Abstract
We have employed a rapid fluorescence-based screen to assess the polyspecificity of several aaRSs against an array of unnatural amino acids. We discovered that a p-cyanophenylalanine specific aminoacyl-tRNA synthetase (pCNF-RS) has high substrate permissivity for unnatural amino acids, while maintaining its ability to discriminate against the canonical twenty amino acids. This orthogonal pCNF-RS, together with its cognate amber nonsense suppressor tRNA is able to selectively incorporate 18 unnatural amino acids into proteins, including trifluoroketone, alkynyl, and hydrazino substituted amino acids. In an attempt to better understand this polyspecificity, the x-ray crystal structure of the aaRS/p-cyanophenylalanine complex was determined. A comparison of this structure with those of other mutant aaRSs showed that both binding site size and other more subtle features control substrate polyspecificitiy.
Keywords: Unnatural amino acids, p-cyanophenylalanine, aminoacyl-tRNA synthetase, polyspecificity
A wide variety of unnatural amino acids (UAAs) have been genetically encoded in bacteria, yeast, and mammalian cells with excellent fidelity and efficiency. These novel amino acids include NMR, IR, fluorescent, and crystallographic probes, orthogonal bioconjugation partners, metal ion chelators, redox active centers, and photocrosslinking and photocaging agents.(1, 2) An orthogonal aminoacyl-tRNA synthetase/suppressor tRNA (aaRS/tRNACUA) pair (one that does not significantly cross-react with host tRNAs or aminoacyl-tRNA synthetases) is used to selectively incorporate the unnatural amino acid of interest in response to a nonsense or frameshift codon. In order to evolve aaRSs that recognize an unnatural amino acid and no endogenous amino acid, a double sieve selection scheme is employed which ties the activity of the aaRS to the viability of E. coli.(3, 4) In the positive selection, large libraries of aaRS active site mutants are screened for their ability to incorporate the unnatural amino acid in response to an amber nonsense codon at a permissive site in an essential protein; in the negative selection, aaRSs that incorporate endogenous amino acids in to a lethal protein in the absence of the unnatural amino acid are removed.
Because no selective pressure is applied against other unnatural amino acids during the selection, evolved aaRSs may also cross-react with other unnatural amino acids, while maintaining their orthogonality to endogenous amino acids.(5–9) This polyspecificity can be exploited to incorporate additional unnatural amino acids without the evolution of new, orthogonal aaRS/tRNA pairs. Aminoacyl-tRNA synthetase promiscuity is not problematic in this regard as the growth media can be supplemented with only the desired unnatural amino acid. Previous experiments have examined the substrate permissivity of the pyrrolysyl, p-benzoylphenylalanine- and napthylalanine-synthetases, and mutations were introduced to broaden their substrate specificity. However, a thorough analysis of multiple aminoacyl-tRNA synthetases with a broad range of unnatural amino acid substrates has yet to be realized.3a Thus, to systematically analyze and better understand the permissivity of multiple evolved aaRSs, we have investigated their ability to incorporate a diverse library of unnatural amino acids using a simple fluorescent reporter-based assay.
ExperimentalProcedures
GFP Expression
GFP expression was carried out as previously described. BL-21 (DE3) E. coli was co-transformed with the appropriate pEVOL plasmid and pET-GFPY151X plasmid(1), grown in 2xYT media at 37 °C to saturation with chloramphenicol (40 μg/mL) and ampicillin (100 μg/mL) and diluted in 2YT to an OD600 of 0.2. Diluted cultures were grown at 37 °C to an OD600 of 0.7 – 1.0, and induced with IPTG (1 mM) and arabinose (0.02%). The culture was immediately aliquoted (100 μL/well) into a 96 well plate containing 10 μL of 10 mM unnatural amino acid per well and background fluorescence was measured on a Spectramax Gemini EM (Ex/Em= 395/509 nm; Molecular Probes). After 14 h at 30 °C fluorescence was measured again to determine the level of GFP expression. Amino acids that afforded a significantly measurable signal were then employed in similar expression experiments in triplicate. Each sample was normalized to cell density using the OD600 and fluorescence was measured and compared to wild type expression (Table 2), confirming the UAA incorporation.
Table 2.
Unnatural amino acid incorporation by pCNF-RS.
| aaRS | UAA | Expected[a] | Observed | Relative % Expression [b] |
|---|---|---|---|---|
| WT[c] | Phe[c] | 18352 | 18353 | 100 ± 3.9 |
| pCNF | 1 | 18377 | 18376 | 79 ± 2.9 |
| pCNF | 2 | 18478 | 18477 | 87 ± 2.2 |
| pCNF | 3 | 18431 | 18431 | 96 ± 1.4 |
| pCNF | 4 | 18386 | 18386 | 91 ± 5.2 |
| pCNF | 5 | 18370 | 18368 | 45 ± 4.9 |
| pCNF | 6 | 18376 | 18376 | 84 ± 1.4 |
| pCNF | 7 | 18394 | 18396 | 60 ± 3.0 |
| pCNF | 8 | 18393 | 18393 | 95 ± 2.3 |
| pCNF | 9 | 18450 | 18450 | 43 ± 6.1 |
| pCNF | 10 | 18394 | 18393 | 82 ± 3.9 |
| pCNF | 11 | 18396 | 18395 | 75 ± 3.7 |
| pCNF | 12 | 18429 | 18326 | 81 ± 1.2 |
| pCNF | 13 | 18396 | 18394 | 61 ± 1.8 |
| pCNF | 14 | 18406 | 18405 | 72 ± 3.0 |
| pCNF | 15 | 18382 | 18382 | 45 ± 4.2 |
| pCNF | 16 | 18383 | 18383 | 94 ± 1.4 |
| pCNF | 17 | 18407 | 18407 | 77 ± 3.2 |
| pCNF | 18 | 18422 | 18422 | 61 ± 4.6 |
LC/MS expected mass for UAA incorporated into F107TAG myoglobin
Based on GFP-Y151TAG expression, and normalized to WT GFP expression (16 mg/L)
Mass for wild type Myo with Phe at residue.
