Abstract
Transcription and mRNA export are linked processes. However, the molecular mechanisms of this coordination are not clear. Sus1 (hENY2) participates in this coordination as part of two protein complexes: SAGA, a transcriptional co-activator; TREX-2, which functions in mRNA biogenesis and export. Here, we investigate the coordinated action of SAGA and TREX-2 required for gene expression. We demonstrate that TREX-2 subunit Sem1 also participates in transcription activation. Like Sus1, Sem1 is required for the induction of ARG1 and GAL1, these being SAGA-regulated genes. Chromatin immunoprecipitations show that proper recruitment of certain SAGA subunits to the GAL1 promoter depends on Sem1. Notably, both in vivo and in vitro analyses reveal that Sem1 influences SAGA-dependent histone H2B deubiquitylation. Most of these phenotypes are also found to depend on another TREX-2 subunit, Thp1. These results unveil a new role for Sem1 in the activation of the SAGA-dependent gene GAL1 and influencing H2B deubiquitylation. Our work provides insights into a novel functional relationship between Sem1 and the SAGA complex.
INTRODUCTION
Gene expression is a complicated multistep process that is essential for all cells. The synthesis and transport of messenger RNA from the transcription site to the translation site in the cytoplasm involves many interconnected steps, including transcription, mRNA processing and export. A major focal point in the field has been to uncover the functional links among the different steps of the gene expression pathway (1–4). Great progress has been made to identify the factors coordinating this functional coupling, and they have provided a solid base to investigate the molecular mechanisms controlling this process (5–8).
Early analyses suggested a role for NPC (nuclear pore complex) in promoting gene expression by means of a gene-to-pore recruitment mechanism (9). Current studies support this view by showing that specific loci are targeted to the vicinity of NPCs on activation (10). One example of an inducible gene regulated by this mechanism is GAL1, the gene that encodes galactokinase, which has served as a model for many in-depth studies into transcription regulation (11,12). GAL1 is a SAGA-dependent gene recruited to the nuclear periphery on induction, and this depends on the coordinated action of the SAGA and TREX-2 complexes (13). SAGA (Spt-Ada-Gcn5-acetyltransferase) is a histone-modifier complex that binds to its target promoters facilitating transcriptional activation via histone acetylation and/or deubiquitylation (14,15). TREX-2 (transcription and export complex-2) is an NPC-associated complex, which plays roles in mRNA biogenesis and export (16). An intriguing aspect is that SAGA and TREX-2 share a component, the conserved small protein Sus1, which links transcription to mRNA export (7).
One model, which has emerged from several studies, proposes that a cascade of events is functionally and spatially linked through the action of overlapping factors along the gene expression pathway (10,17–19). In this cascade, the gene might be recruited to the NPC, which subsequent SAGA recruitment would strengthen this interaction via Sus1 and other SAGA/TREX-2 factors (10,17). Several reports, including structural studies, have shed light on TREX-2 and SAGA coordination, but the precise mechanism remains uncertain (18,20–22).
Sus1 is required for the SAGA-dependent histone H2B deubiquitylation of its target genes (20,23). Along these lines, it has been well established that H2B ubiquitylation increases early on during GAL1 activation to then decrease, which triggers full GAL1 induction (24–27). As Sus1 participates in this process, it is conceivable that the histone H2B deubiquitylation of GAL1 promoter is regulated by the coordinated action of TREX-2 and SAGA.
Here, we show that Sem1 plays a role in SAGA recruitment, and in its dependent H2B deubiquitylation. The results show that the TREX-2 subunit Sem1 is a crucial factor to maintain the functional linkage between SAGA and TREX-2. Sem1 affects TREX-2 stability and is necessary for transcription of SAGA-dependent genes GAL1 and ARG1. In the absence of Sem1, some SAGA factors are not efficiently recruited to the GAL1 promoter. Moreover, SAGA-mediated DUB activity is enhanced by Sem1. Strikingly, lack of TREX-2 subunit Thp1, which directly binds to Sem1, also prevents in vitro SAGA-mediated deubiquitylation activity.
MATERIALS AND METHODS
Yeast strains, DNA recombinant work and microbiological techniques
The yeast strains used in this study are listed in Supplementary Table S1, along with the quantitative polymerase chain reaction (qPCR) primers and antibodies (Supplementary Tables S2 and S3). Microbiological techniques and yeast plasmid transformation were essentially done as described previously (7). The chromosomal integration of TAP (URA3 marker), MYC (HIS3 marker) and C-terminal tags was performed as previously described (28,29). For gene disruptions, the indicated gene was deleted by high-efficiency transformation using a PCR product amplified from either the KanMX4 plasmid pRS400 or the HIS3 plasmid pFA6a. All the deletions and genomically tagged strains were confirmed by PCR analysis and/or western blot analysis. Strains were grown under standard conditions. For the growth analysis, yeast cells were diluted to 0.5 OD600, and serial dilutions (1:10) were spotted onto YP + glucose and incubated at various temperatures.
TAP purifications, immunoprecipitations and western blot analysis
Purification of Sus1-tandem affinity purification (TAP), Ada2-TAP and Ubp8-TAP in wild-type (BY4741) and mutant strains was performed as previously described (7). TAP fusion proteins and associated proteins were recovered from cell extracts by affinity selection in an IgG matrix. After washing, the Tobacco Etch Virus (TEV) protease was added to release the bound material. The eluate was incubated with calmodulin-coated beads in the presence of calcium. This second affinity step was required to remove not only the TEV protease but also traces of contaminants remaining after first affinity selection. After washing, the bound material was released with ethylene glycol tetraacetic acid (EGTA). This enriched fraction was called calmoduline eluate. The calmoduline eluates from the TAP-purified complexes were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). The western analysis of calmoduline eluates was performed using anti-MYC, anti-Sem1 polyclonal or anti-TAP according to standard procedures.
To analyze the TAP-purified protein complexes, trichloroacetic acid (TCA)-precipitation, LysC/trypsin digestion and multidimensional protein identification technology (MudPIT) analyses by mass spectrometry were performed as described elsewhere (30). The immunoprecipitation of Sus1-MYC, Ubp8-TAP and Ada2-TAP in wild-type and mutants was performed as follows: 50 ml of yeast cultures were grown on rich medium to a 0.5 OD600. Cells were harvested, washed with water and resuspended in 250 μl of lysis buffer [50 mM HEPES–KOH at pH 7.5, 140 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 10% glycerol, 0.5% NP-40, 1 mM PMSF and protease inhibitors]. An equal volume of glass beads was added. Breakage was achieved by four pulses of vortexing at 4°C lasting 1 min. The clarified extracts were immunoprecipitated for 60 min at 4°C using anti-MYC antibodies or IgG Sepharose six Fast Flow resin (GE Healthcare). Immunoprecipitates were washed three times with 1 ml of lysis buffer for 10 min and were subsequently resuspended in 50 μl of SDS–PAGE sample buffer. Western analysis was performed using anti-MYC, anti-Sem1 and anti-TAP according to standard procedures.
Yeast whole-cell extracts (WCEs) were prepared as described elsewhere. Typically, 7 μl of WCE were loaded on SDS–PAGE gels. After electrophoresis and transfer to nitrocellulose membranes, samples were subjected to immunoblot analyses using procedures and detection reagents from ECL Amersham®.
