Abstract
Since first imaged by electron microscopy, much effort has been placed into determining the structure and mechanism of the 26S proteasome. While the proteolytic core is understood in atomic detail, how substrates are engaged and transported to this core remains elusive. Substrate delivery is accomplished by a 19-subunit regulatory particle that binds to ubiquitinated substrates, detaches ubiquitin tags, unfolds the substrate, and translocates it into the peptidase in an ATP-dependent fashion. Recently, several labs have determined subnanometer cryoEM structures of the 26S proteasome, shedding light on the architecture of the regulatory complex. We discuss the biological insights into substrate processing provided by these structures, and the technical hurdles ahead to achieve an atomic resolution structure of the 26 proteasome.
Introduction
Within the cell are a myriad of proteins, some of which are turned over at an astonishing rate. In eukaryotes, this turnover is almost entirely accomplished by a single enzyme, the 26S proteasome. A structural description of proteasomal function brings with it a mechanistic understanding of one of the most fundamental proteome regulators in the cell. The proteasome structure can be subdivided into two main components – the proteolytic 20S core particle (CP), which houses the destructive sites of proteolysis, and the 19S regulatory particle (RP), which includes ubiquitin receptors, a deubiquitinase, and a ring of AAA+ ATPases that caps the CP. The RP functions as a selective gateway to the CP proteases, granting passage only to proteins that have been covalently tagged with specific polyubiquitin chains. After engagement of an ubiquitinated substrate by the RP, the deubiquitinase detaches the ubiquitin chain, and the ATPase ring actively unfolds the protein and translocates the polypeptide to the proteolytic core. Previous preconceptions of the RP's architecture have in the recent years been upturned with a burst of new structural studies. By blending crystallography, molecular modeling, novel expression systems, and subnanometer cryoEM reconstructions, the proteasome community has made great strides in elucidating the structure of the 19S RP, revealing intriguing and unexpected features of this multifaceted module. These findings have answered some of the questions surrounding many aspects of the proteasome function, but have also given rise to new questions.
The 19S RP has been a target of study for molecular and structural biologists for more than two decades, and during this time we have learned much about the RP's requirements for recognizing and deubiquitinating polyubiquitinated substrates, as well as for unfolding and translocating the substrate polypeptide into the CP. In order to fully describe the mechanisms that govern these observations, it is crucial to place them in a structural context. While atomic structures for several isolated RP subunits have been determined by NMR and crystallography1–7, all attempts to produce an atomic structure of the complete 19S RP by crystallographic methods have so far failed, likely due to the sheer size and inherent flexibility of this dynamic assembly.
Low-resolution electron microscopy (EM) provided the first glimpses of the RP's three-dimensional organization, offering key insights into the architecture of the RP and its relationship to the CP8–11. In 1998 it was shown that the RP itself could be further dissociated into two subcomponents, and EM analysis was used to ascribe these subcomponents to two large stacked densities capping the CP, naming the proximal mass the “base”, and the distal mass the “lid”12. The base contains six AAA+ ATPase subunits (Rpt1–6), two large non-ATPase scaffolding subunits (Rpn1 and Rpn2), and an intrinsic ubiquitin receptor (Rpn13). The lid, meanwhile, is made up of eight non-ATPase subunits that are one-to-one paralogs of the core proteins within the eukaryotic translation initiation factor eIF3 and the COP9 signalosome (CSN) particle (Figure 1D). Six of the lid subunits (Rpn3, Rpn5, Rpn6, Rpn7, Rpn9, Rpn12) contain a C-terminal winged-helix fold flanked by a helical segment, together known as the PCI (Proteasome-CSN-Initiation factor 3) motif, while the remaining two subunits (Rpn8 and Rpn11) each contain an Mpr1-Pad1 N-terminal (MPN) domain. Interestingly, the MPN domain of Rpn11 contains catalytic residues that endow the subunit with deubiquitinase (DUB) activity, whereas Rpn8's MPN domain appears to be purely structural13. The proteasome's second intrinsic ubiquitin receptor, Rpn10, binds to an arm of lid and is situated at the interface of the two RP subcomplexes (Figure 1A). Significant improvements in single particle cryoEM instrumentation, data collection software, and image processing methodologies have given rise to several subnanometer reconstructions of the proteasome in recent years, and continued development of cryoEM technologies holds the promise of future atomic-resolution reconstructions. This is evidenced by recent work from Yifan Chen's lab, which has obtained a 3.3 Å structure of the archaeal CP using cryo-EM (personal communication).
