Abstract
The BAR (Bin/Amphiphysin/Rvs) domain proteins arfaptin1 and arfaptin2 are localized to the trans-Golgi network (TGN) and, by virtue of their ability to sense and/or generate membrane curvature, could play an important role in the biogenesis of transport carriers. We report that arfaptins contain an amphipathic helix (AH) preceding the BAR domain, which is essential for their binding to phosphatidylinositol 4-phosphate (PI(4)P)-containing liposomes and the TGN of mammalian cells. The binding of arfaptin1, but not arfaptin2, to PI(4)P is regulated by protein kinase D (PKD) mediated phosphorylation at Ser100 within the AH. We also found that only arfaptin1 is required for the PKD-dependent trafficking of chromogranin A by the regulated secretory pathway. Altogether, these findings reveal the importance of PI(4)P and PKD in the recruitment of arfaptins at the TGN and their requirement in the events leading to the biogenesis of secretory storage granules.
Keywords: amphipathic helix, arfaptins, PI(4)P, PKD, protein secretion
Introduction
A large number of carriers form at the trans-Golgi network (TGN) that include clathrin-coated vesicles for trafficking to the endosomes, COPI for retrograde transport to the preceding Golgi cisternae and to the endoplasmic reticulum (ER), and carriers called CARTS for the trafficking of specific cargoes to the cell surface (De Matteis and Luini, 2008; Emr et al, 2009; Campelo and Malhotra, 2012; Valente et al, 2012; Wakana et al, 2012). The TGN is also the source of secretory storage granules in secretory cells and for the production of apical- and basolateral-specific carriers in polarized cells (Mellman and Nelson, 2008). How is the biogenesis of these different classes of transport carriers regulated at the TGN? For example, phosphatidylinositol 4-phosphate (PI(4)P) and the small GTPase Arf1 are required for the formation of clathrin-coated vesicles and CARTS but protein kinase D (PKD), which promotes the production of PI(4)P at the TGN, is required only for the biogenesis of the CARTS. In other words, there are components shared by processes that generate carriers destined for different membranes of the cell. How is this compartmentation or spatial segregation of transport machinery achieved at the TGN? It is also not known whether different carriers, mentioned above, form continuously or if there is a competition among different classes depending on the cargo. For example, does increase in secretory cargo affect the biogenesis of clathrin-coated or COPI vesicles and favour the formation of carriers destined for the cell surface?
We are interested in membrane fission regulated by PKD, which is necessary for the generation of a specific class of transport carriers at the TGN. Two lipids diacylglycerol (DAG) and PI(4)P are essential for the PKD-dependent transport carrier biogenesis. DAG and Arf1 recruit PKD to the TGN, PKD is then activated by PKCη and the trimeric G protein subunits Gβγ, and promotes the production of PI(4)P by activating phosphatidylinositol 4-kinase IIIβ (PI(4)KIIIβ) (Baron and Malhotra, 2002; Diaz Anel and Malhotra, 2005; Hausser et al, 2005; Pusapati et al, 2010). As a result of the PI(4)P synthesis, the ceramide transfer protein (CERT), oxysterol binding protein (OSBP), and FAPP1 and FAPP2 are recruited to the TGN by virtue of their Pleckstrin homology (PH) domains (Levine and Munro, 1998, 2002; Godi et al, 2004). PKD phosphorylates CERT and OSBP and they detach from the TGN (Fugmann et al, 2007; Nhek et al, 2010). Based on these findings, it has been proposed that the PKD-dependent PI(4)P production, CERT and OSBP recruitment, and control of the binding of those proteins are essential for events leading to membrane fission to generate specific transport carriers (Graham and Burd, 2011; Bankaitis et al, 2012; Campelo and Malhotra, 2012). More recently, it has been shown that PKD and CtBP1-S/BARS are linked by 14-3-3γ to recruit and control a number of other proteins, which together regulate membrane fission (Valente et al, 2012). Strangely, thus far there is no evidence of a coat (like clathrin, COPI or COPII) or a BAR (Bin/Amphiphysin/Rvs) domain-containing protein in the PKD-dependent biogenesis of transport carriers.
We have made a surprising finding that BAR domain-containing proteins arfaptin1 and arfaptin2 are recruited to the TGN by a PI(4)P-dependent reaction. Moreover, the binding of arfaptin1 to PI(4)P is regulated by PKD whereas the binding of arfaptin2 to PI(4)P is insensitive to PKD activity. We also report the specific requirement of arfaptin1 in the trafficking of chromogranin A (Cg A); the description of our findings follows.
Results
Arfaptins bind PI(4)P
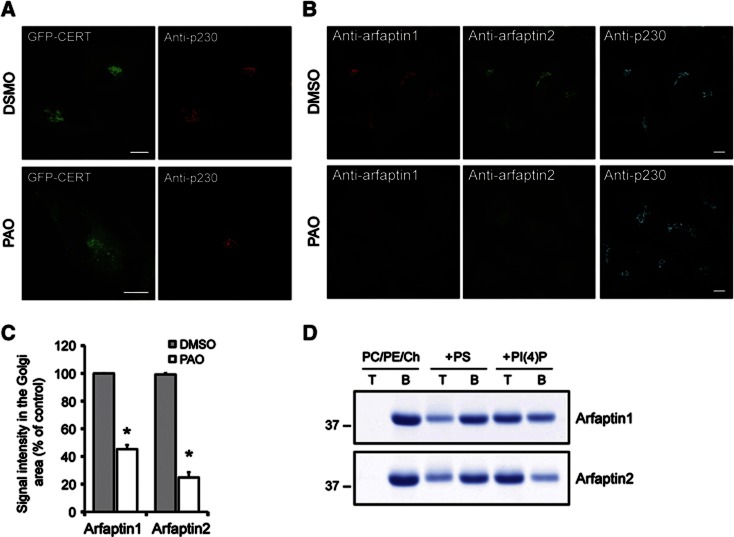
We have found that ceramide levels affect events leading to the biogenesis of transport carriers (Duran et al, 2012). The ceramide transport protein CERT binds PI(4)P at the TGN and thus controls the amount of ceramide imported into the TGN (Hanada et al, 2003). To study the connection between PI(4)P and CERT, we tested the effect of phenylarsine oxide (PAO), a chemical inhibitor of PI(4)P synthesis, on the localization of CERT at the TGN (Wiedemann et al, 1996). PAO treatment induced a partial dissociation of GFP-CERT from the TGN in HeLa cells (Figure 1A). Surprisingly, however, PAO treatment also dissociated arfaptin1 and arfaptin2 from the TGN used as controls in this experiment because they lack a typical PH domain for binding to PI(4)P (Figures 1B and C). The localization of β-COP and p230, two Golgi membrane peripheral proteins, was unaffected by PAO treatment (Figure 1B; Supplementary Figure S1). Arfaptins are metazoan-specific proteins that contain a BAR domain (Peter et al, 2004). Arfaptin1 was identified as a class I Arf-binding protein and reported to control Arf-dependent phospholipase D activation (Kanoh et al, 1997; Tsai et al, 1998; Williger et al, 1999). Subsequent studies revealed that arfaptin1 and the related protein arfaptin2 bind to both class I Arfs and Arl1 (Lu et al, 2001; Shin and Exton, 2001; Man et al, 2011). It has also been shown that the knockdown of Arl1 dissociates arfaptins from the TGN in HeLa cells (Man et al, 2011). Importantly, Arl1 is reported to recruit a guanidine nucleotide exchange factor (GEF) for Arf1 and thus activates Arf1 at the TGN (Christis and Munro, 2012). The question then arises whether arfaptins bind directly to Arf1 and Arl1, or to PI(4)P produced by the enzyme PI(4)KIIIβ, which is an effector of Arf1 at the TGN (Godi et al, 1999).
Figure 1.
Arfaptin1 and 2 bind to PI(4)P-containing membranes. (A) HeLa cells expressing GFP-CERT were treated for 5 min with 0.1% DMSO or 10 μM PAO and then visualized by fluorescence microscopy with anti-p230 antibody. (B) Non-transfected HeLa cells processed as in (A) were visualized with anti-arfaptin1, anti-arfaptin2 and anti-p230 antibodies, respectively. Scale bars, 10 μm. (C) Quantification of the immunofluorescence signal for arfaptin1 and arfaptin2 at the Golgi membranes is shown. Results are shown as the mean±s.e.m. of three experiments. The data were analysed using a paired Student’s t-test (*, P<0.01 versus DMSO-treated cells). (D) Recombinant arfaptin1 and arfaptin2 were incubated with PC/PE/Ch liposomes containing or lacking PS or PI(4)P at 10 mol%. Liposomes (T, top fraction) were separated from the unbound proteins (B, bottom fraction) by flotation and analysed by SDS–PAGE followed by staining of the proteins by Coomassie blue.
Source data for this figure is available on the online supplementary information page.
We then tested the binding of arfaptins to phosphoinositides and other phospholipids by using a protein-lipid overlay assay and recombinant non-tagged arfaptin1 and 2. This analysis revealed that arfaptin1 binds to PI(3)P, PI(5)P and to less extent to PI(4)P, phosphatidylserine (PS) and PI(3,5)P2. Arfaptin2, on the other hand, showed a preference for binding to PI(3)P, PI(4)P, PI(5)P and PS (Supplementary Figure S2). It is known that PI(3)P is not contained in the TGN (van Meer et al, 2008). Moreover, little is known about the location and the function of PI(5)P (Grainger et al, 2012). PS is found at the cytoplasmic leaflets of the plasma membrane, TGN and endo-lysosomal compartments (Fairn et al, 2011). However, the arfaptins do not localize to compartments other than the TGN. Based on these reasons, we suggest that arfaptins bind anionic lipids and we provide further test of this binding with specific focus on PI(4)P.
