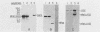Abstract
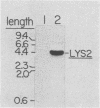
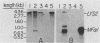
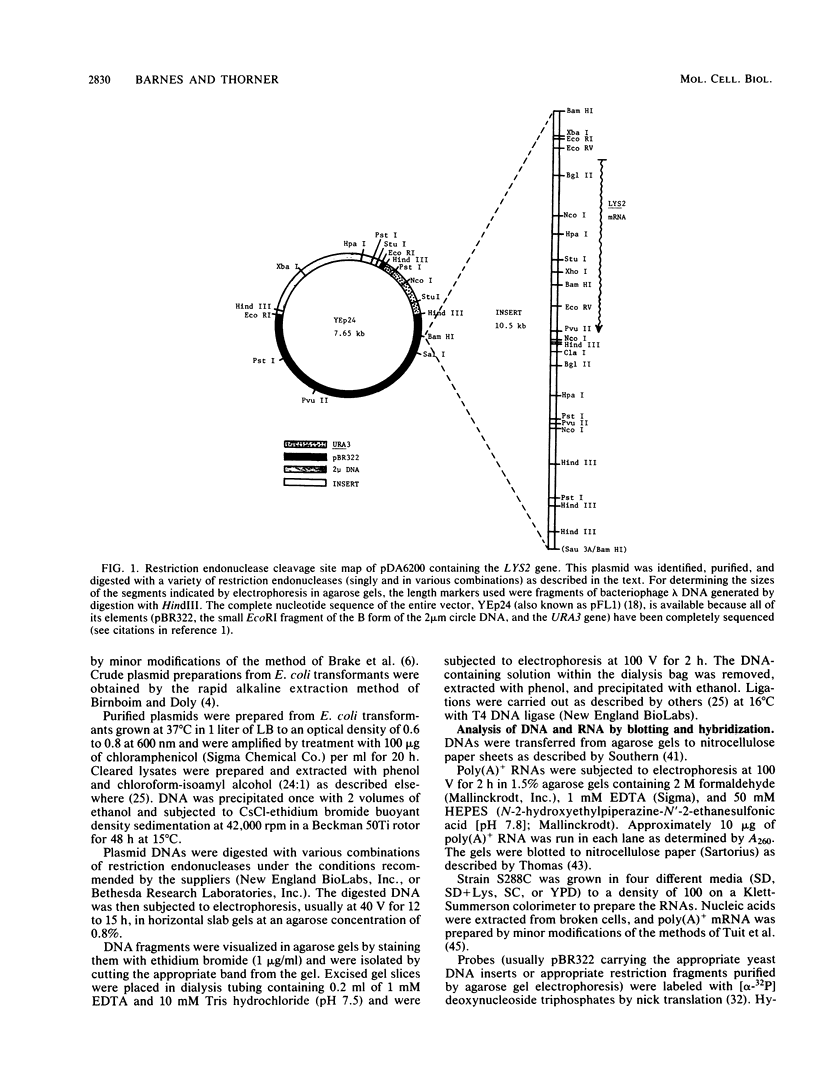
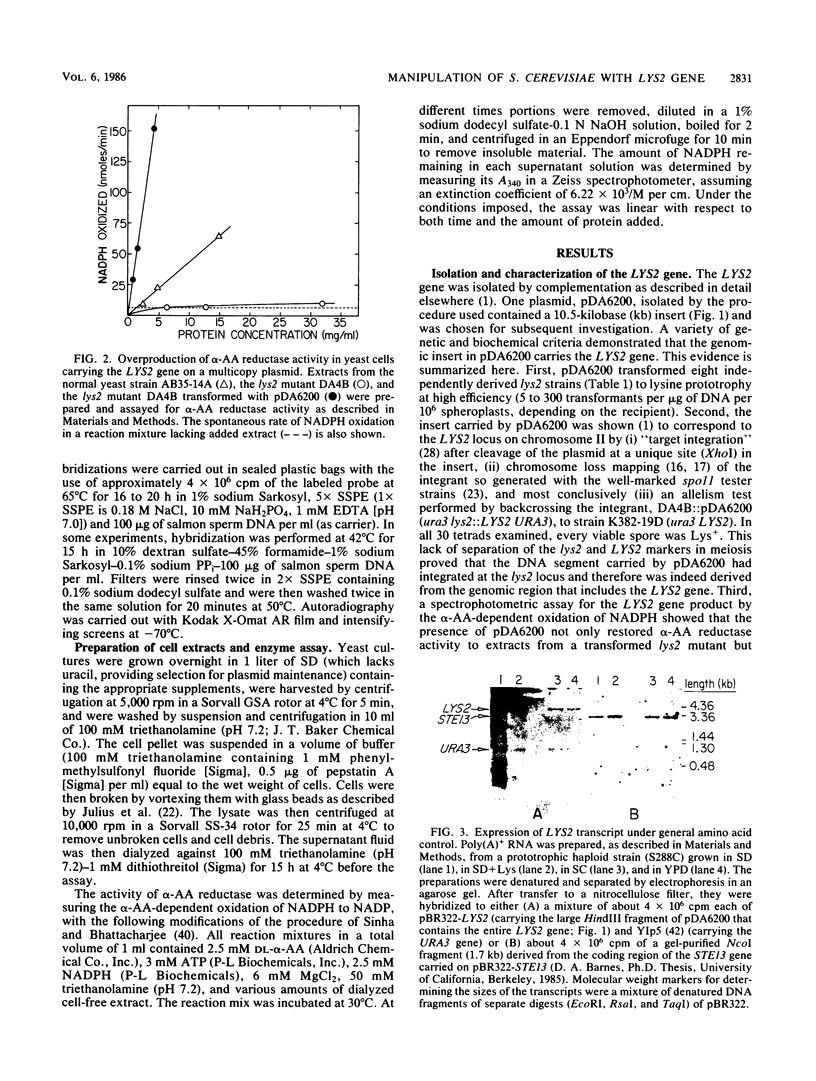
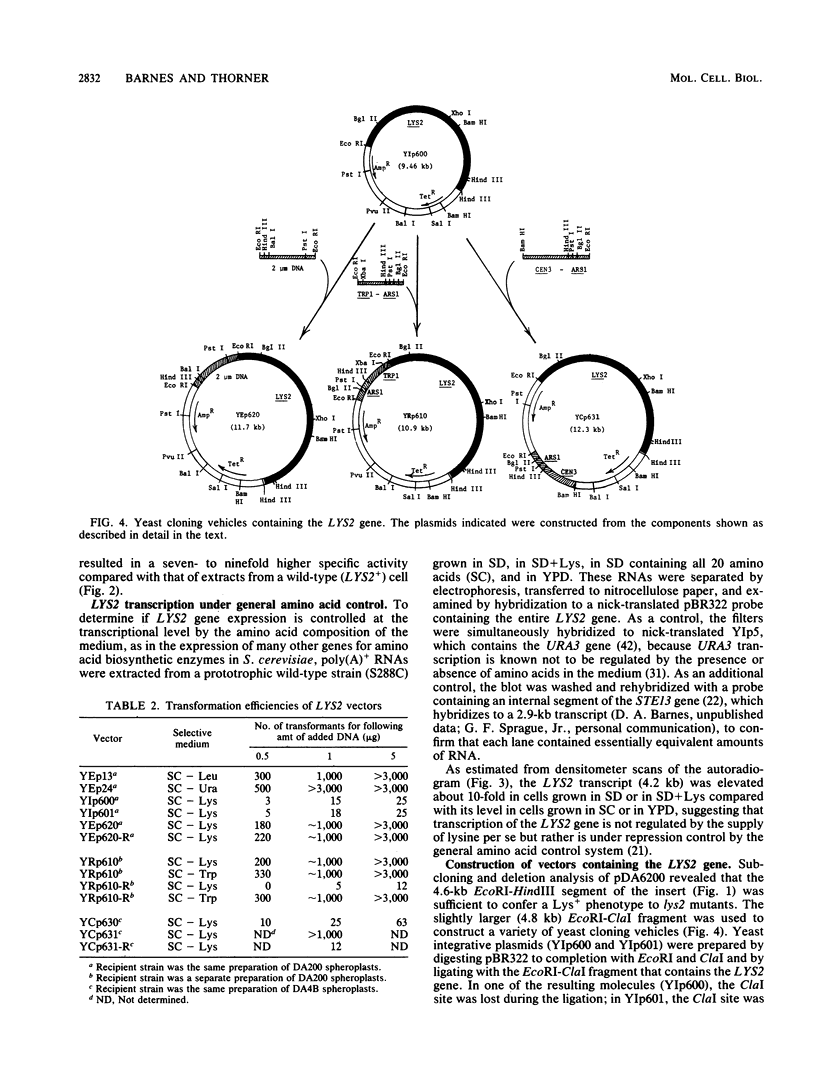
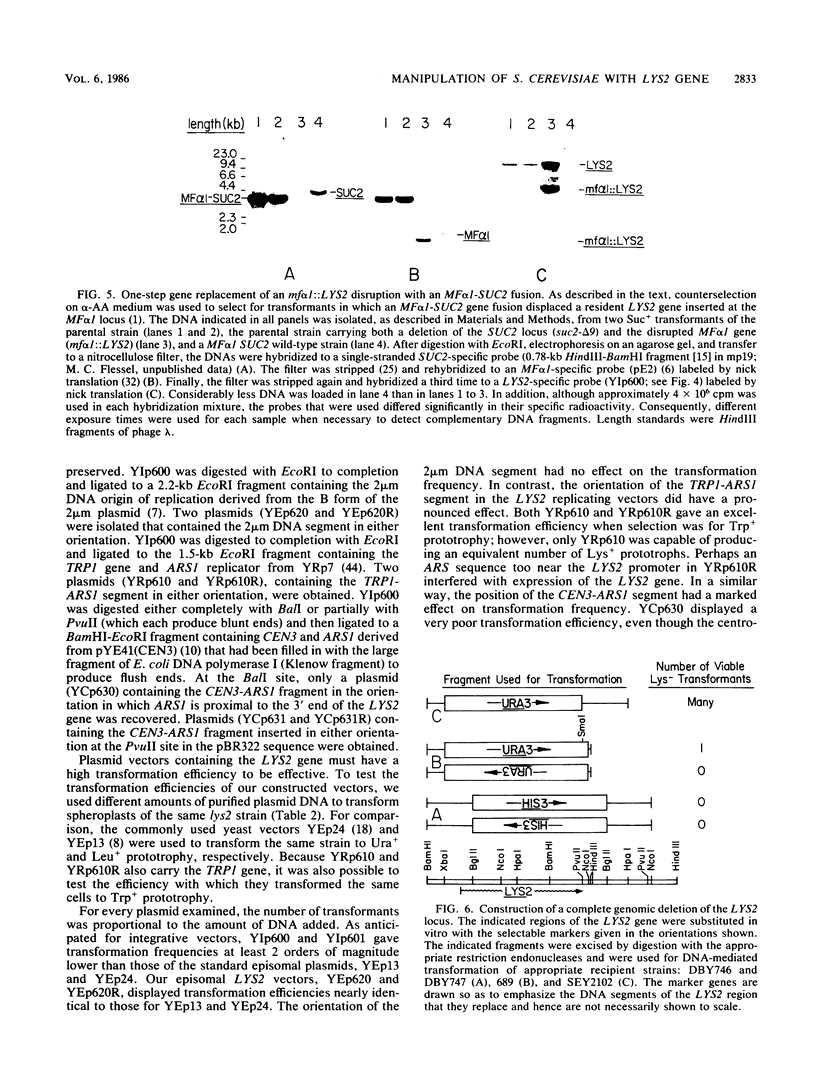
The structural gene for alpha-aminoadipate reductase (LYS2) was isolated from a Saccharomyces cerevisiae genomic DNA library by complementation of a lys2 mutant. Both genetic and biochemical criteria confirmed that the DNA obtained corresponds to the LYS2 locus on chromosome II. Subcloning and deletion analysis showed that a functional LYS2 gene is contained within a 4.6-kilobase (kb) EcoRI-HindIII fragment of the original insert, and the slightly larger EcoRI-ClaI segment (4.8 kb) was used to construct a series of cloning vehicles, including integrating, episomal, replicative, and centromeric vectors. The cloned DNA was also used to generate a genomic deletion that lacks all LYS2 coding sequences on chromosome II. The level of the LYS2 transcript (4.2 kb) was 10-fold higher in cells grown on minimal medium than in cells grown on complete medium and was not repressed by the presence of lysine alone. Gene disruption, gene replacement, and promoter analysis of the major alpha-factor structural gene (MF alpha 1) were performed to illustrate the utility of the LYS2 gene for the genetic manipulation of yeasts. Because all fungi synthesize lysine via the alpha-aminoadipate pathway, the techniques developed here for using the S. cerevisiae LYS2 gene should be directly applicable to other fungal systems.