SUMMARY
TET proteins oxidize 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC). 5fC and 5caC are excised by mammalian DNA glycosylase TDG, implicating 5mC oxidation in DNA demethylation. Here we show that the genomic locations of 5fC can be determined by coupling chemical reduction with biotin tagging. Genome-wide mapping of 5fC in mouse embryonic stem cells (mESCs) reveals that 5fC preferentially occurs at poised enhancers among other gene regulatory elements. Application to Tdg null mESCs further suggests that 5fC production coordinates with p300 in remodeling epigenetic states of enhancers. This process, which is not influenced by 5hmC, appears to be associated with further oxidation of 5hmC and commitment to demethylation through 5fC. Finally, we resolved 5fC at base-resolution by hydroxylamine-based protection from bisulfite-mediated deamination, thereby confirming sites of 5fC accumulation. Our results reveal roles of active 5mC/5hmC oxidation and TDG-mediated demethylation in epigenetic tuning at regulatory elements.
INTRODUCTION
Epigenetic information encoded by 5-methylcytosine (5mC) has a profound influence on mammalian development and human disease (Klose and Bird, 2006). However, one of the most fundamental areas of interest, the active demethylation of 5mC in mammalian cells, has only recently been unveiled (Bhutani et al., 2011). Recently, 5mC was discovered to be further oxidized to 5-hydroxymethylcytosine (5hmC) by the TET family dioxygenases in mammalian cells (Kriaucionis and Heintz, 2009; Tahiliani et al., 2009). TET family dioxygenases can further oxidize 5hmC to 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) in a stepwise manner (He et al., 2011; Ito et al., 2011; Pfaffeneder et al., 2011). The later oxidation products 5fC and 5caC are recognized and excised by mammalian DNA glycosylase, TDG, and subsequently converted to cytosine through base excision repair (BER) (Cortazar et al., 2011; Cortellino et al., 2011; He et al., 2011; Maiti and Drohat, 2011; Zhang et al., 2012), resulting in an active DNA demethylation pathway in mammals.
Genomic profiling of 5hmC has revealed its association with genes and gene regulatory elements in particular, where 5hmC is most abundant and 5mC is depleted (Ficz et al., 2011; Khare et al., 2012; Pastor et al., 2011; Song et al., 2011; Stadler et al., 2011; Stroud et al., 2011; Szulwach et al., 2011; Williams et al., 2011; Wu et al., 2011a; Xu et al., 2011; Yu et al., 2012). Meanwhile, roles for 5hmC and TET-family proteins in normal development (Dawlaty et al., 2011; Doege et al., 2012; Gu et al., 2011; Ito et al., 2010; Koh et al., 2011; Williams et al., 2011; Xu et al., 2011) and disease (Ko et al., 2010; Lian et al., 2012; Moran-Crusio et al., 2011; Tan and Shi, 2012) continue to emerge. As a result, it is becoming clear that although present at low levels, oxidized forms of 5mC represent important dynamic epigenetic states at functional genomic elements, serving to modulate transcriptional programs. However, despite these emerging paradigms, accurate methods for studying 5mC oxidation beyond 5hmC are lacking.
To further understand how 5mC oxidation dynamics shape patterns of DNA methylation the genomic distribution of 5fC and/or 5caC must be determined because these modifications are “committed” to demethylation through BER. Unfortunately, 5fC and 5caC behave similarly to cytosine in bisulfite sequencing-based methods (Booth et al., 2012; Yu et al., 2012), and their low abundance in mammalian genomic DNA (only ppm of total cytosines in mESC (Ito et al., 2011)) make it challenging to effectively apply antibody-based immunoprecipitation, which typically works well with dense modifications (Pastor et al., 2011). Here we present two methods for the distinction of 5fC in genomic DNA. We first introduced a 5-formylcytosine selective chemical labeling (fC-Seal) approach for genome-wide profiling of 5fC. Second, we developed a chemically assisted bisulfite sequencing (fCAB-Seq) method for the base-resolution detection of 5fC. Application of these methods to mESCs, as well as Tdg null mESCs, revealed the genomic distribution and TDG-dependent regulation of 5fC. Genome-wide 5fC profiling further revealed distinct properties of 5mC/5hmC oxidation at various gene regulatory elements, beyond that afforded by 5hmC profiling alone. Our results show that 5fC is enriched at poised and active enhancers, but exhibits a preference to poised enhancers, suggesting a role for 5mC/5hmC oxidation to 5fC in the epigenetic priming of enhancers. Finally, in support of this role, we find that accumulation of 5fC in the absence of TDG correlates with increased binding of the transcriptional co-activator p300 at poised enhancers. Therefore, active 5mC/5hmC oxidation and TDG-coupled BER serve to dynamically regulate epigenetic states at functional regulatory elements in mammalian genome.
RESULTS
fC-Seal for selective chemical labeling and capture of 5fC
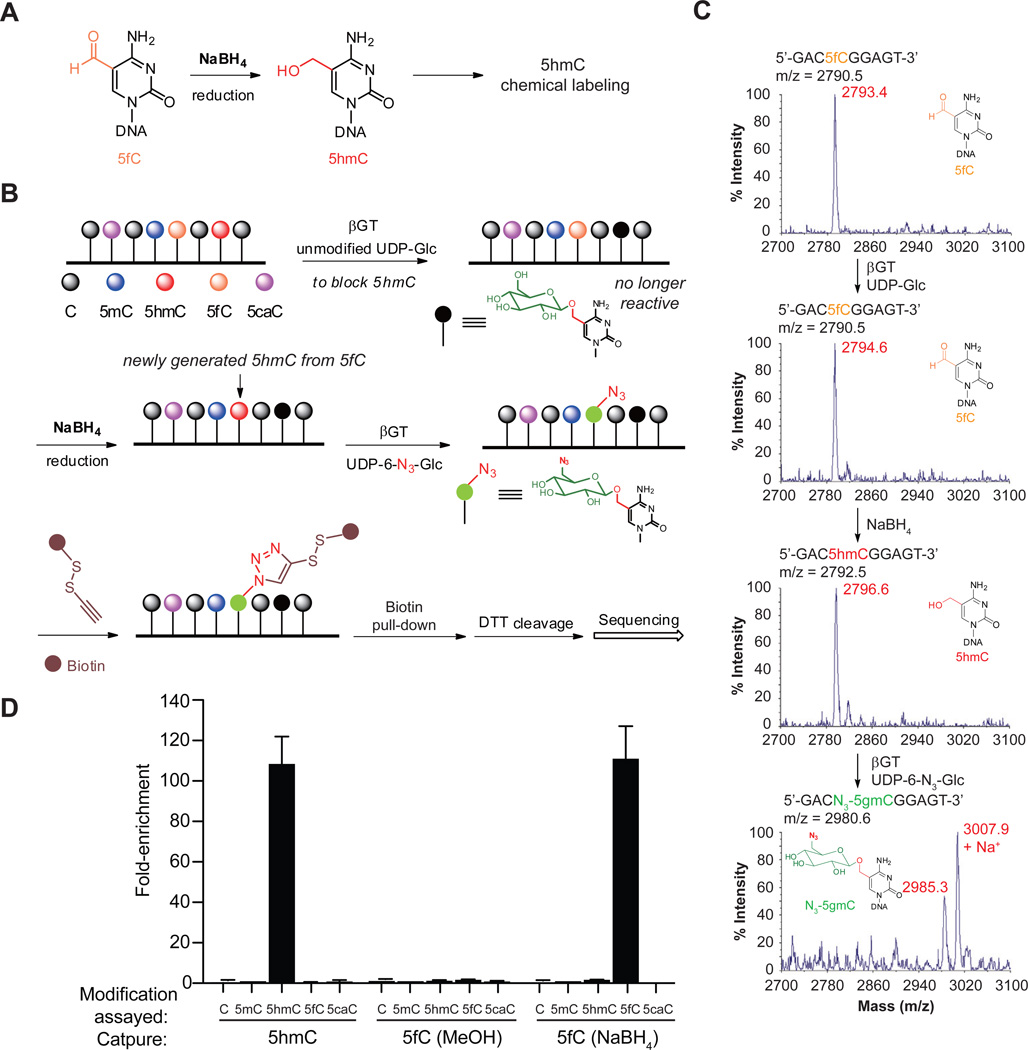
We and others previously have developed selective chemical labeling of 5hmC with biotin for genome-wide profiling that is highly sensitive and specific without density bias (Matarese et al., 2011; Pastor et al., 2011; Song et al., 2011). In duplex DNA, 5fC can be selectively reduced by sodium borohydride (NaBH4) to 5hmC (Figure 1A) (Dai and He, 2011), which prompted us to develop fC-Seal. In this strategy, 5hmC is first blocked with unmodified glucose using β-glucosyltransferase (βGT). We then reduce 5fC to 5hmC using NaBH4 and label the newly generated 5hmC (derived from 5fC) with an azide-modified glucose. The challenge of selectively capturing and profiling 5fC is therefore solved by employing the 5hmC-selective chemical labeling method (hMe-Seal) that we developed previously (Figure 1B).