Chromatin immunoprecipitation and double chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) and chromatin double immunoprecipitation (ChDIP) were performed as formerly described (31,32) using 100 ml (ChDIP) or 50 ml (ChIP) of early-log-phase cultures grown in YP + glucose (repressing), YP + raffinose (de-repressing) or YP + galactose (inducing) for inductions of 25 (ChIP) or 90 min (ChDIP). Cultures were cross-linked with 1% of formaldehyde solution (Sigma) for 20 min at room temperature with intermittent shaking and were then quenched with 125 mM glycine. Cells were collected by centrifugation and washed four times with 25 ml cold Tris–saline buffer (150 mM NaCl and 20 mM Tris–HCl at pH 7.5). Cell breakage was performed in 300 ml of lysis buffer (50 mM HEPES–KOH at pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% Tergitol-type NP-40 (NP-40), 1 mM phenylmethanesulfonylfluoride or phenylmethylsulfonyl fluoride (PMSF) and protease inhibitors) with glass beads, and cell extracts were sonicated in a Bioruptor sonicator (Diagenode) for 30 min in 30 s on/30 s off cycles (chromatin was sheared into an average size of 300 bp). Ten microliters of extract was reserved as the input. The rest was used for immunoprecipitation with magnetic beads (Dynabeads©) coated with polyclonal goat anti-mouse IgG antibodies and was incubated O/N with 2.5 μl of mouse α-MYC antibodies (Sus1-MYC, Gcn5-MYC, Ada2-MYC, Spt8-MYC and Taf9-MYC in wild-type and mutants), or with magnetic beads coated with monoclonal human anti-mouse IgG antibodies (Rpb3-TAP) or by adding 90 μl of M2 agarose slurry (Sigma A2220; pre-washed three times in 1 ml of 125 mM glycine and equilibrated in lysis buffer for 60 min at 4°C) to extracts (wild-type and mutant sem1Δ). Immunoprecipitations were conducted overnight at 4°C, and the immune complexes were washed twice with lysis buffer, twice with lysis buffer supplemented with 360 mM NaCl, twice with wash buffer (10 mM Tris–HCl at pH 8.0, 250 mM LiCl, 0.5% NP-40, 125 mg of nadeoxycholol and 1 mM EDTA) and once with TE 1×. For ChDIP, Flag-H2B was eluted with 25 μl of 3× Flag peptide (Sigma F4799; 4 mg/ml of stock concentration) and incubated overnight at 4°C. Twenty microliters was separated as the input, and a second purification of Flag-H2B pool was performed using 2.5 μl of the anti-hyaluronic acid (HA) antibody (Roche 12CA5, 5 mg/ml), previously incubated with 50 μl of magnetic beads coated with polyclonal goat anti-mouse IgG antibodies (pre-washed three times in 1 ml of phosphate-buffered saline–bovine serum albumin and resuspended in 47.5 μl) and shaken overnight at 4°C. Immunoprecipitation was washed as previously described, and samples were eluted at 65°C for 15 min with 100 μl of elution buffer (50 mM Tris–HCl at pH 8, 10 mM EDTA and 1% SDS). Inputs and immunoprecipitation (IP) samples were incubated overnight at 65°C to reverse the cross-link. Samples were then treated with proteinase K (Ambion), at 100 μg/250 μl of chromatin for 2 h, and DNA was extracted twice with phenol:chloroform:isoamyl alcohol (25:24:1) and once with chloroform:isoamyl alcohol (24:1), ethanol precipitated and was resuspended in 40 μl TE. DNA was used for the qPCR reaction by using the appropriate primers listed in Supplementary Table S2. In all cases, at least three biological replicates were performed to obtain the standard errors of each experiment.
RNA isolation reverse transcriptase–PCR quantitative PCR
Total RNA was harvested from the yeast early-log-phase cultures grown in YP + glucose (repressing), YP + raffinose (de-repressing) or YP + galactose (inducing) by phenol/chloroform extraction (33). RNA was quantified, and quality was checked using Nanodrop®. The reverse transcriptase (RT)–PCR analysis was performed using 1 μg of total RNA. After DNase-I treatment, RNAs were purified by phenol/chloroform extraction. Reverse transcription was performed by following standard procedures with random hexamers and M-MLV reverse transcriptase (Invitrogen®).
Specific pairs of primers were used to amplify the qRT–PCR products of transcripts SCR1 or GAL1 using 3 μl of cDNA as a template (previously diluted at 1/10). For qPCR, SYBR® Green qPCR MM no ROX (Fermentas) was used in 10 μl of the final volume. Each sample was analyzed in triplicate, and qPCR was performed in a LightCycler® Roche 480. An activation step of 10 min at 95°C, followed by 40 cycles of 10 s at 94°C, 15 s at 57°C and 15 s at 72°C, was used for the GAL1 Open Reading Frame (ORF), ARG1 Promoter (PR) and SCR1 primer pairs. For the GAL1 PR pair, the Tm was set at 57°C. Real-time PCR amplification efficiency was calculated from the given slopes in the LightCycler® Roche 480. All the calculations were done by taking into account the real primer efficiencies. See Supplementary Table S2 for a list of these primers. The GAL1 and ARG1 mRNA fold-change (FC) levels were normalized to the reference gene SCR1 and were expressed in relation to the transcript level for each strain at the zero time point, defined as 1.0 (34,35). The ChDIP data were calculated as the signal ratio of IP samples in relation to the input signal minus the H2BK123R and H2BK123R sem1Δ mutant strain signal, set as a background control. The ChIP data were calculated as the signal ratio of IP samples in relation to the input signal minus the background of a no-antibody control. For the H3K79me3 signal, normalization was done in relation to total histone H3 levels. In all cases, at least three biological replicates were performed to obtain the standard errors of each experiment.
In vitro deubiquitylation assay
The Flag-tagged H2B substrate (containing ubH2B and unmodified H2B) was obtained by purifying N-terminally Flag-tagged histone H2B using an M2 agarose slurry (Sigma A2220). This substrate was quantified, and equal amounts (500 ng) were divided and incubated at room temperature for 30 min in deubiquitylation (DUB) buffer (100 mM Tris–HCl at pH 8.0, 1 mM EDTA, 1 mM DTT, 5% glycerol, 1 µM PMSF and 1% protease inhibitor) with the calmoduline eluate of each purification (Sus1-TAP and Ubp8-TAP) from wild-type (BY4741) and mutant strains (see Supplementary Figure S3A for the description of the experimental set-up). As a negative control, histone H2B was incubated solely in DUB buffer (which corresponds with the input of the assay) or with the calmoduline eluates of the Sus1-TAP sgf73Δ mutant, where the DUB activity associated with SAGA was impaired. The reaction was stopped by freezing on dry ice, followed by boiling in 1 volume of 2× SDS sample buffer for 2 min and running in a 15% SDS–PAGE gel. Gels were transferred to nitrocellulose, and a western blot analysis was performed using anti-HA antibodies to detect ubiquitin. The detection with this antibody allows us to identify specifically ubiquitylated histone H2B species, which makes possible accurate band quantification in each strain. Using anti-Flag antibodies allows us to identify all histone H2B species, taking into account that ubiquitylated histone H2B exists in low amounts within the cell; this is not a truthful method for ubH2B quantification (36). In all cases, at least three biological replicates were performed to obtain the standard errors of each experiment.
Fluorescence in situ hybridization
Fluorescence in situ hybridization against poly(A)+ RNA was done by growing yeast cells in 50 ml of YP + glucose medium at 30°C or 37°C at 0.5 OD600. Cells were immediately fixed by adding 10% of formaldehyde for 60 min at room temperature. The fixative was removed by two rounds of centrifugation and was washed with 0.1 M potassium phosphate (pH 6.4). Cells were resuspended in ice-cold washing buffer (1.2 M sorbitol and 0.1 M potassium phosphate, pH 6.4), and, subsequently, the cell wall was digested with 0.5 mg/ml of zymolyase 100 T, whereas samples were applied on poly-l-lysine-coated slide wells. Non-adhering cells were removed by aspiration, cells were rehydrated with 2× SSC (0.15 M NaCl and 0.015 M sodium citrate) and were hybridized overnight at 37°C in 20 µl of pre-hybridization buffer (formamide 50%, dextran sulfate 10%, 125 µg/ml of Escherichia coli tRNA, 4× SSC, 1× Denhardt’s solution and 500 µg/ml of herring sperm DNA) with 0.8 pmol of Cy3-end-labeled oligo(dT) in a humid chamber. After hybridization, slides were washed with 1× SSC at room temperature, air-dried and mounted using VECTASHIELD® Mounting Medium with DAPI. Cy3-oligo(dT) was detected under a Leica DM600B fluorescence microscope. In all cases, at least three biological replicates were performed to obtain the standard errors of each experiment.
Image analysis
The nuclear/cytoplasmic ratio of fluorescence intensity was quantified using the Image J public domain Java image processing program (http://rsb.info.nih.gov/ij/download.htm). To determine the poly(A)+ RNA retention phenotype, the fluorescence intensity of the nucleus and cytoplasm in the wild-type and mutant strains images was measured, and the ratio was calculated by dividing the average nuclear intensity by the average cytoplasmic intensity. Data were then normalized to the mex67-5 control sample set as 100% of poly(A)+ RNA retention. A minimum of 100 cells per image were analyzed per strain (37). Error bars represent SE for at least three independent experiments.
DNA microarray expression profiling
Each mutant strain was profiled four times from two independently inoculated cultures and was harvested in early mid-log phase in synthetic complete medium with 2% glucose. Dual-channel 70mer oligonucleotide arrays were used, with a common reference wild-type RNA, hybridized in dye-swap. Wild-type cultures grown alongside mutants were processed in parallel and also hybridized in dye-swap versus the common reference wild-type RNA sample. After quality control, normalization and dye-bias correction (38), the replicate mutant profiles were averaged and statistically compared with a collection of 200 wild-type cultures, using Limma and Benjamini–Hochberg correction as described in Lenstra et al. (39). The reported FC is the average of the four replicate mutant profiles versus the average of all the wild-types. Shown in the figures are all genes that change at least once significantly (P < 0.05, FC > 1.7) in any single mutant.