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beggs J. D. Transformation of yeast by a replicating hybrid plasmid. Nature. 1978 Sep 14;275(5676):104–109. doi: 10.1038/275104a0. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Brake A. J., Julius D. J., Thorner J. A functional prepro-alpha-factor gene in Saccharomyces yeasts can contain three, four, or five repeats of the mature pheromone sequence. Mol Cell Biol. 1983 Aug;3(8):1440–1450. doi: 10.1128/mcb.3.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Chattoo B. B., Sherman F., Azubalis D. A., Fjellstedt T. A., Mehnert D., Ogur M. Selection of lys2 Mutants of the Yeast SACCHAROMYCES CEREVISIAE by the Utilization of alpha-AMINOADIPATE. Genetics. 1979 Sep;93(1):51–65. doi: 10.1093/genetics/93.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980 Oct 9;287(5782):504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- Donahue T. F., Daves R. S., Lucchini G., Fink G. R. A short nucleotide sequence required for regulation of HIS4 by the general control system of yeast. Cell. 1983 Jan;32(1):89–98. doi: 10.1016/0092-8674(83)90499-3. [DOI] [PubMed] [Google Scholar]
- Eibel H., Philippsen P. Identification of the cloned S. cerevisiae LYS2 gene by an integrative transformation approach. Mol Gen Genet. 1983;191(1):66–73. doi: 10.1007/BF00330891. [DOI] [PubMed] [Google Scholar]
- Eibel H., Philippsen P. Preferential integration of yeast transposable element Ty into a promoter region. 1984 Jan 26-Feb 1Nature. 307(5949):386–388. doi: 10.1038/307386a0. [DOI] [PubMed] [Google Scholar]
