organized using the Sequence Ontology to define their role within the cell (e.g., genome, gene, transcript, polypeptide), this is uninformative regarding antibiotic resistance. As such, at the heart of the CARD is a new Antibiotic Resistance Ontology (ARO). This ontology is a common language that can be used to link gene product function across disparate organisms. Such a controlled vocabulary is essential in the area of antibiotic resistance given the multiple sources of antibiotic resistance elements, antibiotics, and associated phenotypes. The ARO reflects an ontological description of the genes, drugs, and mechanisms involved in antibiotic resistance and also includes antibiotic targets. Like the use of the Sequence Ontology tool, all molecular sequences within the CARD are classified and organized using the ARO, allowing individual antibiotic resistance genes to be placed into a broader functional context (Fig. 2). Core to this organization of molecular data is the relation of drugs to targets and antibiotic resistance genes within the ARO (Fig. 3). As such, the CARD has been designed to use the ARO as its primary organizing principle and all sequence, citation, and protein/chemical structure data are accessible via terms within the ARO (Fig. 4).
Fig 2.
Classification of aminocoumarin-resistant gyrase B in the Antibiotic Resistance Ontology (ARO), illustrating the use of ontological relationships to describe knowledge about the gene (see Table 3). The is_a relationships are depicted by solid arrows labeled with “i” and generally denote classification hierarchies within the major branches of the ARO (mechanism, determinant, antibiotic, target), while dashed arrows labeled with “p” reflect part_of relationships between genes and mechanisms. Dashed arrows labeled with “d” depict derived_from relationships between antibiotic-sensitive precursors and antibiotic-resistant forms of the gene, while those labeled with “t” reflect targeted_by relationships between antibiotic-sensitive forms and antibiotic molecules. Dashed arrows labeled with “r” depict confers_resistance relationships between antibiotic resistance genes and antibiotic molecules. Asterisks denote a derived_from relationship between antibiotic-resistant and -sensitive DNA topoisomerase subunits.
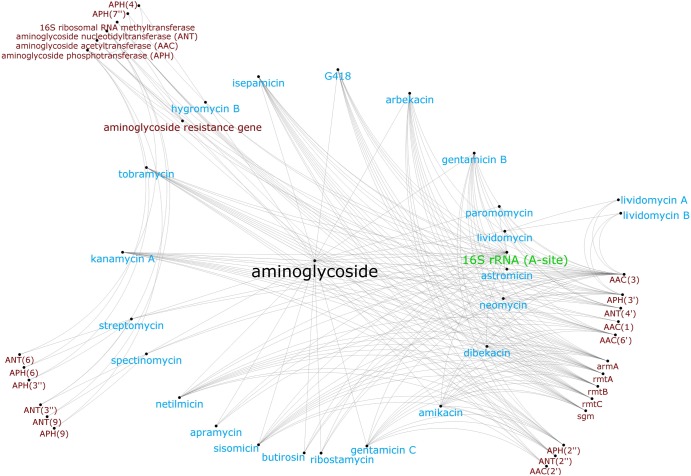
Fig 3.
Organization of aminoglycoside antibiotics (blue), their target (green), and aminoglycoside resistance genes (red) in the CARD's Antibiotic Resistance Ontology, illustrating the diversity of genes providing resistance to single or multiple aminoglycosides. Nodes represent ontology terms, while edges represent relationships between ontology terms.
Fig 4.
An ontology term Web page for tetracycline resistance gene tetX (ARO:3000205) in the CARD, providing descriptive, ontological classification, sequence, protein structure, publication, taxonomic distribution, and bioinformatics data/model information. By providing an ontology-centered interface, the CARD offers a clearinghouse of information on antibiotic resistance genes, mechanisms, drugs, etc. The left column reflects ontological cross-referencing (see Materials and Methods).
We benefited significantly from an initial effort to establish an ARO by Liu and Pop (11), and we have greatly expanded this ARO to include branches that describe antibiotics, biosynthesis, mechanisms, targets, inhibitors, and, of course, the antibiotic resistance genes themselves (Table 2). In addition, we have developed novel relationship types to reflect the biological processes involved in antibiotic resistance (Table 3). In the context of the major branches of the ARO, is_a is used exclusively within branches (e.g., TEM is_a ß-lactamase) whereas part_of is used to bridge the determinant and mechanism branches (e.g., ß-lactamases are part_of antibiotic degradation) or to reflect protein assemblages (e.g., MexA is part_of the efflux pump MexAB-OprM). The “regulates” term connects regulators with the genes they control (e.g., MexR regulates MexAB-OprM). The confers_resistance_to relations bridge the determinant branch and the antibiotic molecule branch (e.g., ß-lactamases confer_resistance_to ß-lactams), while the targeted_by relations bridge the drug target branch and antibiotic molecule branch (e.g., elongation factor Tu is targeted_by_drug pulvomycin). The derives_from relation bridges the drug target branch and the determinant branch (e.g., antibiotic-resistant embB derives_from antibiotic-sensitive embB).