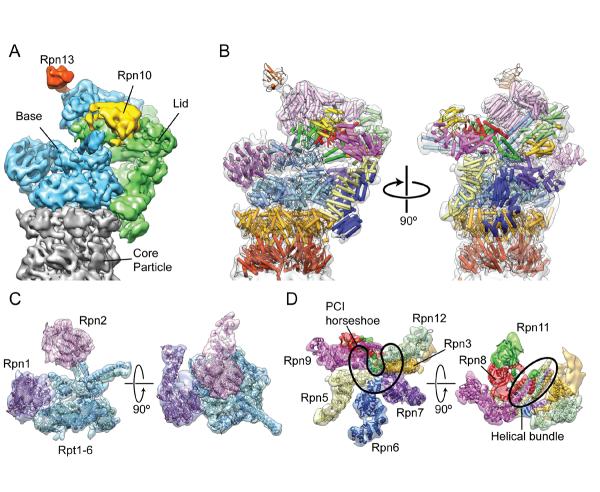
Figure 1.
Proteasome architecture. A. Locations of the base (blue), lid (green), and ubiquitin receptors Rpn10 and Rpn13 within the proteasomal RP. While Rpn13 is considered to be part of the base subcomplex, Rpn10 attaches primarily to the lid and stabilizes the lid-base interaction. B. Atomic models of the RP and the CP fit into the subnanometer reconstruction, shown on the left in the same orientation as A, and facing the lid component on the right (PDB ID 4b4t, except for Rpn1 and Rpt1–6, which were provided by the Pablo Chacón lab). C. The interactions between the Rpts (blue) and Rpn1 and Rpn2 (purple) subunits of the base subcomplex of the RP are shown. D. Architecture of the lid subcomplex. On the left, the horseshoe arrangement of the PCI domains is highlighted in black. On the right, the lid is viewed from the top down, showing the MPN heterodimer (red and green), and the bundle of C-terminal helices (outlined by a black oval). The reconstruction accession number used for this figure is EMD-199215.
The Regulatory Particle at Subnanometer Resolution
The first proteasome reconstruction to achieve subnanometer resolution was obtained in 2010, and was combined with cross-linking/mass spectrometry (MS) data to discriminate the orientation and register of the ATPase subunits relative to the CP14. Cross-linking and MS were also used in conjunction with antibody labeling to localize the DUB Rpn11 to a region of density above the ATPase ring. In the absence of additional external structural information, further architectural details of the RP were based on electron density variance and known stoichiometry of subunits in the holoenzyme. These analyses mistakenly localized the ubiquitin receptor Rpn10 to the side of the RP, while the remainder of the subunit topology remained elusive.
By August of 2012, several labs had determined the complete subunit organization of the S. cerevisiae RP using sub-nanometer cryoEM reconstructions of the 26S proteasome (Figure 1). Two separate studies used EM difference maps of reconstructed deletion mutants to consistently show that the ubiquitin receptors Rpn10 and Rpn13 were flexibly attached to the periphery of the RP15,16 (Figure 1A). The remaining RP subunits were localized using different techniques. The work of Lander et al. used a novel heterologous expression system of the lid subcomplex to label each of the constitutive subunits with a molecular tag, which was localized by negative stain EM analysis. The locations of the non-ATPase subunits of the base were then identified using a combination of antibody and GST-fusion labeling15. Concurrently, a study by Lasker et al. used established protein-protein interactions, known atomic structures, and comparative models to computationally generate a description of the RP17. Particularly significant was the realization that the given name for the “lid” is misleading. This subcomplex attaches to the side of the regulatory particle (Figure 1A), surrounding the ATPase and even contacting the CP at two locations.
A subsequent study of the human proteasome by da Fonseca et al. proposed a revised organization of the RP, based on rigid-body fitting of crystal structures and homology models into a subnanometer resolution cryoEM reconstruction18. Subunits Rpn8, Rpn11, and Rpn12 were localized to positions that contradicted previous structural studies. The redefined position of Rpn12 by da Fonseca et al. was shown to be incorrect upon the crystallization of this subunit, whose atomic structure fit into the previously attributed density with high fidelity7. The ambiguity surrounding the precise locations of Rpn8 and Rpn11 disappeared with the most recent structural work by Beck et al., which has solidified the architectural organization of the RP19.
A colossal dataset of nearly 2.5 million proteasome particles was used to obtain a reconstruction of the 26S proteasome with a reported resolution of 7.4Å (at a Fourier Shell Correlation (FSC) cutoff of 0.5, and a resolution of 6.7Å at a cutoff of 0.3), producing the highest-resolution structure of the proteasome to date. Flexible fitting of atomic structures and homology models into the density generated a quasi-atomic molecular model of the intact RP19(Figure 1B–D). Importantly, this model corrects the Rpn8/Rpn11 architecture proposed by Lander et al., who likely misinterpreted the C-shaped density above the ATPase. Whereas Lander et al. presented a model in which Rpn8 and Rpn11 each occupied one half of this C-shaped density15, Beck et al. suggest that the MPN domains of Rpn8 and Rpn11 dimerize to occupy one half of the C-density, and that the other half corresponds to a large coiled-coil bundle, made up of the C-terminal helices of the eight lid subunits (Figure 1D). The misinterpretation of the Rpn8/Rpn11 heterodimer by Lander et al. exemplifies the limits of molecular tagging, given that flexible linkers are typically used to separate molecular markers from proteins of interest in order to avoid a destabilization of the complex by steric hindrance. Also, these labeling studies are usually carried out using negative stain methods, which provide relatively low-resolution data. Before describing the biological insights provided by recent structural studies, we will review some additional technical aspects of these cryoEM reconstructions for the 26S proteasome.