- Emr S. D., Schekman R., Flessel M. C., Thorner J. An MF alpha 1-SUC2 (alpha-factor-invertase) gene fusion for study of protein localization and gene expression in yeast. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7080–7084. doi: 10.1073/pnas.80.23.7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco S. C., Botstein D. A rapid chromosome-mapping method for cloned fragments of yeast DNA. Genetics. 1983 Dec;105(4):857–872. doi: 10.1093/genetics/105.4.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco S. C., Li Y., Broach J. R., Botstein D. Genetic properties of chromosomally integrated 2 mu plasmid DNA in yeast. Cell. 1982 Jun;29(2):573–584. doi: 10.1016/0092-8674(82)90173-8. [DOI] [PubMed] [Google Scholar]
- Fasiolo F., Bonnet J., Lacroute F. Cloning of the yeast methionyl-tRNA synthetase gene. J Biol Chem. 1981 Mar 10;256(5):2324–2328. [PubMed] [Google Scholar]
- Julius D., Blair L., Brake A., Sprague G., Thorner J. Yeast alpha factor is processed from a larger precursor polypeptide: the essential role of a membrane-bound dipeptidyl aminopeptidase. Cell. 1983 Mar;32(3):839–852. doi: 10.1016/0092-8674(83)90070-3. [DOI] [PubMed] [Google Scholar]
- Klapholz S., Esposito R. E. A new mapping method employing a meiotic rec-mutant of yeast. Genetics. 1982 Mar;100(3):387–412. doi: 10.1093/genetics/100.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter R., Yanofsky C. Attenuation in amino acid biosynthetic operons. Annu Rev Genet. 1982;16:113–134. doi: 10.1146/annurev.ge.16.120182.000553. [DOI] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Mortimer R. K., Schild D. Genetic map of Saccharomyces cerevisiae, edition 9. Microbiol Rev. 1985 Sep;49(3):181–213. doi: 10.1128/mr.49.3.181-213.