RESULTS
Sem1 influences physical interaction of Sus1 with TREX-2
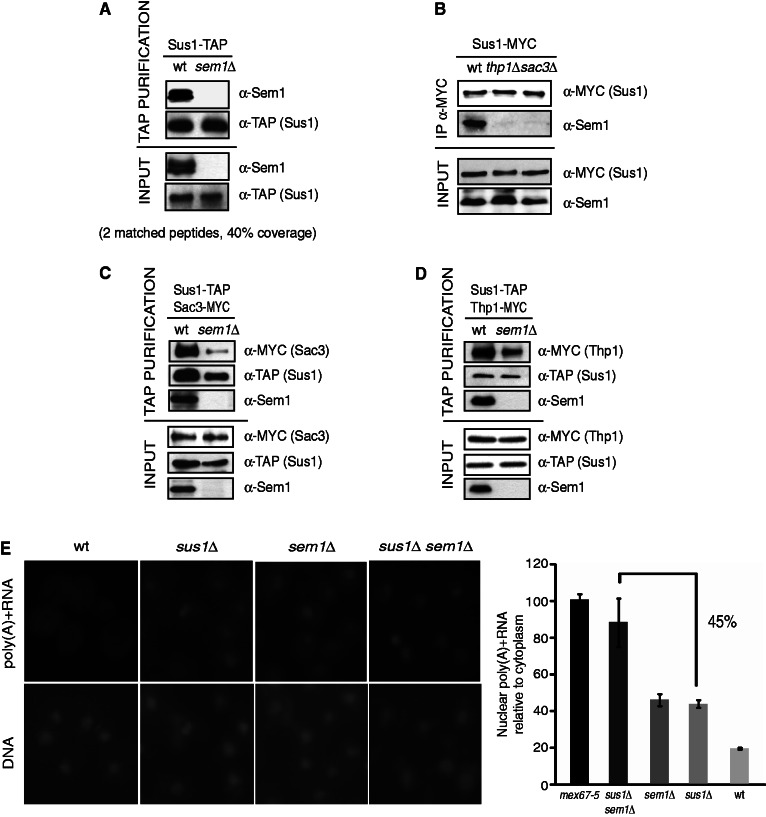
Sem1 is a component of both the proteasome (40) and the COP9 signalosome (41,42). It has also been found to interact with TREX-2 components Sac3 and Thp1 (42,43). More recently, the crystal structure of the Sac3–Thp1–Sem1 segment of the Saccharomyces cerevisiae TREX-2 complex has been solved (18). The small Sem1 protein wraps around the surface of Thp1 and makes few contacts with Sac3 (44). To investigate whether Sem1 interacts with Sus1, which happens for other TREX-2 subunits, Sus1 was TAP-purified, and the presence of Sem1 was analyzed. The western blot analysis using anti-Sem1 antibodies and the analysis of Sus1-TAP eluates by MudPIT reveal that Sem1 co-purifies with Sus1 via standard TAP purification (Figure 1A). To better characterize the Sus1–Sem1 interaction, its dependency on the presence of TREX-2 factors was also investigated. Figure 1B shows that both TREX-2 subunits Sac3 and Thp1 are required for the Sus1–Sem1 association. These results suggest that the interaction between Sus1 and Sem1 might be indirect and could be mediated by other TREX-2 components. Although considerable biochemical and structural information on the TREX-2 complex is available, the relevance of Sem1 in the Sus1 association with TREX-2 has not yet been investigated. Interestingly, although Sem1 and Sus1 have been found to interact with different regions of the complex, absence of Sem1 destabilizes the Sus1 association to TREX-2. Figure1C and D, respectively, illustrates how Sus1 binding to Sac3 and Thp1 is significantly reduced on Sem1 deletion. Thus, Sem1 influences the association of Sus1 with TREX-2, probably by stabilizing the complex or by promoting the assembly of the different subunits. The influence of Sem1 in Sus1 binding to TREX-2 prompted us to test whether Sem1 contributes to the role of Sus1 in mRNA export. Quantitation of the nuclear accumulation of poly(A)+ RNA in relation to cytoplasm concentration showed a 45% enrichment when SUS1 deletion was combined with SEM1 deletion (Figure 1E). Thus, the absence of SEM1 enhances the mRNA export defect provoked by SUS1 deletion, suggesting that they function independently to regulate mRNA export.
Figure 1.
Sem1 interacts physically with Sus1 and overlaps functionally during mRNA export. Sem1 alters Sus1 physical interaction with TREX-2 but not with SAGA. (A) TAP purification of Sus1 TAP-tagged wild-type and sem1Δ cells was performed. Presence of Sem1 within purified eluates was monitored by western blot using anti-Sem1 antibodies (top). Input levels of each protein before TAP purification were detected using anti-Sem1 and anti-TAP antibodies (bottom). Analyses of Sus1-TAP eluates by MudPIT identified two matched Sem1 peptides with 40% sequence coverage. (B) Exponentially growing Sus1-MYC (wild-type), Sus1-MYC thp1Δ and sac3Δ cells were lysed, and MYC-tagged Sus1 was immunoprecipitated from WCEs. Presence of Sem1 and levels of Sus1-MYC bait protein were monitored in immunoprecipitated eluates (top) and inputs (bottom) using the anti-Sem1 and anti-MYC antibodies. (C) The TAP purification of Sus1-TAP Sac3-MYC (wild-type) and Sus1-TAP Sac3-MYC sem1Δ cells was performed. Presence of Sac3 and Sus1 within the purified eluates was revealed using anti-MYC and anti-TAP antibodies (top). The input levels of each protein before TAP purification were detected using the aforementioned antibodies (bottom). (D) The TAP purification of Sus1-TAP Thp1-MYC (wild-type) and Sus1-TAP Thp1-MYC sem1Δ cells was performed. Presence of Thp1 and Sus1 within the purified eluates was revealed using the anti-MYC and anti-TAP antibodies (top). The input levels of each protein were detected using the aforementioned antibodies (bottom). (E) Analysis of nuclear mRNA export in sus1Δ, sem1Δ and sus1Δsem1Δ cells grown at 30°C in YP + glucose. The localization of poly(A)+ RNA was assessed by in situ hybridization using Cy3-labeled oligo(dT) probes. Histogram representation shows the quantitative analysis of the mRNA export defect. Quantification of the cytoplasmic/nuclear fluorescence intensity ratio was performed. Values were plotted as an FC normalized with the mex67-5 ratio, and they were set a 100% export defect. Error bars represent standard error (SE) for at least three independent experiments.
Sem1 interacts functionally with SAGA
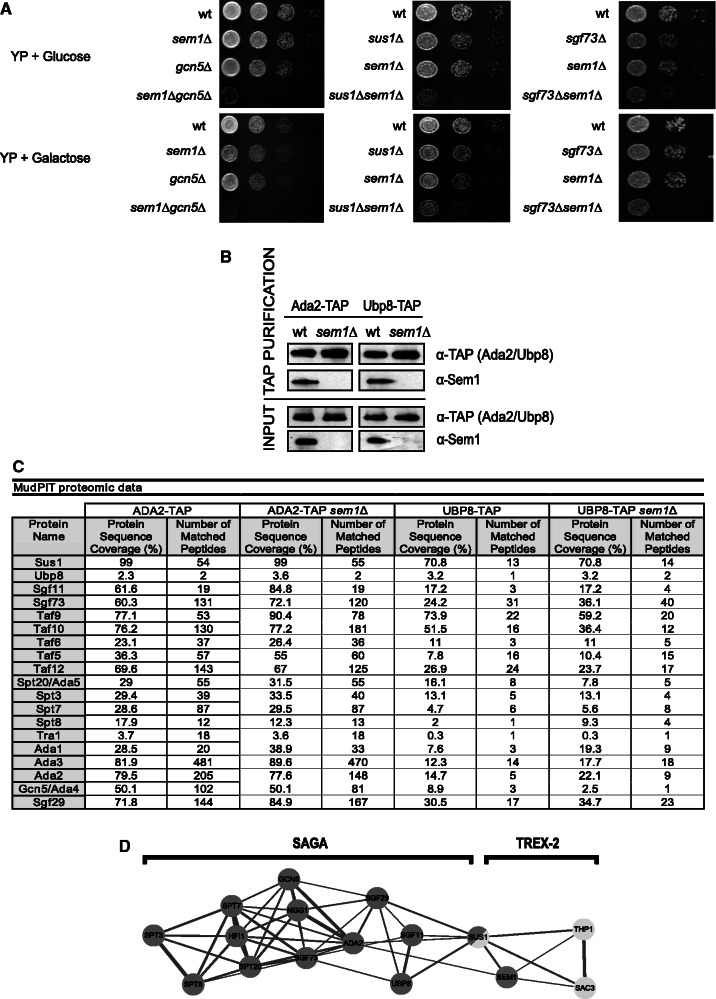
Genetic and physical interactions between components of TREX-2 and SAGA in addition to Sus1 have been shown (45). For instance, mutants of SAC3 and THP1 are synthetic lethal with the deletion of SGF73 (46). To test whether SEM1 is also genetically related to SAGA, double mutants bearing SEM1 deletion were constructed in cells lacking GCN5, SGF73 and SUS1, respectively. As shown in Figure 2A, absence of Sem1 leads to an enhancement of the poor growth exhibit by each single mutant, suggesting that these factors control the same biological function. Both Sac3 and Thp1 co-purify with Ada2 (7,46). To answer whether Sem1 also co-purifies with SAGA, Ada2-TAP and UBP8-TAP were affinity purified from wild-type (wt) and sem1Δ strains. As shown in Figure 2B, Sem1 co-enriches with SAGA subunits. Nevertheless, the purification of the SAGA complex in cells lacking SEM1 did not show significant differences in the composition of the SAGA complex from wild-type cells (Figure 2C). Thus, Sem1 interacts genetically and physically with SAGA, but it is dispensable for its biochemical integrity.