Figure 1. Selective labeling of 5fC in genomic DNA.
(A) 5fC is selectively reduced to 5hmC.
(B) General procedure for fC-Seal. Endogenous 5hmC is blocked by a regular glucose through βGT-catalyzed glucosylation. 5fC is then reduced to 5hmC by NaBH4. The newly generated 5hmC from 5fC can be specifically enriched for sequencing using the 5hmC-selective chemical labeling method (hMe-Seal).
(C) MALDI-TOF characterization of 5fC-containing 9mer duplex DNA in βGT-catalyzed blocking with unmodified glucose, NaBH4-based reduction, and βGT-catalyzed azide-glucose labeling. Calculated MS shown in black, observed MS shown in Red.
(D) Enrichment tests of a single pool of spike-in amplicons containing C, 5mC, 5hmC, 5fC, or 5caC, separately, using hMe-Seal and fC-Seal with NaBH4 or a control with methanol only. Values shown are fold-enrichment over input, normalized to 5mC-modified DNA (n = 3, mean ± s.e.m). See also Figure S1.
We used a 9mer duplex model DNA to confirm that NaBH4-based reduction of 5fC to 5hmC can be carried out in aqueous solution, and that 5fC can be labeled with azide-modified glucose only after reduction using mass spectrometry (MS) (Figure 1C). The glucosylation protection of 5hmC is quantitative (Yu et al., 2012) and protected 5hmC, as well as 5caC and 5mC, cannot be reduced by NaBH4 under our reaction conditions (Figure S1). We further confirmed the specificity of the NaBH4-based 5fC reduction on synthetic DNA by HPLC (Figure S1D). In addition, we performed pull-down assays from genomic DNA spiked with a pool of 2-kb amplicons bearing C, 5mC, 5hmC, 5fC, or 5caC. Quantitative PCR analyses confirmed that fC-Seal only enriched 5fC-containing DNA, and that enrichment is NaBH4 dependent (Figure 1D). In comparison to a hydroxylamine-based method for the labeling of 5fC (Raiber et al., 2012), our method significantly reduced the non-specific capture of DNA (Table S1).
Parallel genome-wide profiling of 5fC and 5hmC in wild-type mESCs
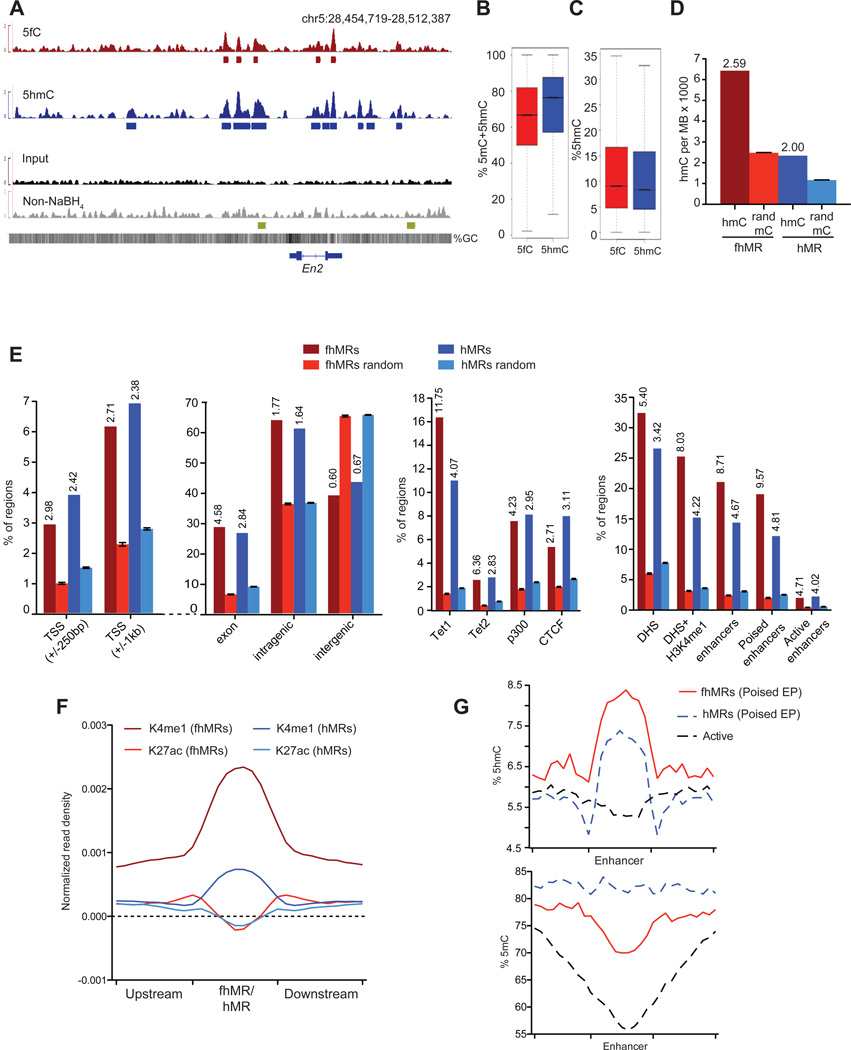
Using hMe-Seal and fC-Seal, we first performed parallel 5fC and 5hmC profiling in wild-type mESCs (Tdgfl/fl) (Figure 2). When sequencing to comparable depths, biological replicates of 5fC and 5hmC profiling were highly reproducible (Figure S2A, Tables S2 and S3). We defined high-confidence 5fC- and 5hmC-containing regions in Tdgfl/fl mESCs (see Extended Experimental Procedures) using a Poisson-based method (Zhang et al., 2008) (Table S4, p ≤ 1e–5, FDR ≤ 1%) (Figure 2A). Sequencing of glucose-blocked, non-NaBH4-treated DNA subjected to biotin labeling and capture confirmed complete blocking of 5hmC (Figure 2A, S2B, and Table S4). Each set of regions was first compared to base-resolution maps of 5hmC and 5mC+5hmC in mESCs (Stadler et al., 2011; Yu et al., 2012). On average, 5mC+5hmC abundance is 6.4% lower at regions marked with 5fC compared to 5hmC-enriched regions (Figure 2B), while 5hmC abundance is slightly higher (0.8%) at 5fC-marked regions (Figure 2C), suggesting a localized increase in 5mC oxidation at 5fC-marked loci. As 5fC-marked loci have comparatively higher 5hmC levels, likely due to an increase in localized Tet-mediated 5mC oxidation, we subsequently separated 5fC+5hmC regions (formyl- and hydroxymethyl-marked regions, fhMRs) and regions marked only by 5hmC but not 5fC (5-hydroxymethylated only regions, hMRs). Within hMRs, there are 2.00X more 5hmC bases than expected by chance (Figure 2D, Z-score = 385) and 92.0% have at least one 5hmC base (Figure S2C). Yet, at fhMRs there are 2.59X more 5hmC bases than expected (Figure 2D, Z-score = 413) and 89.2% of the 5fC-enriched regions contain at least one 5hmC base (Figure S2C), consistent with the observation that 5fC-marked regions contain an overall increase in oxidized 5mC in comparison to 5hmC-marked regions. These data demonstrate a relative decrease of 5mC occurring concomitant with an increased frequency and abundance of 5hmC at fhMRs compared to hMRs, indicating further refinement of genomic elements through mapping 5fC.
Figure 2. Annotation and comparison of 5hmC- and 5fC-containing regions in the wild-type (Tdgfl/fl) mESCs.