Myoglobin Expression
BL-21(DE3) E. coli was co-transformed with the appropriate pEVOL plasmid and pET-MyoF107X, grown in 2xYT media at 37 °C to saturation with chloramphenicol (40 μg/mL) and ampicillin (100 μg/mL) and diluted in 2YT to an OD600 of 0.2. Diluted cultures were grown at 37 °C to an OD600 of 0.7, induced with IPTG (1 mM), arabinose (0.2%) and the appropriate unnatural amino acid (1 mM) and grown at 37 °C for 16 h. The cultures were pelletted and lysed using Bug Buster (Novagen) and the protein was purified on Ni-NTA spin columns (Qiagen) according to the manufacturer’s protocol. Myoglobin expression was analyzed on a 4–20% Gly-Tris polyacrylamide gel (see Supporting Information) and the mass of purified protein was determined by LC/MS on an Agilent 1100 Series LC/MSD. The chromatographic peak corresponding to myoglobin (between 6.1 min and 6.5 min) was charge deconvoluted using Agilent LC/MSD ChemStation software (Rev. B.03.02). Deconvolution parameters were set to high Mr= 17,000 and low Mr= 21,000, maximum charge= 50, and minimum peaks in set = 3 –8. Error± 0.02%, as determined from control samples. Liquid chromatography mass spectrometry results for MyoY107X expression experiments are listed in the Supporting Information.
Aminoacyl-tRNA synthetase Crystallization
Protein expression and crystallization was carried out as previously described(10–12). The pCNF-RS DNA was amplified from the pBK-CN vector(13) (Fwd Primer: 5′ GCAAGCGCATATGGACGAATTTGAAATGA 3′; Rev Primer: 5′ GTTCGGCTCGAGTAATCTCTTTCTAATTG 3′), inserted into the pET-22b(+) vector using the XhoI and NdeI restriction sites, and transformed into BL21(DE3) cells. A 1L 2X YT (Amp) expression culture was inoculated to OD600 0.2 and grown at 37 °C to an OD600 of 0.6 and induced with IPTG (1 mM). The culture was grown at 37 °C for 16 h, pelleted and lysed (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 10 mM BME, pH 8.0) by sonication. The sample was then centrifuged (18,000 rpm; 20 min) and the supernatant was incubated with Ni NTA resin (2 mL; Qiagen) for 1 hour at 4 °C. The resin was then washed (100 mL; 50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, 10 mM BME, pH 8.0) and eluted (5 mL; 50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, 10 mM BME, pH 8.0), followed by dialysis (3 × 1h rt; 25 mM Tris, 50 mM NaCl, 1 mM EDTA, 10 mM BME, pH 8.0). The aaRS was purified by FPLC on a MonoQ column (Buffer A: 25 mM Tris, 25 mM NaCl, 1 mM EDTA, 10 mM BME, pH 8.5; Buffer B: 25 mM Tris, 1 M NaCl, 1 mM EDTA, 10 mM BME, pH 8.5), concentrated to 2 mL and dialyzed into crystallization buffer (20 mM Tris, 50 mM NaCl, 10 mM BME, pH 8.0). Crystals were grown by the sitting-drop-vapor-diffusion method using a 1:1 mixture of concentrated protein (15 mg/mL, with 2 mM pCNF) and mother liquor (0.2M (NH4) Tartrate, 20% PEG-3350, pH 6.6) at 20 °C.
Data Collection, Structure Determination and Refinement
Data for the pCNF co-crystals were collected at beamline 5.0.3 of the Advanced Light source (ALS) at a wavelength of 0.9778Å to a maximum Bragg spacing of 2.3Å (Table 1). All data were reduced and scaled using the HKL2000 package(14). The crystal belonged to the monoclinic crystal system with the β angle approaching 90º. A strong peak in the native Patterson map indicated the presence of pseudo-translational symmetry and that the crystal was pseudo-orthorhombic. This was confirmed by the inability of the unreduced data to scale reasonably in the equivalent orthorhombic spacegroup. The presence of non-crystallographic translational symmetry induces considerable deviation from Wilson Statistics. The Wilson Ratio (<I2>/<I>2) for the data set calculated from phenix xtriage is 2.752 and 2.776 for acentric and centric reflections respectively, whereas perfect data should have values of 2.0 and 3.0 respectively. The Wilson distribution, which is based on the assumption of a random distribution of atoms in the cell, is therefore considerably skewed by this translational symmetry and statistics such as the R-factor and procedures such as the calculation of normalized amplitudes which are based on this assumption can be affected. Despite this the structure was easily solved using Phaser(15) using all data between 50 and 2.3Å resolution using the wild type Tyrosyl tRNA synthetase(16) (PDB code 1J1U) as a probe. The molecular replacement solution confirmed the presence of the pseudo-translational symmetry between the two molecules in the asymmetric unit. Electron density produced from an initial refinement with Buster clearly showed the mutated residues between the probe model and the structure as well as electron density for pCNF in both molecules, confirming the validity of the molecular replacement solution. Model building and refinement were performed by iterative building and refinement with Coot and Buster.(17, 18) All other crystallographic manipulations were carried out with the CCP4 program suite.(19) Convergence of the refinement was checked throughout, with weights between geometric and X-ray terms optimized on the basis of the R-free in combination with a criterion for reasonable geometry. The Local Structure Symmetry Restraints (LSSR) procedure was also applied throughout refinement(20), also incorportated based on its effect on the R-free. Validity of the final structure was checked using a combination of the validation tools in Coot (17), including inspection of the Ramachandran plot, real space correlation, rotamer and geometry analysis. Using these refinement and geometrical critia final model converged with Rcryst and Rfree of 23.0 % and 30.0 % respectively, with excellent geometry, root mean squared deviation (rmsd) of bonds and angles 0.013Å2 and 1.3º respectively, no residues in disallowed regions of the Ramachandran plot (Table 1) and all of the mutations in the structure in a preferred rotamer conformation
Table 1.
