Abstract
Background
Colorectal cancer (CRC) is one of the most commonly diagnosed cancers. Circulating microRNAs (miRNAs) have been suggested as potentially promising markers for early detection of CRC. We aimed to identify and evaluate a panel of miRNAs that might be suitable for CRC early detection.
Methods
MiRNAs were profiled by TaqMan MicroRNA Array and screened for differential expression in 5 pools of plasma samples of CRC patients (N = 50) and 5 pools of neoplasm-free controls (N = 50). Additional miRNAs were selected from a literature review. Identified candidates were evaluated in independent validation samples with respect to discrimination of CRC patients (N = 80) or advanced adenoma patients (N = 50) and neoplasm-free controls (N = 194). Diagnostic performance of the panel of miRNAs was assessed by multiple logistic regression, using bootstrap analysis to correct for over-optimism.
Results
Five miRNAs identified to be differentially expressed from TaqMan MicroRNA Array (miR-29a, -106b, -133a, -342-3p, -532-3p), and seven miRNAs reported to be differentially expressed in the literature (miR-18a, -20a, -21, -92a, -143, -145, -181b) were selected for validation. Nine of the twelve miRNAs (miR-18a, -20a, -21, -29a, -92a, -106b, -133a, -143, -145) were found to be differentially expressed in CRC patients and controls in the validation samples. The optimism-corrected area under the curve was 0.745 (95% confidence interval: 0.708–0.846). None of the selected miRNAs showed significant differential expression between advanced adenoma patients and neoplasm-free controls.
Conclusion
The identified panel of miRNAs could be of potential use in the development of a multi-marker blood based test for early detection of CRC. Impact: The study underscores the high potential of plasma miRNAs for the improvement of current offers of non-invasive CRC screening.
Introduction
With over 1.2 million new cases and 608,700 deaths per year, colorectal cancer (CRC) is the third most commonly diagnosed cancer in males and the second in females, and the fourth most common cause of death from cancer worldwide [1]. Due to its typically very slow development over many years, perspectives for early detection are much better than for many other forms of cancer. It has been estimated that over 95% of cases of CRC would benefit from curative surgery if diagnosis was made at an early or premalignant polyp stage [2], [3]. A number of early detection procedures have been developed and are increasingly applied, including endoscopic examinations, stool- and blood-based tests. Blood based tests would appear to be particularly attractive as they are minimally invasive and might receive high levels of adherence when applied as primary screening tests in population based screening. A large number of blood markers have been proposed and evaluated, including protein, cytological, mRNA, and DNA markers [4], [5], but diagnostic performance has mostly been insufficient for application as a primary tool in population-based screening. Furthermore, most studies relied on small convenience samples from clinical settings, and rather promising results from small studies have often not been replicated in subsequent larger scale validations.
MicroRNAs (miRNA) are 18∼22 nucleotide non-coding RNAs that post-transcriptionally regulate gene expression and control various cellular mechanisms [6]. There is increasing evidence that miRNAs are widely dysregulated in cancer and may have potential application for cancer diagnosis, prognosis and treatment [7]. Furthermore, recent development of miRNA microarrays has made large profiling studies in cancer patients possible.
The stability of cell-free miRNAs in body fluids enables circulating miRNAs to be potential biomarkers for noninvasive diagnosis of cancer and other disease [8]. Several recent studies found some circulating miRNAs, such as miR-29a, miR-92a and miR-221, to be differentially expressed in CRC patients and therefore to be of potential use as non-invasive biomarkers for CRC screening [9]–[11].
The aim of this study was to identify and evaluate a panel of plasma miRNAs that might serve as biomarkers for the early detection of CRC.
Materials and Methods
Study Design and Study Population
Cases with sporadic CRC were recruited prior to initiation of therapy at the University Clinic of Heidelberg in the context of the DACHS+ study, which is a satellite study to DACHS, an ongoing case-control study conducted in the Rhine-Neckar region of Germany [12], [13].
Patients with advanced colorectal adenomas and controls free of colorectal neoplasms were randomly selected from participants of screening colonoscopy recruited in the BLITZ study, an ongoing study designed to evaluate novel promising markers for early detection of CRC and previously described in detail elsewhere [14]–[16]. Briefly, patients were recruited, and blood samples were taken at gastroenterologists’ offices at a preparatory visit, typically about one week prior to screening colonoscopy.