Figure 2.
Sem1 interacts genetically and physically with some SAGA subunits. (A) Wild-type, gcn5Δ, sus1Δ and sgf73Δ single mutant cells combined with SEM1 deletion were diluted in 10−1 steps, and equivalent amount of cells were spotted onto YP + glucose and YP + galactose plates. Cells were grown for 48 h at 30°C. (B) The TAP purification of Ada2 and Ubp8 TAP-tagged wild-type and sem1Δ cells was performed. The presence of Sem1 and levels of Ada2-TAP and Ubp8-TAP bait proteins were monitored in the purified eluates (top) and inputs (bottom) using the anti-Sem1 and anti-TAP antibodies. (C) The TAP immunoprecipitation of Ada2 and Ubp8 TAP-tagged wild-type and sem1Δ cells was performed. TAP-purified protein complexes were TCA-precipitated, LysC/trypsin-digested and subjected to MudPIT by (Matrix-assisted laser desorption/ionisation-time of flight mass spectrometry) MALDI-TOF-MS. All the SAGA subunits were identified. The number of matched peptides indicates the number of peptides that match the full-length protein, whereas protein sequence coverage indicates the percentage of total protein covered by the matched peptides. (D) A network of the relationships among Sem1, SAGA and TREX-2. Nodes represent mutated components. Edges between nodes are drawn if the correlation between the two signatures was >0.3. The thickness of the edge corresponds to the strength of the correlation. The network was generated using an edge-weighted, spring-embedded network algorithm (47).
In light of these genetic and physical interactions between Sem1 and SAGA, we further investigated the role of Sem1 in transcription. Thus, DNA microarray gene expression profile was generated for sem1Δ and was compared with the profiles of the mutants in the components of TREX-2 and SAGA, all of which were generated identically (39). Supplementary Figure S1A shows a hierarchical cluster diagram of the color-coded profiles according to complex composition. The dendrogram of similarities indicates that the gene expression changes observed in sem1Δ correspond most closely to those of sus1Δ. These profiles are also similar to the expression changes observed in the mutation of TREX-2 components THP1 and SAC3 (see Supplementary Figure S1B for all pairwise correlations). An unsupervised network recapitulates all these correlations and positions Sus1 as playing an intermediary role between SAGA and TREX-2 (Figure 2D). Interestingly, the sem1Δ transcriptional profile also correlates with ada2Δ to a lesser extent. Moreover, our analyses show that the expression changes of sem1Δ are enriched by genes with TATA-containing promoters with a P-value of <0.0025, these generally being SAGA-dominated genes (48,49). In summary, these results suggest that Sem1 plays a role in transcription in a similar manner to Sus1.
Sem1 is required for the induction of SAGA-regulated genes GAL1 and ARG1
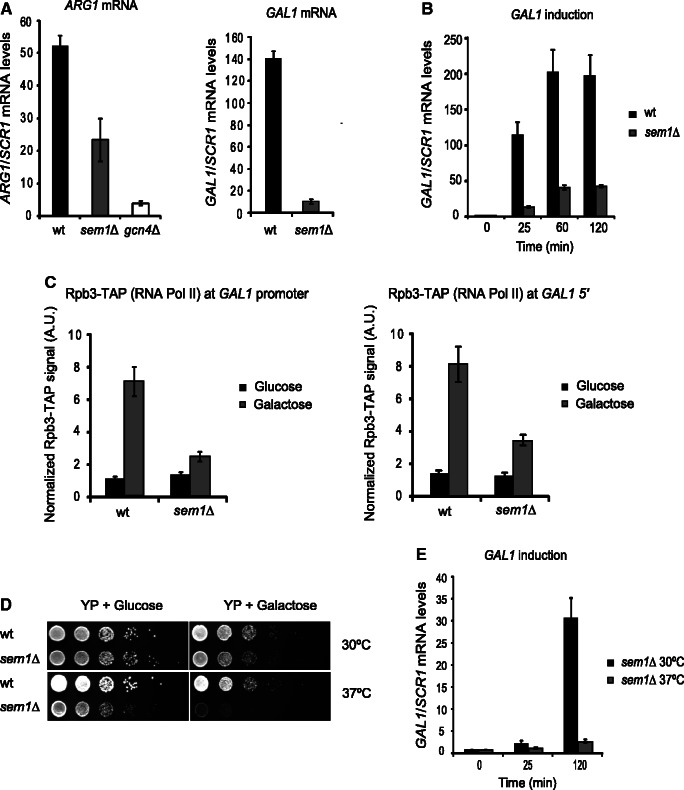
Our results suggest that Sem1 could play a role in the transcription of SAGA-dependent genes. Induction of SAGA-regulated genes, such as GAL1 and ARG1, has been shown to depend on Sus1 (7,50); thus, the requirement of Sem1 for the induction of both genes was also investigated. Figure 3A shows that the induction of ARG1 by the addition of sulfometuron (SM) or GAL1 on a 25-min shift to galactose-containing medium does not reach the wild-type expression levels in cells lacking SEM1. To better follow the effect of loss of Sem1 on the GAL1 expression, GAL1 mRNA abundance was monitored during galactose induction at different time points. The absence of SEM1 reduces GAL1 induction after a shift to galactose (Figure 3B). In accordance with poor GAL1 induction, the cells lacking SEM1 displayed a 3-fold depletion of RNA pol II at the GAL1 promoter and in coding regions on the addition of galactose (Figure 3C). Thus, RNA pol II recruitment to GAL1 diminished in those cells lacking SEM1. As expected with inefficient GAL1 induction, sem1Δ mutant cells grew poorly on galactose-containing plates (Figure 3D). Interestingly, this phenotype was greatly enhanced by raising the growth temperature to 37°C (Figure 3D lower right), which correlated with a downregulation of the GAL1 mRNA expression levels in sem1Δ at that temperature (Figure 3E).
Figure 3.
Sem1 is necessary for the induction of GAL1 and ARG1. (A) ARG1 induction in wild-type, sem1Δ and gcn4Δ cells was performed with addition of SM for 25 min. GAL1 induction in wild-type and sem1Δ cells was performed by a shift from 2% raffinose to 2% galactose medium for 25 min. Relative GAL1 and ARG1 mRNA levels were determined by qPCR and normalized to the SCR1 mRNA levels. Values were referred to each strain at the zero time point. Error bars represent SE for at least three independent experiments. (B) The time course analysis of GAL1 induction in wild-type and sem1Δ cells for 120 min in the presence of 2% galactose. The relative GAL1 mRNA levels were normalized to SCR1. Values were referred to each strain at the zero time point. Error bars represent SE for at least three independent experiments. (C) GAL1 promoter and 5′-end RNA pol II occupancy was monitored by ChIP analysis in Rpb3-TAP (wild-type) and Rpb3-TAP sem1Δ cells after 25 min of galactose induction. The RNA pol II occupancy level was calculated as the signal ratio from IP samples in relation to the input signal minus the background of a no-antibody control. The resulting normalized ratios were plotted. Error bars represent SE for at least three independent experiments. The qPCR primer sequences are provided in Supplementary Table S2. (D) Serial dilutions of wild-type and sem1Δ strains were grown in glucose- and galactose-containing plates. Plates were incubated for 48 h at 30°C and 37°C. (E) Time course analysis of GAL1 induction in sem1Δ cells at 30°C and 37°C for 120 min in the presence of 2% galactose. The relative GAL1 mRNA levels were normalized to SCR1. Values were referred to the 30°C or the 37°C condition at the zero time point. Error bars represent SE for at least three independent experiments.