Data and refinement statistics
| Space Group | P21 |
|---|---|
| Unit Cell Å | a=52.52Å,b=68.93Å,c=82.09Å β=90.59 |
| Wavelength Å | 0.9778 |
| Resolution Ǻ(highest resolution shell) | 2.3 (2.34–2.3) |
| Rmerge % (highest resolution shell) | 0.066 (0.25) |
| Unique Refs | 46732 |
| Completeness % (highest resolution shell) | 96.2 (76.6) |
| I/sigma (Highest resolution shell) | 15.0 (2.1) |
| Redundancy (highest resolution shell) | 3.0(2.1) |
| Refinement | |
| Rfactor (Rfree *) % † | 0.23 (0.30) |
| No. protein atoms | 4913 |
| No. water atoms | 172 |
| No. Hetero Atoms | 28 |
| rmsd bonds Å | 0.013 |
| rmsd angles º | 1.37 |
| Mean B factor Å2 | 47.9 |
Rfactor = Σ Σ|Ii-<Ii>| |/Σ|Ii| where Ii is the scaled intensity of the ith measurement, and <Ii> is the mean intensity for that reflection.
Rfree = as for Rcryst, but for 5.0% of the total reflections chosen at random and omitted from refinement.
Estimated overall coordinate error
Results and Discussion
Assay Development
To develop a rapid assay to analyze the incorporation of unnatural amino acids by an evolved aaRS, a fluorescence-based GFP reporter was used. A GFP gene with an amber mutation at Tyr151 (GFPY151X) was placed under the control of the T7 promoter in a pET101 vector as previously described.(21) Tyrosine 151 is a surface residue that can be substituted with a range of structurally diverse amino acids without affecting protein stability or fluorescence.(21) This reporter affords fluorescence only when the TAG codon is used as a sense codon for the acylated suppressor tRNACUA (and not a stop codon). Mutant Methanocaldococcus jannaschii tyrosyl-RS/tRNACUA (MjTyrRS/tRNACUA) pairs encoded in the efficient pEVOL system(21) were co-transformed into BL-21(DE3) cells with the pET-GFPY151X construct to enable rapid screening for substrate permissivity. Aminoacyl-tRNA synthetases specific for pAcF (p-acetylphenylalanine, 7),(22) pAzF (p-azidophenylalanine, 8),(23) pCNF (p-cyanophenylalanine, 1),(13) pIF (p-iodophenylalanine, 2), (24) pBoF (p-borophenylalanine),(25) pPrF (p-propargylphenylalanine, 14),(26) pCMF (p-carboxymethyl-phenylalanine),(27) pBpF (p-benzoylphenylalanine),(28) NapA (napthylalanine),(29) CouA (7-hydroxycoumarin-4-yl ethylglycine),(30) PLA (p-hydroxyL-phenyllactic acid),(31) BipyA (bipyridylalanine),(32) and HQA (8-hydroxyquinolin-3-yl alanine),(33) in addition to the wild type tyrosyl-tRNA synthease, were analyzed.(1, 34) A 96-well format was used to screen 72 unnatural amino acids including para and meta substituted phenylalanine analogues, histidine and alanine derivatives, amine, biaryl, and thiol containing amino acids (see Supporting Information). While fluorescent amino acids were included in the plate, other than CouA (well E7), the wavelengths of excitation and emission were nonoverlapping with GFP, and a background subtraction prior to expression was sufficient to allow for assessment of incorporation. The fluorescence of each well supplemented with 1 mM amino acid was measured after 14 hours of expression in rich media (Figure 1). In order to rapidly assess amino acid incorporation fluorescence readings were taken on the cultures, as opposed to lysed cells, and potential hits were later confirmed by protein isolation and mass spectrometry.
Figure 1.
