Abstract
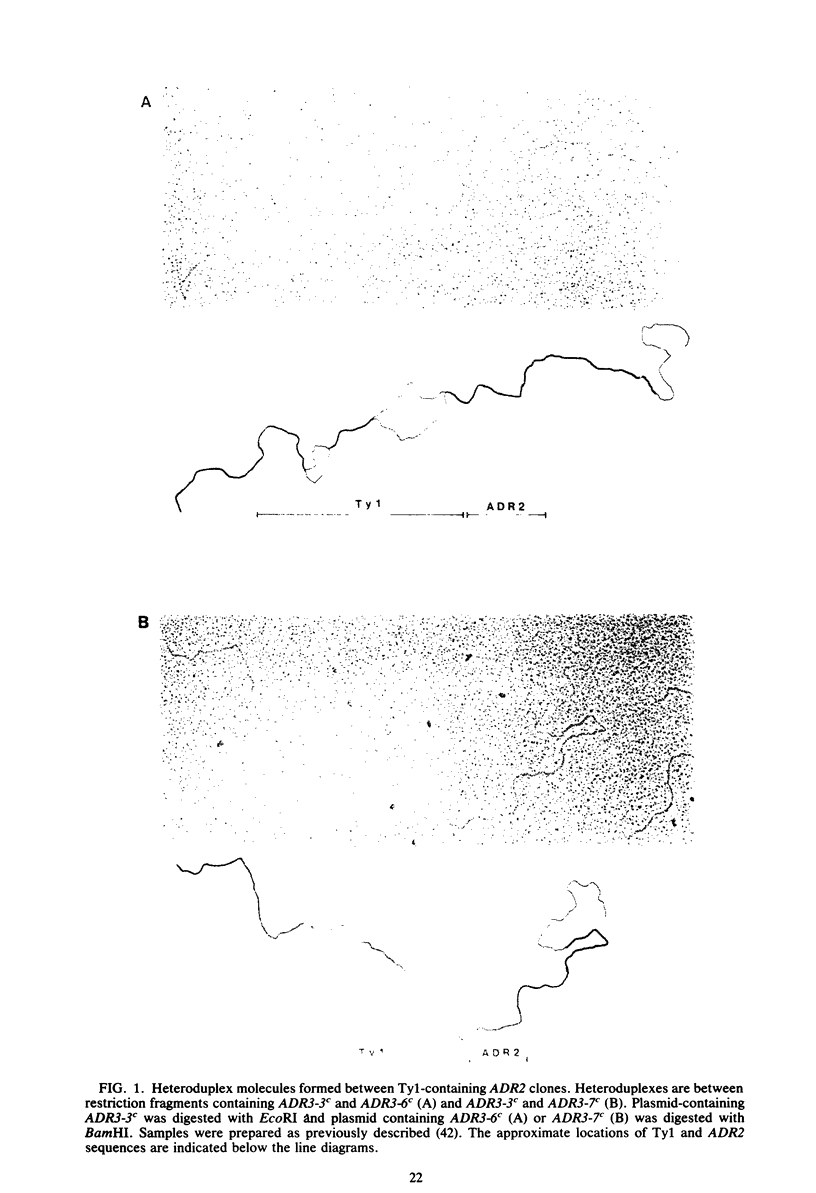
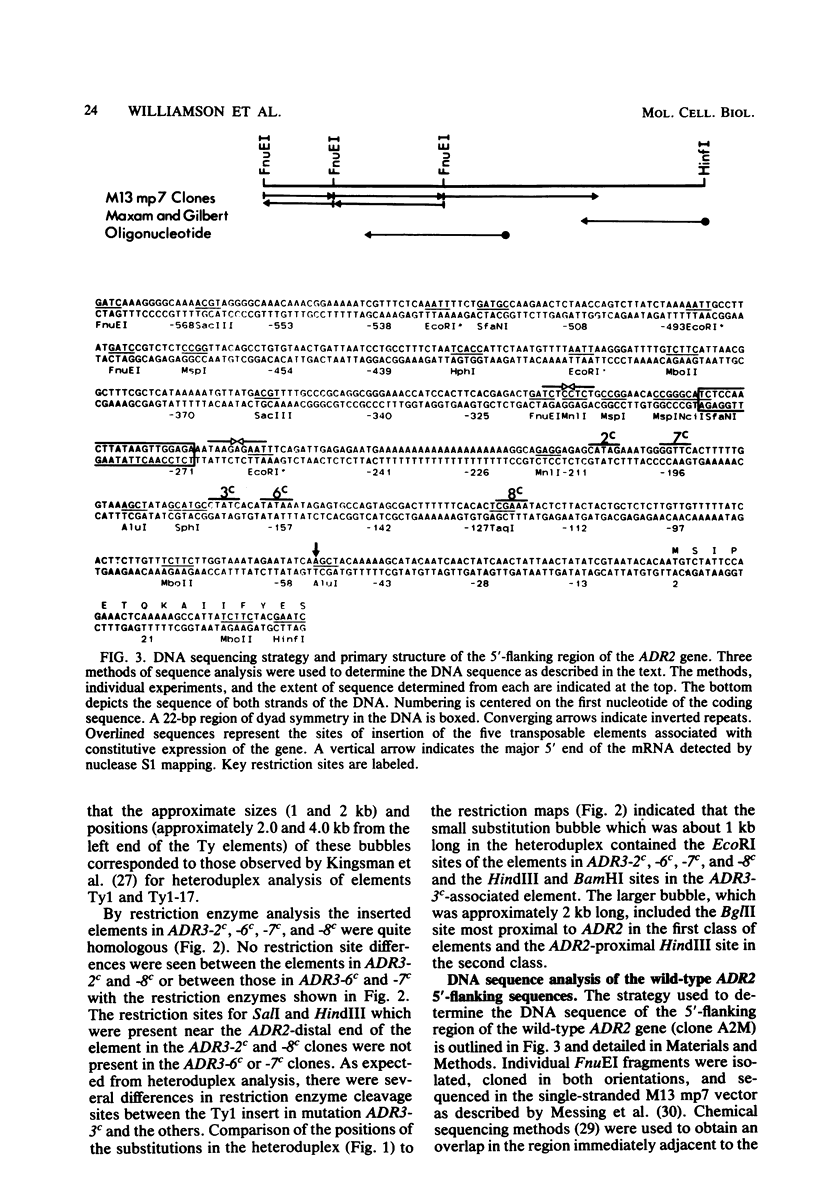
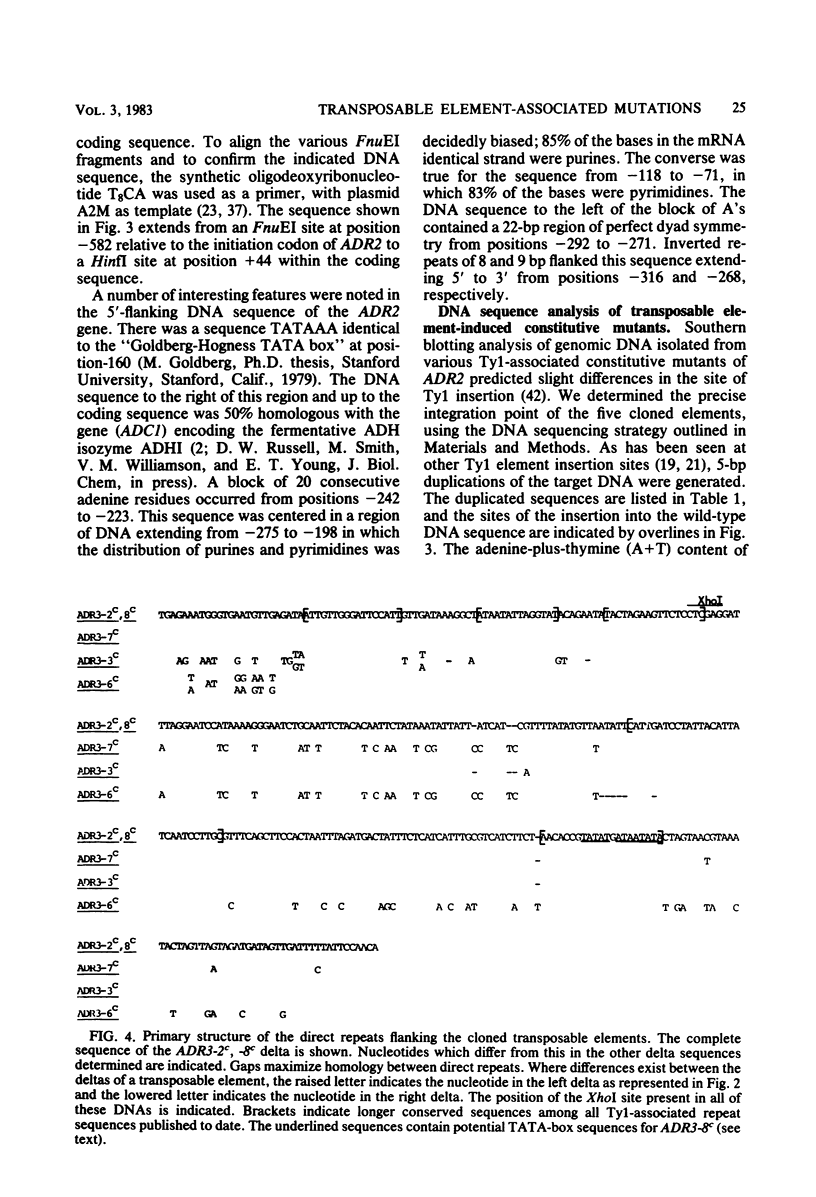
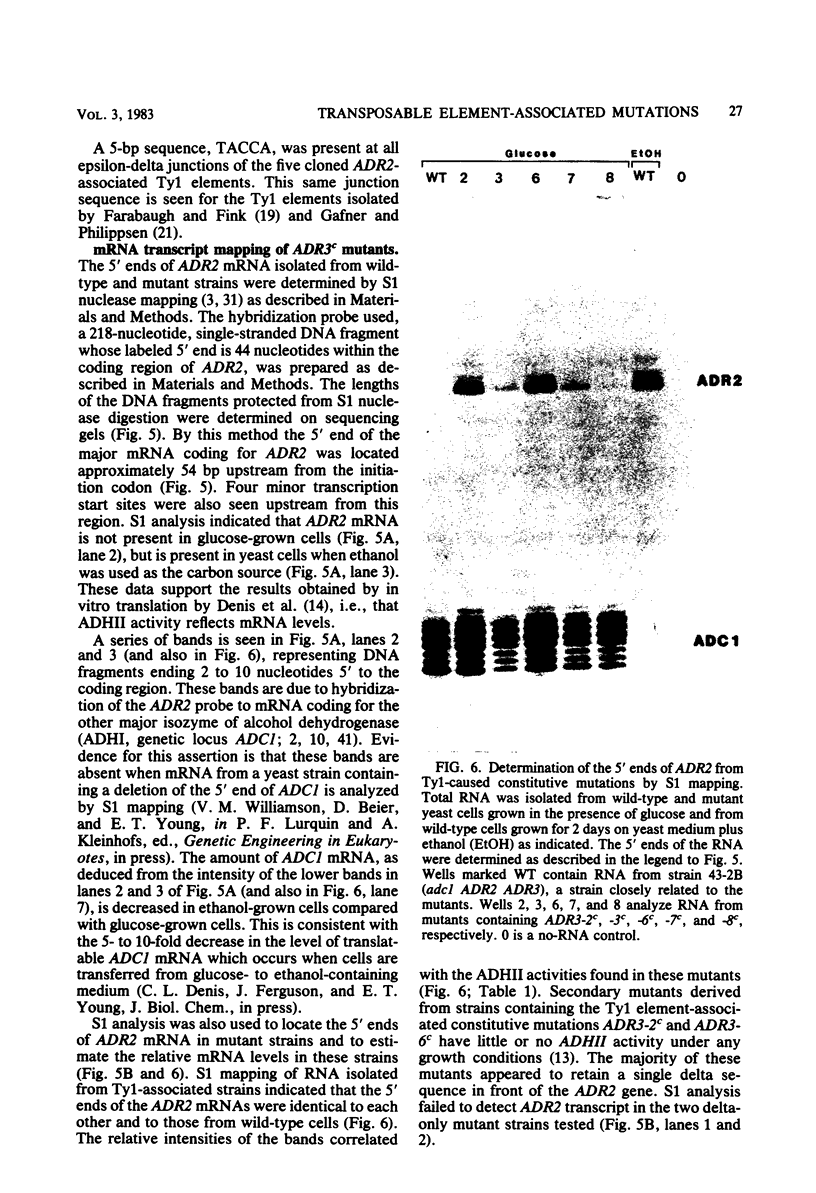
Seven cis-dominant, constitutively expressed mutations of the normally glucose-repressible isozyme of alcohol dehydrogenase (ADHII) from the yeast Saccharomyces cerevisiae are caused by insertion of transposable elements from the Ty1 family in front of the ADHII structural gene (ADR2) (V. M. Williamson, E. T. Young, and M. Ciriacy, Cell 23:605-614, 1981). We cloned ADR2 with its associated Ty1 element from five S. cerevisiae strains carrying these mutations. Comparison of the Ty1 elements by heteroduplex studies and restriction enzyme analyses indicated that four were very similar; the fifth, although the same size as the others (about 5.6 kilobases), differed by the presence of two large substitutions of approximately 1 and 2 kilobases. The DNA sequences of the terminal direct repeats (deltas) were very homologous but not identical and were similar to previously reported Ty1 element direct repeats. We determined the 5'-flanking sequences of the ADR2 gene isolated from a wild-type strain and from five Ty1-associated mutations. The 5-base pair target sequence at the site of Ty1 insertion was present at both ends of each Ty1 element. The sites of insertion of the elements were all different and occurred from 125 to 210 base pairs in front of the coding region of ADR2. The 5' end of the major transcript as determined by S1 mapping was the same in wild-type cells and in Ty1-associated constitutive mutants and was approximately 54 base pairs upstream from the coding region. ADR2 transcripts were not detected when a solo delta sequence was present in the 5'-flanking region of this gene.