Sem1 is required for SAGA integrity at the GAL1 promoter
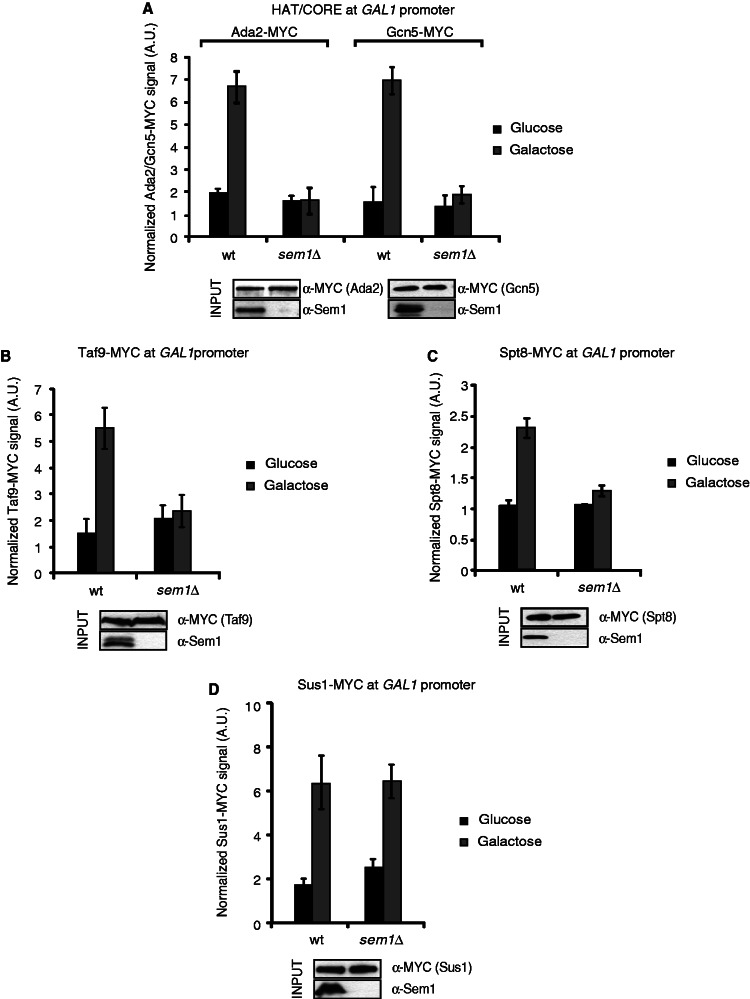
As shown earlier in the text, Sem1 is required for the expression of some SAGA-dependent genes. A straightforward explanation would be that SAGA components are not efficiently recruited to the GAL1 gene in sem1Δ cells. To investigate this notion, the recruitment of several SAGA subunits, which are the components of distinct modules, was monitored. Recruitment of the components of the HAT/CORE (Ada2 and Gcn5), SA_TAF (Taf9), SA_SPT (Spt8) and the DUB (Sus1) modules was analyzed. As shown in Figure 4A, the presence of HAT/CORE subunits Ada2 and Gcn5 is significantly reduced in the absence of Sem1. In addition to the poor recruitment of the HAT/CORE subunits, the presence of Taf9 (Figure 4B) and Spt8 (Figure 4C) from SA_TAF and SA_SPT, respectively, also diminished at GAL1 in sem1Δ cells. However, the recruitment of Sus1, to the same DNA region, was not significantly affected under similar conditions (Figure 4D). In summary, the ChIP results shown herein suggest that Sem1 influences SAGA integrity at the GAL1 promoter, but not Sus1 association.
Figure 4.
Recruitment of certain SAGA subunits to GAL1 promoter depends on Sem1. (A) Ada2 and Gcn5 (HAT/CORE) occupancy at the GAL1 promoter was monitored by the ChIP analysis in wild-type and sem1Δ cells after 25 min of galactose induction. The occupancy level was calculated as the signal ratio of IP samples in relation to the input signal minus the background of a no-antibody control. The resulting normalized ratios were plotted. The input level of each protein was detected from the inputs by western blot using the indicated antibodies (bottom). (B) The Taf9 (SA_TAF) occupancy at the GAL1 promoter was monitored by ChIP analysis in wild-type and sem1Δ cells after 25 min of galactose induction. The occupancy level was calculated as in A. The resulting normalized ratios were plotted. The input level of each protein was detected from the lysates as in A (bottom). (C) The Spt8 (SA_SPT) occupancy at the GAL1 promoter was monitored by ChIP analysis in wild-type and sem1Δ cells after 25 min of galactose induction. The occupancy level was calculated as in A. The resulting normalized ratios were plotted. The input level of each protein was detected as in A (bottom). (D) The Sus1 (DUB) occupancy at the GAL1 promoter was monitored by ChIP analysis in wild-type and sem1Δ cells after 25 min of galactose induction. The occupancy level was calculated as in A. The resulting normalized ratios were plotted. The input level of each protein was detected as in A (bottom). Error bars represent SE for at least three independent experiments. All the qPCR primer sequences are provided in Supplementary Table S2.
Sem1 influences SAGA-dependent deubiquitylation activity
As shown earlier in the text, Sus1 was associated with GAL1 on the shift to galactose in sem1Δ, whereas other SAGA subunits are absent. However, under our experimental conditions, we were unable to demonstrate whether Ubp8 recruitment to the GAL1 promoter was affected in absence of SEM1. Thus, we ought to investigate whether SAGA-mediated DUB activity at GAL1 was compromised. To monitor the in vivo H2B ubiquitylation status at the GAL1 promoter, real-time qPCR after ChDIP was performed. Figure 5A shows that the cells lacking SEM1 displayed a 3-fold enrichment of ubiquitylated H2B at the GAL1 promoter. Levels of ubiquitylated histone H2B correlated with the H3K79 trimethylation status at the GAL1 promoter (51–54). Accordingly, H3K79 trimethylation levels abnormally increased at the GAL1 promoter on gene activation in sem1Δ cells (Supplementary Figure S2). Hence, Sem1 is required for in vivo SAGA-mediated histone H2B deubiquitylation and for H3K79 trimethylation at the GAL1 promoter.
Figure 5.
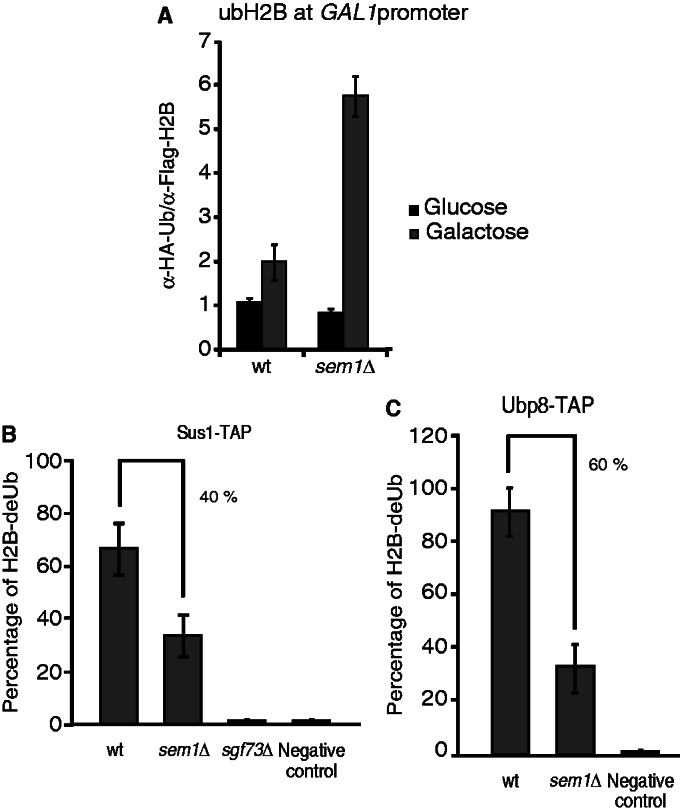
Sem1 enhances SAGA-mediated histone H2B deubiquitylation activity. (A) ChDIP of ubH2B under galactose induction is shown. Wild-type and sem1Δ strains bearing Flag-tagged H2B and HA-tagged ubiquitin were grown in raffinose and shifted to a galactose-containing medium for 90 min. Eluted DNA was analyzed by qPCR. The ubiquitylated H2B occupancy at the GAL1 promoter was calculated as the signal ratio of IP samples in relation to the input signal minus the H2BK123R and H2BK123R sem1Δ mutant strain signal, set as a background control. Error bars represent SE for at least three independent experiments. All the qPCR primer sequences are provided in Supplementary Table S2. (B) Sus1-TAP wild-type and sem1Δ cells were subjected to an in vitro deubiquitylation assay. The TAP purification of Sus1 TAP-tagged cells was performed, and native calmoduline eluates were incubated with purified histone H2B. Sus1-TAP sgf73Δ was established as a negative control. (C) Ubp8-TAP wild-type and sem1Δ cells were subjected to an in vitro deubiquitylation assay as in B. Sole purified histone H2B was incubated and used as a negative control. Error bars represent SE for at least three independent experiments. Error bars represent SE for at least three independent experiments.
The biochemical analyses shown in Figure 2C indicate that Sem1 is not necessary for SAGA integrity. However, at GAL1 promoter, Sem1 is required for SAGA-mediated deubiquitylation of H2B (Figure 5A). To further characterize whether the DUB module is partially inactive in sem1Δ cells, in vitro H2B deubiquitylation assays, with the SAGA complex purified via Sus1-TAP from wild-type and sem1Δ cells, were carried out. As Sgf73 is required for SAGA DUB activity, Sus1 purified via TAP in sgf73Δ was used as a negative control (20,46,55,56). Figure 5B shows that SEM1 deletion reduced in vitro H2B Sus1-dependent DUB activity by decreasing in 40% level of H2B-deUB. Moreover, the level of H2B-deUB is reduced in 60% when the complex was purified by Ubp8-TAP from sem1Δ (Figure 5C and Supplementary Figure S3). Thus, these results indicate that Sem1 enhances SAGA-mediated histone H2B deubiquitylation activity in vitro.