Table 2.
Major branches of the AROa
| Major ARO branch | Scope |
|---|---|
| Determinant of Antibiotic Resistance (ARO:3000000) | Antibiotic resistance genes, SNPs, or other molecular entities organized by target (e.g., aminocoumarin, glycopeptides, etc.) and mode of action (e.g., antibiotic inactivation, molecular bypass, etc.) |
| Mechanism of Antibiotic Resistance (ARO:1000002) | Target alteration, target replacement, antibiotic inactivation, antibiotic efflux, antibiotic target protection, reduced permeability to antibiotic |
| Antibiotic Target (ARO:3000708) | Targeted cell membrane components, protein or nucleotide synthesis machinery, enzymes, etc. |
| Antibiotic Molecule (ARO:1000003) | Hierarchical classification of antibiotics (e.g., sulfonamide, β-lactam, glycopeptide antibiotics, etc.) |
| Inhibitor of Antibiotic Resistance (ARO:0000076) | β-Lactamase and other inhibitors |
| Antibiotic Biosynthesis (ARO:3000082) | Phosphonoacetaldehyde methyltransfererase, glycopeptide biosynthesis, macrolide biosynthesis, streptogramin biosynthesis, fosfomycin biosynthesis, aminocoumarin biosynthesis, phosphoenolpyruvate (PEP) mutase, phosphonopyruvate decarboxylase |
| Antibiotic Resistance Terminology (ARO:3000045) | Ontological relationship types, bioinformatic model types, reference molecular sequence types, etc. |
ARO, Antibiotic Resistance Ontology. All major branches are part_of ARO:1000001, “process or component of antibiotic biology or chemistry.”
Table 3.
Relationship types used within the AROa
| Relationship type | Description | Source |
|---|---|---|
| is_a | An axiomatic relationship ontology term in which the subject is placed into a higher order classification | RO |
| part_of | A relationship ontology term in which the subject is but part of the object | RO |
| derives_from | A relationship ontology term in which the subject has its origins in the object | RO |
| regulates | A relationship ontology term in which the subject regulates expression of the object | ARO |
| confers_resistance_to | A relationship ontology term in which the subject confers antibiotic resistance to the object | ARO |
| confers_resistance_to_drug | A relationship ontology term in which the subject (usually a gene) confers clinically relevant resistance to a specific antibiotic | ARO |
| targeted_by | A relationship ontology term in which the subject is targeted by the object (usually a class of antibiotics) | ARO |
| targeted_by_drug | A relationship ontology term in which the subject is targeted by a specific antibiotic | ARO |
ARO, Antibiotic Resistance Ontology; RO, Relation Ontology, a part of the Open Biological and Biomedical Ontologies resource (27). Descriptions are paraphrased.
Horizontal gene transfer and pathogen diversity.
GMOD's Chado relational database schema was designed with a discrete number of organisms in mind, but this is not reflective of the reality of antibiotic resistance, where the same or similar genes may be found in numerous bacterial genomes. Antibiotic resistance can encompass any number of bacterial strains, and horizontal gene transfer (HGT) is common. Unlike eukaryotic organism databases, where the underlying genome sequence often represents a static sample, the CARD is a dynamic database that must handle an ever-changing landscape of antibiotic resistance. For example, a plasmid recorded in CARD for a specific bacterial pathogen may quickly be discovered in additional pathogens and strains. Due to promiscuous plasmids, the metallo-ß-lactamase NDM-1 alone has been recorded in Acinetobacter baumannii, Klebsiella oxytoca, Proteus mirabilis, Enterobacter cloacae, Citrobacter freundii, Providencia spp., Shigella spp., Pseudomonas spp., Stenotrophomonas spp., Aeromonas spp., and Vibrio cholerae since its initial discovery in Klebsiella pneumoniae and Escherichia coli in 2008 (21). To handle this biological diversity, the CARD employs an ontological approach to recoding the organismal origin of molecular sequences, via a custom NCBI Taxonomy ontology, such that each bacterial strain has its own ontology term and all such terms are placed in a taxonomic hierarchy. This allows a single plasmid sequence stored in the CARD to be associated with multiple bacterial strains, via assignment of ontology terms, without the sequence itself being stored multiple times in the CARD. The use of a taxonomic ontology was also explicitly developed to allow searching of data for specific clinical strains (e.g., list all β-lactamases found in Salmonella enterica subsp. enterica serovar Typhi strain CT18) or at broader taxonomic levels (e.g., list all β-lactamases found in all Salmonella pathogens).