(A) Genome browser view of the En2 locus in 5fC- and 5hmC-specific profiling, along with the input as well as the glucose-blocked, non-NaBH4-treated control. Below each track are regions defined as marked with each respective mark. The gold track at the bottom corresponds to known poised enhancers at En2 (Shen et al., 2012).
(B) Quantification of %5mC+5hmC at 5fC- and 5hmC-marked regions in Tdgfl/fl mESCs.
(C) Quantification of %5hmC at 5fC- and 5hmC-marked regions in Tdgfl/fl mESCs.
(D) The relative enrichment of single-base 5hmC calls from (Yu et al., 2012) within fhMRs and hMRs Tdgfl/fl mESCs. mC random are randomly sampled 5mC bases defined by conventional bisulfite sequencing (Stadler et al., 2011) (10 iterations, mean ± s.d.). 5hmC base call counts are normalized in the genomic space covered by each set of enriched/random regions in megabases (MB) and divided by 103. Values above bars indicate the o/e ratios.
(E) Percentage of fhMRs and hMRs overlapping a given genomic/epigenomic annotation compared to the average percent overlap of 10 randomized sets of equal number and length (mean ± s.d). Vertical values above bars indicate the o/e ratios with significant enrichment (p < 1e–15, Fisher’s exact). Genomic annotations are listed in the left panel and epigenomic annotations are listed on the right.
(F) H3K4me1 and H3K27ac normalized read densities at fhMRs (red) and hMRs (blue).
(G) %5hmC (top panel) and %5mC (bottom panel) at fhMRs (red) and hMRs (blue) associated with poised Enhancer-Promoters (Poised EP) and active enhancers (black). See also Figure S2.
fhMRs further refine diverse gene regulatory elements
Previous efforts have localized 5hmC to TSSs, gene-bodies, enhancers, as well as CTCF-binding sites, with the overall abundance highest at promoter distal regulatory elements (Ficz et al., 2011; Jin et al., 2011; Pastor et al., 2011; Song et al., 2011; Stroud et al., 2011; Szulwach et al., 2011; Williams et al., 2011; Wu et al., 2011a; Xu et al., 2011; Yu et al., 2012), which may indicate distinct mechanisms for the regulation of 5mC oxidation at diverse gene regulatory elements. We therefore associated fhMRs and hMRs with genomic and epigenomic annotations, comparing them to equal numbers of equal length fragments randomized throughout the genome (Figure 2E). Notably, both fhMRs and hMRs are enriched at Tet1-binding sites (Williams et al., 2011; Wu et al., 2011b) (Figure 2E, observed to expected, o/e = 11.75, and o/e = 4.07) as well as at Tet2-binding sites (Figure 2E, o/e = 6.36 and o/e = 2.83) (Chen et al., 2012), corresponding to a 2.89- and 2.25-fold preference for fhMRs versus hMRs (fhMR:hMR), respectively (Figure S2D). fhMRs are enriched intragenically, particularly within exons, (Figure 2E and S2D, o/e = 4.58, fhMR:hMR = 1.61), strongly enriched at enhancers (Figure 2E and S2D o/e = 8.71, fhMR:hMR = 1.87), but are depleted at intergenic regions (Figure 2E and S2D, o/e = 0.60, fhMR:hMR = 0.89). Enrichment of fhMRs is further increased at enhancers predicted as linked to promoters on the basis of correlated chromatin state and RNAPII occupancy (Figure S2E, o/e = 9.61, fhMR:hMR = 1.92) (Shen et al., 2012), suggesting that promoter-linked enhancers may be more prone to 5mC/5hmC oxidation.
We also found depletion of fhMRs at repeat element classes including LINEs, LTRs, and DNA repeats (Figure S2F). Although repeat elements are generally depleted of fhMRs, among these repeat classes, fhMRs most frequently associate with SINEs (Figure S2F and S2G). fhMRs occur more frequently than expected at p300-binding sites (o/e = 4.23, fhMR:hMR = 1.44), DNaseI hypersensitive sites (DHSs) (o/e = 5.40, fhMR:hMR = 1.58), and are further enriched at H3K4me1-positive DHSs (o/e = 8.03, fhMR:hMR = 1.90), thus supporting the strong association of 5mC oxidation with enhancers (Figure 2E and S2D). Furthermore, fhMRs are enriched at poised enhancers (H3K4me1[+] H3K27ac[−], o/e = 9.57, fhMR:hMR = 1.99) in comparison to active enhancers (H3K4me1[+] H3K27ac[+], o/e = 4.71, fhMR:hMR = 1.17) (Creyghton et al., 2010; Rada-Iglesias et al., 2011; Zentner et al., 2011), implicating 5mC oxidation to 5fC in the preferential marking of these elements (Figure 2E and S2D). CTCF-bound regions are also associated with fhMRs (o/e = 2.67), although at a reduced frequency relative to hMRs (fhMR:hMR = 0.87) (Figure 2E and S2D). In contrast to Tet1, Tet2, p300, and CTCF sites, measurement of normalized 5fC read densities at 18 additional sets of diverse transcription factors showed that 5fC is not strongly enriched at these elements (Figure S2H). As p300 and CTCF interact with various transcription factors, this observation may support a role for 5mC oxidation and TDG-coupled removal of 5fC at regulatory elements “organized” by these factors. These results suggest that some regulatory elements, such as enhancers, may be more prone to 5mC/5hmC oxidation, while others, such as CTCF-bound loci, may contain more stable 5hmC.
5fC is preferentially enriched at poised enhancers and LMRs
Overall, we found that 21.1% of fhMRs are associated with an enhancer, significantly more than observed for hMRs (14.4%) (Figure 2E, p ≤ 2.2e−16, Fisher’s Exact). Among enhancer subtypes, we also observed a significant increase in the frequency of fhMRs at poised (H3K4me1[+] H3K27ac[−], o/e = 9.57) versus active (H3K4me1[+] H3K27ac[+], o/e = 4.71) enhancers (Figure 2E, p ≤ 2.2e−16, Fisher’s Exact). The association between fhMRs and poised enhancers is further strengthened with those predicted as functionally linked to promoters, approaching that observed for Tet1 (Figure S2E, o/e = 10.95, fhMR:hMR = 2.14) (Shen et al., 2012). Comparison between fhMRs and hMRs also revealed that fhMRs occur more frequently than hMRs at poised versus active enhancers (Figure 2E, p ≤ 2.2e−16, Fisher’s Exact). Consistent with the strong link between fhMRs and enhancers, particularly poised enhancers, quantification of H3K4me1- and H3K27ac-normalized ChIP-Seq read densities at fhMRs and hMRs demonstrated a clear distinction in H3K4me1-signal, but not H3K27ac (Figure 2F).
We next measured methylation levels, as defined by conventional whole genome bisulfite sequencing (WGBS) and Tet-assisted bisulfite sequencing (TAB-Seq) of 5hmC in normal mESCs (Stadler et al., 2011; Yu et al., 2012) at enhancer associated fhMRs and hMRs. At poised enhancers predicted as linked to promoters (poised enhancer-promoter, EP), there is a higher average abundance of 5hmC within fhMRs compared to hMRs, consistent with the overall increase in 5mC oxidation at fhMRs (Figure 2G). Conversely, there is a depletion of 5mC only at fhMRs, but not hMRs. On the other hand, active enhancers (H3K4me1[+] H3K27ac[+]) display a 20% reduction in 5mC from an average 74% down to 53% (Figure 2G). Active enhancers also show a lack of 5hmC compared to poised enhancers at fhMRs or hMRs (Figure 2G). These results indicate that within regions defined as poised EPs on the basis of histone modifications, the presence of 5fC and 5hmC correlates with a reduced methylation state relative to the presence of 5hmC alone. This distinction among poised EPs may reflect the link between TDG-mediated removal of 5fC and dynamic active demethylation at this particular subset of enhancers.
Measurements of 5fC and 5hmC signals at segments of the mESC methylome defined on the basis of DNA methylation (5mC+5hmC) using conventional WGBS (Stadler et al., 2011) (Un-, Low-, and Fully-methylated regions; UMRs, LMRs, and FMRs, respectively) revealed that 5fC and 5hmC strongly accumulate specifically at the LMR fraction of the mouse methylome (Figure S2I and S2J). Notably, LMRs, harboring reduced 5mC+5hmC levels (~30% abundance), are frequently present at promoter distal regulatory elements and contain binding sites/motifs for diverse transcription factors (Stadler et al., 2011). It is also interesting to note the small amount of 5fC captured at the previously assigned UMRs (Stadler et al., 2011); 5fC behaves as C under conventional bisulfite conditions. The presence of 5fC suggests that these sites could represent a subclass of UMRs that are undergoing active 5mC/5hmC oxidation in the presence of Tet1 and/or Tet2.