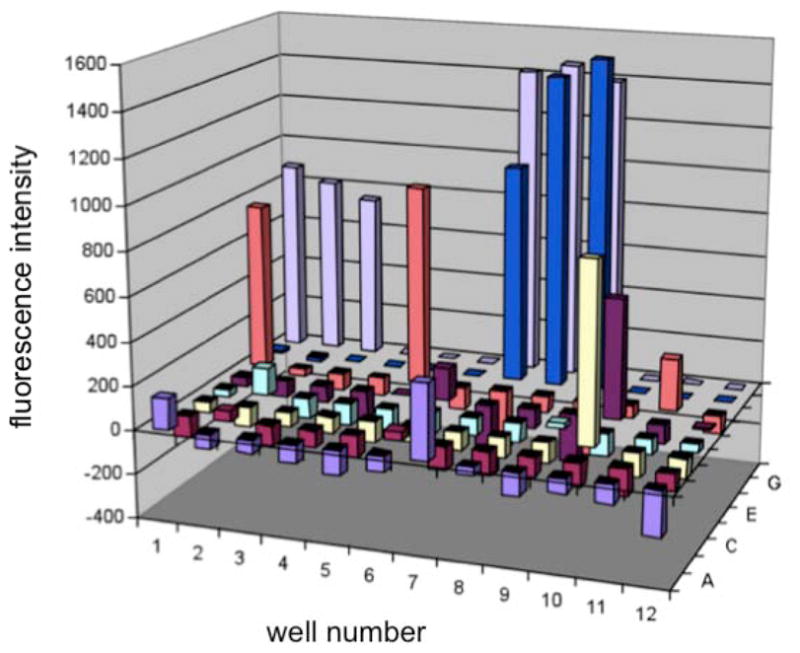
GFP fluorescence assay for pCNF-RS dependent incorporation of unnatural amino acids into GFPY151X. A 96-well plate contains different non-native amino acids (1 mM, see supporting information) incubated with BL-21(DE3) cells harboring the GFPY151X reporter and the pEVOL-pCNF in 2YT media. Fluorescence was measured after 14 h at 30 °C of expression and corrected for background by a non-induced culture.
Many of the aaRSs failed to exhibit a significant degree of substrate promiscuity with respect to the UAA in our screen. Not surprisingly, the wild type MjTyrRS showed no detectable incorporation of any unnatural amino acid in the screen. Additionally, the pBoF, pPrF, pCMF, HQA, CouA, PLA, NapA, and BipyA specific aaRSs displayed a very low propensity for incorporation of unnatural amino acids beyond the specific unnatural amino acid for which they were evolved. The pAcF, pIF and pAzF synthetases were able to incorporate several para-substituted phenylalanine analogs included in the screen, but only to a limited extent (see Supporting Information). Most interestingly, pCNF-RS(13) exhibited a substantial degree of polyspecificity as it afforded increased GFPY151X fluorescence in the presence of p-chlorophenylalanine (2), p-fluorophenylalanine (5), p-acetylphenylalanine (7), p-alkynylphenylalanine (6), benzylserine (15), and p-phenyl phenylalanine (12) (Figure 1). Screen hits were validated by incorporation of the unnatural amino acid into a his-tagged myoglobin mutant containing an F107TAG amber mutation (MyoF107X).(21) The protein was purified using a Ni-NTA resin and analyzed by SDS-PAGE. The calculated mass of purified mutant protein was confirmed by liquid chromatography mass spectroscopy (LCMS). In the absence of any unnatural amino acid low levels of phenylalanine incorporation in response to the amber codon could be detected (< 20%); however, in the presence of any of these amino acids (>1mM), only incorporation of the unnatural amino acid was observed within the detection limits of LCMS.(21) The behavior of pCNF-RS in this assay was unique both with respect to incorporation efficiency and substrate diversity relative to the other 13 aminoacyl-tRNA synthetases.
Determination of Substrate Scope for pCNF-RS Polyspecificity
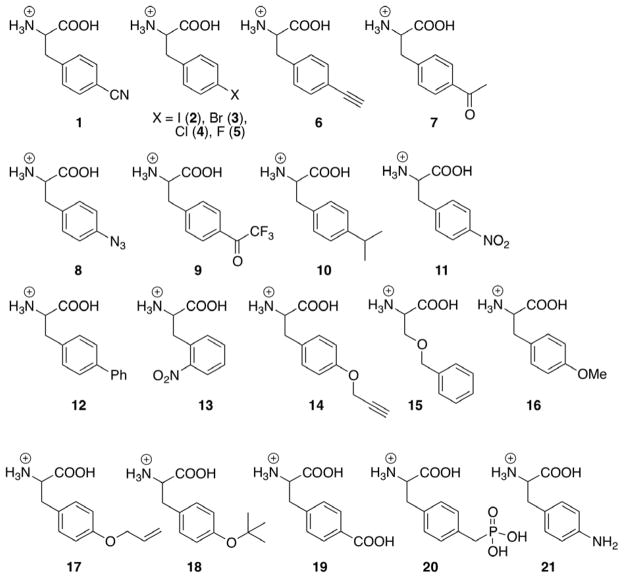
The pCNF-RS appears to be highly tolerant of para-substituted phenylalanine derivatives. Consequently, the pCNF-RS was further screened against additional para-substituted aromatic amino acids (Figure 2). The majority of these amino acids (1–18) were incorporated into GFPY151X by the pCN-RS (relative incorporation was assessed in triplicate and normalized to cell density, Table 2); incorporation was further validated by incorporation into MyoF107X with subsequent mass spectral analysis (Table 2). pCNF-RS substrates include UAAs that have already been genetically encoded in bacteria, e.g., pIF (2), pBrF (p-bromophenylalanine 3), pAcF (7), pAzF (8), pPrF (14), pNO2F (p-nitrophenylalanine 11), oNO2F (o-nitrophenylalanine 13), O-methyl Tyr (16), and O-allyl Tyr (17).(2)p CNF-RS substrates also include unnatural amino acids which have not previously been encoded in bacteria, e.g., pClF (4), pFF (5), p-ethynyl phenylalanine (6), p-trifluoromethylacetyl phenylalanine (9), p-phenyl phenylalanine (12), benzylserine (15), and O-tert-butyl tyrosine (18). Based on GFP expression levels, many of the amino acids are good substrates for pCNF-RS, affording >80% protein expression relative to wild type GFP. Interestingly, several amino acids (pIF (2), pBrF (3), pClF (4), p-alkynylF (6), pAzF (8), p-isopropylF (10), and OMeY (16)) afford higher protein yields than did the pCNF substrate (1), the UAA against which the pCNF-RS was originally selected. Only amino acids 19–21 (p-carboxyphenylalanine, p-methyl-phosphonic acid phenylalanine, and p-aminophenylalanine), which have hydrogen bond donating substituents, were not incorporated. It is interesting to note that despite the apparent polysubstrate specificity of the pCNF-RS, 21 (p-aminophenylalanine) was not incorporated while other significantly larger substituents were tolerated, indicating the pCNF-RS has some degree of specificity (21 has been previously shown to be taken up efficiently by E. coli and is stable in the cytoplasm).(35) In total, the pCNF-RS was found to incorporate 18 unnatural amino acids, seven of which had not been previously encoded. A number of the latter UAAs, e.g., p-alkynylphenylalanine (6) and p-trifloroacetylphenylalanine (9), may be useful as orthogonal bioconjugation handles or as warheads for the selective inactivation of enzymes.