Integration with NCBI, PDB, and other resources.
Terms in the ARO and the NCBI Taxonomy ontology have been associated with useful supplementary information via active curation, such as key publications in PubMed, chemical structures in PubChem, and three-dimensional (3D) protein structures in the Protein Data Bank (PDB). Both PubChem and PDB structures can be viewed graphically within the CARD. PDB structures have similarly been associated with the underlying gene sequences. PubMed entries are integrated among genes and other molecular sequences, ARO terms, and pathogen strains. The CARD automatically updates citations as new information becomes available in PubMed.
The CARD also provides cross-references to other ontologies and databases, such as the Sequence Ontology, the Gene Ontology, and NCBI's Taxonomy section. Efforts are under way to link the ARO to higher-order ontologies, such as the Gene Ontology and the developing Infectious Disease Ontology (22), to provide a broader context for the antibiotic resistance biology than that covered by the ARO.
Searching the CARD.
The entire contents of the CARD can be queried via a text search box, which searches all gene annotations, ontology terms, publications, and linkages to PubChem or PDB structure annotations. ARO terms in the CARD include curated lists of synonyms (e.g., Amicacin as a synonym for the drug amikacin, or meticilin for methicillin), allowing searching of the CARD using multiple terminologies. The search box itself suggests solutions in the form of a drop-down box of suggested ARO or NCBI Taxonomy ontology terms based on the supplied text. As the CARD is organized as an ontology-centered interface, selection of a suggested ontology term provides the user with a curated catalog on antibiotic resistance genes, mechanisms, drugs, and pathogens recorded in the CARD for the term selected. In fact, examination of ARO terms is often the most powerful form of search, as ontology term Web pages include precomputed molecular sequence lists within the context of the ARO and the NCBI Taxonomic ontology, e.g., “how many aminoglycoside acetyltransferase genes are recorded in the CARD for any strain of A. baumannii?” These cross-references allow the user to browse lists of antibiotic resistance genes, browse to specific gene sequences, and generate multiple sequence alignments and are the product of active curation of sequences available in GenBank associated with peer-reviewed publications.
In addition to extensive browsing tools, integrated within the CARD and available as tools on the website are BLAST databases for all genes stored in the CARD. BLAST searches can include all genes or subsets of CARD reflecting antibiotic resistance genes, antibiotic targets, and antibiotic biosynthesis genes.
The Resistance Gene Identifier (RGI).
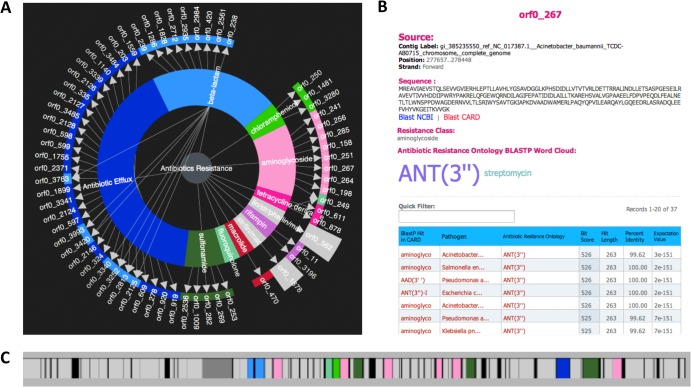
In addition to extensive search tools, the CARD provides a novel analytical tool in the form of the Resistance Gene Identifier (RGI). The RGI provides a preliminary annotation of the submitted DNA sequence(s) based upon the data available in the CARD. RGI can accept GenBank accession or GI numbers, pasted sequences, or uploaded nucleotide sequence files in FASTA format. Data with two or more FASTA sequences, such as whole-genome-sequencing (WGS) assembly contigs, can be accepted (maximum size, 20 Mb). The RGI analyzes the submitted sequences and provides a detailed output of predicted antibiotic resistance genes and targeted drug classes. This includes resistance to antibiotics via mutations in their targets or via dedicated antibiotic resistance gene products (enzymes, protective proteins, and efflux systems). RGI results are summarized using a “resistance wheel,” with overall antibiotic resistance in the center, antibiotic resistance classes in the middle, and individual antibiotic resistance genes on the outer ring (Fig. 5). Clicking on an individual gene designation brings up the annotation details for that gene. A toolbox panel allows export of results for all genes or individual genes in tab-delimited format plus a downloadable image of the resistance wheel. Analysis results can be saved and loaded on the CARD server or shared by email with collaborators. The panel providing gene details (Fig. 5B) includes the coordinates and sequence of the antibiotic resistance gene, browsable BLASTP results, and a weighted word cloud of the antibiotic resistance evidence. A clickable gene map (Fig. 5C) is provided for identification of gene clusters, with color coding based on the resistance wheel.