To test whether arfaptins can directly bind to PI(4)P-containing membranes, we carried out liposome flotation assays with recombinant arfaptin1 and 2 and liposomes containing or lacking PI(4)P. The amount of arfaptins found in the liposome fraction was barely detectable when the assay was performed with liposomes composed only of phosphatidylcholine (PC)/phosphatidylethanolamine (PE)/cholesterol (Ch) (Figure 1D). The Golgi membranes are known to contain the negatively charged phospholipid PS (van Meer et al, 2008; Fairn et al, 2011), we therefore assayed the binding of arfaptins to liposomes containing 10 mol% PS. Liposomes made of PC/PE/Ch/PS recruited 31 and 34% of arfaptin1 and 2, respectively, from the total amounts of these proteins included in the reaction mixture (Figure 1D). The binding increased to 58% for arfaptin1 and to 74% for arfaptin2 when the assay was performed with liposomes containing 10 mol% PI(4)P instead of PS (Figure 1D). These results indicate that although arfaptin1 and 2 can bind to membranes containing negatively charged phospholipids, such as PS, they bind more efficiently to membranes with PI(4)P.
PI(4)P is required for the targeting of arfaptins to the Golgi complex
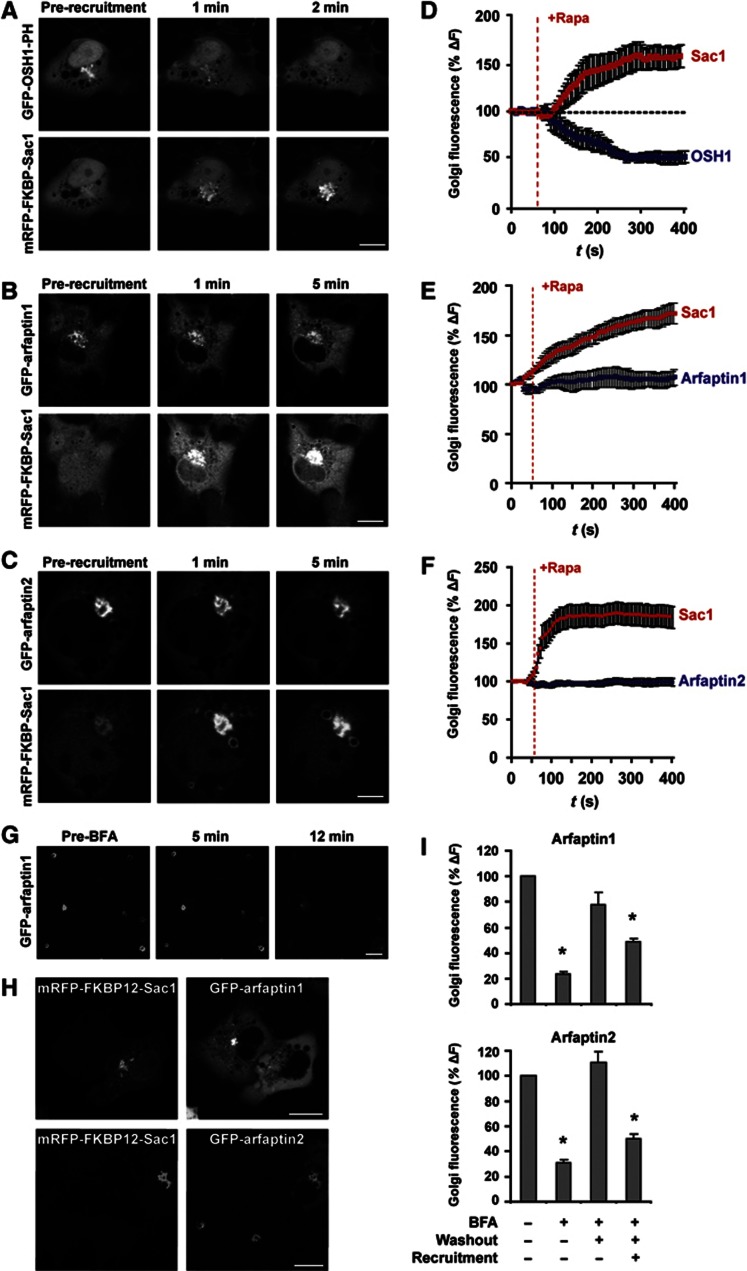
We have taken advantage of a recently developed approach to acutely deplete PI(4)P by the rapamycin-induced recruitment of the Sac1 phosphatase to the Golgi membranes (Szentpetery et al, 2010). COS-7 cells were transfected with plasmids coding for the Golgi-associated TGN38-FRB-CFP recruiter together with the cytosolic mRFP-FKBP12-Sac1 and either GFP-arfaptin1 or GFP-arfaptin2. The localization of the PH domain of OSH1 fused to GFP was used as a control. The Golgi membrane-associated levels of Sac1 and arfaptins/OSH1 were monitored by live-cell confocal microscopy prior to and after adding rapamycin for up to 10 min (Figures 2A–C). The recruitment of Sac1 to the Golgi membranes rapidly dissociated OSH1, without any obvious effect on the localization of the arfaptins (Figures 2D–F). We then tested the requirement of PI(4)P in the recruitment of arfaptins by the following procedure. The cells were treated with brefeldin A (BFA) for 12 min, which is known to prevent Arf1-dependent binding of proteins to the Golgi membranes. Arfaptins dissociated from the TGN under these experimental conditions (Figure 2G). The cells were then washed to remove BFA and incubated in the absence or the presence of rapamycin for 1 h to allow the recruitment of Sac1 to the Golgi membranes. Quantification of the Golgi complex-associated levels of GFP-arfaptin1 and 2 after this procedure revealed that the Sac1 recruitment reduced the amount of both arfaptin1 and 2 that can re-associate with the Golgi membranes after BFA wash-out (Figures 2H and I). It is therefore clear that an Arf or Arf-like GTPase is required for the binding of arfaptins to the Golgi membranes. Importantly, these results reveal that PI(4)P is also necessary for the recruitment of arfaptins to the Golgi membranes.
Figure 2.
PI(4)P depletion at the Golgi reduces the rate of arfaptin1 and 2 association with the Golgi membranes. (A–F) COS-7 cells co-expressing mRFP-FKBP12-Sac1, TGN38-FRB-CFP and GFP-OSH1-PH (A, D), GFP-arfaptin1 (B, E) or GFP-arfaptin2 (C, F) were imaged by live-cell confocal microscopy and treated with 100 nM rapamycin to induce the recruitment of the cytosolic FKBP12-Sac1 to the Golgi complex. Time-lapse images of individual cells were recorded for 400 s and representative images are shown for GFP-OSH1-PH (A), GFP-arfaptin1 (B) and GFP-arfaptin2 (C) at 0 min (pre-recruitment) and at 1 and 2 or 5 min after recruitment. Graphs represent normalized Golgi fluorescence intensity for Sac1 and OSH1-PH (D) (n=6), arfaptin1 (E) (n=9) or arfaptin2 (F) (n=9) in cells with effective Sac1 recruitment. Values are shown as the mean±s.e.m. (G) COS-7 cells transfected with GFP-arfaptin1 were treated with BFA (5 μg/ml) for 12 min. Confocal microscopy images are shown at 0 min (pre-BFA) and at 5 and 12 min after treatment. (H) Confocal images of COS-7 cells co-expressing mRFP-FKBP12-Sac1, TGN38-FRB-CFP, and either GFP-arfaptin1 (upper panel) or GFP-arfaptin2 (lower panel), treated with BFA for 12 min, washed out and incubated with 100 nM rapamycin for 1 h. (I) Bar graphs depict normalized Golgi fluorescence intensities of arfaptin1 (upper graph) or arfaptin2 (lower graph) for untreated cells (arfaptin1 n=21, arfaptin2 n=18), or cells treated with BFA for 12 min (arfaptin1 n=26, arfaptin2 n=28), washed out and incubated in complete medium with DMSO (arfaptin1 n=23, arfaptin2 n=18) or 100 nM rapamycin (arfaptin1 n=41, arfaptin2 n=37) for 1 h. Values are shown as the mean±s.e.m. The data were analysed using the one-way ANOVA (*, P<0.01 versus untreated cells). Scale bars, 10 μm.