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 1983;101:228–245. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn M. D., Galgoci B., Greer H. Identification of AAS genes and their regulatory role in general control of amino acid biosynthesis in yeast. Proc Natl Acad Sci U S A. 1983 May;80(9):2704–2708. doi: 10.1073/pnas.80.9.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn M. D., Thireos G., Greer H. Temporal analysis of general control of amino acid biosynthesis in Saccharomyces cerevisiae: role of positive regulatory genes in initiation and maintenance of mRNA derepression. Mol Cell Biol. 1984 Mar;4(3):520–528. doi: 10.1128/mcb.4.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rose M., Winston F. Identification of a Ty insertion within the coding sequence of the S. cerevisiae URA3 gene. Mol Gen Genet. 1984;193(3):557–560. doi: 10.1007/BF00382100. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Schaefler S. Inducible system for the utilization of beta-glucosides in Escherichia coli. I. Active transport and utilization of beta-glucosides. J Bacteriol. 1967 Jan;93(1):254–263. doi: 10.1128/jb.93.1.254-263.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S., Davis R. W. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle D., DiMaio D., Nathans D. Directed mutagenesis. Annu Rev Genet. 1981;15:265–294. doi: 10.1146/annurev.ge.15.120181.001405. [DOI] [PubMed] [Google Scholar]