Fig 5.
Analysis of the whole genome of Acinetobacter baumannii strain TCDC-AB0715 by the Resistance Gene Identifier (RGI). The A. baumannii strain TCDC-AB0715 is a clinical isolate with resistance to carbapenems, fluoroquinolones, and cephalosporins (26). (A) “Resistance wheel” for A. baumannii strain TCDC-AB0715, predicting resistance to a broad range of antibiotic classes. (B) Details screen of orf0_267, illustrating detection of the aminoglycoside nucleotidyltransferase ANT(3″). (C) Open reading frame (ORF) map of a region of the A. baumannii strain TCDC-AB0715 chromosome, with prediction of β-lactamases TEM-1 and TEM-33 (light blue), aminoglycoside phosphotransferases, nucleotidyltransferases, and acetyltransferases (pink), chloramphenicol acetyltransferase (bright green), sulfonamide-resistant dihydropteroate synthase sul1 (dark green), tetracycline efflux pump tetB (dark pink), and genes implicated in general efflux (dark blue). ORFs unrelated to antibiotic resistance are presented in gray, while non-protein-coding regions are presented in black. The resistance genes identified are consistent with the reported resistance phenotype (26).
Public release of CARD data.
Both the Antibiotic Resistance Ontology (ARO) and curated antibiotic resistance, target, and biosynthesis gene and polypeptide sequences are available for download (http://arpcard.mcmaster.ca/download). The ARO download is available in Open Biological and Biomedical Ontologies (OBO) format version 1.2. Sequence downloads are available in FASTA format and include original GenBank annotation and curated ARO term assignments.
Social media.
The CARD developers post updates to Twitter (@arpcard) and host an Antibiotic Resistance discussion group for the research community on LinkedIn (http://arpcard.mcmaster.ca/linkedin). Users can post questions, initiate discussions, and submit bug reports at SourceForge (http://arpcard.mcmaster.ca/discussion). The CARD collaboratively develops the Timeline of Antibiotic Resistance with members of the research community using Dipity (http://arpcard.mcmaster.ca/timeline).
DISCUSSION
Antibiotic resistance research spans the clinic, the laboratory, the environment, human and animal populations, and the pharmaceutical sector. While disparate in their objectives, nevertheless, all these areas are based on antibiotic resistance genes and their associated phenotypes. What is missing from the arsenal of tools for researchers, clinicians, drug discoverers, and regulators is a comprehensive accessible data platform that integrates genomic and molecular data across the entire sequence space of bacterial genomes and metagenomes with respect to antibiotics, antibiotic resistance, and drug targets. The field of antibiotic resistance lacks unifying informational tools that serve to inform all aspects of antibiotic resistance research. Such applications have proven to be foundational, enabling the design of platforms for the study of numerous organisms (yeast, Pseudomonas, C. elegans, Drosophila, etc.) and systems (e.g., host defense 9). There are a few antibiotic resistance databases available, but none provide a comprehensive accessible data platform: Can-R (www.can-r.com) (23) has a surveillance focus, the Bush-Jacoby ß-lactamase list (www.lahey.org/Studies/) (24) is exclusively focused on compiling an inventory of a functional subset of antibiotic resistance genes, ResFinder (cge.cbs.dtu.dk/services/ResFinder/) (25) is limited to BLAST analysis of a subset of antibiotic resistance genes and lacks a unifying ontology, and ARDB (ardb.cbcb.umd.edu) (11) has not been updated since 2009. In contrast, the CARD provides a first step in bringing together genomic data and tools specific to antibiotics, antibiotic resistance, and antibiotic targets.