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baltimore D. Gene conversion: some implications for immunoglobulin genes. Cell. 1981 Jun;24(3):592–594. doi: 10.1016/0092-8674(81)90082-9. [DOI] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J Biol Chem. 1982 Mar 25;257(6):3018–3025. [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Spliced early mRNAs of simian virus 40. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1274–1278. doi: 10.1073/pnas.75.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham P. M., Judd B. H. A copy of the copia transposable element is very tightly linked to the Wa allele at the white locus of D. melanogaster. Cell. 1981 Sep;25(3):705–711. doi: 10.1016/0092-8674(81)90177-x. [DOI] [PubMed] [Google Scholar]
- Bingham P. M., Levis R., Rubin G. M. Cloning of DNA sequences from the white locus of D. melanogaster by a novel and general method. Cell. 1981 Sep;25(3):693–704. doi: 10.1016/0092-8674(81)90176-8. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Transposable elements. Cell. 1980 Jul;20(3):579–595. doi: 10.1016/0092-8674(80)90305-0. [DOI] [PubMed] [Google Scholar]
- Cameron J. R., Loh E. Y., Davis R. W. Evidence for transposition of dispersed repetitive DNA families in yeast. Cell. 1979 Apr;16(4):739–751. doi: 10.1016/0092-8674(79)90090-4. [DOI] [PubMed] [Google Scholar]
- Chaleff D. T., Fink G. R. Genetic events associated with an insertion mutation in yeast. Cell. 1980 Aug;21(1):227–237. doi: 10.1016/0092-8674(80)90130-0. [DOI] [PubMed] [Google Scholar]