Technical Points Concerning Proteasome Reconstructions
The proteasome is, in many respects, an ideal sample for cryoEM studies. The large size of the 26S particle makes alignment of individual particle images quite robust, and a richness of atomic structures for individual components can now be used for the interpretation (and validation) of cryoEM structures of the whole complex. Additionally, the inherent two-fold symmetry of the holoenzyme helps to increase the signal to noise ratio of a three dimensional reconstruction. This C2 symmetry was not imposed during the processing of the recent structure presented by Beck et al., achieving a high signal-to-noise ratio through particle number rather than symmetry, and it is suggested that there is a certain degree of asymmetry in the proteasome. However, Beck et al. only note that secondary structural elements are better resolved in one regulatory particle than the other, and do not describe any observable conformational differences between the RP caps. In light of these findings, it is likely that preferential alignment of the better-ordered or well-defined RP within doubly-capped complexes gave rise to this apparent asymmetry, rather than functionally relevant structural differences between the 19S complexes of the proteasome.
For reasons such as this, cryoEM studies of the 26S are not without challenges. A possible source of artifacts in an EM reconstruction concerns a tendency of some macromolecules, including the proteasome, to adopt a preferred orientation on an EM grid15,20. Treating the carbon support of an EM grid with polylysine before adsorbing proteasomes to the surface18,21 or adding small amounts of detergent to the buffer before vitrifying particles over open holes15 are experimental measures that can help obtain a random distribution of particle orientations. Importantly, if an isotropic distribution of particle views around at least one full axis of rotation is not achieved, significant loss of detail and distortions in the reconstruction can occur. Such artifacts may not be negatively reflected in the FSC curve used for estimating resolution, especially in the case of very large datasets, making it important to examine the distribution of Euler angles assigned to the dataset.
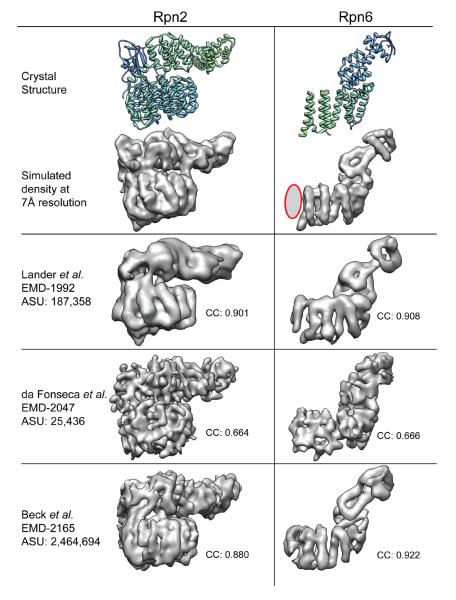
In order to assess the validity and degree of isotropy of the recent 26S cryo-EM reconstrutions15,18,19, we focused on a couple of subunits known in atomic detail. We used crystal structures to simulate the electron density maps of Rpn2 and Rpn6 at 7Å resolution, and compared them to corresponding segmented densities from the three reported proteasome reconstructions (Figure 2). These two subunits contain alpha helical segments that run in approximately orthogonal directions within the complex (Figure 1C,D), and thus are good reporters of the isotropy within the 26S proteasome reconstructions. The segmented subunits from Lander et al. show that the level of resolvable detail is internally consistent within these two regions of the reconstructions, indicative of evenly distributed particle orientations. The reconstruction by Beck et al. shows significant differences in resolvable details between the Rpn2 and Rpn6 densities. While the Rpn6 density exhibits the highest observable resolution of all the reconstructions, as evidenced by a clear delineation of all the secondary structural elements, the helices of the Rpn2 solenoid are considerably less well-resolved. A non-isotropic distribution of particle orientations may explain the noticeable differences in these structural details. It is difficult to assess isotropy of the reconstruction from da Fonseca et al, since these subunits lack any resolvable secondary structure. Such limited detail is not surprising given the low number of particles used in the reconstruction, and indicates an overestimation of resolution, at least for these segments of the structure (see below).
Figure 2.
Comparison of segmented subunits from three subnanometer reconstructions. Density maps for two RP subunits, Saccharomyces cerevisiae Rpn2 and Drosophila melanogaster Rpn6, were generated from their crystal structures (PDB IDs: 4ady and 3txn, respectively) and filtered to 7Å resolution. The first 37 residues of Rpn6, which are predicted to form two short alpha helices, are absent from the crystal structure and thus do not appear in the simulated 7Å density. A gray ellipse (outlined in red) is used to represent these N-terminal helices. The densities corresponding to the Rpn2 and Rpn6 subunits were segmented from the subnanometer-resolution reconstructions of proteasomes from Saccharomyces cerevisiae (EMD-199215 and EMD-216519) and Homo sapiens (EMD-204718). The segmented densities were aligned to the simulated EM density and a cross-correlation value calculated.