Together, the results support a model of relatively strong DNA demethylation at promoter distal regulatory regions and selected transcription factor binding sites, as has been previously proposed (Stadler et al., 2011; Yu et al., 2012). The particularly strong link between H3K4me1 and fhMRs in comparison to hMRs indicates that further oxidation of 5hmC towards demethylation occurs at the subsets of 5hmC-containing regions associated with enhancers.
TDG affects 5fC deposition in mESCs
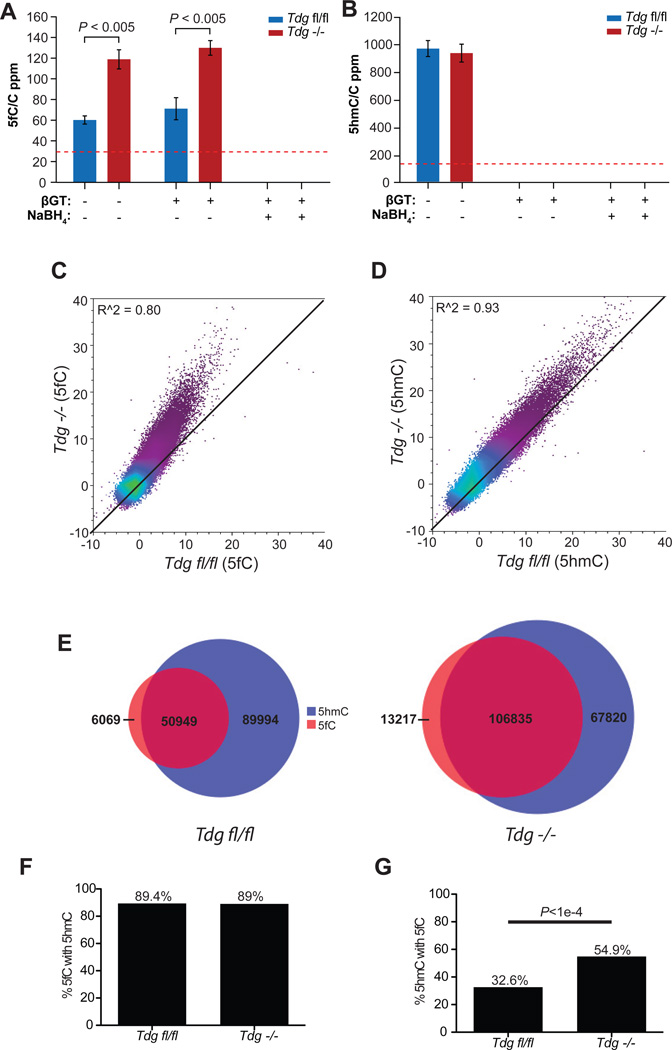
We next compared the genome-wide distributions of 5fC and 5hmC between wild-type and Tdg−/− mESCs. Detailed pluripotency and self-renewal characterization on the Tdgfl/fl and Tdg−/− mESCs found no evidence for altered self-renewal or pluripotency between floxed and Tdg−/− cell lines (Figure S3A–E) (Cortazar et al., 2011). LC-MS/MS quantification of 5fC and 5hmC showed that Tdg knockout leads to ~2-fold increase of 5fC in genomic DNA with no significant change of the 5hmC level (Figure 3A and 3B).
Figure 3. Comparison of 5fC and 5hmC signals in Tdgfl/fl and Tdg−/− mESCs.
(A–B) Mass spectrometry quantification of the genomic content of 5fC (A) and 5hmC (B) relative to cytosine in Tdgfl/fl and Tdg−/− mESCs in fC-Seal. Error bars indicate s.e.m. for n = 4 experiments. The red dotted lines indicate the detection limits under the assay conditions.
(C–D) Scatter plot of input-normalized 5fC (C) and 5hmC (D) read counts (reads/million) in 10kb bins genome-wide in Tdgfl/fl and Tdg−/− mESCs. Read counts per 10kb bin are normalized to the total number of reads in millions and similarly normalized values from input control genomic DNA subtracted. R2 values are denoted in the upper left-hand corner and the black diagonal is provided for reference.
(E) Venn diagram of the number of 5fC-marked regions (red) overlapping 5hmC-enriched regions (blue) in Tdgfl/fl (top) and Tdg−/− (bottom) mESCs.
(F–G) The percentage of the genomic regions with 5fC also marked with 5hmC (F) and the percentage of 5hmC-enriched regions also containing 5fC (G). See also Figure S3.
On a genomic scale, sequence reads derived from the DNA containing 5fC also indicate accumulation of 5fC in Tdg−/− mESCs (Figure 3C, r2=0.80) with little difference in the 5hmC pattern (Figure 3D, r2=0.93), consistent with the fact that TDG lacks 5hmC glycosylase activity (Cortellino et al., 2011; Maiti and Drohat, 2011). As 5fC is derived from the iterative oxidation of 5mC/5hmC, we found that ≥89% of 5fC-marked regions could be explained by 5hmC enrichment regardless of genotype (Figure 3E and 3F, Figure S3F, Table S4). However, while only 32.6% of 5hmC-enriched regions contain 5fC in Tdgfl/fl mESCs, in Tdg−/− mESCs, the fraction of 5hmC-enriched regions also harboring 5fC increases significantly to 54.9% as expected based on the elevated level of 5fC (Figure 3E and Figure 3G, Table S4, p<1.e–4).
The absence of TDG in mESCs causes alterations in DNA methylation states upon differentiation and during embryonic development (Cortazar et al., 2011; Cortellino et al., 2011). We therefore measured 5fC and 5hmC levels in Tdgfl/fl and Tdg−/− mESCs differentiated to embryoid bodies (mEBs). In mEBs the 5hmC level decreased by ~50% (Figure S4A) while the 5fC level was further decreased to ~15% of that in mESCs (Figure S4B). The depletion of 5hmC and 5fC in mEBs agrees with a previous report (Pfaffeneder et al., 2011) and the observation that Tet1/Tet2 expression is reduced upon differentiation (Koh et al., 2011). Since the 5fC levels in Tdgfl/fl and Tdg−/− mEBs are similar, we performed hMe-Seal and fC-Seal in Tdgfl/fl mEBs. In accordance with the quantification data, there is a clear reduction in the total number of 5fC-marked regions in mEBs (Figure S4C) as well as in mEB-normalized 5fC read densities at regions marked by 5fC in mESCs (Figure S4D).