Key to the development of CARD has been the establishment of an Antibiotic Resistance Ontology (ARO). Ontologies, also known as controlled vocabularies, form the foundation of genomic bioinformatics; they provide rigorous vocabularies for genes and their products that link them to their activities and enable robust investigation of molecular data (12). The ARO thus provides a unifying language, specific to antibiotics, enabling codification of antibiotic resistance and target genes, compounds, and molecular activities germane to the field. The ARO has been built by taking into consideration a first attempt published by Liu and Pop several years ago (11). We have greatly expanded this effort and have a functional ontology that links well to others such as GO (Gene Ontology) (17), GenBank's organism taxonomy, SO (Sequence Ontology) (18), and the emerging IDO (Infectious Disease Ontology) (22).
The Resistance Gene Identifier (RGI) illustrates the power provided by an integrated use of ontologies, bioinformatic models, and molecular sequence data. Resistance to antibiotics can occur via mutations in their targets or by orthogonally evolved antibiotic resistance enzymes, protective proteins, and efflux systems. Furthermore, antibiotic resistance elements can be acquired through horizontal gene transfer (HGT) of genetic elements. HGT can result in the broad spread of evolutionary successful antibiotic resistance elements throughout microbial populations, both among pathogens and among benign environmental and host microbiome-associated bacteria, with devastating effect. Monitoring, classifying, and biochemically investigating such elements moving through bacterial populations, including components of the microbiome, by HGT is vital. By incorporating information at all levels of antibiotic resistance, the RGI provides a powerful new tool for broad analysis of antibiotic resistance at the genome level.
The use of the GMOD software components by the CARD ensures robust and proven technology and the opportunity to expand the CARD with new open source modules as they become available from the GMOD community. The GMOD community is very collaborative, and the CARD is an active participant, particularly in the handling of complex prokaryotic data. The CARD is unique in that development of the ARO is paired with development of models and algorithms for high-throughput assignment of ontology terms to molecular and other data. CARD stores and provides seed alignments, model data, and text-mining terms to its users, uniquely allowing users to view genes and their annotations alongside the models and criteria used for their annotation. Hidden Markov models (HMMs) and other search terms are designed to be downloaded from CARD to be used for independent research projects and wide dissemination by the research community.
The CARD addresses critical unmet needs in the antibiotic resistance and discovery communities. As genome sequencing of pathogens and microbial communities becomes ever more prevalent and feasible even in diagnostic settings, tools such as the CARD will become even more significant. The challenge of implementing such an approach with respect to antibiotics and antibiotic resistance is that it requires the integration of data linked to not just one genome or organism but to hundreds if not thousands of organisms, species, and individual isolates. This is in addition to associated genetic elements such as plasmids, transposons, and integrons that provide individual organisms with additional genetic information often associated with pathogenesis and antibiotic resistance. At the same time, the compounds that are themselves antibiotics or that somehow modulate antibiotic activity (adjuvants, inhibitors, etc.) add to the complexity of an informatic resource that would be useful to researchers, clinicians, and other interested parties, including patients, farmers, etc. Despite these challenges, an electronic resource such as the CARD is absolutely essential to help unify data and make it easily accessible to all stakeholders to ensure that growing genomic information relevant to antibiotics and antibiotic resistance can be easily mined. The CARD is envisioned to be a living resource to serve investigators over the long term; therefore, the developers encourage new entries as new resistance elements are identified as well as the contribution of any additional information deemed relevant through a clickable “contribute/corrections” button. With this electronic resource and continued contributions and updates by stakeholders, the result will be more informed clinical, research, public health, and drug discovery communities.
ACKNOWLEDGMENTS
We thank members of the Antibiotic Resistance Pipeline, a Canada-United Kingdom Joint Health Research Pilot Program on Antibiotic Resistance, for discussion of the targets and scope of the Comprehensive Antibiotic Resistance Database during its development. Mihai Pop (Center for Bioinformatics and Computational Biology, University of Maryland) provided insight into the bioinformatic organization of antibiotic resistance data and kindly allowed us to expand upon his set of ontology terms for antibiotic resistance (http://ardb.cbcb.umd.edu). Members of the Generic Model Organism Database (GMOD) community, particularly Scott Cain (Ontario Institute for Cancer Research, Toronto, Canada), provided help in the application of GMOD's Chado database schema to antibiotic resistance data.
This work was supported by the Canada Research Chairs program, a joint initiative between the Canadian Institutes of Health Research and the Medical Research Council (United Kingdom), and a Killam Research Fellowship awarded to G.D.W.