Another important fact that must be taken into consideration during the analysis and interpretation of proteasome reconstructions is that large macromolecular complexes, such as the 26S proteasome, often contain mobile regions for which the resolution drops below that of the rest of the structure. The proteasomal CP is the most stable component of the proteasome and thus contains the highest resolution information within a reconstructed density, while the RP includes components that exhibit varying degrees of flexibility and will accordingly vary in resolution. Indeed, it has been proposed that the entirety of the RP complex does not remain in a fixed position relative to the CP9,19. For this reason, a local resolution assessment should be calculated for proteasome reconstructions, or any similarly complex macromolecular assembly, and conclusions drawn only at the level of detail dictated by the resolution at the region of interest.
Among the most dynamic subunits of the RP are the ubiquitin receptors Rp10 and Rp13, as well as Rpn1. The flexibility of these subunits is likely due to their role as the proteasome's main interactors with the cellular environment. Rpn10 and Rpn13 accommodate interactions with a variety of polyubiquitin chain linkages in vivo and in vitro22,23, while Rpn1 serves as a docking platform for shuttle factors and the intrinsic DUB Ubp624,25. Higher resolution structures of these domains in the context of the holoenzyme will only be possible by rigidifying these subunits biochemically, perhaps by specific crosslinking, by the addition of stabilizing cofactors, and/or through extensive three-dimensional sorting and focused sub-classification of large datasets.
Lid Assembly and Incorporation into the Holoenzyme
The recent structural studies not only reveal the organization of the subunits within the RP, but also offer key insights into the lid's assembly and interactions with the base to form the 19S RP. Six of the eight lid subunits contain PCI domains that likely serve as scaffolding motifs26–28, organizing subunits Rpn3, Rpn5, Rpn6, Rpn7, Rpn9, and Rpn12 into a horseshoe-shaped density with the N-termini of these subunits radiating outward like the fingers of a hand15,17–19 (Figure 1D). An additional feature determined by Beck et al. is the organization of the C-terminal helices of the lid subunits into a bundle extending away from the six PCI subunits19(Figure 1D). It was shown that the Rpn8/Rpn11 dimer is very flexible in the isolated lid, suggesting that the interaction between the MPN domains and the PCI domains is minimal. Rpn8 and Rpn11, which do not contain the PCI motif, likely associate with the six PCI subunits primarily via interaction with this helical bundle before the lid is incorporated into the RP. Integration of the lid into the RP stabilizes the position of the MPN heterodimer above the N-ring of the ATPases through interactions with the large scaffolding subunit Rpn2 of the base15,19.
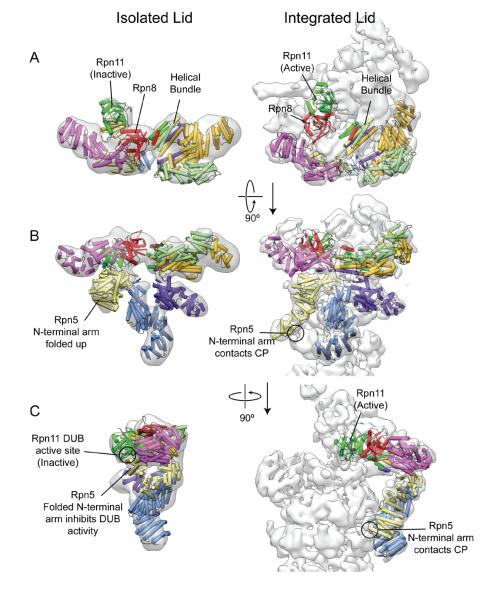
The helical bundle of the lid C-termini, together with the PCI horseshoe, likely serve as a scaffolding anchor that allows a conformational change of the lid as it attaches to the base. A negative-stain reconstruction of the isolated lid shows that many of the subunits are in slightly different positions before and after RP assembly, and that the N-terminal domain of Rpn5 undergoes a dramatic switch in conformation15 (Figure 3). This movement may be directly related to regulation of Rpn11 DUB activity, a potential explanation for the observation that the lid does not exhibit DUB activity in isolation29. Before integration of the lid into the RP, the N-terminal domain of Rpn5 likely interacts with Rpn11 to block DUB activity. As the lid is incorporated into the holoenzyme, the N-terminus of Rpn5 swings down to make contact with the alpha-1 subunit of the CP. This may free and catalytically activate Rpn11, which then assumes a position directly above the pore of the ATPases, next to Rpn2. It is also proposed that the C-terminal helices of Rpn11 may play an additional role in regulating its activity by blocking the active site in the isolated lid19. Movement of these helices with the C-terminal bundle would free the site for catalysis upon holoenzyme assembly. Such structural autoregulation of the lid's DUB activity would elegantly prevent indiscriminate removal of polyubiquitin chains from substrates prior to holoenzyme assembly. Due to the flattening effect that negative stain has on protein complexes, there is a possibility that the proposed motions of Rpn11 and the C-terminal bundle during RP-incorporation are not as dramatic as those shown in Figure 3A,C. Future cryoEM studies of the lid complex will provide a better understanding of the motions of these subunits and their relationship to allosteric activation of the DUB.