Figure 2.
Unnatural amino acids screened for incorporation by pCNF-RS
X-ray Crystal Structure of pCNF-RS
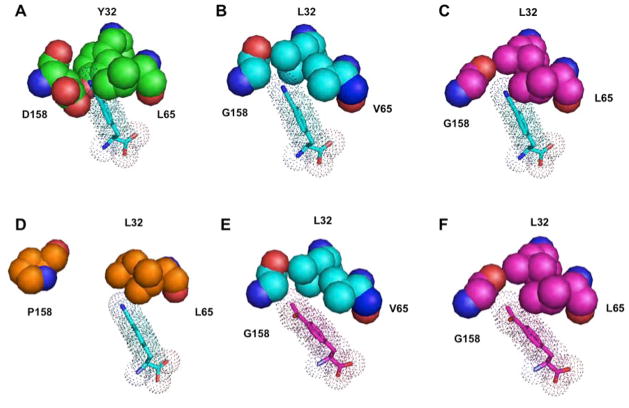
To better understand the molecular basis of pCNF-RS’s polysubstrate specificity (which in contrast to polyspecific enzymes like p450s, is not a solvent exposed surface cavity), the aaRS was overexpressed and crystallized for x-ray structure analysis (10–12, 36). The crystal structure of the pCNF-RS pCNF complex was solved to 2.3 Å with an Rfactor = 0.23 (Table 1 and Supporting Information). The pCNF-RS structure superimposes well onto the WT-TyrRS-Tyr structure (PDB 1J1U) except in the amino acid binding site. The amino acid binding site for the pCNF-RS is relatively hydrophobic containing the following mutations relative to the wild type synthetase: Y32L, L65V, F108W, N109M, D158G, and I159A. A comparison of the WT-TyrRS structure to the pCNF bound pCNF-RS structure (Figure 4A vs. Figure 4B) reveals that mutations create a larger binding pocket which allows the enzyme to accept the nitrile group. In the WT-TyrRS structure, residues Y32 and D158 are involved in hydrogen bonding with the tyrosine hydroxyl group. These residues in the pCNF-RS are mutated to leucine and glycine, respectively, which removes hydrogen bonding potential and likely explains the inability of pCNF-RS to accept tyrosine and other phenylalanine derivatives with hydrogen bond donor substituents in the para position. Additionally, the mutation L65V creates a larger binding pocket and appears to alter the angular position of the bound amino acid. This mutation is unique to the pCNF-RS, as no other previously evolved aminoacyl-tRNA synthetases possess the valine residue at this position. Although the binding pocket is larger in pCNF-RS, key residues (Leu 32, Val65 and Gly158) are still able to stabilize the pCNF (1) through van Der Waals interactions (Figure 4B).
Figure 4.
Structural analysis of 3 key residues (32, 65, and 158) for amino acid recognition in various synthetases. A) Wild-type tyrosyl RS (green, PDB 1J1U) modeled with pCNF. B) pCNF-RS (cyan) modeled with pCNF. C) pAcF-RS (magenta, PDB 1ZH6) modeled with pCNF. D) NapA-RS (orange, PDB 1ZH0) modeled with pCNF. E) pCNF-RS (cyan) modeled with pAcF. F) pAcF-RS (magenta, PDB 1ZH6) modeled with pAcF.