TDG-dependent 5fC at selective regulatory elements of mESCs
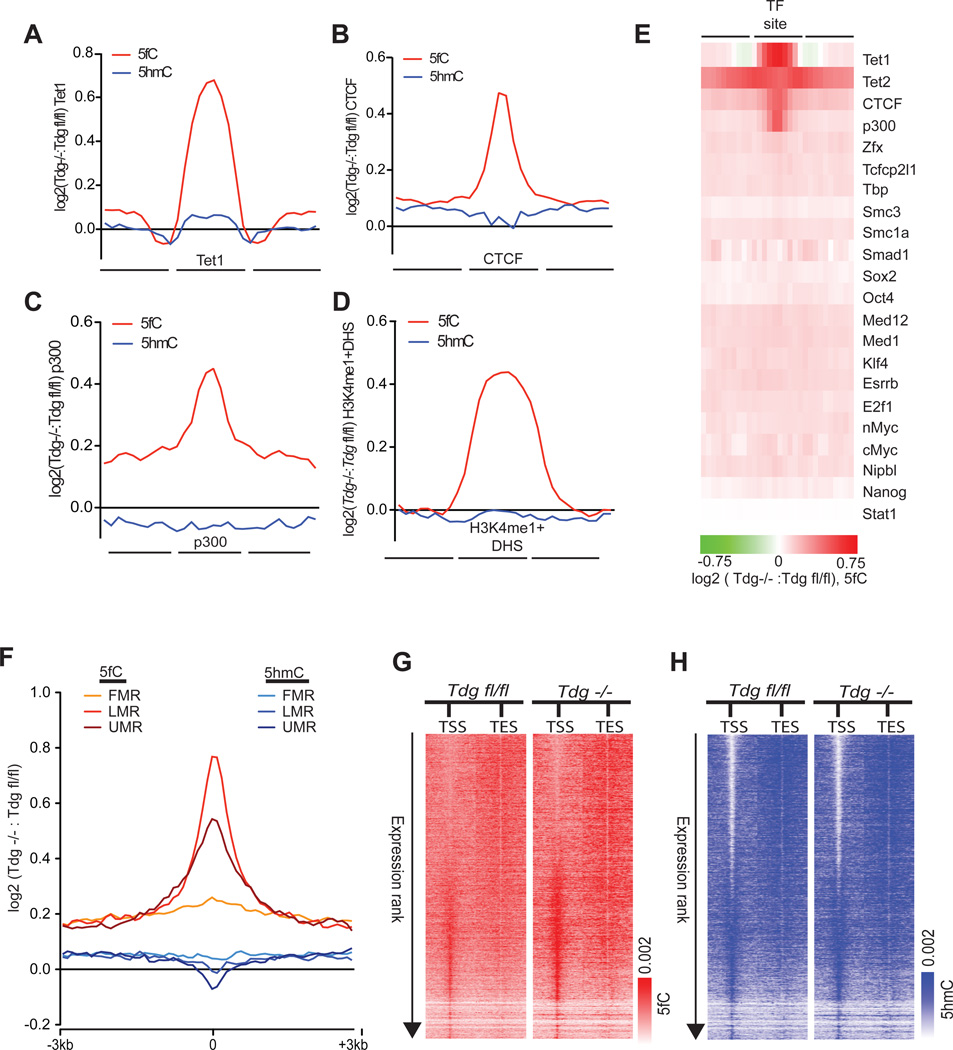
Assessment of normalized 5fC signals also reveals a clear accumulation of 5fC at Tet1-bound loci in Tdg−/− mESCs, as well as at other genomic elements normally enriched for 5fC but no difference between genotypes in immediately adjacent regions (Figure 4A–E). Comparison to 18 additional sets of transcription factor-binding sites also indicated that TDG-dependent regulation occurs preferentially at Tet1-, Tet2-, p300-, and CTCF-binding sites, but not uniformly across all types of binding sites (Figure 4E). Furthermore, among 5fC sites that are specifically found in Tdg−/− mESCs we observed more frequent associations with DHS and p300-and CTCF-bound loci as well as TSSs as compared to 5fC sites in Tdgfl/fl mESCs (Figure S4E). At UMRs, LMRs, and FMRs, we found preferential acquisition of 5fC within LMRs relative to UMRs and FMRs (Figure 4F). Gains of 5fC at both LMRs and UMRs occur without concomitant decreases in 5hmC (Figure 4F).
Figure 4. TDG-dependent 5fC regulation at defined gene regulatory elements.
(A–D) Log2 ratios of 5fC- and 5hmC-normalized read densities (reads/million/base, Tdg−/−: Tdgfl/fl) at genomic elements enriched for 5fC in Tdgfl/fl and Tdg−/− mESCs. (A) Tet1, (B) CTCF, (C) p300, and (D) H3K4me1+DHS. Each region of interest, denoted as the central portion of the x-axis, was divided into bins of 10 equal portions and reads were intersected to 10 bins within, upstream, and downstream of each region.
(E) Heatmap representation of the Log2 ratios of 5fC-read densities (reads/million/base, Tdg−/−: Tdgfl/fl) at 22 distinct sets of transcription factor (TF)-binding sites.
(F) Log2 ratio of the normalized 5fC and 5hmC read densities (reads/million/base, Tdg−/−: Tdgfl/fl) at FMRs, LMRs, and UMRs. Normalized read densities are plotted ± 3kb from the center of each segment as log2 fold-enrichment over input normalized read densities.
(G) Heatmap representations of 5fC-normalized read densities (reads/million/base) at RefSeq TSSs/TESs (± 5kb). 5fC signals at genes that are ranked by RPKM in descending order. Heatmap scales correspond to normalized read densities.
(H) Heatmap representations of 5hmC-normalized read densities (reads/million/base) at RefSeq TSSs/TESs (± 5kb). 5hmC signals at genes that ranked by RPKM in descending order. Heatmap scales correspond to normalized read densities. See also Figure S4.
A previous report suggested that 5fC is present and regulated by TDG at the TSS-associated CpG islands of transcribed genes, which are also depleted of 5hmC (Raiber et al., 2012). In contrast to this, we found that in Tdgfl/fl mESCs, 5fC is most enriched at the TSSs of genes with low expression (Figure 4G), where 5hmC is highest, consistent with the fact that 5fC is derived from 5hmC. Through further examination of TSSs ranked by expression level (RNA-Seq RPKM) in Tdg−/− mESCs, we found that the absence of TDG leads to accumulation of 5fC at the promoter regions of genes with low to intermediate expression while 5hmC remained unchanged (Figure 4G, 4H, and Table S5). This observation indicates that TDG-mediated regulation of 5fC occurs at “poised” genes in mESCs, similar to that described for 5hmC (Pastor et al., 2011; Williams et al., 2011; Wu et al., 2011b). While these various gene regulatory regions are each marked with 5hmC under normal conditions, the increased frequency of 5fC without a change of 5hmC in Tdg−/− mESCs indicate that these regions are more likely to be undergoing TDG-dependent removal of 5fC in a demethylation process that couples TET oxidation with TDG-based BER.
TDG-dependent regulation of 5fC correlates with p300 binding
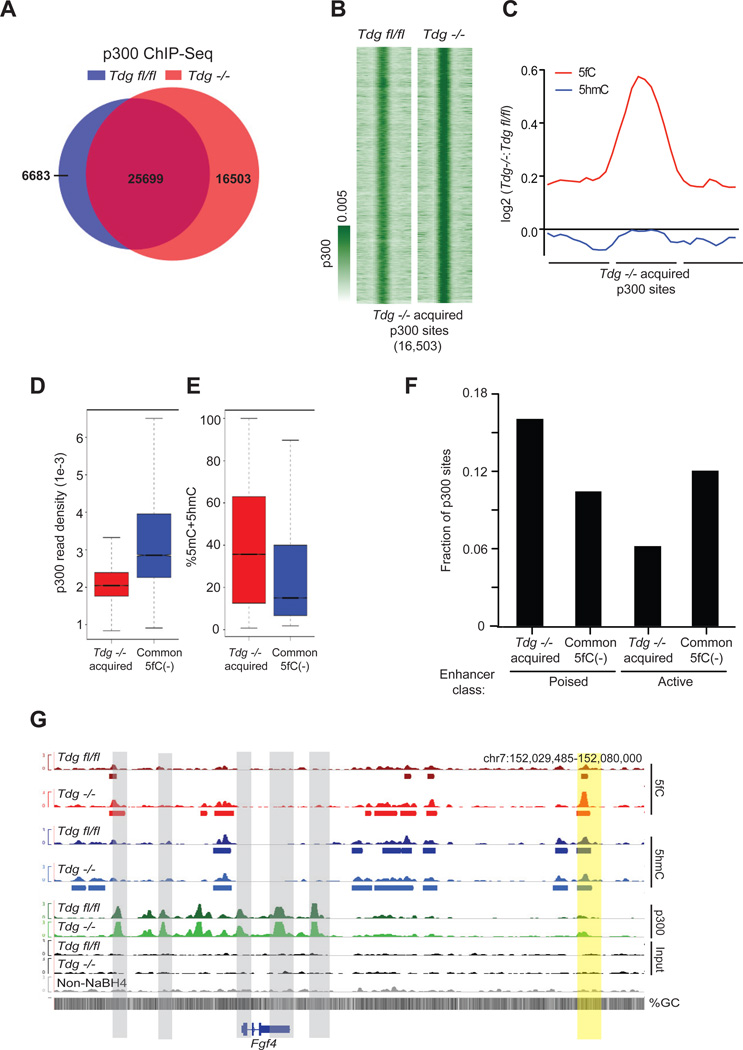
We took advantage of the altered 5fC content in Tdg−/− mESCs to further explore the impact of 5fC accumulation at sites of transcription factor binding. To do so, we focused on the transcriptional co-activator p300 because TDG and p300/CBP are known to interact, providing a link between DNA demethylation and p300 localization (Cortellino et al., 2011; Tini et al., 2002). By mapping p300 genome-wide in Tdgfl/fl and Tdg−/− mESCs, we found that the vast majority of high-confidence p300 sites in Tdgfl/fl mESCs (79.2%) remained in Tdg−/− mESCs (Figure 5A, S5A, S5B, and Table S4), indicating that the loss of TDG does not widely disrupt p300 localization to chromatin. However, examination of p300 binding in Tdg−/− mESCs identified a 31.2% increase in the total number of high-confidence p300-binding sites (Figure 5A and Table S4) with 43% marked by 5fC (Figure S5C), significantly more than that observed in Tdgfl/fl mESCs (12.9%, p<2.2e−16, Fisher’s exact). In the absence of TDG, a total of 16,503 unique p300 sites were acquired, as opposed to only 6,683 sites unique to Tdgfl/fl mESCs (Figures 5A and 5B), and an increased proportion of these of p300 sites were also marked with 5fC in comparison to Tdgfl/fl mESCs (Figure S5D and S5E). Neither p300 nor CBP displayed significantly altered expression (p300: Tdgfl/fl = 25.887, Tdg−/− = 27.431, RPKM, p-value = 0.22; CBP: Tdgfl/fl = 7.274, Tdg−/− = 7.571, RPKM, p-value = 0.41). Subsequent quantification of normalized 5fC read densities at p300 sites acquired in Tdg−/− demonstrated a strong gain in 5fC without changes in 5hmC (Figure 5C). Yet, at sites consistently bound by p300 in each genotype, and at which 5fC was not detected, we did not observe as significant an increase in p300 binding, nor did we observe accumulation of 5fC in the absence of TDG (Figure S5F and S5G). These common p300 sites exhibit significantly stronger p300 binding (Figure 5D, p<10−16 Welch’s two-tailed t-test) and reduced levels of 5mC+5hmC as compared to sites of Tdg−/−-specific p300 acquisition (Figure 5E).