Figure 3.
Rearrangements of the lid subunits upon incorporation into the RP. The atomic models of the lid subunits (PDB ID: 4b4t)19 were docked into reconstructions of the isolated lid (EMD-1993) and holoenzyme (EMD-1992)15. The N-terminal region of Rpn9 (purple) was duplicated to occupy the N-terminal region of Rpn3 (dark yellow), for which there is no atomic model. The structures are shown from the top (A), front (B), and side (C). Several subunits, in particular the N-terminal arm of Rpn5, undergo considerable movements between the isolated and integrated states. In the isolated form, the Rpn5 N-terminal helices are folded up against Rpn11, potentially blocking the DUB active site, which is located at the bottom of Rpn11 and facing Rpn5. Upon lid binding, the Rpn5 N-terminal arm swings down to interact with the CP, and the Rpn8/Rpn11 heterodimer (red and green) extends toward the center of the RP.
Two subunits that have been strongly implicated in the incorporation of the lid into the RP are Rpn10 and Rpn12. It is well established that addition of Rpn10 stabilizes the lid-base interaction, as its deletion led to the discovery of the lid and base subcomplexes12. In all proteasome reconstructions, Rpn10's globular Von Willebrand factor type A (VWA) domain is shown to make extensive contacts with the N-terminal domain of Rpn9 at the lid-base boundary15–19. While there is little to no density depicting a robust interaction with the base complex, the ubiquitin-interacting motif (UIM) of Rpn10 is attached to the VWA by a long flexible linker and may interact with the N-terminal portions of the Rpt4/5 coiled coil. In contrast, Rpn12 (or Nin1), which is located opposite to Rpn10's binding site on the lid, appears to make a series of contacts with the Rpn2 subunit of the base. This localization supports evidence that Rpn12 is crucial for stable attachment of the lid to the base.7,26,30,31. Together, Rpn10 and Rpn12 may function as structural staples that bind the lid and base together.
A 3D Model for Substrate Engagement and Translocation
The RP model described by these EM studies provides a structural basis for previously established requirements of proteasomal degradation of ubiquitinated substrates (Figure 4A). Deubiquitination of the substrate is an obligatory step in degradation29,32, and it is known that the polyubiquitin chain attached to a targeted substrate must contain at least four ubiquitin monomers for efficient substrate delivery33. Both Rpn10 and Rpn13 are known to bind polyubiquitin chains between ubiquitin moieties2,4, requiring that the ubiquitin chain be, at least locally, in an open conformation. Both Rpn10 and Rpn13's ubiquitin-binding domains are approximately 75Å away from the Rpn11 DUB active site15,19 and thus spatially arranged to accommodate simultaneous binding and DUB cleavage of a fully extended tetraubiquitin chain.
Figure 4.
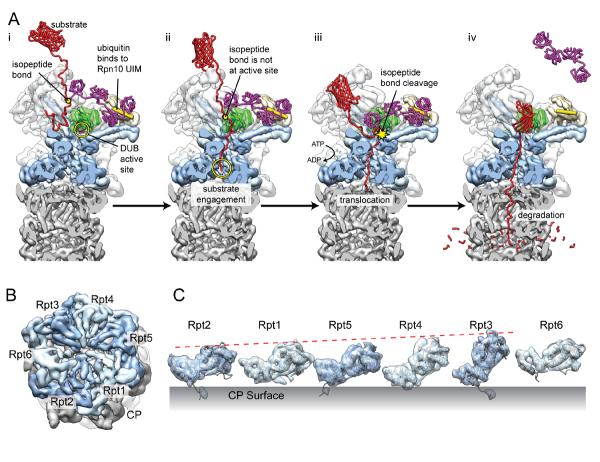
Model for substrate degradation and staircase arrangement of the ATPase. A. Putative model for ATP-dependent substrate deubiquitination and degradation. (i) Binding of the Rpn10 UIM (yellow cylinder) between two ubiquitin moieties of a tetraubiquitin chain (purple). (ii) The unfolded tail of the substrate (red) is threaded through the ATPase pore and becomes engaged. At this point, the isopeptide bond between the substrate and the tetraubiquitin chain is not in the vicinity of the DUB active site (pink, circled in yellow), which is located at the bottom of Rpn11 (green) and faces the ATPase pore. (iii) Translocation of the substrate tail progresses in an ATP-dependent fashion, leading to the positioning of the isopeptide linkage between the ATPase pore and the DUB active site, and the ubiquitin chain is cleaved off. (iv) As the tetraubiquitin dissociates, the remainder of the substrate is unfolded and translocated into the peptidase for degradation.