- Simchen G., Winston F., Styles C. A., Fink G. R. Ty-mediated gene expression of the LYS2 and HIS4 genes of Saccharomyces cerevisiae is controlled by the same SPT genes. Proc Natl Acad Sci U S A. 1984 Apr;81(8):2431–2434. doi: 10.1073/pnas.81.8.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha A. K., Bhattacharjee J. K. Lysine biosynthesis in Saccharomyces. Conversion of -aminoadipate into -aminoadipic -semialdehyde. Biochem J. 1971 Dec;125(3):743–749. doi: 10.1042/bj1250743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschumper G., Carbon J. Sequence of a yeast DNA fragment containing a chromosomal replicator and the TRP1 gene. Gene. 1980 Jul;10(2):157–166. doi: 10.1016/0378-1119(80)90133-x. [DOI] [PubMed] [Google Scholar]
- Tuite M. F., Plesset J., Moldave K., McLaughlin C. S. Faithful and efficient translation of homologous and heterologous mRNAs in an mRNA-dependent cell-free system from Saccharomyces cerevisiae. J Biol Chem. 1980 Sep 25;255(18):8761–8766. [PubMed] [Google Scholar]
- Winston F., Chumley F., Fink G. R. Eviction and transplacement of mutant genes in yeast. Methods Enzymol. 1983;101:211–228. doi: 10.1016/0076-6879(83)01016-2. [DOI] [PubMed] [Google Scholar]
- Wolfner M., Yep D., Messenguy F., Fink G. R. Integration of amino acid biosynthesis into the cell cycle of Saccharomyces cerevisiae. J Mol Biol. 1975 Aug 5;96(2):273–290. doi: 10.1016/0022-2836(75)90348-4. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. alpha-Aminoadipate as a primary nitrogen source for Saccharomyces cerevisiae mutants. J Bacteriol. 1985 May;162(2):579–583. doi: 10.1128/jb.162.2.579-583.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]