A highly conserved region preceding the arfaptin BAR domain is required for TGN localization and PI(4)P binding
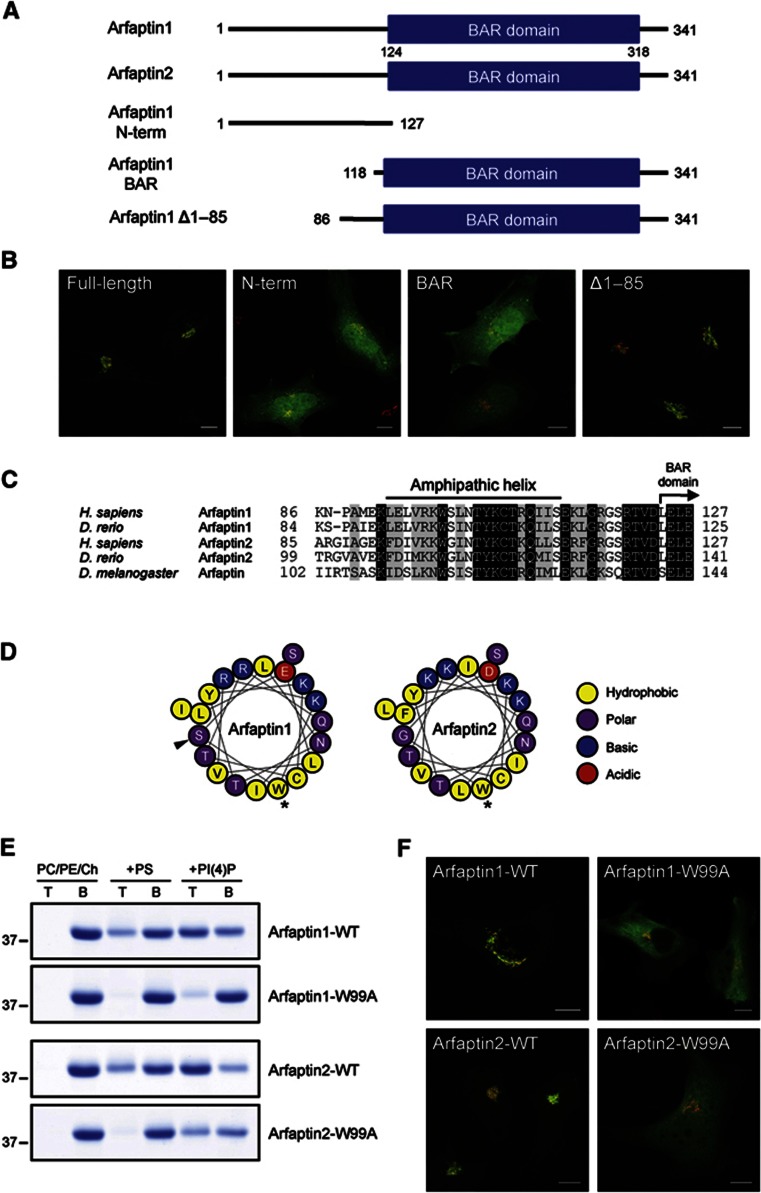
We generated several deletion mutants of arfaptin1 tagged with GFP, which were expressed in HeLa cells to monitor the localization of cognate polypeptides by fluorescence microscopy (Figure 3A). Western blotting the lysates of the cells expressing the deletion mutants revealed the predicted sizes allowing us to use them for further analyses (Supplementary Figure S3). As shown in Figure 3B, the full-length arfaptin1 was located at the TGN, whereas the N-terminal half (aa 1–127) and the BAR domain-containing C-terminal half (aa 118–341) of arfaptin1 were predominantly cytoplasmic and nuclear. A mutant lacking aa 1–85 of arfaptin1 was then expressed in HeLa cells and a significant amount of the expressed protein was localized at the TGN (Figure 3B). These results indicate that the region comprising residues 86–118 is required for the localization of the arfaptin1 BAR domain to the TGN. Sequence alignment of the 40-aa region (aa 86–127) preceding the arfaptin BAR domain showed a high level of conservation across species (Figure 3C). Computational analysis of this region with Heliquest (Gautier et al, 2008) revealed the existence, in both arfaptins, of a putative amphipathic helix (AH) from residues 93 to 112 (Figure 3D). The polar face of this AH contains four conserved basic residues that could participate, by means of their interaction with negatively charged phospholipids, in the binding of arfaptins to the outer leaflet of the TGN. To test whether the predicted AH is required for the binding of arfaptins to membranes, we perturbed the formation of the AH by mutating the most bulky hydrophobic residue in the predicted AH, Trp99, to alanine and analysed the binding of the cognate proteins to liposomes. Recombinant full-length arfaptin1-W99A and arfaptin2-W99A were incubated with liposomes made of PC/PE/Ch containing or lacking PS or PI(4)P. The liposomes were collected from the reaction mixture and analysed by SDS–PAGE. The results show that replacing Trp99 with alanine reduced by 80% and 55% the binding of arfaptin1 and 2, respectively, to PI(4)P-containing liposomes as compared with that observed for the wild-type proteins (Figure 3E). In addition, the W99A mutation strongly decreased the binding of both arfaptins to PC/PE/Ch/PS liposomes (Figure 3E). To test the significance of the AH in the recruitment of arfaptins to the TGN in intact cells, HeLa cells were transfected with wild-type or the W99A mutant of arfaptin1 and 2 tagged with GFP and visualized by fluorescence microscopy. As shown in Figure 3F, the wild-type forms but not the mutants were recruited to the TGN. These results reveal the importance of the AH in the binding of arfaptins to PI(4)P-containing membranes and in their localization to the TGN in intact cells. Thus, PI(4)P-dependent binding of arfaptins to the TGN, and their insertion into the outer leaflet through the AH could play an important role in the sorting and the export of cargo by specific carriers from the TGN.
Figure 3.
An amphipathic helix is required for the localization of arfaptin1 and 2 at the TGN. (A) Schematic representation of full-length arfaptin1 and arfaptin2, and the truncated forms of arfaptin1 tested. Numbers indicate amino-acid position. (B) HeLa cells were transfected with constructs coding for either the wild-type full-length arfaptin1 or the indicated deletion mutants of arfaptin1 fused to GFP (green) and visualized by fluorescence microscopy along with anti-p230 antibodies (red). (C) Amino-acid sequence alignment of the region preceding the BAR domain of human and zebrafish arfaptin1 (GenBank accession Nos. NP_001020764 and NP_998533, respectively), human and zebrafish arfaptin2 (NP_036534 and NP_001017649, respectively), and Drosophila melanogaster arfaptin (NP_650058). Residues in the sequences that are conserved and similar are shown in the black and shaded background, respectively. (D) Schematic representation of the predicted AH of arfaptin1 and 2. Asterisks indicate mutations to alanine, and black triangle marks the PKD phosphorylation site. (E) Flotation assays with the wild-type forms and the W99A mutants of arfaptin1 and 2 using PC/PE/Ch liposomes containing or lacking PS or PI(4)P at 10 mol%. T, top fraction; B, bottom fraction. (F) HeLa cells expressing arfaptin1-WT, -W99A, arfaptin2-WT or -W99A tagged with GFP (green) were fixed, stained with anti-p230 antibodies (red) and examined by confocal microscopy. Scale bars, 10 μm.
Source data for this figure is available on the online supplementary information page.
PKD phosphorylated arfaptin1 does not bind the TGN
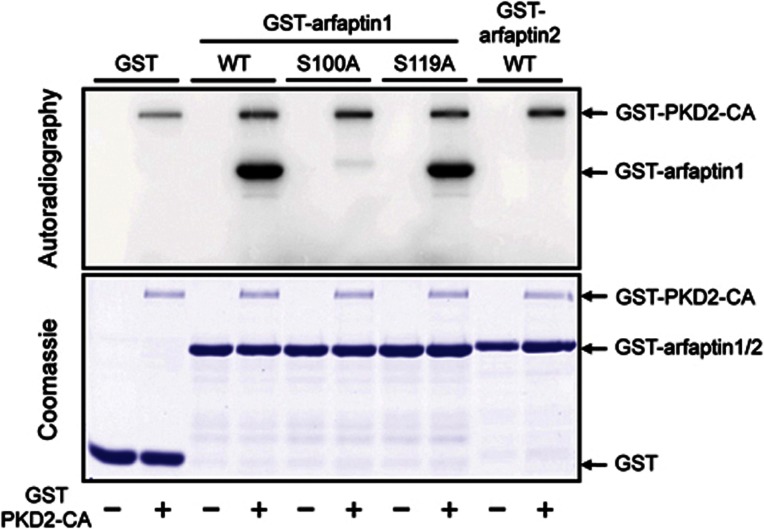
The 40-aa region preceding the BAR domain of arfaptin1, but not arfaptin2, contains two potential phosphorylation sites for PKD (Ser100 and Ser119, Figure 3C) fitting the consensus PKD phosphorylation sequence [LIV]-X-[KR]-X(1,2)-pS (where X is any amino acid and pS is phosphoserine). To test the significance of these serines in the recruitment of arfaptin1 to the PI(4)P-containing membranes, recombinant GST-tagged arfaptin1-wild type (WT), arfaptin1-S100A, arfaptin1-S119A and arfaptin2-WT variants were incubated with a constitutively activated GST-PKD2 (PKD2-CA) and [γ-32P]ATP. The proteins were then analysed by SDS–PAGE followed by autoradiography (Figure 4). Activated PKD2 phosphorylated arfaptin1-WT but not arfaptin2-WT. Moreover, replacing Ser100 with alanine (arfaptin1-S100A) inhibited the PKD2-dependent phosphorylation of arfaptin1. Arfaptin1-S119A, on the other hand, was phosphorylated by PKD2-CA (Figure 4). These results confirm that Ser100 of arfaptin1 is phosphorylated by PKD in vitro.
Figure 4.
PKD2 phosphorylates arfaptin1 at Ser100. Recombinant GST, GST-arfaptin1-WT, -S100A, -S119A and GST-arfaptin2-WT proteins were incubated in kinase buffer containing [γ-32P]ATP in the absence (−) or presence (+) of purified GST-PKD2-CA. Proteins were analysed by SDS–PAGE and stained with Coomassie blue (bottom) followed by autoradiography (top).
Source data for this figure is available on the online supplementary information page.