To determine which mutations might be determinants of polyspecificity, the pCNF bound pCNF-RS amino acid binding site structure was overlaid with the substrate-bound pAcF-RS (Y32L, D158G, I159C, L162R; PDB 1ZH6) and NapA-RS (Y32L, D158P, I159A, L162Q, A167V; PDB 1ZH0) structures. The former aaRS accepts relatively few other unnatural amino acids, while the latter displays no polyspecificity (Figure 3). As previously noted, the Y32L mutation increases the amino acid binding site size (especially at the para-position of phenylalanine derivatives); however, this mutation is also found in the pAcF-RS and NapA-RS (among others), which do not exhibit a comparable degree of polyspecificity to pCNF-RS. The unique L65V mutation appears to create unoccupied space at the back of the amino acid binding that is filled by other residues in pAcF-RS. This mutation may enable pCNF-RS to accept a diverse group of para-substituted phenylalanine or tyrosine derivatives (Figure 3). Interestingly, when pCNF is modeled into the pAcF-RS binding pocket, the leucine at residue 65 interferes with the cyano substituent (Figure 4C; the pAcF-RS complex is shown for comparison in Figure 4F). In contrast, pAcF (7) can readily be docked into the active site of the pCNF-RS (Figure 4E). Additionally, mutation of leucine 65 to smaller amino acids has been shown to allow the incorporation of larger unnatural amino acids in other aaRSs. For example, the o-nitrobenzyl tyrosine (ONBY) synthetase contains a L65G mutation, facilitating the incorporation of the sterically bulky bicyclic ONBY amino acid.(37)
Figure 3.
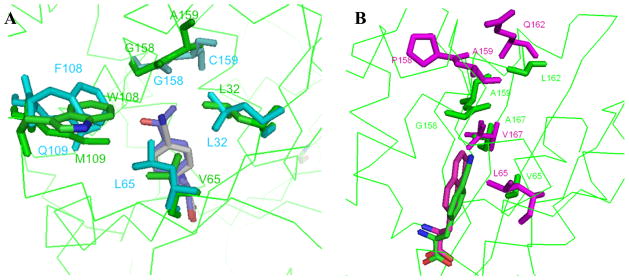
A) Overlay of the key active site residues for substrate bound pAcF-RS·pAcF (cyan; PDB 1ZH6) and pCNF-RS·pCNF (green) complexes. The pAcF (blue) and pCNF (gray) substrates are show at the center of the structure. B) Overlay of the substrate bound pNapA-RS·NapA (magenta, PDB 1ZH0) and pCNF-RS·pCNF (green) complexes. The NapA (magenta)and pCNF (green) substrates are showat the bottom of the structure.
While an increased substrate binding site size may be partially responsible for increased polyspecificity, other factors are involved. This is apparent when comparing the pCNF-RS and NapA-RS structures. Although the D158P mutation in the NapA-RS gives it a significantly larger binding pocket than the pCNF-RS, this aaRS exhibits virtually no permissivity. When pCNF is modeled into the amino acid binding site of NapA-RS, proline 158 prevents side chain Van der Waals interactions that might stabilize the bound and amino acid (Figure 4D). Moreover, a number of the other aminoacyl-tRNA synthetases examined with larger binding sites (e.g., that accommodate bipyridyl, 8-hydroxyquinolinyl and benzoylphenyl side chains) do not show significant polyspecificity, suggesting that there is a plasticity unique to pCNF-RS active site.
The uniqueness of the pCNF-RS is also evident when examining backbone perturbations in the mutant aminoacyl-tRNA synthetases. As previously reported, the D158P and I159A mutations terminate the α8-helix in NapA-RS; a similar backbone alteration is observed in the pAcF-RS (albeit to a lesser extent).(12) Interestingly, this perturbation is not observed in the pCNF-RS, despite having the same D158G mutation as the pAcF-RS and the same I159A mutation as the NapA-RS. In fact, the pCNF-RS aligns much more closely to the wild-type tyrosyl-tRNA synthetase than with the other aminoacyl-tRNA synthetases in this region (Figure 5A). Another distinctive difference is apparent in the pCNF-RS when examining the helical residues 108 and 109. Relative to both the WT-RS and the pAcF-RS, the helix is moderately displaced and is slightly truncated (Figure 5B). This may afford a greater degree of space and plasticity for the accommodation of larger para-substituted amino acids.
Figure 5.
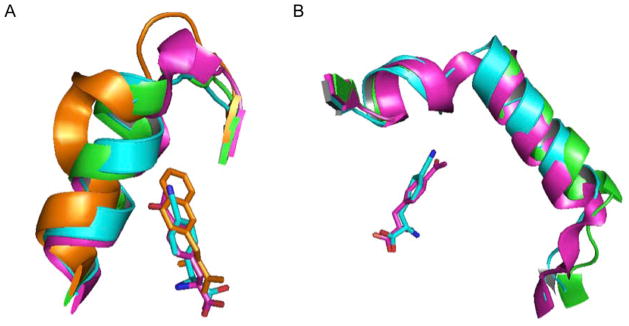
Backbone pertubations in various aminoacyl-tRNA synthetases. A) Overlay of the helices containing mutated residues 158 and 159 in NapA-RS (orange; PDB 1ZH0), WT-TyrRS (green; PDB 1J1U), pAcF-RS (magenta; PDB 1ZH6) and pCNF-RS (cyan). The termination of the helix which accommodates large amino acids is especially apparent in NapA-RS B) Overlay of the helices containing mutated residues 108 and 109 in WT-TyrRS (green; PDB 1J1U), pAcF-RS (magenta; PDB 1ZH6) and pCNF-RS (cyan). pCNF = cyan, pAcF = magenta, NapA = orange.
Importantly, the pCNF-RS amino acid binding site still highly discriminates against Phe, Tyr and p-aminophenylalanine, while it accepts similar amino acid such as O-methyltyrosine (16), p-chlorophenylalanine (4) and p-fluorophenylalanine (5). Clearly, mutations in a relatively small number of active site residues result in the ability of this aaRS to discriminate subtle changes in substrate structure. This result indicates that the negative selection step in the evolution of pCNF-RS is quite robust in its ability to remove polyspecific mutants that accept endogenous amino acids. Work is currently underway to co-crystallize pCNF-RS and various mutants with additional common and unnatural amino acids in an effort to further decipher this unique enzyme’s specificity.