Figure 5. TDG-dependent p300 binding in Tdgfl/fl and Tdg−/− mESCs.
(A) Venn diagram summarizing the total number of p300 ChIP-Seq peaks identified in Tdgfl/fl (32,160) and Tdg−/− mESCs (42,202), the number of Tdg−/− p300 sites overlapping with Tdgfl/fl sites (25,699), and the number of p300 sites unique to Tdgfl/fl (6,683) and Tdg−/− mESCs (16,503).
(B) p300 ChIP-Seq signals (reads/million/base) at the Tdg−/− specific p300-binding sites (16,503).
(C) Log2-fold-change in 5fC and 5hmC signals at the Tdg−/− specific p300-binding sites (16,503).
(D–E) p300 ChIP-Seq signals (reads/million/base, Tdg−/− mESCs) (D) and percent 5mC+5hmC (E) at p300 sites specific to Tdg−/− (16,503) and the 5fC negative p300-binding sites common to Tdgfl/fl and Tdg−/− (16,323).
(F) The fraction of Tdg−/− specific p300-binding sites (16,503) and 5fC negative p300-binding sites common to Tdgfl/fl and Tdg−/−(16,323) that occur at active and poised enhancers.
(G) Genome browser view of the Fgf4 locus at which multiple strong p300 sites lacking 5fC and 5hmC occur surrounding Fgf4 (gray), with a downstream poised enhancer (Shen et al., 2012) displaying a gain in 5fC and p300 in Tdg−/− (yellow). Shown below each track are the regions defined as marked for each respective mark. See also Figure S5.
The acquisition of a relatively large number of p300-binding sites in the absence of TDG, which concomitantly accumulate 5fC, suggests that the active oxidation of 5mC/5hmC to 5fC may serve as an initial step to counteract 5mC and to facilitate p300 binding; however, we cannot rule out the possibility that a more open chromatin state correlates with increased p300 binding and 5mC/5mC oxidation. Further oxidation and removal of 5fC could facilitate p300 binding on chromatin by generating or maintaining a reduced methylation state, as indicated by both the decreased 5mC+5hmC signal at sites with stronger p300 binding (Figure 5E) and the overall negative correlation between p300 binding strength and 5fC/5hmC levels (Figure S5H). Distinct chromatin states could exist at regions that acquire p300 binding in Tdg−/− mESCs when compared to p300 sites lacking 5fC that are common between both genotypes. Indeed, Tdg−/− acquired p300 sites occur preferentially at regions normally marked by histone modifications defining poised enhancers (H3K4me1[+]H3K27ac[−]) in comparison to 5fC-negative p300 sites consistently identified in each genotype, which occur preferentially at active enhancers (H3K4me1[+]H3K27ac[+]) (Figure 5F, p < 2.2e−16, Fisher’s Exact). These effects are apparent at Fgf4 (Figure 5G), a key regulator of ES cell differentiation (Wilder et al., 1997). Our findings suggest that the active oxidation of 5mC/5hmC to 5fC could serve as an epigenetic priming mechanism at poised enhancers.
Single-base resolution detection of 5fC
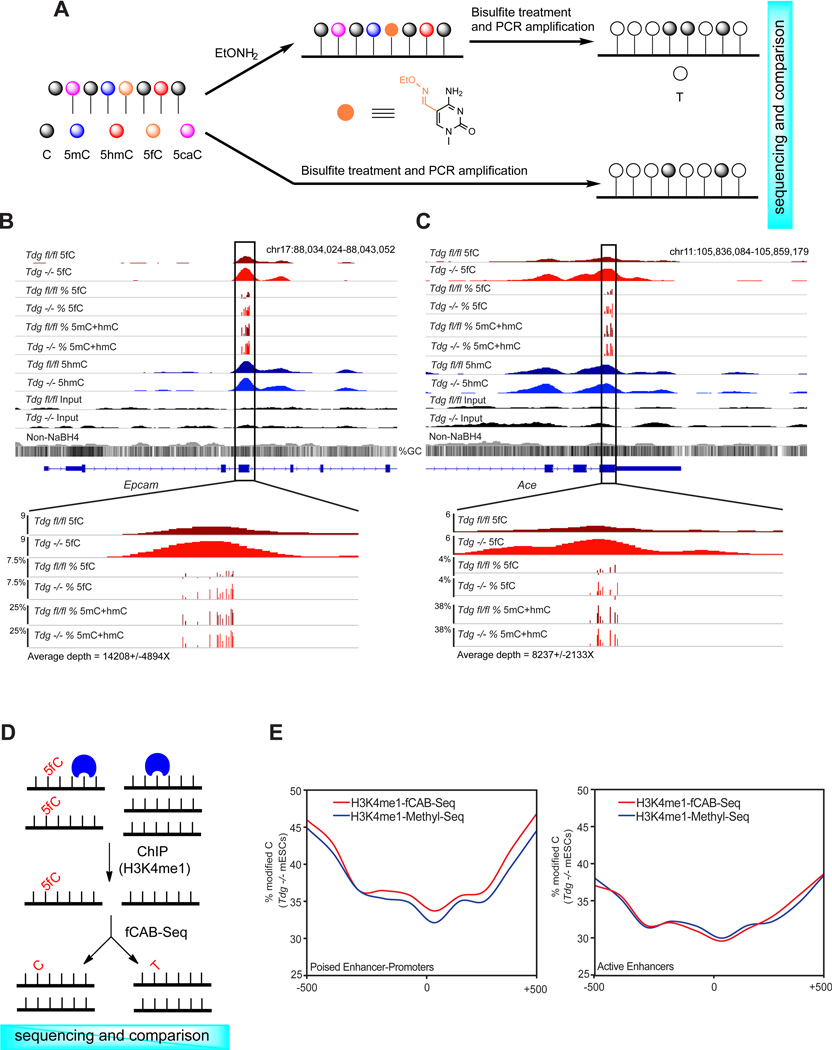
We next sought to develop an independent method for detecting 5fC at base resolution to further confirm the presence and accumulation of 5fC at defined sites. 5fC can be converted to uracil in bisulfite sequencing (Booth et al., 2012). If a specific chemical treatment prevents 5fC from bisulfite-mediated deamination, we can determine 5fC at base resolution through a chemically assisted bisulfite sequencing method (fCAB-Seq). Both hydroxylamine-protected 5fC (Figure 6A, Figure S6A and S6B) and reduction of 5fC to 5hmC could protect 5fC from bisulfite-mediated deamination; we found that the O-ethylhydroxylamine (EtONH2)-based protection of 5fC against bisulfite-mediated deamination is more effective (Figure S6B). Through comparison of EtONH2-treated bisulfite sequencing and traditional bisulfite sequencing of the same sample, we can determine the genomic locations of 5fC at single-base level (Figure 6A). Using a 76mer DNA model, we determined a working curve for 5fC conversion in fCAB-Seq with a linear correlation up to 50% 5fC abundance (Figure S6C and S6D), which is sufficient to analyze almost all potential 5fC sites as they are expected to exist in low abundance.