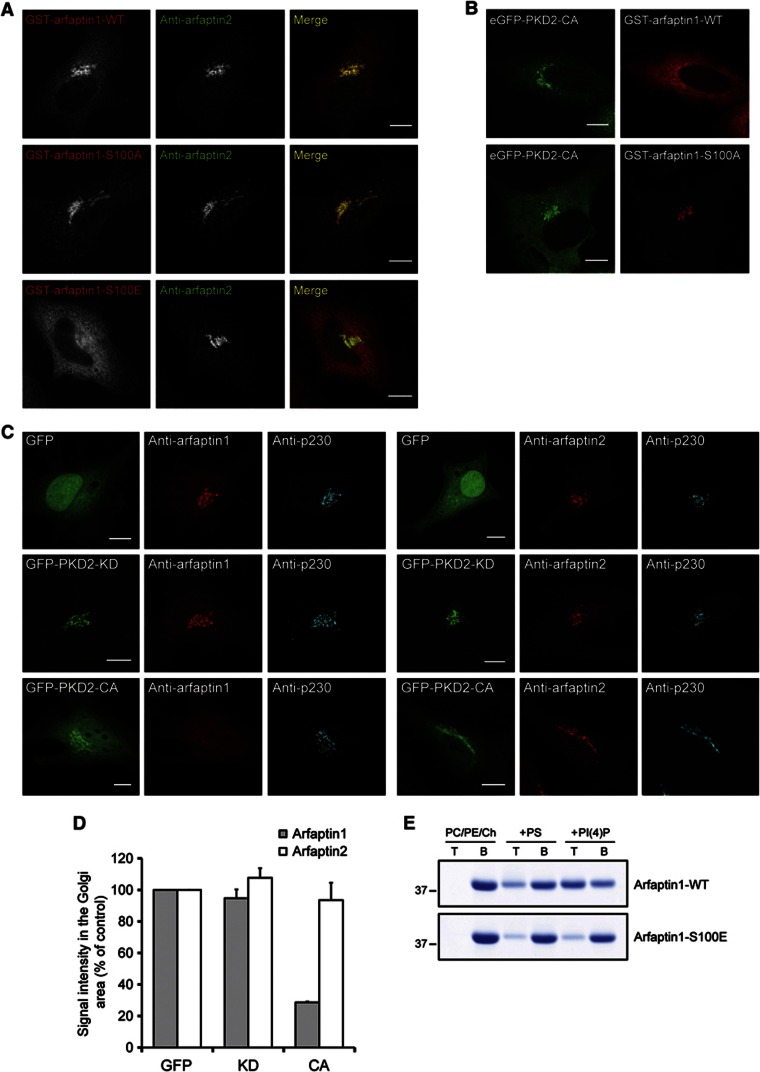
We then tested the role of Ser100 in the localization of arfaptin1 in HeLa cells. HeLa cells were transfected with the GST-tagged wild type, the non-phosphorylatable (S100A), or the phosphomimetic (S100E) forms of arfaptin1 and visualized by fluorescence microscopy. The wild-type and the non-phosphorylatable forms were found to colocalize with arfaptin2 at the TGN; however, the phosphomimetic S100E mutant was entirely cytoplasmic (Figure 5A). In another experiment, PKD2-CA tagged with GFP was co-expressed in HeLa cells with either GST-arfaptin1-WT or GST-arfaptin1-S100A. The cells were then visualized by fluorescence microscopy using an anti-GST antibody. Expression of PKD2-CA induced a partial translocation of GST-arfaptin1-WT from the TGN to the cytoplasm, whereas GST-arfaptin1-S100A (the non-phosphorylatable form) remained at the TGN under these conditions (Figure 5B). Expression of constitutively activated PKD2 also decreased the Golgi localization of endogenous arfaptin1, but not of endogenous arfaptin2, when analysed by immunofluorescence (Figure 5C). Quantification of this effect showed that the amount of arfaptin1 at the Golgi complex is reduced by 75% in cells expressing GFP-PKD2-CA compared with cells expressing only GFP or a kinase-dead form of PKD2 (GFP-PKD2-KD) (Figure 5D). On the other hand, the levels of arfaptin2 at the Golgi complex were unaffected under such experimental conditions (Figures 5C and D). Taken together, these results indicate that PKD-mediated phosphorylation of Ser100 dissociates arfaptin1 from the TGN.
Figure 5.
Ser100 phosphorylation inhibits the localization of arfaptin1 at the TGN. (A) HeLa cells were transfected with vectors coding for GST-arfaptin1-WT, -S100A or -S100E. After 24 h, cells were fixed and visualized with anti-GST and anti-arfaptin2 antibodies, respectively. (B) HeLa cells co-expressing GFP-PKD2-CA and either GST-arfaptin1-WT or -S100A were visualized with an anti-GST antibody by fluorescence microscopy. (C) HeLa cells were transfected with constructs encoding GFP, GFP-PKD2-KD or GFP-PKD2-CA. After 24 h, cells were fixed and visualized by fluorescence microscopy with anti-p230 antibody in combination with either arfaptin1- (left panel) or arfaptin2- (right panel) specific antibodies. Scale bars, 10 μm. (D) Quantification of the immunofluorescence signal for arfaptin1 or 2 at the Golgi area in (C). Results are shown as the mean±s.e.m. of two experiments. (E) Liposome flotation assays with recombinant arfaptin1-WT and -S100E, and PC/PE/Ch liposomes containing or lacking 10 mol% of either PS or PI(4)P. T, top fraction; B, bottom fraction.
Source data for this figure is available on the online supplementary information page.
Ser100 in arfaptin1 is contained in the AH that is required for the anchoring to PI(4)P-containing membrane and TGN localization; we hypothesized that phosphorylation of Ser100 could control the ability of the AH to be assembled and, therefore, the binding of arfaptin1 to PI(4)P. To test this, the wild-type and the phosphomimetic forms of arfaptin1 were incubated with liposomes containing or lacking PS or PI(4)P, the liposomes were isolated and analysed by SDS–PAGE. As shown in Figure 5E, the binding of arfaptin1-S100E to PS- or PI(4)P-containing liposomes was significantly reduced compared with arfaptin1-WT. Taken together, these results reveal that PKD phosphorylates Ser100 of arfaptin1 inhibiting its binding to PI(4)P-containing membranes, likely by interfering with the assembly of the AH.
Arfaptin1 is required for the export of regulated secretory cargo from the TGN
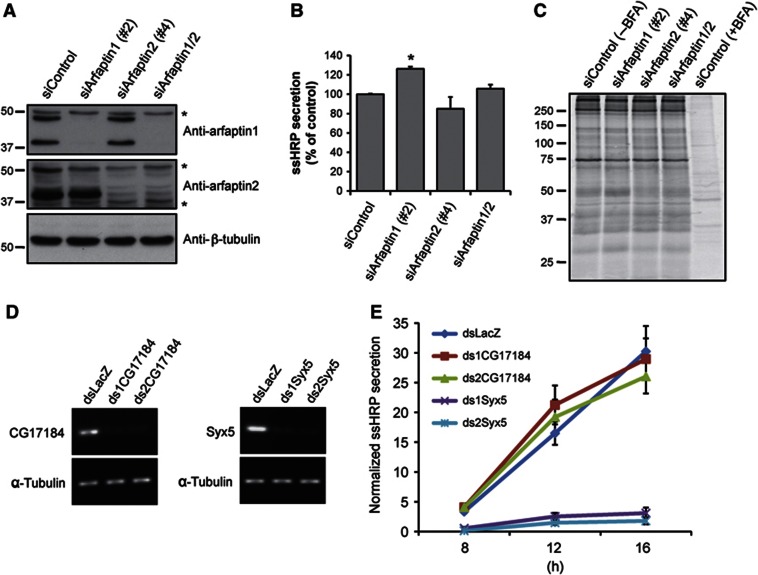
At the TGN, PKD is required for the trafficking of cargoes that contain sorting information for trafficking to the basolateral surface (Yeaman et al, 2004). PKD is also required for the biogenesis of CARTS and secretory storage granules (von Wichert et al, 2008; Wakana et al, 2012). Because PKD phosphorylates and dissociates arfaptin1, we tested the requirement of arfaptins in the PKD-dependent export routes from the TGN. HeLa cells express two splicing variants of arfaptin1 with apparent molecular weights of 38 and 43 kDa (Figure 6A; Supplementary Figure S4). Although the data presented thus far only describe the binding of the shorter version, the longer isoform also contains the same PI(4)P-binding site, it can be phosphorylated in vitro by PKD and its phosphomimetic mutant displays a cytoplasmic localization (Supplementary Figure S4). To test the functional significance of arfaptin1 and arfaptin2 in protein secretion, both proteins were individually or simultaneously knocked down in HeLa cells by RNA interference. As shown in Figure 6A, transfection with specific siRNA oligonucleotides reduced the levels of both arfaptin1 isoforms and the levels of arfaptin2 to >85% compared with their levels in HeLa cells transfected with a control siRNA. Co-transfection with both siRNA for arfaptin1 and 2 induced similar knockdown levels as observed with the single transfection (Figure 6A). HeLa cells stably expressing horseradish peroxidase containing a signal sequence (HeLa-ssHRP) transfected with arfaptin1 and/or arfaptin2 siRNA oligonucleotides were then used to monitor effects on the secretion of ssHRP as described previously (Bossard et al, 2007; von Blume et al, 2009). Individual knockdown of arfaptin1, arfaptin2 or double knockdown did not strongly affect ssHRP secretion, although arfaptin1 knockdown induced a slight increase in the secretion of this protein (Figure 6B). We also found that the individual knockdown of arfaptin1 and 2 did not affect the secretion of PAUF, an endogenous secretory cargo that is transported by specific vesicles called CARTS (Supplementary Figure S5). To further ascertain the role of arfaptins in general protein secretion, we performed the following experiment. HeLa cells were transfected with arfaptin1 and/or arfaptin2 siRNA oligonucleotides for 72 h. The cells were then labelled with 35S-methionine for 15 min and chased for 2.5 h in medium containing unlabelled methionine. Medium from the cells was collected to precipitate secreted proteins and analysed by SDS–PAGE and autoradiography. Knockdown of arfaptins had no effect on the secretion of newly synthesized proteins (Figure 6C). Treatment with BFA, as expected, severely inhibited protein secretion under these experimental conditions (Figure 6C). To further address this issue, we tested the role of arfaptins in the secretion of ssHRP in Drosophila S2 cells. The Drosophila melanogaster genome encodes a single arfaptin-like protein, which has the predicted AH found in human arfaptins and the serine that it is phosphorylated by PKD in arfaptin1. Flag-tagged Drosophila arfaptin was expressed in S2 cells stably expressing mannosidase II-GFP and its intracellular distribution was examined by immunofluorescence microscopy. Drosophila arfaptin was localized in close proximity to the mannosidase II-containing Golgi membrane in S2 cells (Supplementary Figure S6). S2 cells stably transfected with a vector encoding ssHRP under the control of a Cu2+-inducible promoter were incubated for 5 days with specific dsRNA for Drosophila arfaptin, syntaxin5, or LacZ as a negative control. The knockdown efficiency was monitored by RT–PCR (Figure 6D). The same knockdown procedure was repeated and after 5 days the cells were incubated with Cu2+ to promote the synthesis of ssHRP. The medium and the cell lysates were tested for HRP activity by chemiluminescence. Knockdown of syntaxin5, which is required for ER to Golgi transport significantly inhibited ssHRP secretion, whereas knockdown of Drosophila arfaptin did not affect the secretion of this cargo (Figure 6E). Taken together, our results indicate that arfaptins are not required for constitutive protein secretion.
Figure 6.