Conclusion
The use of previously evolved aaRSs for the incorporation of additional unnatural amino acids will facilitate the expansion of the genetic repertoire to include novel chemical functionalities. Seven amino acids which had previously not been genetically encoded in bacteria were selectively incorporated into proteins using the pCNF-RS synthetase. These include p-chlorophenylalanine, p-fluorophenylalanine, p-alkynylphenylalanine (4-6), p-trifluoromethylacetylphenylalanine (9), p-phenyl phenylalanine (12), benzylserine (15), and O-tert-butyl tyrosine (18). Future screens of additional amino acids and aaRSs may yields aaRSs that incorporate classes of unnatural amino acids rather than a single residue.
Supplementary Material
Acknowledgments
This work was funded by grant DE-FG03-00ER46051 from the Division of Materials Sciences, DOE (P.G.S.). This is manuscript 20691 of the Scripps Research Institute.
D.D.Y. is grateful for a NIH fellowship IF32CA144213 and T.S.Y. is grateful for an Achievement Rewards for College Scientists Scholarship. The work in this paper is based on experiments conducted at beamline 5.0.3 of the advanced light source (ALS). The ALS is supported by the Director, Office of Science, Office of Basic Energy Sciences, Material Sciences Division of the U.S. Department of Energy under contract No. DE-AC03-76SF00098 at Lawrence Berkeley National Laboratory. We would like to thank all of the staff of these beamlines for their continued support.
Footnotes
Supporting Information Available: Unnatural amino acids screened and 96-well plate format, myoglobin mass spectral data, and crystal refinement statistics. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Young TS, Schultz PG. Beyond the Canonical 20 Amino Acids: Expanding the Genetic Lexicon. J Biol Chem. 2010 doi: 10.1074/jbc.R109.091306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu CC, Schultz PG. An Expanding Genetic Code. Annual Review of Biochemistry. 2010:79. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 3.Wang L, Schultz PG. A general approach for the generation of orthogonal tRNAs. Chem Biol. 2001;8:883–890. doi: 10.1016/s1074-5521(01)00063-1. [DOI] [PubMed] [Google Scholar]
- 4.Santoro SW, Wang L, Herberich B, King DS, Schultz PG. An efficient system for the evolution of aminoacyl-tRNA synthetase specificity. Nat Biotechnol. 2002;20:1044–1048. doi: 10.1038/nbt742. [DOI] [PubMed] [Google Scholar]
- 5.Stokes AL, Miyake-Stoner SJ, Peeler JC, Nguyen DP, Hammer RP, Mehl RA. Enhancing the utility of unnatural amino acid synthetases by manipulating broad substrate specificity. Mol Biosyst. 2009;5:1032–1038. doi: 10.1039/b904032c. [DOI] [PubMed] [Google Scholar]
- 6.Hartman MC, Josephson K, Lin CW, Szostak JW. An expanded set of amino acid analogs for the ribosomal translation of unnatural peptides. PLoS One. 2007;2:e972. doi: 10.1371/journal.pone.0000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyake-Stoner SJ, Refakis CA, Hammill JT, Lusic H, Hazen JL, Deiters A, Mehl RA. Generating permissive site-specific unnatural aminoacyl-tRNA synthetases. Biochemistry. 2010;49:1667–1677. doi: 10.1021/bi901947r. [DOI] [PubMed] [Google Scholar]
- 8.Kirshenbaum K, Carrico IS, Tirrell DA. Biosynthesis of proteins incorporating a versatile set of phenylalanine analogues. Chembiochem. 2002;3:235–237. doi: 10.1002/1439-7633(20020301)3:2/3<235::AID-CBIC235>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 9.Munier R, Cohen GN. Incorporation of structural analogues of amino acids into bacterial proteins during their synthesis in vivo. Biochim Biophys Acta. 1959;31:378–391. doi: 10.1016/0006-3002(59)90011-3. [DOI] [PubMed] [Google Scholar]
- 10.Liu W, Alfonta L, Mack AV, Schultz PG. Structural basis for the recognition of para-benzoyl-L-phenylalanine by evolved aminoacyl-tRNA synthetases. Angew Chem Int Ed Eng. 2007;146:6073–6075. doi: 10.1002/anie.200701990. [DOI] [PubMed] [Google Scholar]
- 11.Turner JM, Graziano J, Spraggon G, Schultz PG. Structural characterization of a p-acetylphenylalanyl aminoacyl-tRNA synthetase. J Am Chem Soc. 2005;127:14976–14977. doi: 10.1021/ja0549042. [DOI] [PubMed] [Google Scholar]
- 12.Turner JM, Graziano J, Spraggon G, Schultz PG. Structural plasticity of an aminoacyl-tRNA synthetase active site. Proc Natl Acad Sci U S A. 2006;103:6483–6488. doi: 10.1073/pnas.0601756103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schultz KC, Supekova L, Ryu Y, Xie J, Perera R, Schultz PG. A genetically encoded infrared probe. J Am Chem Soc. 2006;128:13984–13985. doi: 10.1021/ja0636690. [DOI] [PubMed] [Google Scholar]