B. The heterohexameric arrangement of the ATPase catalytic domains (light and dark blue) are shown atop the CP (grey), looking down the central pore (EMD-2165)19. C. The segmented densities corresponding to the catalytic domains are lined up with their exterior surface facing the reader. The large AAA+ subdomains become progressively more upright, as indicated by the red dashed line, producing a staircase-like arrangement in the closed ATPase ring. Interestingly, Rpt6 is suspended above the CP surface at an intermediate height between Rpt3 and Rpt2.
A polyubiquitin tag alone, however, is not sufficient for proteasomal degradation of a protein. Substrates must contain also an unstructured initiation site (or “tail”) at least thirty residues in length to permit engagement by the ATPase ring to initiate translocation34–37. This engagement may additionally prevent dissociation of the substrate after removal of the polyubiquitin chain. Interestingly, it has been reported that DUB activity occurs in an ATP-dependent fashion29,32,38, suggesting a link between ubiquitin cleavage by Rpn11 and ATP hydrolysis of the unfoldase. It is possible that early engagement of the unstructured initiation site by the ATPase plays an active role in positioning the substrate for deubiquitination (Figure 4A). The commencement of ATP-dependent translocation of the polypeptide toward the CP protease could pull the isopeptide bond of the tethered proximal ubiquitin into position at the Rpn11 DUB active site. The active site's enclosed position at the bottom of Rpn11, inaccessible to the globular proteins of the cytosol, ensures that the DUB only cleaves off ubiquitin chains from committed substrates (Figure 4A).
After deubiquitination, the substrate is unfolded and delivered to the proteolytic core by the heterohexameric ring of ATPases. Encircling the entrance to the CP's central pore, the large domains of the ATPase subunits are arranged in a staircase-like configuration with Rpt3 assuming the uppermost position, descending through Rpt4, Rpt5, and Rpt1, with Rpt2 in the lowest position (Figure 4B,C). Rpt6 is situated in an intermediary orientation between Rpt3 and Rpt215,19. A spiraling organization of subunits has been observed in viral, prokaryotic, and eukaryotic DNA helicases, and were suggested to indicate a translocation mechanism that involves a sequential progression of each subunit through the various conformational registers within the spiral39–42. The asymmetric organization of the closed heterohexamer revealed by the proteasome reconstructions directly contradicts this mechanism, unless this observed configuration is a low-energy state assumed by the ATPase in the absence of substrate. It is possible that this asymmetric arrangement persists during translocation, and that local small-scale motions propel substrate through the central pore. This static asymmetric model for translocation is reinforced by the fact that lid subunits Rpn5–7 collectively make extensive contacts with the catalytic domains of four of the ATPase subunits15,17,19. Interestingly, these four ATPase subunits occupy the uppermost portion of the spiral staircase, suggesting that lid incorporation into the RP organizes the ATPase into its staircase configuration. Because all current reconstructions of the proteasome were generated in the absence of substrate, details concerning the substrate pathway through the ATPase remain unknown.
Conclusion
The comprehensive subunit architecture of the proteasomal RP described by recent cryoEM studies provides a structural context for putative models of substrate recognition, deubiquitination, and translocation. Future structural work that takes advantage of novel expression systems, high-throughput cryoEM data collection, and integrative methodologies will be necessary to describe the biochemical mechanisms responsible for each of these steps. Although the fickle RP remains refractory to high-resolution cryoEM studies, as evidenced by the fact that a 2.5 million-particle dataset was unable to break the 6Å barrier, large-scale datasets will prove invaluable in the development of localized 3D variance and classification algorithms, as well as techniques for construction of homology-based atomic models and flexible fitting methodology19. Such algorithms and techniques will be critical not only for detailed investigations of the movements and dynamics of the proteasome, but also benefit the global cryoEM community and our ability to explore the structures of other dynamic macromolecules.
Highlights
-
-
The complete subunit architecture of the proteasome regulatory particle is now known
-
-
Technical challenges still impede cryoEM structure determination of the proteasome
-
-
The proteasomal lid undergoes dramatic conformational movements during assembly
-
-
A model for substrate processing emerges, based on recent cryoEM reconstructions
Acknowledgements
We are grateful to Dr. Mary Matyskiela for helpful discussion and comments during the preparation of this manuscript. G.C.L. acknowledges support from the Damon Runyon Cancer Research Foundation. Work was funded by the Human Frontiers in Science program (E.N.), the Searle Scholars Program (A.M.), and NIH grant R01-GM094497-01A1 (A.M.). E.N. is a Howard Hughes Medical Institute Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Husnjak K, et al. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. doi:10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schreiner P, et al. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature. 2008;453:548–552. doi: 10.1038/nature06924. doi:10.1038/nature06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanches M, Alves BS, Zanchin NI, Guimaraes BG. The crystal structure of the human Mov34 MPN domain reveals a metal-free dimer. Journal of molecular biology. 2007;370:846–855. doi: 10.1016/j.jmb.2007.04.084. doi:10.1016/j.jmb.2007.04.084. [DOI] [PubMed] [Google Scholar]
- 4.Riedinger C, et al. Structure of Rpn10 and its interactions with polyubiquitin chains and the proteasome subunit Rpn12. The Journal of biological chemistry. 2010;285:33992–34003. doi: 10.1074/jbc.M110.134510. doi:10.1074/jbc.M110.134510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pathare GR, et al. The proteasomal subunit Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:149–154. doi: 10.1073/pnas.1117648108. doi:10.1073/pnas.1117648108. [DOI] [PMC free article] [PubMed] [Google Scholar]; * The first crystal structure of a subunit within the lid subcomplex is described and docked into a subnanometer resolution cryoEM reconstruction of the proteasome. The Rpn6 subunit contacts the core particle, as well as an ATPase subunit, suggesting that Rpn6 stabilizes the regulatory particle-core particle interaction.