- Ciriacy M. Cis-dominant regulatory mutations affecting the formation of glucose-repressible alcohol dehydrogenase (ADHII) in Saccharomyces cerevisiae. Mol Gen Genet. 1976 Jun 15;145(3):327–333. doi: 10.1007/BF00325831. [DOI] [PubMed] [Google Scholar]
- Ciriacy M., Williamson V. M. Analysis of mutations affecting Ty-mediated gene expression in Saccharomyces cerevisiae. Mol Gen Genet. 1981;182(1):159–163. doi: 10.1007/BF00422784. [DOI] [PubMed] [Google Scholar]
- Denis C. L., Ciriacy M., Young E. T. A positive regulatory gene is required for accumulation of the functional messenger RNA for the glucose-repressible alcohol dehydrogenase from Saccharomyces cerevisiae. J Mol Biol. 1981 Jun 5;148(4):355–368. doi: 10.1016/0022-2836(81)90181-9. [DOI] [PubMed] [Google Scholar]
- Eibel H., Gafner J., Stotz A., Philippsen P. Characterization of the yeast mobile element Ty1. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):609–617. doi: 10.1101/sqb.1981.045.01.079. [DOI] [PubMed] [Google Scholar]
- Elder R. T., St John T. P., Stinchcomb D. T., Davis R. W., Scherer S., Davis R. W. Studies on the transposable element Ty1 of yeast. I. RNA homologous to Ty1. II. Recombination and expression of Ty1 and adjacent sequences. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):581–591. doi: 10.1101/sqb.1981.045.01.075. [DOI] [PubMed] [Google Scholar]
- Errede B., Cardillo T. S., Sherman F., Dubois E., Deschamps J., Wiame J. M. Mating signals control expression of mutations resulting from insertion of a transposable repetitive element adjacent to diverse yeast genes. Cell. 1980 Nov;22(2 Pt 2):427–436. doi: 10.1016/0092-8674(80)90353-0. [DOI] [PubMed] [Google Scholar]