- 14.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymo. 1997;1276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 15.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi T, Nureki O, Ishitani R, Yaremchuk A, Tukalo M, Cusack S, Sakamoto K, Yokoyama S. Structural basis for orthogonal tRNA specificities of tyrosyl-tRNA synthetases for genetic code expansion. Nat Struct Biol. 2003;10:425–432. doi: 10.1038/nsb934. [DOI] [PubMed] [Google Scholar]
- 17.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 18.Bricogne G, Blanc E, Brandl M, Flensburg C, Keller P, Paciorek P, Roversi P, Sharff A, Smart O, Vonrhein C, Womack T. BUSTER version 2.9. Global Phasing Ltd; Cambridge, United Kingdom: 2010. [Google Scholar]
- 19.Bailey S. The Ccp4 Suite - Programs for Protein Crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 20.Smart O, Brandl M, Flensburg C, Keller P, Paciorek W, Wonrhein C, Womack T, Bricogne G. Refinement with Local Structure Similarity Restraints (LSSR) Enables Exploitation of Information from Related Structures and Facilitates use of NCS. Abstr Annu Meet Am Crystallogr Assoc. 2008:117. [Google Scholar]
- 21.Young TS, Ahmad I, Yin JA, Schultz PG. An Enhanced System for Unnatural Amino Acid Mutagenesis in E. coli. J Mol Biol. 2009 doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 22.Wang L, Zhang ZW, Brock A, Schultz PG. Addition of the keto functional group to the genetic code of Escherichia coli. P Natl Acad Sci USA. 2003;100:56–61. doi: 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chin JW, Santoro SW, Martin AB, King DS, Wang L, Schultz PG. Addition of p-azido-L-phenylaianine to the genetic code of Escherichia coli. Journal of the American Chemical Society. 2002;124:9026–9027. doi: 10.1021/ja027007w. [DOI] [PubMed] [Google Scholar]
- 24.Xie JM, Wang L, Wu N, Brock A, Spraggon G, Schultz PG. The site-specific incorporation of p-iodo-L-phenylalanine into proteins for structure determination. Nature Biotechnology. 2004;22:1297–1301. doi: 10.1038/nbt1013. [DOI] [PubMed] [Google Scholar]
- 25.Brustad E, Bushey ML, Lee JW, Groff D, Liu W, Schultz PG. A Genetically Encoded Boronate-Containing Amino Acid. Angew Chem Int Edit. 2008;47:8220–8223. doi: 10.1002/anie.200803240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deiters A, Schultz PG. In vivo incorporation of an alkyne into proteins in Escherichia coli. Bioorg Med Chem Lett. 2005;15:1521–1524. doi: 10.1016/j.bmcl.2004.12.065. [DOI] [PubMed] [Google Scholar]
- 27.Xie JM, Supekova L, Schultz PG. A genetically encoded metabolically stable analogue of phosphotyrosine in Escherichia coli. Acs Chemical Biology. 2007;2:474–478. doi: 10.1021/cb700083w. [DOI] [PubMed] [Google Scholar]
- 28.Chin JW, Martin AB, King DS, Wang L, Schultz PG. Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. P Natl Acad Sci USA. 2002;99:11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Brock A, Schultz PG. Adding L-3-(2-naphthyl)alanine to the genetic code of E-coli. Journal of the American Chemical Society. 2002;124:1836–1837. doi: 10.1021/ja012307j. [DOI] [PubMed] [Google Scholar]
- 30.Wang JY, Xie JM, Schultz PG. A genetically encoded fluorescent amino acid. Journal of the American Chemical Society. 2006;128:8738–8739. doi: 10.1021/ja062666k. [DOI] [PubMed] [Google Scholar]
- 31.Guo JT, Wang JY, Anderson JC, Schultz PG. Addition of an alpha-hydroxy acid to the genetic code of bacteria. Angew Chem Int Edit. 2008;47:722–725. doi: 10.1002/anie.200704074. [DOI] [PubMed] [Google Scholar]
- 32.Xie JM, Liu WS, Schultz PG. A genetically encoded bidentate, metal-binding amino acid. Angew Chem Int Edit. 2007;46:9239–9242. doi: 10.1002/anie.200703397. [DOI] [PubMed] [Google Scholar]
- 33.Lee HS, Spraggon G, Schultz PG, Wang F. Genetic Incorporation of a Metal-Ion Chelating Amino Acid into Proteins as a Biophysical Probe. Journal of the American Chemical Society. 2009;131:2481. doi: 10.1021/ja808340b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo J, Wang J, Lee JS, Schultz PG. Site-specific incorporation of methyl- and acetyl-lysine analogues into recombinant proteins. Angew Chem Int Ed Eng. 2008;147:6399–6401. doi: 10.1002/anie.200802336. [DOI] [PubMed] [Google Scholar]
- 35.Mehl RA, Anderson JC, Santoro SW, Wang L, Martin AB, King DS, Horn DM, Schultz PG. Generation of a bacterium with a 21 amino acid genetic code. Journal of the American Chemical Society. 2003;125:935–939. doi: 10.1021/ja0284153. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Wang L, Schultz PG, Wilson IA. Crystal structures of apo wild-type M. jannaschii tyrosyl-tRNA synthetase (TyrRS) and an engineered TyrRS specific for O-methyl-L-tyrosine. Protein Sci. 2005;14:1340–1349. doi: 10.1110/ps.041239305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deiters A, Groff D, Ryu YH, Xie JM, Schultz PG. A genetically encoded photocaged tyrosine. Angew Chem Int Edit. 2006;45:2728–2731. doi: 10.1002/anie.200600264. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.