- 6.He J, et al. The structure of the 26S proteasome subunit Rpn2 reveals its PC repeat domain as a closed toroid of two concentric alpha-helical rings. Structure. 2012;20:513–521. doi: 10.1016/j.str.2011.12.015. doi:10.1016/j.str.2011.12.015. [DOI] [PubMed] [Google Scholar]; * The crystal structure of Rpn2 was solved, confirming that it contains a closed toroid of helical repeats, as well as rod-like arm extending away from the solenoid. An EM reconstruction of Rpn1 shows that these subunits share a similar structural morphology.
- 7.Boehringer J, et al. Structural and functional characterization of Rpn12 identifies residues required for Rpn10 proteasome incorporation. The Biochemical journal. 2012;448:55–65. doi: 10.1042/BJ20120542. doi:10.1042/BJ20120542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikai A, Nishigai M, Tanaka K, Ichihara A. Electron microscopy of 26 S complex containing 20 S proteasome. FEBS letters. 1991;292:21–24. doi: 10.1016/0014-5793(91)80824-m. [DOI] [PubMed] [Google Scholar]
- 9.Walz J, et al. 26S proteasome structure revealed by three-dimensional electron microscopy. Journal of structural biology. 1998;121:19–29. doi: 10.1006/jsbi.1998.3958. doi:10.1006/jsbi.1998.3958. [DOI] [PubMed] [Google Scholar]
- 10.Nickell S, et al. Automated cryoelectron microscopy of “single particles” applied to the 26S proteasome. FEBS letters. 2007;581:2751–2756. doi: 10.1016/j.febslet.2007.05.028. doi:10.1016/j.febslet.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 11.da Fonseca PC, Morris EP. Structure of the human 26S proteasome: subunit radial displacements open the gate into the proteolytic core. The Journal of biological chemistry. 2008;283:23305–23314. doi: 10.1074/jbc.M802716200. doi:10.1074/jbc.M802716200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glickman MH, et al. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 13.Maytal-Kivity V, Reis N, Hofmann K, Glickman MH. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC biochemistry. 2002;3:28. doi: 10.1186/1471-2091-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bohn S, et al. Structure of the 26S proteasome from Schizosaccharomyces pombe at subnanometer resolution. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:20992–20997. doi: 10.1073/pnas.1015530107. doi:10.1073/pnas.1015530107. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This publication introduces the first reconstruction of the 26S proteasome to be solved at subnanometer resolution. Importantly, the organization of the ATPase subunits relative to the base is defined by combining this structural work with cross-linking and mass spectrometry.
- 15.Lander GC, et al. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. doi:10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This paper describes a novel heterologous expression system of the lid subcomplex, which was used to localize the 8 constitutive subunits. Combined with additional molecular labeling or deletions of subunits, the overall architecture of the regulatory particle is described.
- 16.Sakata E, et al. Localization of the proteasomal ubiquitin receptors Rpn10 and Rpn13 by electron cryomicroscopy. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1479–1484. doi: 10.1073/pnas.1119394109. doi:10.1073/pnas.1119394109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lasker K, et al. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1380–1387. doi: 10.1073/pnas.1120559109. doi:10.1073/pnas.1120559109. [DOI] [PMC free article] [PubMed] [Google Scholar]; * A subnanometer-resolution cryoEM reconstruction is combined with crystal structures, cross-linking/mass spectrometry experiments, and known protein-protein interactions to computationally generate a model of the regulatory particle subunit organization.
- 18.da Fonseca PC, He J, Morris EP. Molecular model of the human 26S proteasome. Molecular cell. 2012;46:54–66. doi: 10.1016/j.molcel.2012.03.026. doi:10.1016/j.molcel.2012.03.026. [DOI] [PubMed] [Google Scholar]
- 19.Beck F, et al. Near-atomic resolution structural model of the yeast 26S proteasome. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:14870–14875. doi: 10.1073/pnas.1213333109. doi:10.1073/pnas.1213333109. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** The highest resolution proteasome reconstruction to date, it provides the basis for a complete atomic model of the regulatory particle. The Rpn8/Rpn11 subunits form a heterodimer within the lid, and the C-termini of the 8 lid subunits form a coiled coil bundle.