Arfaptins are not required for constitutive protein secretion. (A) HeLa cells stably expressing ssHRP (HeLa-ssHRP) were transfected with non-targeting siRNA (siControl), arfaptin1 siRNA (siArfaptin1 #2), arfaptin2 siRNA (siArfaptin2 #4), or both arfaptin1 and 2 siRNAs (siArfaptin1/2). After 72 h, cells were lysed and the lysates were analysed by western blotting with anti-arfaptin1, anti-arfaptin2 and anti-β-tubulin antibodies, respectively. Asterisks indicate non-specific bands recognized by the anti-arfaptin antibodies. (B) HeLa-ssHRP cells transfected as in A were incubated with fresh medium for 4 h and the HRP activity monitored in the media and the cell lysates by chemiluminescence. Results are shown as the mean±s.e.m. of three experiments. The data were analysed using the one-way ANOVA with Dunnett’s multiple comparison post-testing (*, P<0.05 versus siControl-transfected cells). (C) Normal HeLa cells transfected with non-targeting siRNA (siControl), arfaptin1 siRNA (siArfaptin1 #2), arfaptin2 siRNA (siArfaptin2 #4), or both arfaptin1 and 2 siRNAs (siArfaptin1/2) were pulsed with 35S-methionine for 15 min and chased for 2.5 h in medium containing unlabelled methionine. The medium was collected and analysed by SDS–PAGE and autoradiography. Brefeldin A (BFA) was added at 5 μg/ml to one well of the siControl-transfected cells for the last 15 min of incubation with [35S]-methionine and kept throughout the chase. (D) Drosophila S2 cells stably transfected with the pMT-ssHRP-V5 vector were treated with dsRNA oligonucleotides for LacZ (dsLacZ), Drosophila arfaptin (ds1CG17184 and ds2CG17184), or Drosophila syntaxin5 (ds1Syx5 and ds2Syx5). After 5 days, total RNA was extracted and the knockdown efficiencies were monitored by RT–PCR. (E) Drosophila S2 cells stably transfected with the pMT-ssHRP-V5 vector were transfected with dsRNAs as in (D). After 5 days, the cells were incubated with fresh medium containing 0.5 mM CuSO4 for the indicated times and the HRP activity monitored in the media and the cell lysates by chemiluminescence. Results are shown as the mean±s.e.m. of three experiments.
Source data for this figure is available on the online supplementary information page.
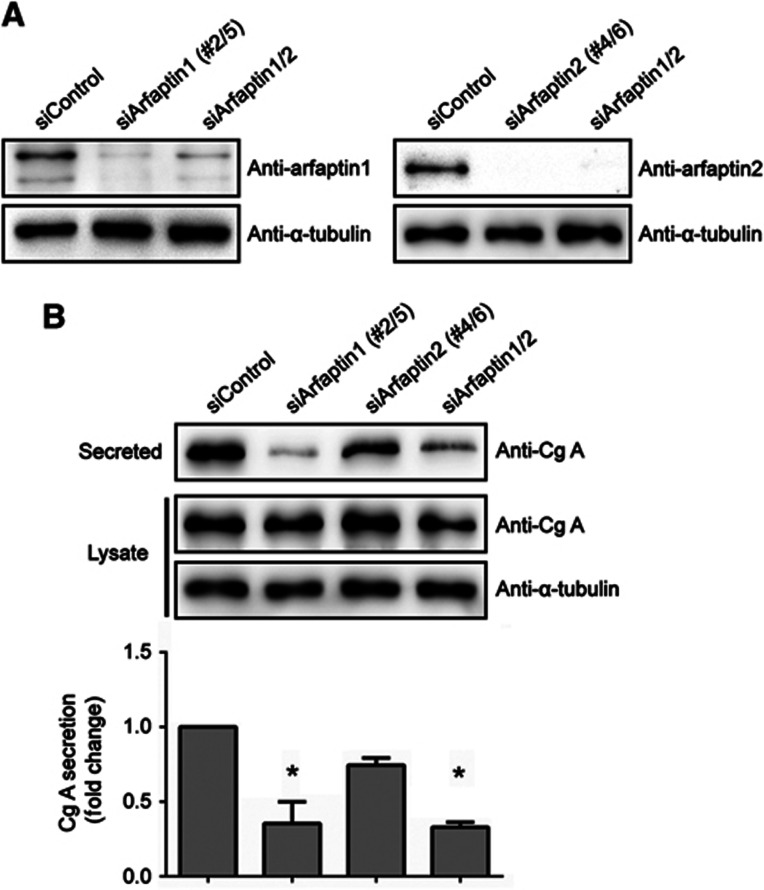
It has recently been reported that arfaptin1 is required for the formation of insulin-containing secretory granules from the TGN in rat INS1 cells (Gehart et al, 2012). Human BON cells express and secrete Cg A via the regulated secretory pathway in a PKD-dependent manner (von Wichert et al, 2008) and were used to test the requirement of arfaptins in this process. BON cells were transfected with arfaptin1 and/or arfaptin2 siRNA oligonucleotides and, after 48 h, the knockdown efficiencies were monitored by western blotting (Figure 7A). Under these experimental conditions, the amount of secreted Cg A during a 2-h incubation period was measured by western blotting with an anti-Cg A antibody. This analysis revealed that single knockdown of arfaptin1 and 2 inhibited Cg A secretion by 65 and 25%, respectively, compared with control cells (Figure 7B). Simultaneous knockdown of both arfaptins showed a 65% reduction in the secretion of Cg A (Figure 7B). Unfortunately, we have not been able to express siRNA-resistant forms of arfaptin1 in BON cells, and therefore we could not rescue the defect in secretion of Cg A observed upon knockdown of arfaptin1. However, our findings nicely complement the observations of Ricci and colleagues on the involvement of arfaptin1 in the regulated trafficking of insulin in rat INS1 cells (Gehart et al, 2012) and together these findings suggest that arfaptin1 is required for the secretion of cargoes exported from the TGN by the regulated secretory pathway, whereas arfaptin2 does not have a major role in this process.
Figure 7.
Involvement of arfaptins in the secretion of Cg A in BON cells. (A) BON cells were transfected with non-targeting siRNA (siControl), or co-transfected with either two siRNAs for arfaptin1 (siArfaptin1 #2/5), two siRNAs for arfaptin2 (siArfaptin2 #4/6), or the four siRNAs for arfaptin1 and 2 (siArfaptin1/2). After 48 h, cells were lysed and the lysates were analysed by western blotting with anti-arfaptin1, anti-arfaptin2 and anti-α-tubulin antibodies. (B) Non-targeting, arfaptin1, arfaptin2, and arfaptin1 and 2 siRNAs were transfected into BON cells as indicated. Forty-eight hours after transfection, cells were incubated with serum-free medium for 2 h and the Cg A levels in the conditioned medium and in the cell lysates were measured by western blotting. Secreted Cg A levels were normalized to Cg A levels in the lysates. Results are shown as the mean±s.e.m. of three experiments. The data were analysed using the one-way ANOVA with Dunnett’s multiple comparison post-testing (*, P<0.001 versus siControl-transfected cells).
Source data for this figure is available on the online supplementary information page.
Discussion
Our findings provide insights into the mechanism by which the BAR domain-containing proteins arfaptins are recruited to the TGN and their requirement in the biogenesis of secretory storage granules.
PI(4)P mediated recruitment of arfaptins and their function at the TGN
The BAR domain-containing proteins employ the positively charged amino-acid residues at the concave face of the BAR domain to bind the negatively charged lipids in the membranes. The specificity of the binding to a specific intracellular membrane is provided by a phosphoinositide-binding PH or PX domain contained in some BAR domain-containing proteins (Peter et al, 2004; Qualmann et al, 2011; Rao and Haucke, 2011). Our results reveal that the phosphoinositide PI(4)P is required for the localization and recruitment of arfaptins to the TGN. Arfaptins lack a canonical PH domain but contain a highly conserved region preceding the BAR domain that is predicted to form an AH. The AH-containing region is necessary for the localization of arfaptins at the TGN. Moreover, a conserved tryptophan at position 99 within this region is required for the binding of arfaptins to PI(4)P-containing membranes. We suggest that the interaction of the relatively high number of basic residues in the polar face of the predicted AH with the negatively charged phospholipids promotes the binding of arfaptins to the PI(4)P-enriched TGN membranes. A similar mode of recruitment has been proposed for the anchoring of the AH of the BAR domain-containing proteins endophilin and amphiphysin to the cytoplasmic face of the plasma membrane (Drin and Antonny, 2010).
Phosphorylation at Ser100, which is predicted to be localized at the hydrophobic-polar interface of the AH, regulates the binding of arfaptin1 to PI(4)P-containing membranes. The phosphorylation of Ser100 could block the assembly of the AH, by electrostatic repulsion with the negative charges of the anionic phospholipids, and thus inhibit the binding of arfaptin1 to the TGN. Ricci and colleagues have also reported recently that PKD1 phosphorylates arfaptin1 at Ser100 to control the attachment of arfaptin1 with the TGN (Gehart et al, 2012). Together, these findings provide strong evidence for the involvement of PKD in regulating the recruitment of arfaptin1 to the TGN.
The arfaptin2 BAR domain dimer bound to Arl1 has recently been crystallized (Nakamura et al, 2012). Mutation of a single phenylalanine (F285A) inhibits the binding of arfaptin2 to Arl1 and the localization of full-length arfaptin2 to the TGN in HeLa cells (Nakamura et al, 2012). However, as shown here, the BAR domain alone of arfaptin1, which is similar to the BAR domain of arfaptin2, and contains the same Phe285, does not localize at the TGN in intact cells. Whereas, the BAR domain of arfaptin1 appended to the AH-containing fragment (Δ1–85 mutant) localizes to the TGN in HeLa cells. Based on the cumulative data, we suggest that the binding of arfaptins to the TGN requires both the AH, which binds PI(4)P, and the BAR domain, which binds the small GTPase. Arfaptins are therefore similar to proteins such as AP1, the GGAs, and the FAPPs, which require both PI(4)P and Arf1 for their recruitment to the TGN (Wang et al, 2003, 2007; Godi et al, 2004).