Footnotes
Published ahead of print 6 May 2013
REFERENCES
- 1. Cooper MA, Shlaes D. 2011. Fix the antibiotics pipeline. Nature 472:32. 10.1038/472032a [DOI] [PubMed] [Google Scholar]
- 2. D'Costa VM, King CE, Kalan L, Morar M, Sung WWL, Schwarz C, Froese D, Zazula G, Calmels F, Debruyne R, Golding GB, Poinar HN, Wright GD. 2011. Antibiotic resistance is ancient. Nature 477:457–461 [DOI] [PubMed] [Google Scholar]
- 3. Wright GD. 2010. The antibiotic resistome. Expert Opin. Drug Discov. 5:779–788 [DOI] [PubMed] [Google Scholar]
- 4. Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Köser CU, Ellington MJ, Cartwright EJP, Gillespie SH, Brown NM, Farrington M, Holden MTG, Dougan G, Bentley SD, Parkhill J, Peacock SJ. 2012. Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathog. 8:e1002824. 10.1371/journal.ppat.1002824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wright GD, Poinar H. 2012. Antibiotic resistance is ancient: implications for drug discovery. Trends Microbiol. 20:157–159 [DOI] [PubMed] [Google Scholar]
- 7. Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MOA, Dantas G. 2012. The shared antibiotic resistome of soil bacteria and human pathogens. Science 337:1107–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lynn DJ, Winsor GL, Chan C, Richard N, Laird MR, Barsky A, Gardy JL, Roche FM, Chan THW, Shah N, Lo R, Naseer M, Que J, Yau M, Acab M, Tulpan D, Whiteside MD, Chikatamarla A, Mah B, Munzner T, Hokamp K, Hancock REW, Brinkman FSL. 2008. InnateDB: facilitating systems-level analyses of the mammalian innate immune response. Mol. Syst. Biol. 4:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tsafnat G, Copty J, Partridge SR. 2011. RAC: Repository of Antibiotic resistance Cassettes. Database (Oxford) 2011:bar054. 10.1093/database/bar054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu B, Pop M. 2009. ARDB–Antibiotic Resistance Genes Database. Nucleic Acids Res. 37:D443–D447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Antezana E, Kuiper M, Mironov V. 2009. Biological knowledge management: the emerging role of the Semantic Web technologies. Brief. Bioinform. 10:392–407 [DOI] [PubMed] [Google Scholar]
- 13. Mungall CJ, Emmert DB, FlyBase Consortium 2007. A Chado case study: an ontology-based modular schema for representing genome-associated biological information. Bioinformatics 23:i337–i346 [DOI] [PubMed] [Google Scholar]
- 14. Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, Lewis S. 2002. The generic genome browser: a building block for a model organism system database. Genome Res. 12:1599–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Federhen S. 2012. The NCBI taxonomy database. Nucleic Acids Res. 40:D136–D143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25:25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eilbeck K, Lewis SE, Mungall CJ, Yandell M, Stein L, Durbin R, Ashburner M. 2005. The Sequence Ontology: a tool for the unification of genome annotations. Genome Biol. 6:R44. 10.1186/gb-2005-6-5-r44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eddy SR. 2011. Accelerated Profile HMM searches. PLoS Comput. Biol. 7:e1002195. 10.1371/journal.pcbi.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lukashin AV, Borodovsky M. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26:1107–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nordmann P, Poirel L, Walsh TR, Livermore DM. 2011. The emerging NDM carbapenemases. Trends Microbiol. 19:588–595 [DOI] [PubMed] [Google Scholar]
- 22. Cowell LG, Smith B. 2010. Infectious disease ontology, p 373–395 In Sintchenko V. (ed), Infectious disease informatics. Springer, New York, NY [Google Scholar]
- 23. Zhanel GG, Low DE. 2007. Launching of the CAN-R Web site—the official Web site of the Canadian Antimicrobial Resistance Alliance. Can. J. Infect. Dis. Med. Microbiol. 18:151–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bush K, Jacoby GA. 2010. Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 54:969–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67:2640–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen C-C, Lin Y-C, Sheng W-H, Chen Y-C, Chang S-C, Hsia K-C, Liao M-H, Li S-Y. 2011. Genome sequence of a dominant, multidrug-resistant Acinetobacter baumannii strain, TCDC-AB0715. J. Bacteriol. 193:2361–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, The OBI Consortium, Leontis N, Rocca-Serra P, Ruttenberg A, Sansone S-A, Scheuermann RH, Shah N, Whetzel PL, Lewis S. 2007. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol. 25:1251–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]