TREX-2 is involved in enhancing SAGA-dependent deubiquitylation activity
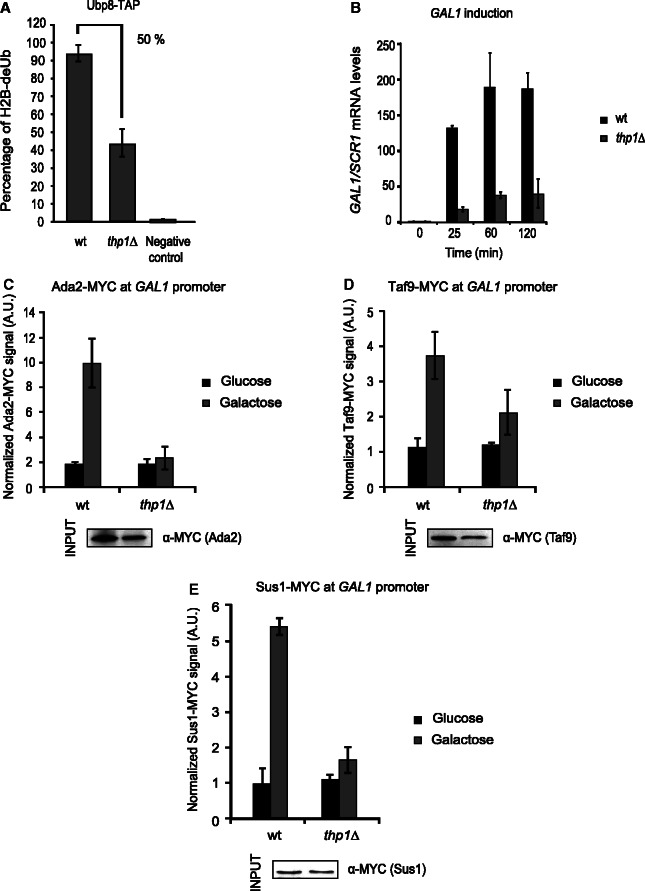
Sem1 affects the recruitment of different SAGA subunits and enhances SAGA-mediated histone H2B deubiquitylation. Whether this is related to its role in stabilizing TREX-2 is unknown. To test whether the mutation in other TREX-2 subunit also lead to similar phenotypes, in vitro deubiquitylation activity, GAL1 induction and SAGA recruitment were investigated in a thp1Δ strain. As shown in Figure 6A, the absence of Thp1 leads to a significant reduction of Ubp8-mediated DUB activity, similarly to that observed for Sem1 deletion (Figure 5C). Moreover, as observed for sem1Δ, the absence of THP1 impairs GAL1 induction (Figure 6B). Finally, the ChIP experiments done to monitor recruitment of different SAGA factors to the GAL1 promoter in the wild-type and thp1Δ on the shift to galactose reveal that THP1 deletion decreased the correct recruitment of distinct SAGA subunits to the GAL1 promoter (Figure 6C and D). As it happens for sac3Δ (50), the presence of Sus1 is also compromised in the thp1Δ under the same conditions (Figure 6E). In conclusion, TREX-2 subunit Thp1 is needed for a precise SAGA recruitment to GAL1. All in all, the data herein suggest that Sem1 influences the GAL1 expression by a mechanism that interferes with SAGA-DUB-mediated activity and by stabilizing the TREX-2 complex.
Figure 6.
The TREX-2 subunit Thp1 influences SAGA–DUB-mediated activity. (A) Ubp8-TAP wild-type and thp1Δ cells were subjected to an in vitro deubiquitylation assay. The TAP purification of Ubp8 TAP-tagged cells was performed, and native calmoduline eluates were incubated with purified histone H2B. Sole purified histone H2B was incubated and used as a negative control. Error bars represent SE for at least three independent experiments. (B) The time course analysis of GAL1 induction in wild-type and thp1Δ cells for 120 min in the presence of 2% galactose. The relative GAL1 mRNA levels were normalized to SCR1. Values were referred to each strain at the zero time point. Error bars represent SE for at least three independent experiments. (C) The Ada2 (HAT/CORE) occupancy at the GAL1 promoter was monitored by ChIP analysis in wild-type and thp1Δ cells after 25 min of galactose induction. The occupancy level was calculated as the signal ratio of IP samples in relation to the input signal minus the background of a no-antibody control. The resulting normalized ratios were plotted. Levels of the Ada2-MYC bait protein were detected from the inputs by western blot using the anti-MYC antibodies (bottom). (D) The Taf9 (SA_TAF) occupancy at the GAL1 promoter was monitored by ChIP analysis in wild-type and thp1Δ cells after 25 min of galactose induction. The occupancy level was calculated as in C. The resulting normalized ratios were plotted. Levels of the Taf9-MYC bait protein were detected from the inputs as in C (bottom). (E) The Sus1 (DUB) occupancy at the GAL1 promoter was monitored and calculated as in C. The resulting normalized ratios were plotted. Levels of the Sus1-MYC bait protein were detected from the inputs as in C (bottom). Error bars represent SE for at least three independent experiments. All the qPCR primer sequences are provided in Supplementary Table S2.
DISCUSSION
Sem1 is a versatile protein that functions in different cellular processes and interacts with several protein complexes, including the proteasome and TREX-2 (41–43,57). However, its role in transcription regulation remains unknown. In this study, we demonstrate a novel role for Sem1 in the transcription of certain SAGA-dependent genes via their impact on SAGA recruitment and histone H2B deubiquitylation activity.
It has been proposed that Sem1 serves as a molecular adhesive of small subcomplexes in larger molecular assemblies (41). Previous studies have shown that Sem1 forms part of TREX-2 and contributes to mRNA export (42,43,58). More recently, the work by Stewart and colleagues (18) has reported the structural basis for the in vitro assembly of a portion of TREX-2-containing Sem1. The structures solved to date show that Sus1 binds to Sac3, whereas Sem1 binds to Thp1 (18,22,59,60). As expected from these data, our in vivo biochemical results confirm that the Sus1 association with Sem1 depends on both Sac3 and Thp1 (Figure 1). Surprisingly, the absence of Sem1 leads to a reduction in Sus1 association with TREX-2, which suggests a more complicated pattern of interactions among different TREX-2 components than inferred from the structural analysis. Despite our knowledge having substantially improved in recent years, solving the TREX-2 complex structure, including all its subunits, to gain a better understanding of its nature in vivo would be of much interest.
TREX-2 is functionally connected to SAGA through its shared subunit, Sus1, which plays roles in both mRNA export and transcription (7). The transcriptional profile data presented herein further support an intermediary role for Sus1 between SAGA and TREX-2. Along these lines, Sem1 is functionally closer to Sus1 than to other TREX-2 factors. The correlation between the transcriptional profiles of sem1Δ as compared with the sus1Δ, sac3Δ and thp1Δ profiles is 0.41, 0.32 and 0.32, respectively. Hence, it is conceivable that Sem1 and Sus1 are also linked in its function in transcription. The different analyses presented in the present work support a novel role for Sem1 in the transcription of SAGA-dependent genes. First, the DNA microarray data indicate an enrichment of the SAGA-dependent genes for sem1Δ. Second, the induction of SAGA-regulated genes ARG1 and GAL1 in the absence of SEM1 is poor. Third, the GAL1 ChIP experiments demonstrate a poor recruitment of RNA pol II and several SAGA factors dependent on SEM1. A vast number of studies describe how GAL1 and ARG1 are regulated (61–64). The SAGA complex is targeted to the promoter of these genes, and they are induced through a series of distinct histone modifications (14,65). It is known that the control of histone H2B ubiquitylation levels is crucial for GAL1 regulation (24). In previous studies, we demonstrated that Sus1 is recruited to GAL1 and ARG1, and that it influences their expression by the role it plays in SAGA and TREX-2 (23,50). Here, we find that Sem1 is required for the assembly of certain SAGA subunits at the GAL1 promoter and for correct H2B deubiquitylation. In the light of these findings, an important question to answer is raised: whether this reduction in enzymatic activity is due to a direct effect of Sem1 on SAGA integrity or is the result of Sem1’s role in TREX-2. On one hand, Sem1 might have a direct role in influencing DUB activity by stabilizing the structural features within SAGA. In this sense, the structural similarities between the DUB module of SAGA and the TREX-2 complex have been reported (66). On the other hand, by stabilizing TREX-2, Sem1 might influence the SAGA–DUB interaction which, in turn, activates Ubp8. To support this model, the direct partner of Sem1 in TREX-2, Thp1, is also required for the transcription of GAL1, the recruitment of certain SAGA subunits to chromatin and for in vitro H2B DUB activity (Figure 6). All in all, we cannot exclude the possibility that Sem1 controls other(s) factor(s) or signal(s) that influence the cross-talk between SAGA and TREX-2 to activate GAL1 expression. It is worth to note that the ChIP analyses shown here indicate that the HAT/Core components Gcn5 and Ada2 are poorly recruited in the absence of Sem1. Thus, the hypothesis that Sem1 could affect the expression of GAL1 not only by decreasing the deubiquitylating activity of SAGA but also by strongly affecting the recruitment of the acetylation module is an interesting open question that requires further work.