Figure 6. fCAB-Seq for base-resolution detection of 5fC.
(A) Schematic diagram of EtONH2-modified bisulfite sequencing for base-resolution detection of 5fC in genomic DNA (fCAB-Seq).
(B) fCAB-Seq validation of TDG-dependent 5fC in genomic DNA. An example of 5fC detection by fCAB-Seq amplicon deep sequencing at a region of Epcam. Sequencing depth = 14,208 ± 4894.
(C) fCAB-Seq validation of TDG-dependent 5fC in genomic DNA. An example of 5fC detection by fCAB-Seq amplicon deep sequencing at a region of Ace. Sequencing depth = 8,237 ± 2,133. For (B) and (C) 5fC track is equivalent to the signal from EtONH2 treatment minus (5mC+hmC) signal. All 5fC bases shown have p ≤ 0.005, Fisher’s exact.
(D) Schematic diagram of ChIP-fCAB-Seq. DNA fragments associated with H3K4me1 are enriched in ChIP and then subjected to fCAB-Seq for the determination of 5fC at base resolution.
(E) H3K4me1-ChIP-fCAB (Red) and H3K4m1-ChIP-Methyl-Seq (Blue) signals at 5fC-positive poised enhancers predicted as linked to promoters (left) and at all active enhancers (right) in Tdg− /− mESCs. Plotted are the weighted methylation signals in 100bp bins within the 1kb enhancer region. See also Figure S6.
We applied fCAB-Seq to Tdgfl/fl and Tdg−/− mESCs genomic DNA, and subjected the bisulfite amplicons to high-throughput sequencing in order to achieve sequencing depths sufficient to distinguish low abundance hydroxylamine-protected 5fC from the conventional bisulfite signals (~1,000X or higher coverage). Using this approach we were able to validate the presence and accumulation of specific 5fC sites in genomic DNA within five 5fC-marked endogenous loci (from fC-Seal) displaying TDG-dependent accumulation of 5fC (Figures 6B, 6C, Figures S6E–G, and Table S6, p < 0.005). We also confirmed that hydroxylamine does not alter the behavior of cytosine in bisulfite sequencing (Figure S6H).
We next employed a ChIP-fCAB-Seq approach by capturing H3K4me1-bound DNA via chromatin immunoprecipitation, in which 5fC-marked regions defined by fC-Seal are enriched, and then subjected the captured DNA to either conventional bisulfite (Brinkman et al., 2012; Statham et al., 2012) (H3K4me1-ChIP-Methyl-Seq) or fCAB (H3K4me1-ChIP-fCAB-Seq) treatments, followed by sequencing (Figure 6D). We then quantified the percentage of cytosine bases protected from deamination in each treatment within poised and active enhancers predicted to be linked to promoters (Shen et al., 2012), as such enhancers display the most significant enrichment of 5fC-marked regions (Figure S2E) in normal mESCs. We found that within these poised enhancers, the fCAB-Seq treatment resulted in an increase in the fraction of cytosines protected from deamination in the absence of TDG (Tdg−/− mESCs, 0.98% higher weighted average H3K4me1-ChIP-fCAB signal, p = 5.25−5, Fisher’s Exact), consistent with the occurrence and TDG-dependent regulation of 5fC at poised enhancers (Figure 6E). At active enhancers H3K4me-ChIP-Methyl-Seq and H3K4me1-ChIP-fCAB-Seq signals were very similar (33.04% and 33.03%, respectively, p = 1.69−3) (Figure 6E). In Tdgfl/fl mESCs, although we observed more variability in H3K4me1-ChIP-Methyl-Seq and H3K4me1-ChIP-fCAB-Seq signals, the increases in H3K4me1-ChIP-fCAB-Seq relative to H3K4me-ChIP-Methyl-Seq were reduced in comparison to those in Tdg−/− mESCs (Figure S6I and S6J).
DISCUSSION
Understanding TET-mediated 5mC oxidation has led to the identification of three additional DNA modifications in mammalian genomes, 5hmC, 5fC, and 5caC. The study of 5hmC has benefited greatly from the recent development of 5hmC-selective methods for affinity enrichment (Ficz et al., 2011; Pastor et al., 2011; Song et al., 2011; Wu et al., 2011a) and baseresolution detection (Booth et al., 2012; Yu et al., 2012). In particular, base-resolution maps of 5hmC have demonstrated that the relative abundance is greatest at distal enhancers for mESCs (Yu et al., 2012), suggesting a preference for active demethylation at such regulatory elements. There are also indications that rather than serving only as an intermediate in an active DNA demethylation pathway (Guo et al., 2011; He et al., 2011; Ito et al., 2011), 5hmC may itself serve as a distinct and stable epigenetic mark recognized by putative 5hmC-binding proteins (Frauer et al., 2011; Mellen et al., 2012; Spruijt et al., 2013; Yildirim et al., 2011). However, the means by which 5hmC may be dynamically regulated or stably maintained at distinct genomic elements remains a central challenge. To meet this challenge, it is essential to be able to accurately detect 5fC and 5caC in genomic DNA. Here we present a pair of methods, fC-Seal and fCAB-Seq, which employ 5fC-selective chemical manipulation to enable its affinity enrichment and base-resolution detection.
fC-Seal is developed as a highly selective chemical labeling approach for the affinity purification and genome-wide profiling of 5fC. By profiling 5hmC and 5fC in parallel using analogous methods, we observed a relative decrease of 5mC that occurs concomitant with an increased frequency and abundance of 5hmC at 5fC-marked regions when compared to 5hmC-marked regions. We show that fhMRs occur more frequently than hMRs at poised versus active enhancers, whereas other elements, such as CTCF-binding sites, are more frequently marked with 5hmC in comparison to 5fC. Furthermore, when classifying poised enhancers defined by chromatin state as associated with fhMRs or hMRs, we found that the presence of 5fC correlates with an increased abundance of 5hmC concomitant with a decreased 5mC levels as compared to hMRs. This observation supports a role for 5mC/5hmC oxidation to 5fC, and likely TDG-dependent removal of 5fC, in dynamic DNA demethylation at a subclass of poised enhancers. These results also indicate that, in addition to chromatin modifications, DNA methylation states can be informative in classification of gene regulatory elements.
Comparison of 5hmC and 5fC profiles between wild-type and Tdg KO mESCs further indicates distinct regulation of 5fC, but not 5hmC, by TDG. However, we found that the effect of TDG is greatest at regions of the genome that generally harbor intermediate levels of DNA methylation (LMRs and poised enhancers), as opposed to regions that can be classified as fully methylated (FMRs) or unmethylated (UMRs, TSSs of highly expressed genes, and active enhancers). These results indicate distinct role(s) for 5fC and 5hmC in influencing the epigenetic state and function of gene regulatory elements. To further examine this possibility, we generated TDG-dependent binding profiles of p300 as TDG and p300 are known to interact. We found that the loss of TDG leads to a relatively large gain in the overall number of p300-binding sites, and that these sites also accumulate 5fC. Intriguingly, the p300-binding sites acquired in the absence TDG, which also accumulate 5fC, occur preferentially at poised enhancers, whereas p300 sites common to Tdgfl/fl and Tdg−/− mESCs, which are not marked by 5fC, occur preferentially at active enhancers. These results indicate that 5fC production coordinates with the binding of p300 at poised enhancers, thus indicating functional roles for 5fC-based DNA demethylation in the epigenetic priming of regulatory elements. As a large fraction of sites marked by 5fC only in Tdg−/− mESCs do not exhibit concomitant p300 binding (Figure S5D), and 5fC is enriched at diverse gene regulatory elements, our data also suggest that TDG-dependent regulation of 5fC could influence other regulatory elements in a similar manner. Further assessment of these regions in Tdgfl/fl and Tdg−/− mESCs may yield additional insight into the roles of 5mC/5hmC oxidation in embryonic stem cells.