With respect to the role of the arfaptins at the Golgi complex, our findings indicate that only arfaptin1 is required for the trafficking of Cg A by the regulated secretory pathway. We have not observed a role for arfaptins in the constitutive transport along the secretory pathway. Ricci and colleagues have reported that siRNA-dependent knockdown of arfaptin1 or overexpression of the wild-type or the non-phosphorylatable forms of arfaptin1 reduced glucose-stimulated insulin secretion without affecting the constitutive secretion (Gehart et al, 2012). Together, these findings introduce the role of BAR domain-containing proteins in the production of secretory granules at the TGN.
The role of PKD and its interacting partners in vesicle biogenesis at the TGN
At the TGN, PKD is required for the biogenesis of vesicles that contain cargo destined for the basolateral surface (Yeaman et al, 2004; Campelo and Malhotra, 2012). PKD is also required for the regulated secretion of Cg A and for the biogenesis of a new class of carriers called CARTS (von Wichert et al, 2008; Wakana et al, 2012). PKD is reported to increase the PI(4)P levels at the TGN by activating PI(4)KIIIβ and to regulate the attachment of CERT and OSBP (Hausser et al, 2005; Fugmann et al, 2007; Nhek et al, 2010). CERT controls the delivery of ceramide to the TGN and we have recently shown that perturbing the levels of the ceramide product sphingomyelin (SM)—generated by the action of SM synthase—inhibits the biogenesis of transport carriers at the Golgi complex (Duran et al, 2012). SM assembles with Ch into liquid-ordered domains and therefore the levels of these two lipids have to be tightly regulated to generate transport carriers. The regulation of CERT and OSBP attachment to the TGN via PKD is therefore an important reaction in the overall scheme of transport carrier biogenesis. Our new data reveal the significance of PKD in the location of arfaptin1 at the TGN. Perturbing levels of SM at the TGN affect the biogenesis of transport carriers destined to all export routes from the Golgi membranes, whereas arfaptin1 is required for the biogenesis of only the secretory storage granules. We suggest that PKD-dependent dissociation of arfaptin1 from the TGN is the means to regulate the biogenesis of secretory granules and thus the amount of secretory cargo transported by this pathway without affecting the secretion of cargo exported by the constitutive pathway and CARTS. How the TGN is compartmentalized to generate these different carriers that all require PKD but have different requirements for the BAR domain-containing proteins (arfaptins) is an interesting new challenge.
Materials and methods
Antibodies and reagents
Polyclonal antibodies used in this study were as follows: goat anti-arfaptin1 and rabbit anti-GFP (Santa Cruz Biotechnology, Inc.), rabbit anti-arfaptin2 (Invitrogen), goat anti-GST (GE Healthcare) and rabbit anti-β-COP (a gift of M Tagaya, Tokyo University of Pharmacy and Life Sciences, Tokyo, Japan). Mouse monoclonal antibodies were against arfaptin2 (Abnova), p230 (BD Biosciences), α-tubulin, β-tubulin, and FLAG (Sigma-Aldrich), His (QIAGEN), and Cg A (Dako). PAO, rapamycin and BFA were obtained from Sigma-Aldrich, Calbiochem and Epicenter Technologies, respectively. [γ-32P]ATP and [35S]-methionine label were purchased from Hartmann Analytic. PC, PE, PS and Ch were purchased from Sigma-Aldrich and PI(4)P from Avanti Polar Lipids.
Plasmid expression vectors
The coding sequences of the 341-aa form of human arfaptin1 (GenBank accession No. NM_001025593.1) and the 341-aa form of human arfaptin2 (NM_012402.3) were amplified by RT–PCR from HeLa cells and cloned into the pGEX-6P1 plasmid (GE Healthcare) or the peGFP-C1 plasmid (BD Clontech) generating vectors coding for GST-tagged or GFP-tagged arfaptin1 and arfaptin2, respectively. The coding sequence of human arfaptin1 was inserted into the pMePy-GST plasmid (Maeda et al, 2001) creating a vector encoding arfaptin1 with an N-terminal GST tag for expression in mammalian cells. Mutagenesis resulting in single aa substitution was performed using the QuikChange II site-directed mutagenesis system (Agilent). For deletion mutants, the 277–657, 628–1302 and 532–1302 nucleotides of human arfaptin1 cDNA (NM_001025593.1) were generated by PCR for N-term, BAR and Δ1–85, respectively, and cloned into the peGFP-C1 plasmid. SnapGene software (from GSL Biotech; available at snapgene.com) was used for molecular cloning procedures. All constructs were verified by sequencing. The plasmid encoding the single D. melanogaster arfaptin (CG17184) fused to a C-terminal Flag-HA tag and driven by a metallothionein promoter was purchased from the Drosophila Genomics Resource Center (clone ID FMO02655). The vectors coding for mRFP-FKBP12-Sac1, TGN38-FRB-CFP and GFP-OSH1-PH have been described earlier (Szentpetery et al, 2010). The plasmid coding for GFP-CERT was provided by M Olayioye (University of Stuttgart, Stuttgart, Germany). peGFP-PKD2-D695A, peGFP-PKD2-S706/710E and pGMEX-PKD2-S706/710E plasmids were described previously (Auer et al, 2005; von Blume et al, 2007).
Recombinant proteins
For the purification of GST-arfaptin proteins, pGEX-6P1-arfaptin plasmids were transformed into BL21(DE3) cells. Expression of the GST fusion proteins was induced with 0.2 mM IPTG for 4 h at 25°C. Bacteria were harvested, resuspended in phosphate-buffered saline (PBS) containing 0.2% (v/v) Triton X-100 and protease inhibitors, and lysed by French press. Lysates were incubated overnight with glutathione-Sepharose beads (GE Healthcare) at 4°C. Beads were then washed three times with PBS and the GST fusion proteins were eluted with 20 mM glutathione. Recombinant arfaptins without the GST tag were obtained by treating GST-arfaptin-immobilized glutathione-Sepharose beads with PreScission Protease (GE Healthcare) following manufacturer’s instructions. For expression and purification of a constitutively active form of GST-PKD2, HEK-293 cells were transfected with the pGMEX-PKD2-S706/710E vector. Seventy-two hours after transfection, cells were harvested in lysis buffer (50 mM Tris–HCl, pH 7.4, 1 mM EGTA, 1 mM EDTA, 1 mM sodium orthovanadate, 25 mM NaF, 0.27 M sucrose, 1% (v/v) Triton X-100, 15 mM 2-mercaptoethanol and protease inhibitors), incubated for 20 min at 4°C and centrifuged at 16 000 g for 15 min. Supernatants were incubated overnight with glutathione-Sepharose beads at 4°C. Beads were then washed once with lysis buffer containing 0.5 M NaCl and once with pre-elution buffer (250 mM Tris–HCl, pH 8.5, 0.1 mM EGTA, 10% (v/v) glycerol, 0.27 M sucrose, 0.2 M NaCl, 0.1% (v/v) Triton X-100 and 15 mM 2-mercaptoethanol). The GST fusion protein was eluted by incubating the beads with 20 mM glutathione in pre-elution buffer. Recombinant proteins were dialysed against either kinase assay or liposome buffer and concentrated by centrifugation using Amicon Ultra filters (Millipore).
Cell culture, and plasmid and siRNA transfection
Drosophila S2 cells stably transfected with the plasmids pAc5-MannII-GFP or pMT-ssHRP-V5 (Bard et al, 2006) were cultured in Schneider’s medium (Invitrogen) supplemented with 10% (v/v) fetal calf serum (FCS) at 20°C. Normal HeLa cells, HeLa cells stably expressing ssHRP (a gift of Dr I Stamenkovic, University of Lausanne, Switzerland), COS-7 cells, and HEK-293 cells were grown in DMEM containing 10% (v/v) FCS at 37°C in a 7% CO2 incubator. Human BON carcinoid tumour cells were maintained in DMEM supplemented with 10% (v/v) FCS in a humidified atmosphere of 5% CO2: 95% air at 37°C. HeLa cells, COS-7 cells and HEK-293 cells were transfected with plasmids using FuGENE6 (Roche), Lipofectamine 2000 (Invitrogen), and TransIT-293 transfection reagent (Mirus), respectively. In the case of siRNA oligonucleotides, normal HeLa cells and HeLa cells stably expressing ssHRP were transfected with HiPerfect (QIAGEN) and Lipofectamine 2000, respectively, and analysed after 72 h. BON cells were transfected with TransMessenger transfection reagent (QIAGEN) and analysed 48 h after siRNA transfection. The siRNA oligonucleotides used were non-targeting siControl (5′-AAUUGCGUAGUCUAAGUUAAAGUGG-3′), siArfaptin1 (#2, 5′-GUGUACUCGACAGAUUAUC-3′; #5, 5′-CAAUGCCAUUGCCGCUUAC-3′) and siArfaptin2 (#4, 5′-GCUCAAGUUCCUGGAAGAA-3′; #6, 5′-GACACGCUCAUGACUGUGA-3′).
Protein-lipid overlay assay
Protein-lipid overlay assays were performed with recombinant non-tagged arfaptin1 and arfaptin2 and PIP strips (Echelon) following manufacturer’s instructions. Bound proteins were detected by incubating the strips with goat anti-arfaptin1 or mouse anti-arfaptin2 antibodies followed by horseradish peroxidase-conjugated secondary antibody.
Liposome flotation assay
A lipid film containing either PC:PE:Ch (45:45:10 mol%) or PC:PE:Ch:other (40:40:10:10 mol%, where other is PS or PI(4)P) was obtained by evaporation of the solvent under nitrogen and resuspended in liposome buffer (20 mM HEPES, pH 7.4, 150 mM NaCl, 1.5 mM DTT) at a final concentration of 1.47 mM lipids. Liposomes were generated by sonication in a water-bath sonicator until a homogeneous suspension was formed (20 min). For liposome binding assays, 3 μM of proteins was incubated with 1 mM of liposomes in 85 μl of liposome buffer for 30 min at room temperature. In all, 62% sucrose in liposome buffer was mixed with the sample to adjust the sucrose concentration to 30%. The sample was overlaid with 300 μl of 25% sucrose in liposome buffer and subsequently with 50 μl of liposome buffer. The sample was centrifuged at 240 000 g for 60 min in a Beckman TLS-55 rotor. The top (140 μl) and the bottom (375 μl) fractions were collected and the proteins were precipitated by chloroform/methanol. Proteins were analysed by SDS–PAGE, visualized by Coomassie stain, and quantified using ImageJ 1.45r software.