- Farabaugh P. J., Fink G. R. Insertion of the eukaryotic transposable element Ty1 creates a 5-base pair duplication. Nature. 1980 Jul 24;286(5771):352–356. doi: 10.1038/286352a0. [DOI] [PubMed] [Google Scholar]
- Fink G., Farabaugh P., Roeder G., Chaleff D. Transposable elements (Ty) in yeast. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):575–580. doi: 10.1101/sqb.1981.045.01.074. [DOI] [PubMed] [Google Scholar]
- Gafner J., Philippsen P. The yeast transposon Ty1 generates duplications of target DNA on insertion. Nature. 1980 Jul 24;286(5771):414–418. doi: 10.1038/286414a0. [DOI] [PubMed] [Google Scholar]
- Gehring W. J., Paro R. Isolation of a hybrid plasmid with homologous sequences to a transposing element of Drosophila melanogaster. Cell. 1980 Apr;19(4):897–904. doi: 10.1016/0092-8674(80)90081-1. [DOI] [PubMed] [Google Scholar]
- Gillam S., Smith M. Use of E. coli polynucleotide phosphorylase for the synthesis of oligodeoxyribonucleotides of defined sequence. Methods Enzymol. 1980;65(1):687–701. doi: 10.1016/s0076-6879(80)65067-8. [DOI] [PubMed] [Google Scholar]
- Hayward W. S., Neel B. G., Astrin S. M. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature. 1981 Apr 9;290(5806):475–480. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- Jones C. W., Kafatos F. C. Structure, organization and evolution of developmentally regulated chorion genes in a silkmoth. Cell. 1980 Dec;22(3):855–867. doi: 10.1016/0092-8674(80)90562-0. [DOI] [PubMed] [Google Scholar]