- 20.Hegerl R, Pfeifer G, Puhler G, Dahlmann B, Baumeister W. The three-dimensional structure of proteasomes from Thermoplasma acidophilum as determined by electron microscopy using random conical tilting. FEBS letters. 1991;283:117–121. doi: 10.1016/0014-5793(91)80567-m. [DOI] [PubMed] [Google Scholar]
- 21.Ortega J, et al. The axial channel of the 20S proteasome opens upon binding of the PA200 activator. Journal of molecular biology. 2005;346:1221–1227. doi: 10.1016/j.jmb.2004.12.049. doi:10.1016/j.jmb.2004.12.049. [DOI] [PubMed] [Google Scholar]
- 22.Xu P, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. doi:10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saeki Y, et al. Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. The EMBO journal. 2009;28:359–371. doi: 10.1038/emboj.2008.305. doi:10.1038/emboj.2008.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elsasser S, et al. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nature cell biology. 2002;4:725–730. doi: 10.1038/ncb845. doi:10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 25.Rosenzweig R, Bronner V, Zhang D, Fushman D, Glickman MH. Rpn1 and Rpn2 coordinate ubiquitin processing factors at proteasome. The Journal of biological chemistry. 2012;287:14659–14671. doi: 10.1074/jbc.M111.316323. doi:10.1074/jbc.M111.316323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fukunaga K, Kudo T, Toh-e A, Tanaka K, Saeki Y. Dissection of the assembly pathway of the proteasome lid in Saccharomyces cerevisiae. Biochemical and biophysical research communications. 2010;396:1048–1053. doi: 10.1016/j.bbrc.2010.05.061. doi:10.1016/j.bbrc.2010.05.061. [DOI] [PubMed] [Google Scholar]
- 27.Scheel H, Hofmann K. Prediction of a common structural scaffold for proteasome lid, COP9-signalosome and eIF3 complexes. BMC bioinformatics. 2005;6:71. doi: 10.1186/1471-2105-6-71. doi:10.1186/1471-2105-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu H, Reis N, Lee Y, Glickman MH, Vierstra RD. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. The EMBO journal. 2001;20:7096–7107. doi: 10.1093/emboj/20.24.7096. doi:10.1093/emboj/20.24.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verma R, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. doi:10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 30.Gordon C, McGurk G, Wallace M, Hastie ND. A conditional lethal mutant in the fission yeast 26 S protease subunit mts3+ is defective in metaphase to anaphase transition. The Journal of biological chemistry. 1996;271:5704–5711. doi: 10.1074/jbc.271.10.5704. [DOI] [PubMed] [Google Scholar]
- 31.Tomko RJ, Jr., Hochstrasser M. Incorporation of the Rpn12 subunit couples completion of proteasome regulatory particle lid assembly to lid-base joining. Molecular cell. 2011;44:907–917. doi: 10.1016/j.molcel.2011.11.020. doi:10.1016/j.molcel.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. doi:10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 33.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. The EMBO journal. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. doi:10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inobe T, Fishbain S, Prakash S, Matouschek A. Defining the geometry of the two-component proteasome degron. Nature chemical biology. 2011;7:161–167. doi: 10.1038/nchembio.521. doi:10.1038/nchembio.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prakash S, Inobe T, Hatch AJ, Matouschek A. Substrate selection by the proteasome during degradation of protein complexes. Nature chemical biology. 2009;5:29–36. doi: 10.1038/nchembio.130. doi:10.1038/nchembio.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nature structural & molecular biology. 2004;11:830–837. doi: 10.1038/nsmb814. doi:10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- 37.Shabek N, et al. The size of the proteasomal substrate determines whether its degradation will be mediated by mono- or polyubiquitylation. Molecular cell. 2012;48:87–97. doi: 10.1016/j.molcel.2012.07.011. doi:10.1016/j.molcel.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 38.Eytan E, Armon T, Heller H, Beck S, Hershko A. Ubiquitin C-terminal hydrolase activity associated with the 26 S protease complex. The Journal of biological chemistry. 1993;268:4668–4674. [PubMed] [Google Scholar]
- 39.Costa A, et al. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nature structural & molecular biology. 2011;18:471–477. doi: 10.1038/nsmb.2004. doi:10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Itsathitphaisarn O, Wing RA, Eliason WK, Wang J, Steitz TA. The Hexameric Helicase DnaB Adopts a Nonplanar Conformation during Translocation. Cell. 2012;151:267–277. doi: 10.1016/j.cell.2012.09.014. doi:10.1016/j.cell.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. doi:10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 42.Thomsen ND, Berger JM. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell. 2009;139:523–534. doi: 10.1016/j.cell.2009.08.043. doi:10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]