In vitro kinase assay
In all, 20 μl reactions containing 6 μg of GST-arfaptin and 1 μg of GST-PKD2-S706/710E were incubated for 10 min at 30°C in kinase assay buffer (50 mM Tris–HCl, pH 7.4, 10 mM MgCl2) containing 0.2 mM ATP, 1 mM DTT and 5 μCi [γ-32P]ATP. Reactions were stopped by addition of SDS sample buffer and the samples were processed by SDS–PAGE and autoradiography.
Immunofluorescence and confocal microscopy
For immunofluorescence, cells were fixed with 3.7% (v/v) formaldehyde at room temperature for 15 min and then incubated with indicated primary, and secondary antibodies conjugated to Alexa Fluor 488, 594 and/or 647 (Invitrogen). The samples were visualized with a Leica SPE confocal microscope with the 63 × ACS Apo NA 1.3 objective and LAS-AF software. Image processing and analysis were performed with ImageJ 1.45r software. The immunofluorescence signal intensity in the Golgi area for arfaptin1 or arfaptin2 was measured as follows: the boundaries of each analysed cell were manually traced and converted into a region of interest (ROI cell); using this ROI, a binary threshold (60–255 grey levels) was applied to the z projection image of the anti-p230 signal channel creating an ROI for the Golgi area (ROI Golgi). A third ROI was created by substracting the ROI Golgi to the ROI cell (ROI cytoplasm). The mean grey values were measured in the z projection images of the anti-arfaptin1 or anti-arfaptin2 signal channels using ROI Golgi and ROI cytoplasm. Finally, the value for the cytoplasm was substracted to the value obtained for the ROI Golgi. For the experiments using the rapamycin-induced recruitment of Sac1, COS-7 cells were grown on 25-mm glass coverslips. Sixteen hours after plasmid transfection, COS-7 cells were washed three times with a modified Krebs-Ringer solution (120 mM NaCl, 4.7 mM KCl, 1.2 mM CaCl2, 0.7 mM MgSO4, 10 mM glucose, 10 mM HEPES, pH 7.4) and the coverslip was mounted onto live-cell microscopy chambers. All live-cell experiments were performed on a heated stage with medium and the objective temperature kept at 35°C, using an inverted Zeiss LSM-510 scanning-laser confocal microscope (Carl Zeiss) and an objective heater (Bioptechs).
Western blotting
Protein extracts from cultured cells were prepared by lysis in 1 × RIPA buffer (50 mM mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% (v/v) Triton X-100, 0.1% (w/v) SDS, 1% (w/v) sodium deoxycholate) containing protease inhibitors and dissolved in reducing SDS sample buffer. Proteins were separated by SDS–PAGE and transferred onto PVDF membranes. Membranes were incubated in 5% (w/v) bovine serum albumin, 0.1% (v/v) Tween-20, 150 mM NaCl, and 25 mM Tris (pH 7.4) for 1 h and then with the respective primary antibody diluted in blocking buffer overnight at 4°C. Blots were washed and incubated for 1 h at room temperature with the appropriate horseradish peroxidase-conjugated secondary antibody diluted in blocking buffer. Membranes were washed and polypeptides detected by chemiluminescence. Signal intensity of immunoreactive bands was evaluated with ImageJ 1.45r software.
ssHRP secretion assay
HeLa cells stably expressing ssHRP were transfected with siRNA oligonucleotides. After 72 h, cells were incubated with fresh medium for 4 h. In all, 50 μl of medium was mixed with ECL reagent (Thermo Fisher Scientific) and luminescence was measured with a Victor 3 plate reader (Perkin-Elmer, Waltham, MA). For normalization, cells were lysed with 1 × RIPA buffer, and internal HRP activity was measured.
PAUF secretion assay
Analysis of PAUF secretion was performed as previously described (Wakana et al, 2012). Briefly, HeLa cells were transfected with siRNA oligonucleotides and, after 48 h, cells were transfected with a plasmid coding for PAUF-MycHis. Twenty-four hours later, the medium was replaced with Opti-MEM, the cells were incubated at 20°C for 2 h, and then shifted to 32°C for 1 h to restore protein secretion from the Golgi. Secreted proteins in the medium were precipitated with trichloroacetic acid and resuspended in reducing SDS sample buffer. The levels of PAUF-MycHis in the precipitates and in the cell lysates were measured by western blotting with anti-His antibody.
Metabolic labelling
Normal HeLa cells were transfected with siRNA oligonucleotides. After 72 h, cells were cultured in DMEM without L-methionine and L-cysteine for 1 h. Starting from the last 15 min of starvation until the end of the experiment, the cells were incubated in the absence or presence of 5 μg/ml BFA. The cells were pulsed for 15 min with 200 μCi [35S]-methionine label and then chased for 2.5 h with medium supplemented with 10mM L-methionine. Secreted proteins were collected by precipitation by trichloroacetic acid and analysed by SDS–PAGE and autoradiography. Cells were lysed in 1 × RIPA buffer for 10 min on ice, and [35S] incorporation was determined for normalization.
dsRNA transfection and ssHRP secretion assay in S2 cells
DsRNA oligonucleotides for Drosophila arfaptin (ds1CG17184 and ds2CG17184) and syntaxin5 (ds1Syx5 and ds2Syx5) were purchased from the Drosophila RNAi Screening Center. For transfection of dsRNA, S2 cells stably transfected with the Cu2+-inducible ssHRP coding vector were plated in a 96-well plate with 1 μg of dsRNA in 60 μl of medium without FCS. Serum was added to the cells after 2 h of incubation and, after 5 days, ssHRP expression was induced by incubating the cells with 100 μl of medium containing 0.5 mM CuSO4. After 8, 12 and 16 h, the medium was collected and the cells were lysed with 100 μl of 1 × RIPA containing protease inhibitors and centrifuged at 20 000 g for 15 min. hRP activity was detected at each time point by measuring chemiluminescence in 10 μl of medium as described above, and normalized to the activity in the cell lysate.
Measurement of Cg A secretion
BON cells were incubated 72 h after plating in phenol-free DMEM without FCS for 2 h at 37°C. The medium was collected and the Cg A levels in the conditioned medium and in the cell lysates were determined by western blotting using a specific Cg A antibody. The amount of secreted Cg A was normalized to the intracellular levels of Cg A.
RT–PCR
RNA from dsRNA-transfected S2 cells was extracted using the RNAeasy mini kit (QIAGEN) and cDNAs were synthesized with Cloned AMV First-Strand cDNA Synthesis Kit (Invitrogen) according to manufacturer’s protocol. Drosophila arfaptin (CG17184), syntaxin5 and α-tubulin were amplified with specific primers.
Supplementary Material
Acknowledgments
We thank members of the Malhotra Lab for helpful discussion. We also thank M Tagaya and M Olayioye for kindly providing reagents used in this study. We especially thank Dr Romeo Ricci for disclosing their findings on the PKD-dependent phosphorylation of arfaptin1 prior to publication. V Malhotra is an Institució Catalana de Recerca i Estudis Avançats (ICREA) professor at the Center for Genomic Regulation, and the work in his laboratory is funded by grants from Plan Nacional (BFU2008-00414), Consolider (CSD2009-00016), Agència de Gestió d’Ajuts Universitaris i de Recerca (AGAUR) Grups de Recerca Emergents (SGR2009-1488; AGAUR-Catalan Government), and European Research Council (268692). The research of M Jovic and T Balla was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health. J Villeneuve is funded by a long-term EMBO postdoctoral fellowship.