Both the DACHS+ and the BLITZ study were approved by the ethics committees of the Medical Faculty at the University of Heidelberg and of the Medical Boards of Baden-Wuerttemberg and Rhineland-Palatinate. Written informed consent was obtained from each participant.
Our investigation involved two main phases, a marker identification phase and a marker validation phase:
In the marker identification phase, promising miRNAs were screened by TaqMan MicroRNA Array in pooled plasma samples from 50 CRC patients and 50 neoplasm-free controls, using five pools of plasma samples from ten CRC patients each, and five pools of plasma samples from ten neoplasm-free controls each. In addition, seven miRNAs described to be differentially expressed in CRC cases and controls in previous publications were identified from a systematic literature review (miR-18a, -20a, -21, -92a, -143, -145, -181b) [17].
In the marker validation phase, expression of the identified miRNAs was evaluated in independent samples of (i) 80 CRC cases and 144 controls free of colorectal neoplasms and (ii) 50 patients with advanced adenomas and 50 controls free of colorectal neoplasms.
Laboratory Procedures
(i) Sample preparation and RNA extraction
The blood samples were collected in EDTA tubes. Blood samples from cancer patients were taken before surgery or any other therapy and blood samples from participants who underwent screening colonoscopy were taken prior to colonoscopy. The blood samples were centrifuged at 2123g for 10 min at 4°C and the supernatant was transferred into new tubes. Plasma samples were stored at −80°C until use. Total RNA containing small RNA was extracted from 200 µl (samples used in the identification phase) or 115 µl (samples used in the validation phase) plasma using a combination of Trizol LS reagent (Invitrogen, Carlsbad, California, USA) and miRNeasy Mini Kit (Qiagen, Hilden, Germany) as well as 5 fmol/µl cel-miR-39 as external preparation control as described before [18]. Samples were eluted in a final volume of 30 µl.
(ii) MicroRNA profiling from plasma samples
Profiling was performed using TaqMan MicroRNA Array (Applied Biosystems, Foster City, California, USA) which enables quantification of 667 human microRNAs.(Table S1) Megaplex reverse transcription reaction and pre-amplification reaction were performed by G-Storm GS2 Thermal Cycler (G-Storm, UK). Real-time quantitative polymerase chain reaction (qRT-PCR) was performed using 7900HT Fast Real-Time PCR System (Applied Biosystems). The cycle threshold (Ct) is defined as the number of cycles required for the fluorescent signal to cross the threshold in qRT-PCR. Raw Ct values were calculated using the SDS software version 2.2 applying automatic baseline settings and a threshold of 0.1 was set. Only miRNAs whose Ct value was equal to or below 33 in at least either the case or the control group were taken into account for further data analysis. Data normalization was done as described by Kroh et al. [19].
(iii) Real time quantitative PCR verification
Selected miRNAs were measured using TaqMan MicroRNA Assays (Applied Biosystems) according to the manufacturer’s protocol. Triplicates of qRT-PCR of each sample were performed using LightCycler 480 Real-time PCR system (Roche Applied Science, Germany). ΔCt was calculated by subtracting the Ct values of internal controls from the Ct values of the miRNAs of interest, and mean ΔCt values were compared between cases and controls. Multiplex assays were performed using predefined pools of RT-primers (Table S2). The efficiency of each miRNA’s assay was determined by constructing a standard curve using a series of total RNA dilutions. All assays showed good linearity (R2>0.96) between the Ct values and the log of the starting quantity of total RNA of each dilution (data not shown).
Statistical Analyses
MiRNA expression levels were compared between CRC patients and neoplasm-free controls and between advanced adenoma patients and neoplasm-free controls using the Wilcoxon-Mann-Whitney-Test (hereafter: Wilcoxon test). The comparison of miRNA expression levels between CRC patients and neoplasm-free controls in the marker identification phase was performed using the exact version of the Wilcoxon test. All tests were two-sided and P values of 0.05 or less were considered to be statistically significant.
Multiple logistic regression was employed to assess the joint use of the identified panel of miRNAs in predicting CRC in the marker validation phase. Receiver operating characteristic (ROC) curves were constructed and areas under the ROC curves (AUCs) were calculated both from unadjusted (apparent) and adjusted (“optimism-corrected”) estimates to assess discrimination of the prediction model between patients with and without CRC. The.632+ bootstrap method (with 1000 replicates) was used to adjust for overfitting of the apparent misclassification error and over-estimation of AUCs by the unadjusted estimates [20], [21]. 95% confidence intervals (CI) for the adjusted and unadjusted estimates were obtained by ordinary bootstrap analyses (with 1000 replicates).