- Ju G., Skalka A. M. Nucleotide sequence analysis of the long terminal repeat (LTR) of avian retroviruses: structural similarities with transposable elements. Cell. 1980 Nov;22(2 Pt 2):379–386. doi: 10.1016/0092-8674(80)90348-7. [DOI] [PubMed] [Google Scholar]
- Kingsman A. J., Gimlich R. L., Clarke L., Chinault A. C., Carbon J. Sequence variation in dispersed repetitive sequences in Saccharomyces cerevisiae. J Mol Biol. 1981 Feb 5;145(4):619–632. doi: 10.1016/0022-2836(81)90306-5. [DOI] [PubMed] [Google Scholar]
- Levis R., Dunsmuir P., Rubin G. M. Terminal repeats of the Drosophila transposable element copia: nucleotide sequence and genomic organization. Cell. 1980 Sep;21(2):581–588. doi: 10.1016/0092-8674(80)90496-1. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. A., Tatchell K., Hall B. D., Astell C., Smith M. Physical analysis of mating-type loci in Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):961–981. doi: 10.1101/sqb.1981.045.01.113. [DOI] [PubMed] [Google Scholar]
- Payne G. S., Bishop J. M., Varmus H. E. Multiple arrangements of viral DNA and an activated host oncogene in bursal lymphomas. Nature. 1982 Jan 21;295(5846):209–214. doi: 10.1038/295209a0. [DOI] [PubMed] [Google Scholar]
- Roeder G. S., Farabaugh P. J., Chaleff D. T., Fink G. R. The origins of gene instability in yeast. Science. 1980 Sep 19;209(4463):1375–1380. doi: 10.1126/science.6251544. [DOI] [PubMed] [Google Scholar]
- Rubin G. M., Brorein W. J., Jr, Dunsmuir P., Flavell A. J., Levis R., Strobel E., Toole J. J., Young E. Copia-like transposable elements in the Drosophila genome. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):619–628. doi: 10.1101/sqb.1981.045.01.080. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz L. D. Transcriptional role of yeast deoxyribonucleic acid dependent ribonucleic acid polymerase III. Biochemistry. 1978 Feb 21;17(4):750–758. doi: 10.1021/bi00597a031. [DOI] [PubMed] [Google Scholar]