Author contributions: DCG and VM designed the experiments and analysed the data; DCG, MOB and MS performed most of the experiments; MS produced the recombinant arfaptin proteins; JV performed the experiments with S2 cells; MJ and TB designed, carried out and analysed the experiments with the recruitable Sac1; MP and TS performed the experiments and analysed the data on Cg A secretion; VM wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Auer A, von Blume J, Sturany S, von Wichert G, Van Lint J, Vandenheede J, Adler G, Seufferlein T (2005) Role of the regulatory domain of protein kinase D2 in phorbol ester binding, catalytic activity, and nucleocytoplasmic shuttling. Mol Biol Cell 16: 4375–4385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis VA, Garcia-Mata R, Mousley CJ (2012) Golgi membrane dynamics and lipid metabolism. Curr Biol 22: R414–R424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard F, Casano L, Mallabiabarrena A, Wallace E, Saito K, Kitayama H, Guizzunti G, Hu Y, Wendler F, Dasgupta R, Perrimon N, Malhotra V (2006) Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature 439: 604–607 [DOI] [PubMed] [Google Scholar]
- Baron CL, Malhotra V (2002) Role of diacylglycerol in PKD recruitment to the TGN and protein transport to the plasma membrane. Science 295: 325–328 [DOI] [PubMed] [Google Scholar]
- Bossard C, Bresson D, Polishchuk RS, Malhotra V (2007) Dimeric PKD regulates membrane fission to form transport carriers at the TGN. J Cell Biol 179: 1123–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campelo F, Malhotra V (2012) Membrane fission: the biogenesis of transport carriers. Annu Rev Biochem 81: 407–427 [DOI] [PubMed] [Google Scholar]
- Christis C, Munro S (2012) The small G protein Arl1 directs the trans-Golgi-specific targeting of the Arf1 exchange factors BIG1 and BIG2. J Cell Biol 196: 327–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Matteis MA, Luini A (2008) Exiting the Golgi complex. Nat Rev Mol Cell Biol 9: 273–284 [DOI] [PubMed] [Google Scholar]
- Diaz Anel AM, Malhotra V (2005) PKCeta is required for beta1gamma2/beta3gamma2- and PKD-mediated transport to the cell surface and the organization of the Golgi apparatus. J Cell Biol 169: 83–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G, Antonny B (2010) Amphipathic helices and membrane curvature. FEBS Lett 584: 1840–1847 [DOI] [PubMed] [Google Scholar]
- Duran JM, Campelo F, van Galen J, Sachsenheimer T, Sot J, Egorov MV, Rentero C, Enrich C, Polishchuk RS, Goni FM, Brugger B, Wieland F, Malhotra V (2012) Sphingomyelin organization is required for vesicle biogenesis at the Golgi complex. EMBO J 31: 4535–4546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emr S, Glick BS, Linstedt AD, Lippincott-Schwartz J, Luini A, Malhotra V, Marsh BJ, Nakano A, Pfeffer SR, Rabouille C, Rothman JE, Warren G, Wieland FT (2009) Journeys through the Golgi--taking stock in a new era. J Cell Biol 187: 449–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairn GD, Schieber NL, Ariotti N, Murphy S, Kuerschner L, Webb RI, Grinstein S, Parton RG (2011) High-resolution mapping reveals topologically distinct cellular pools of phosphatidylserine. J Cell Biol 194: 257–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugmann T, Hausser A, Schoffler P, Schmid S, Pfizenmaier K, Olayioye MA (2007) Regulation of secretory transport by protein kinase D-mediated phosphorylation of the ceramide transfer protein. J Cell Biol 178: 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier R, Douguet D, Antonny B, Drin G (2008) HELIQUEST: a web server to screen sequences with specific alpha-helical properties. Bioinformatics 24: 2101–2102 [DOI] [PubMed] [Google Scholar]
- Gehart H, Goginashvili A, Beck R, Morvan J, Erbs E, Formentini I, De Matteis MA, Schwab Y, Wieland FT, Ricci R (2012) The BAR domain protein arfaptin-1 controls secretory granule biogenesis at the trans-Golgi network. Dev Cell 23: 756–768 [DOI] [PubMed] [Google Scholar]
- Godi A, Di Campli A, Konstantakopoulos A, Di Tullio G, Alessi DR, Kular GS, Daniele T, Marra P, Lucocq JM, De Matteis MA (2004) FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat Cell Biol 6: 393–404 [DOI] [PubMed] [Google Scholar]
- Godi A, Pertile P, Meyers R, Marra P, Di Tullio G, Iurisci C, Luini A, Corda D, De Matteis MA (1999) ARF mediates recruitment of PtdIns-4-OH kinase-beta and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat Cell Biol 1: 280–287 [DOI] [PubMed] [Google Scholar]
- Graham TR, Burd CG (2011) Coordination of Golgi functions by phosphatidylinositol 4-kinases. Trends Cell Biol 21: 113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger DL, Tavelis C, Ryan AJ, Hinchliffe KA (2012) The emerging role of PtdIns5P: another signalling phosphoinositide takes its place. Biochem Soc Trans 40: 257–261 [DOI] [PubMed] [Google Scholar]
- Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M (2003) Molecular machinery for non-vesicular trafficking of ceramide. Nature 426: 803–809 [DOI] [PubMed] [Google Scholar]
- Hausser A, Storz P, Martens S, Link G, Toker A, Pfizenmaier K (2005) Protein kinase D regulates vesicular transport by phosphorylating and activating phosphatidylinositol-4 kinase IIIbeta at the Golgi complex. Nat Cell Biol 7: 880–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh H, Williger BT, Exton JH (1997) Arfaptin 1, a putative cytosolic target protein of ADP-ribosylation factor, is recruited to Golgi membranes. J Biol Chem 272: 5421–5429 [DOI] [PubMed] [Google Scholar]
- Levine TP, Munro S (1998) The pleckstrin homology domain of oxysterol-binding protein recognises a determinant specific to Golgi membranes. Curr Biol 8: 729–739 [DOI] [PubMed] [Google Scholar]
- Levine TP, Munro S (2002) Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr Biol 12: 695–704 [DOI] [PubMed] [Google Scholar]
- Lu L, Horstmann H, Ng C, Hong W (2001) Regulation of Golgi structure and function by ARF-like protein 1 (Arl1). J Cell Sci 114(Pt 24): 4543–4555 [DOI] [PubMed] [Google Scholar]
- Maeda Y, Beznoussenko GV, Van Lint J, Mironov AA, Malhotra V (2001) Recruitment of protein kinase D to the trans-Golgi network via the first cysteine-rich domain. EMBO J 20: 5982–5990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man Z, Kondo Y, Koga H, Umino H, Nakayama K, Shin HW (2011) Arfaptins are localized to the trans-Golgi by interaction with Arl1, but not Arfs. J Biol Chem 286: 11569–11578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I, Nelson WJ (2008) Coordinated protein sorting, targeting and distribution in polarized cells. Nat Rev Mol Cell Biol 9: 833–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Man Z, Xie Y, Hanai A, Makyio H, Kawasaki M, Kato R, Shin HW, Nakayama K, Wakatsuki S (2012) Structural basis for membrane binding specificity of the Bin/Amphiphysin/Rvs (BAR) Domain of Arfaptin-2 Determined by Arl1 GTPase. J Biol Chem 287: 25478–25489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nhek S, Ngo M, Yang X, Ng MM, Field SJ, Asara JM, Ridgway ND, Toker A (2010) Regulation of oxysterol-binding protein Golgi localization through protein kinase D-mediated phosphorylation. Mol Biol Cell 21: 2327–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT (2004) BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303: 495–499 [DOI] [PubMed] [Google Scholar]
- Pusapati GV, Krndija D, Armacki M, von Wichert G, von Blume J, Malhotra V, Adler G, Seufferlein T (2010) Role of the second cysteine-rich domain and Pro275 in protein kinase D2 interaction with ADP-ribosylation factor 1, trans-Golgi network recruitment, and protein transport. Mol Biol Cell 21: 1011–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualmann B, Koch D, Kessels MM (2011) Let’s go bananas: revisiting the endocytic BAR code. EMBO J 30: 3501–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao Y, Haucke V (2011) Membrane shaping by the Bin/amphiphysin/Rvs (BAR) domain protein superfamily. Cell Mol Life Sci 68: 3983–3993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin OH, Exton JH (2001) Differential binding of arfaptin 2/POR1 to ADP-ribosylation factors and Rac1. Biochem Biophys Res Commun 285: 1267–1273 [DOI] [PubMed] [Google Scholar]
- Szentpetery Z, Varnai P, Balla T (2010) Acute manipulation of Golgi phosphoinositides to assess their importance in cellular trafficking and signaling. Proc Natl Acad Sci USA 107: 8225–8230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SC, Adamik R, Hong JX, Moss J, Vaughan M, Kanoh H, Exton JH (1998) Effects of arfaptin 1 on guanine nucleotide-dependent activation of phospholipase D and cholera toxin by ADP-ribosylation factor. J Biol Chem 273: 20697–20701 [DOI] [PubMed] [Google Scholar]
- Valente C, Turacchio G, Mariggio S, Pagliuso A, Gaibisso R, Di Tullio G, Santoro M, Formiggini F, Spano S, Piccini D, Polishchuk RS, Colanzi A, Luini A, Corda D (2012) A 14-3-3gamma dimer-based scaffold bridges CtBP1-S/BARS to PI(4)KIIIbeta to regulate post-Golgi carrier formation. Nat Cell Biol 14: 343–354 [DOI] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW (2008) Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9: 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Blume J, Duran JM, Forlanelli E, Alleaume AM, Egorov M, Polishchuk R, Molina H, Malhotra V (2009) Actin remodeling by ADF/cofilin is required for cargo sorting at the trans-Golgi network. J Cell Biol 187: 1055–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Blume J, Knippschild U, Dequiedt F, Giamas G, Beck A, Auer A, Van Lint J, Adler G, Seufferlein T (2007) Phosphorylation at Ser244 by CK1 determines nuclear localization and substrate targeting of PKD2. EMBO J 26: 4619–4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wichert G, Edenfeld T, von Blume J, Krisp H, Krndija D, Schmid H, Oswald F, Lother U, Walther P, Adler G, Seufferlein T (2008) Protein kinase D2 regulates chromogranin A secretion in human BON neuroendocrine tumour cells. Cell Signal 20: 925–934 [DOI] [PubMed] [Google Scholar]
- Wakana Y, van Galen J, Meissner F, Scarpa M, Polishchuk RS, Mann M, Malhotra V (2012) A new class of carriers that transport selective cargo from the trans Golgi network to the cell surface. EMBO J 31: 3976–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sun HQ, Macia E, Kirchhausen T, Watson H, Bonifacino JS, Yin HL (2007) PI4P promotes the recruitment of the GGA adaptor proteins to the trans-Golgi network and regulates their recognition of the ubiquitin sorting signal. Mol Biol Cell 18: 2646–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YJ, Wang J, Sun HQ, Martinez M, Sun YX, Macia E, Kirchhausen T, Albanesi JP, Roth MG, Yin HL (2003) Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell 114: 299–310 [DOI] [PubMed] [Google Scholar]
- Wiedemann C, Schafer T, Burger MM (1996) Chromaffin granule-associated phosphatidylinositol 4-kinase activity is required for stimulated secretion. EMBO J 15: 2094–2101 [PMC free article] [PubMed] [Google Scholar]
- Williger BT, Ostermann J, Exton JH (1999) Arfaptin 1, an ARF-binding protein, inhibits phospholipase D and endoplasmic reticulum/Golgi protein transport. FEBS Lett 443: 197–200 [DOI] [PubMed] [Google Scholar]
- Yeaman C, Ayala MI, Wright JR, Bard F, Bossard C, Ang A, Maeda Y, Seufferlein T, Mellman I, Nelson WJ, Malhotra V (2004) Protein kinase D regulates basolateral membrane protein exit from trans-Golgi network. Nat Cell Biol 6: 106–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.