The correlation of plasma miRNA expression levels across the 324 participants of the validation phase was assessed by Spearman correlation coefficients.
SAS version 9.2 (SAS Institute Inc., Cary, North Carolina, USA) was used to conduct Wilcoxon tests and R version 2.15.0 (R Foundation for Statistical Computing, Vienna, Austria) was used to conduct all other analyses. The R-packages “Daim” and “boot” were employed to perform bootstrap analyses [22], [23].
Results
Study Population
A total of 424 individuals were included (130 CRC patients, 50 advanced adenoma patients and 244 neoplasm-free controls) in the study. Demographic characteristics of the study population and distribution of tumor stages are summarized for the marker identification phase and the marker validation phase in Table 1.
Table 1. Study population.
| Characteristic | Marker identification phase | Marker validation phase | |||||
| (pooled samples) | (individual samples) | ||||||
| CRC patients | Neoplasm-free controls | CRC patients | Neoplasm-free controls | Advanced adenoma patients | Neoplasm-free controls | ||
| n = 50 | n = 50 | n = 80 | n = 144 | n = 50 | n = 50 | ||
| Age (years) | |||||||
| Mean±SD | 67.1±11.0 | 61.7±6.4 | 68.0±10.4 | 62.5±7.5 | 65±7.6 | 61.6±6.3 | |
| Gender | |||||||
| Male | 25 | 25 | 45 | 60 | 32 | 20 | |
| Female | 25 | 25 | 35 | 84 | 18 | 30 | |
| AJCC stage | |||||||
| I | 15 | – | 22 | – | – | – | |
| II | 12 | – | 25 | – | – | – | |
| III | 20 | – | 26 | – | – | – | |
| IV | 3 | – | 5 | – | – | – | |
| Not specified | 0 | – | 2 | – | – | – | |
| Tumor location | |||||||
| Proximal colon | 18 | – | 23 | – | 26 | – | |
| Distal colon and rectum | 32 | – | 56 | – | 22 | – | |
| Not specified | 0 | – | 1 | – | 2 | – | |
Abbreviations: CRC, colorectal cancer; SD, standard deviation.
Identification of Differentially Expressed miRNAs in CRC Patients and Neoplasm-free Controls
In microarray analyses, five miRNAs were found to be statistically significantly over-expressed in the plasma of CRC patients compared to neoplasm-free controls (miR-29a: p = 0.016, miR-106b: p = 0.008, miR-133a: p = 0.032, miR-342-3p: p = 0.008, miR-532-3p: p = 0.016, exact Wilcoxon test).
Evaluation of Internal Controls for Real Time Quantitative PCR
In our microarray data we observed no significant difference in terms of the Ct values of miR-188-5p (p = 0.83, Wilcoxon test) and miR-16 (p = 0.40, Wilcoxon test) between CRC and neoplasm-free controls. RNU6B and miR-16 have been the most widely-used endogenous control miRNA for qRT-PCR in the miRNA studies [8]. Therefore, the named three miRNAs were considered as internal controls for miRNA quantification. However, expression levels of RNU6B and miR-188-5p in the plasma were too low to be quantified by TaqMan MicroRNA Assay (mean Ct values were greater than 35). Hence, miR-16 was selected as the internal control as it showed high abundance and less variability in expression. No significant difference was observed in terms of Ct values of miR-16 (p = 0.40, Wilcoxon test) between CRC and neoplasm-free samples.
Validation in an Independent Sample of CRC Patients and Neoplasm-free Controls
To confirm the five miRNAs (miR-29a, -106b, -133a, -342-3p, -532-3p) identified in the microarray analysis and another seven miRNAs (miR-18a, -20a, -21, -92a, -143, -145, -181b) which were previously reported to be differentially expressed in CRC (except miR-92a, they were all from studies based on tissue samples) [17], qRT-PCR was performed to analyze the expression of the selected miRNAs in our validation set (224 plasma samples in total from 80 CRC patients and 144 neoplasm-free controls).