We also developed a chemically assisted bisulfite sequencing method, fCAB-Seq, to detect the relative abundance of 5fC at base resolution. We demonstrated that fCAB-Seq is capable of detecting low abundance 5fC at endogenous loci at levels down to only a few percent when performed in combination with high-throughput bisulfite amplicon sequencing. Finally, we showed that the 5fC content at specific genomic elements could be studied using a ChIP-fCAB-Seq approach to enrich subsets of genomic elements harboring 5fC. By employing ChIP-fCAB-Seq, we confirmed the presence of 5fC at poised enhancers. The approaches presented here are highly sensitive and selective, and have minimal background noise, which is critical in order to accurately determine the distribution of 5fC given its low abundance in the genome (Table S1).
In summary, we have developed and implemented two methods for detecting and profiling 5-formylcytosine in genomic DNA. Genome-wide maps of 5fC in mouse ESCs generated using our approaches demonstrate the utility of mapping 5fC beyond that afforded by mapping 5hmC alone in order to gain additional insight into the general strategies employed by cells for the regulated access of transcription factors to genetic information. Use of fC-Seal in combination with detailed base-resolution detection of 5fC by fCAB-Seq represents a powerful combination of tools for the future study of 5fC in any biological context.
EXPERIMENTAL PROCEDURES
NaBH4-based selective chemical labeling and capture of 5fC (fC-Seal)
50 µg sonicated mESC genomic DNA (average 400 bp) was incubated in a 100 µL solution containing 50 mM HEPES buffer (pH 7.9), 25 mM MgCl2, 300 µM unmodified UDP-Glc, and 2 µM βGT for 1 h at 37° C. The labeled DNA was purified by Micro Bio-Spin 6 spin columns (Bio-Rad, exchange buffer to H2O first). NaBH4 solution was prepared by adding 1.5 mg of NaBH4 (Aldrich) in 1 mL of anhydrous methanol (Acros) and vortexing until the entire solid dissolved. NaBH4 reduction was performed by adding equal volume of freshly prepared NaBH4 solution to the DNA solution. The reaction mixture was then vortexed and incubated for 15 min at room temperature. The DNA samples were purified by isopropanol precipitation and used for azide-glucosylation, biotionylation and capture (see Extended Experimental Procedures).
Hydroxylamine protection of 5fC for bisulfite sequencing (fCAB-Seq)
Hydroxylamine protection of 5fC was performed in 100 mM MES buffer (pH 5.0), 10 mM O-ethylhydroxylamine (Aldrich, 274992), and 100 ng/µl 76mer double-stranded synthetic DNA or sonicated genomic DNA (average 400 bp), or ChIP’d DNA for 2 h at 37 °C. The DNA substrates were purified by Qiagen nucleotide removal kit and subjected to the sodium bisulfite treatment by using EpiTect Bisulfite Kits (Qiagen) following the manufacturers’ instructions except the bisulfite thermal cycle program was run twice or high-throughput bisulfite amplicon sequencing (see Extended Experimental Procedures).
Supplementary Material
HIGHLIGHTS.
Development of 5fC-selective chemical labeling for enrichment and genomic profiling
Genome-wide profiling reveals TDG-dependent regulation of 5fC, not 5hmC, in mESCs
5fC is preferentially enriched at poised enhancers and correlates with p300 binding
Development of a base-resolution method for the detection of 5fC
ACKNOWLEDGMENTS
This study was supported by National Institutes of Health (HG006827 to C.H., NS079625/NS051630/HD073162/AG025688 to P.J.), grants from the Ministry of Science and Technology of China (2011CB946102 and 2012CB966903 to G. X.), the Emory Genetics Discovery Fund (P.J.), and the Simons Foundation Autism Research Initiative (P.J.). We thank Drs. Bing Ren and Gary Hon for sharing single-base maps of 5mC+5hmC and 5hmC. We thank S.F. Reichard, MA for editing the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Sequence data has been deposited to GEO (accession number GSE41545).
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, six figures, and six tables, all of which can be found with this article online.
REFERENCES
- Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell. 2011;146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- Brinkman AB, Gu H, Bartels SJ, Zhang Y, Matarese F, Simmer F, Marks H, Bock C, Gnirke A, Meissner A, et al. Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Res. 2012;22:1128–1138. doi: 10.1101/gr.133728.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2012 doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortazar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E, Wirz A, Schuermann D, Jacobs AL, Siegrist F, et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 2011;470:419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q, He C. Syntheses of 5-formyl- and 5-carboxyl-dC containing DNA oligos as potential oxidation products of 5-hydroxymethylcytosine in DNA. Org Lett. 2011;13:3446–3449. doi: 10.1021/ol201189n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi SW, Page DC, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9:166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doege CA, Inoue K, Yamashita T, Rhee DB, Travis S, Fujita R, Guarnieri P, Bhagat G, Vanti WB, Shih A, et al. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature. 2012;488:652–655. doi: 10.1038/nature11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Frauer C, Hoffmann T, Bultmann S, Casa V, Cardoso MC, Antes I, Leonhardt H. Recognition of 5-hydroxymethylcytosine by the Uhrf1 SRA domain. PLoS One. 2011;6:e21306. doi: 10.1371/journal.pone.0021306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Guo Junjie U, Su Y, Zhong C, Ming G-l, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Wu X, Li AX, Pfeifer GP. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res. 2011;39:5015–5024. doi: 10.1093/nar/gkr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare T, Pai S, Koncevicius K, Pal M, Kriukiene E, Liutkeviciute Z, Irimia M, Jia P, Ptak C, Xia M, et al. 5-hmC in the brain is abundant in synaptic genes and shows differences at the exon-intron boundary. Nat Struct Mol Biol. 2012;19:1037–1043. doi: 10.1038/nsmb.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian Christine G, Xu Y, Ceol C, Wu F, Larson A, Dresser K, Xu W, Tan L, Hu Y, Zhan Q, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150:1135–1146. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J Biol Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matarese F, Carrillo-de Santa Pau E, Stunnenberg HG. 5-Hydroxymethylcytosine: a new kid on the epigenetic block? Mol Syst Biol. 2011;7:562. doi: 10.1038/msb.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffeneder T, Hackner B, Truss M, Munzel M, Muller M, Deiml CA, Hagemeier C, Carell T. The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew Chem, Int Ed. 2011;50:7008–7012. doi: 10.1002/anie.201103899. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiber EA, Beraldi D, Ficz G, Burgess HE, Branco MR, Murat P, Oxley D, Booth MJ, Reik W, Balasubramanian S. Genome-wide distribution of 5-formylcytosine in embryonic stem cells is associated with transcription and depends on thymine DNA glycosylase. Genome Biol. 2012;13:R69. doi: 10.1186/gb-2012-13-8-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, Munzel M, Wagner M, Muller M, Khan F, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- Statham AL, Robinson MD, Song JZ, Coolen MW, Stirzaker C, Clark SJ. Bisulfite sequencing of chromatin immunoprecipitated DNA (BisChIP-seq) directly informs methylation status of histone-modified DNA. Genome Res. 2012;22:1120–1127. doi: 10.1101/gr.132076.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen S. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12:R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song C-X, Han JW, Kim S, Namburi S, Hermetz K, Kim JJ, Rudd MK, et al. Integrating 5-Hydroxymethylcytosine into the Epigenomic Landscape of Human Embryonic Stem Cells. PLoS Genet. 2011;7:e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139:1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tini M, Benecke A, Um SJ, Torchia J, Evans RM, Chambon P. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol Cell. 2002;9:265–277. doi: 10.1016/s1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- Wilder PJ, Kelly D, Brigman K, Peterson CL, Nowling T, Gao QS, McComb RD, Capecchi MR, Rizzino A. Inactivation of the FGF-4 gene in embryonic stem cells alters the growth and/or the survival of their early differentiated progeny. Dev Biol. 1997;192:614–629. doi: 10.1006/dbio.1997.8777. [DOI] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D'Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011a;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D'Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Eve Sun Y, Zhang Y. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011b;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Rando OJ, Fazzio TG. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lu X, Lu J, Liang H, Dai Q, Xu GL, Luo C, Jiang H, He C. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat Chem Biol. 2012;8:328–330. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.