Sem1/DSS1 has been described as a multitasking organizer of PCI/MPN minicomplexes (41). These minicomplexes could incorporate into larger complexes either with or without the presence of Sem1. Following the idea that Sem1 might serve as a facilitator of larger complexes assembly/interactions, and considering that TREX-2 subunits co-purify with SAGA, it is tempting to hypothesize that Sem1 helps assemble and stabilize the interactions between these two complexes (Figure 7). Remarkably, all the complexes linked to Sem1 are related to deubiquitylating enzymes either directly or indirectly. Future work addressing the intimate link between Sem1 and the ubiquitin system will be of much interest for different research areas.
Figure 7.
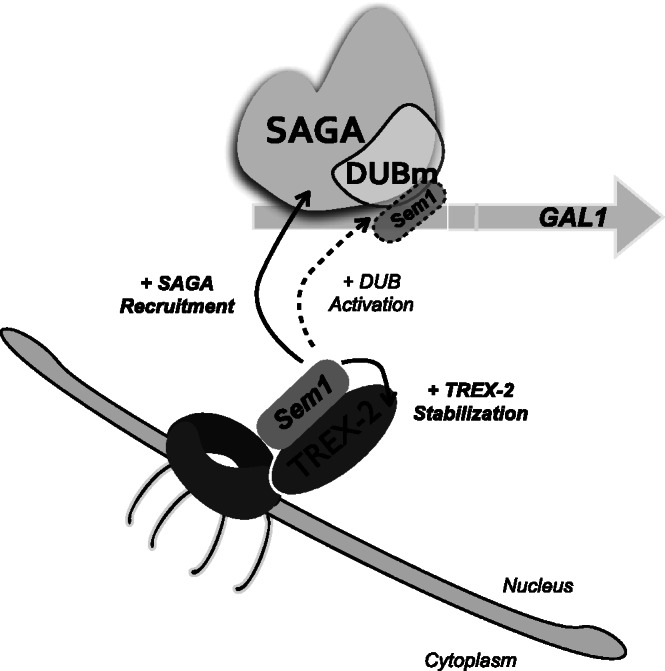
Schematic drawing showing the role of Sem1 in SAGA-dependent transcription. Sem1 interacts with TREX-2 and it is necessary for stabilization of the complex at the NPC. At the GAL1 gene promoter, recruitment of certain SAGA subunits is influenced by the presence of Sem1. Notably, Sem1 enhances the SAGA-dependent deubiquitylation activity, both in vivo (dash line) and in vitro. Whether Sem1 associates to SAGA at the NPC level remains unknown.
In summary, these results support a model of concomitant SAGA recruitment, TREX-2 stabilization and DUB activation to induce SAGA-regulated gene GAL1. A challenge for future studies is to decipher how each complex and their submodules act in concert and how general this scenario is.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3 and Supplementary Figures 1–3.
ACKNOWLEDGEMENTS
The authors are grateful to Dr Martínez-Jimenez (holder of a FEBS postdoctoral fellowship) for her initial contributions to the ChIP of Supplementary Figure S2 and expression experiments, Dr Kobayashi for sending the anti-Sem1 polyclonal antibodies and Dr Chavez and Dr Capelson for the fruitful comments made on our work. Dr Valero’s excellent technical assistance on Mass Spectrometry Service is also acknowledged. They thank all the I-25 laboratory members for scientific comments and A. Llopis for technical assistance.
FUNDING
MINECO [BFU2011-23418 to S.R.N.]; GV [PROMETEO/2012/061 to S.R.N.]; CIPF and FPU [AP2009-0917] to E.G.O. and V.G.M., respectively programs. Funding for open access charge: MINECO [BFU2011-23418 to S.R.N.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Liang Y, Hetzer MW. Functional interactions between nucleoporins and chromatin. Curr. Opin. Cell Biol. 2011;23:65–70. doi: 10.1016/j.ceb.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tutucci E, Stutz F. Keeping mRNPs in check during assembly and nuclear export. Nat. Rev. Mol. Cell Biol. 2011;12:377–384. doi: 10.1038/nrm3119. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez-Navarro S, Hurt E. Linking gene regulation to mRNA production and export. Curr. Opin. Cell Biol. 2011;23:302–309. doi: 10.1016/j.ceb.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Strasser K, Hurt E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 2000;19:410–420. doi: 10.1093/emboj/19.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Del Rosario BC, Pemberton LF. Nap1 links transcription elongation, chromatin assembly, and messenger RNP complex biogenesis. Mol. Cell Biol. 2008;28:2113–2124. doi: 10.1128/MCB.02136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodriguez-Navarro S, Fischer T, Luo MJ, Antunez O, Brettschneider S, Lechner J, Perez-Ortin JE, Reed R, Hurt E. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116:75–86. doi: 10.1016/s0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- 8.Green DM, Marfatia KA, Crafton EB, Zhang X, Cheng X, Corbett AH. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J. Biol. Chem. 2002;277:7752–7760. doi: 10.1074/jbc.M110053200. [DOI] [PubMed] [Google Scholar]
- 9.Blobel G. Gene gating: a hypothesis. Proc. Natl Acad. Sci. USA. 1985;82:8527–8529. doi: 10.1073/pnas.82.24.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dieppois G, Stutz F. Connecting the transcription site to the nuclear pore: a multi-tether process that regulates gene expression. J. Cell Sci. 2010;123:1989–1999. doi: 10.1242/jcs.053694. [DOI] [PubMed] [Google Scholar]
- 11.Schell MA, Wilson DB. Purification and properties of galactokinase from Saccharomyces cerevisiae. J. Biol. Chem. 1977;252:1162–1166. [PubMed] [Google Scholar]
- 12.Maya D, Quintero MJ, de la Cruz Munoz-Centeno M, Chavez S. Systems for applied gene control in Saccharomyces cerevisiae. Biotechnol. Lett. 2008;30:979–987. doi: 10.1007/s10529-008-9647-z. [DOI] [PubMed] [Google Scholar]
- 13.Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin JC, Hurt EC, et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- 14.Koutelou E, Hirsch CL, Dent SY. Multiple faces of the SAGA complex. Curr. Opin. Cell Biol. 2010;22:374–382. doi: 10.1016/j.ceb.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez-Navarro S. Insights into SAGA function during gene expression. EMBO Rep. 2009;10:843–850. doi: 10.1038/embor.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rondon AG, Jimeno S, Aguilera A. The interface between transcription and mRNP export: from THO to THSC/TREX-2. Biochim. Biophys. Acta. 2010;1799:533–538. doi: 10.1016/j.bbagrm.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 17.Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat. Rev. Mol. Cell Biol. 2010;11:490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- 18.Ellisdon AM, Dimitrova L, Hurt E, Stewart M. Structural basis for the assembly and nucleic acid binding of the TREX-2 transcription-export complex. Nat. Struct. Mol. Biol. 2012;19:328–336. doi: 10.1038/nsmb.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallardo M, Luna R, Erdjument-Bromage H, Tempst P, Aguilera A. Nab2p and the Thp1p-Sac3p complex functionally interact at the interface between transcription and mRNA metabolism. J. Biol. Chem. 2003;278:24225–24232. doi: 10.1074/jbc.M302900200. [DOI] [PubMed] [Google Scholar]
- 20.Samara NL, Datta AB, Berndsen CE, Zhang X, Yao T, Cohen RE, Wolberger C. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science. 2010;328:1025–1029. doi: 10.1126/science.1190049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohler A, Zimmerman E, Schneider M, Hurt E, Zheng N. Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell. 2010;141:606–617. doi: 10.1016/j.cell.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilmes GM, Guthrie C. Getting to the gate: crystallization of a Sac3(CID):Sus1:Cdc31 complex. Mol. Cell. 2009;33:671–672. doi: 10.1016/j.molcel.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 23.Kohler A, Pascual-Garcia P, Llopis A, Zapater M, Posas F, Hurt E, Rodriguez-Navarro S. The mRNA export factor Sus1 is involved in Spt/Ada/Gcn5 acetyltransferase-mediated H2B deubiquitinylation through its interaction with Ubp8 and Sgf11. Mol. Biol. Cell. 2006;17:4228–4236. doi: 10.1091/mbc.E06-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chandrasekharan MB, Huang F, Sun ZW. Histone H2B ubiquitination and beyond: regulation of nucleosome stability, chromatin dynamics and the trans-histone H3 methylation. Epigenetics. 2010;5:460–468. doi: 10.4161/epi.5.6.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weake VM, Workman JL. Inducible gene expression: diverse regulatory mechanisms. Nat. Rev. Genet. 2010;11:426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- 27.Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell Biol. 2005;25:637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 29.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 30.Graumann J, Dunipace LA, Seol JH, McDonald WH, Yates JR, III, Wold BJ, Deshaies RJ. Applicability of tandem affinity purification MudPIT to pathway proteomics in yeast. Mol. Cell. Proteomics. 2004;3:226–237. doi: 10.1074/mcp.M300099-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Trujillo KM, Tyler RK, Ye C, Berger SL, Osley MA. A genetic and molecular toolbox for analyzing histone ubiquitylation and sumoylation in yeast. Methods. 