- Smith M., Leung D. W., Gillam S., Astell C. R., Montgomery D. L., Hall B. D. Sequence of the gene for iso-1-cytochrome c in Saccharomyces cerevisiae. Cell. 1979 Apr;16(4):753–761. doi: 10.1016/0092-8674(79)90091-6. [DOI] [PubMed] [Google Scholar]
- St John T. P., Davis R. W. The organization and transcription of the galactose gene cluster of Saccharomyces. J Mol Biol. 1981 Oct 25;152(2):285–315. doi: 10.1016/0022-2836(81)90244-8. [DOI] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temin H. M. Origin of retroviruses from cellular moveable genetic elements. Cell. 1980 Oct;21(3):599–600. doi: 10.1016/0092-8674(80)90420-1. [DOI] [PubMed] [Google Scholar]
- Williamson V. M., Bennetzen J., Young E. T., Nasmyth K., Hall B. D. Isolation of the structural gene for alcohol dehydrogenase by genetic complementation in yeast. Nature. 1980 Jan 10;283(5743):214–216. doi: 10.1038/283214a0. [DOI] [PubMed] [Google Scholar]
- Williamson V. M., Young E. T., Ciriacy M. Transposable elements associated with constitutive expression of yeast alcohol dehydrogenase II. Cell. 1981 Feb;23(2):605–614. doi: 10.1016/0092-8674(81)90156-2. [DOI] [PubMed] [Google Scholar]
- Young T., Williamson V., Taguchi A., Smith M., Sledziewski A., Russell D., Osterman J., Denis C., Cox D., Beier D. The alcohol dehydrogenase genes of the yeast, Saccharomyces cerevisiae: isolation, structure, and regulation. Basic Life Sci. 1982;19:335–361. doi: 10.1007/978-1-4684-4142-0_26. [DOI] [PubMed] [Google Scholar]