2011;54:296–303. doi: 10.1016/j.ymeth.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chandrasekharan MB, Huang F, Sun ZW. Decoding the trans-histone crosstalk: methods to analyze H2B ubiquitination, H3 methylation and their regulatory factors. Methods. 2011;54:304–314. doi: 10.1016/j.ymeth.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Green EM, Jiang Y, Joyner R, Weis K. A negative feedback loop at the nuclear periphery regulates GAL gene expression. Mol. Biol. Cell. 2012;23:1367–1375. doi: 10.1091/mbc.E11-06-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dasgupta A, Juedes SA, Sprouse RO, Auble DT. Mot1-mediated control of transcription complex assembly and activity. EMBO J. 2005;24:1717–1729. doi: 10.1038/sj.emboj.7600646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol. Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 37.Auer A, von Blume J, Sturany S, von Wichert G, Van Lint J, Vandenheede J, Adler G, Seufferlein T. Role of the regulatory domain of protein kinase D2 in phorbol ester binding, catalytic activity, and nucleocytoplasmic shuttling. Mol. Biol. Cell. 2005;16:4375–4385. doi: 10.1091/mbc.E05-03-0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Margaritis T, Lijnzaad P, van Leenen D, Bouwmeester D, Kemmeren P, van Hooff SR, Holstege FC. Adaptable gene-specific dye bias correction for two-channel DNA microarrays. Mol. Syst. Biol. 2009;5:266. doi: 10.1038/msb.2009.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lenstra TL, Benschop JJ, Kim T, Schulze JM, Brabers NA, Margaritis T, van de Pasch LA, van Heesch SA, Brok MO, Groot Koerkamp MJ, et al. The specificity and topology of chromatin interaction pathways in yeast. Mol. Cell. 2011;42:536–549. doi: 10.1016/j.molcel.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Funakoshi M, Li X, Velichutina I, Hochstrasser M, Kobayashi H. Sem1, the yeast ortholog of a human BRCA2-binding protein, is a component of the proteasome regulatory particle that enhances proteasome stability. J. Cell Sci. 2004;117:6447–6454. doi: 10.1242/jcs.01575. [DOI] [PubMed] [Google Scholar]
- 41.Pick E, Hofmann K, Glickman MH. PCI complexes: beyond the proteasome, CSN, and eIF3 Troika. Mol. Cell. 2009;35:260–264. doi: 10.1016/j.molcel.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 42.Wilmes GM, Bergkessel M, Bandyopadhyay S, Shales M, Braberg H, Cagney G, Collins SR, Whitworth GB, Kress TL, Weissman JS, et al. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol. Cell. 2008;32:735–746. doi: 10.1016/j.molcel.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Faza MB, Kemmler S, Jimeno S, Gonzalez-Aguilera C, Aguilera A, Hurt E, Panse VG. Sem1 is a functional component of the nuclear pore complex-associated messenger RNA export machinery. J. Cell Biol. 2009;184:833–846. doi: 10.1083/jcb.200810059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ellisdon AM, Stewart M. Structural biology of the PCI-protein fold. Bioarchitecture. 2012;2 doi: 10.4161/bioa.21131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia-Oliver E, Garcia-Molinero V, Rodriguez-Navarro S. mRNA export and gene expression: the SAGA-TREX-2 connection. Biochim. Biophys. Acta. 2012;1819:555–565. doi: 10.1016/j.bbagrm.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 46.Kohler A, Schneider M, Cabal GG, Nehrbass U, Hurt E. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat. Cell Biol. 2008;10:707–715. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- 47.van Bakel H, Holstege FC. In control: systematic assessment of microarray performance. EMBO Rep. 2004;5:964–969. doi: 10.1038/sj.embor.7400253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Venters BJ, Wachi S, Mavrich TN, Andersen BE, Jena P, Sinnamon AJ, Jain P, Rolleri NS, Jiang C, Hemeryck-Walsh C, et al. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol. Cell. 2011;41:480–492. doi: 10.1016/j.molcel.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- 50.Pascual-García P, Govind CK, Queralt E, Cuenca-Bono B, Llopis A, Chavez S, Hinnebusch AG, Rodríguez-Navarro S. Sus1 is recruited to coding regions and functions during transcription elongation in association with SAGA and TREX2. Genes Dev. 2008;22:2811–2822. doi: 10.1101/gad.483308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakanishi S, Lee JS, Gardner KE, Gardner JM, Takahashi YH, Chandrasekharan MB, Sun ZW, Osley MA, Strahl BD, Jaspersen SL, et al. Histone H2BK123 monoubiquitination is the critical determinant for H3K4 and H3K79 trimethylation by COMPASS and Dot1. J. Cell Biol. 2009;186:371–377. doi: 10.1083/jcb.200906005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- 53.Shahbazian MD, Zhang K, Grunstein M. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol. Cell. 2005;19:271–277. doi: 10.1016/j.molcel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 54.Ezhkova E, Tansey WP. Proteasomal ATPases link ubiquitylation of histone H2B to methylation of histone H3. Mol. Cell. 2004;13:435–442. doi: 10.1016/s1097-2765(04)00026-7. [DOI] [PubMed] [Google Scholar]
- 55.Lee KK, Swanson SK, Florens L, Washburn MP, Workman JL. Yeast Sgf73/Ataxin-7 serves to anchor the deubiquitination module into both SAGA and Slik(SALSA) HAT complexes. Epigenetics Chromatin. 2009;2:2. doi: 10.1186/1756-8935-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Helmlinger D, Hardy S, Sasorith S, Klein F, Robert F, Weber C, Miguet L, Potier N, Van-Dorsselaer A, Wurtz JM, et al. Ataxin-7 is a subunit of GCN5 histone acetyltransferase-containing complexes. Hum. Mol. Genet. 2004;13:1257–1265. doi: 10.1093/hmg/ddh139. [DOI] [PubMed] [Google Scholar]
- 57.Faza MB, Kemmler S, Panse VG. Sem1: a versatile “molecular glue”? Nucleus. 2010;1:12–17. doi: 10.4161/nucl.1.1.10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mannen T, Andoh T, Tani T. Dss1 associating with the proteasome functions in selective nuclear mRNA export in yeast. Biochem. Biophys. Res. Commun. 2008;365:664–671. doi: 10.1016/j.bbrc.2007.11.024. [DOI] [PubMed] [Google Scholar]
- 59.Valkov E, Dean JC, Jani D, Kuhlmann SI, Stewart M. Structural basis for the assembly and disassembly of mRNA nuclear export complexes. Biochim. Biophys. Acta. 2012;1819:578–592. doi: 10.1016/j.bbagrm.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 60.Jani D, Lutz S, Marshall NJ, Fischer T, Köhler A, Ellisdon AM, Hurt E, Stewart M. Sus1, Cdc31, and the Sac3 CID region form a conserved interaction platform that promotes nuclear pore association and mRNA export. Mol. Cell. 2009;33:727–737. doi: 10.1016/j.molcel.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhaumik SR, Green MR. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 2001;15:1935–1945. doi: 10.1101/gad.911401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Govind CK, Yoon S, Qiu H, Govind S, Hinnebusch AG. Simultaneous recruitment of coactivators by Gcn4p stimulates multiple steps of transcription in vivo. Mol. Cell Biol. 2005;25:5626–5638. doi: 10.1128/MCB.25.13.5626-5638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ricci AR, Genereaux J, Brandl CJ. Components of the SAGA histone acetyltransferase complex are required for repressed transcription of ARG1 in rich medium. Mol. Cell Biol. 2002;22:4033–4042. doi: 10.1128/MCB.22.12.4033-4042.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Flick JS, Johnston M. Two systems of glucose repression of the GAL1 promoter in Saccharomyces cerevisiae. Mol. Cell Biol. 1990;10:4757–4769. doi: 10.1128/mcb.10.9.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Atanassov BS, Koutelou E, Dent SY. The role of deubiquitinating enzymes in chromatin regulation. FEBS Lett. 2010;585:2016–2023. doi: 10.1016/j.febslet.2010.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ellisdon AM, Jani D, Kohler A, Hurt E, Stewart M. Structural basis for the interaction between yeast Spt-Ada-Gcn5 acetyltransferase (SAGA) complex components Sgf11 and Sus1. J. Biol. Chem. 2010;285:3850–3856. doi: 10.1074/jbc.M109.070839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.