(i) Expression of miRNAs
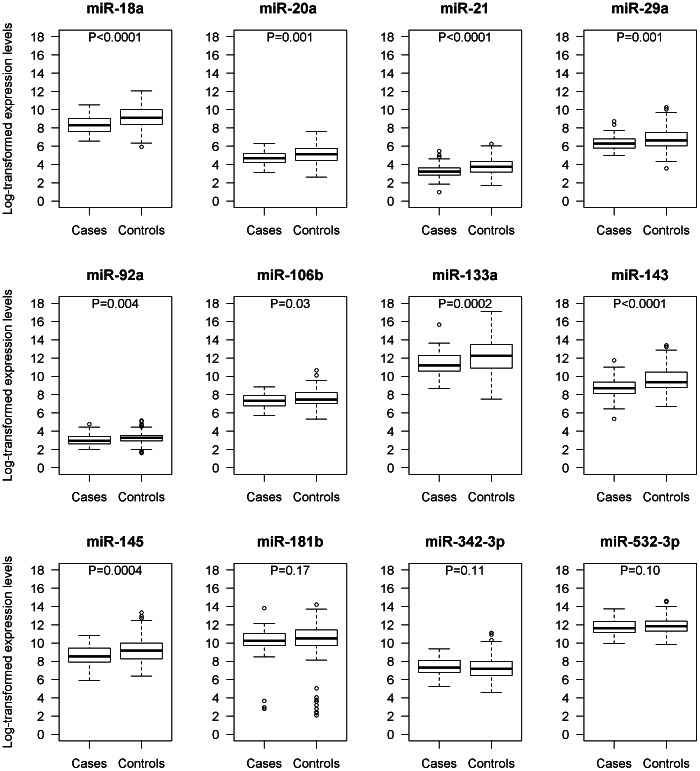
Nine miRNAs (miR-18a, -20a, -21, -29a, -92a, -106b, -133a, -143, -145) showed higher expression in the plasma of CRC patients than in neoplasm-free controls (miR-18a: p<0.0001, miR-20a: p = 0.001, miR-21: p<0.0001, miR-29a: p = 0.001, miR-92a: p = 0.004, miR-106b: p = 0.028, miR-133a: p = 0.0002, miR-143<0.0001, miR-145: p = 0.0004, Wilcoxon test). No miRNA was found to be downregulated. The corresponding expression levels in plasma of CRC patients and neoplasm-free controls (normalized to miR-16) are shown in Figure 1.
Figure 1. Expression levels of microRNAs in 80 colorectal cancer patients and 144 neoplasm-free controls (normalized to miR-16, log2 scale at y-axis).
Box plots with smallest observation, lower quartile, median, upper quartile and largest observation are shown. The whiskers extend to the observations which are no more than 1.5 times the length of the box (interquartile range) away from the box. More extreme observations are considered outliers. P values are based on Wilcoxon tests.
(ii) Relationship between miRNAs and clinicopathological features of CRC patients
We examined the association between miRNA expression levels and some clinicopathological features. MiR-181b’s expression level was higher in patients with lymph node metastasis than in patients without lymph node metastasis (p = 0.024, Wilcoxon test). None of the miRNAs showed any association with tumor site or tumor stage. Likewise, no associations were seen with age, neither in cases nor in neoplasm-free controls (data not shown).
To evaluate whether the expression levels of the investigated miRNAs are associated with the development of CRC, patients were stratified by AJCC stages. In general, expression levels were very similar in early and late stage cancers. MiR-18a, 20a, 21, 29a, 133a, 143 and 145 were found to be over-expressed in plasma of CRC patients at early stages compared with neoplasm-free controls. MiR-18a, -20a, -21, -92a, -133a, -143, -145 and -181b showed higher expression levels in plasma of CRC patients at late stages compared with neoplasm-free controls. MiR-181b showed higher expression in the plasma of late stage CRC patients than early stage CRC patients (Table 2).
Table 2. MicroRNAs differently expressed between stage stratified colorectal cancer patients and neoplasm-free controls.
| Mean ΔCt* ofCRC patients atearly stages(stage I and II) | Mean ΔCt* ofCRC patients atlate stages(stage III and IV) | Mean ΔCt* ofneoplasm-freecontrols | P values† | |||
| CRC at earlystage vs controls(47 vs 144) | CRC at latestage vs controls(31 vs 144) | CRC at early stage vsCRC at late stage(47 vs 31) | ||||
| miR-18a | 8.36 | 8.48 | 9.17 | <0.001 | 0.001 | 0.491 |
| miR-20a | 4.73 | 4.73 | 5.11 | 0.012 | 0.022 | 0.984 |
| miR-21 | 3.24 | 3.27 | 3.80 | 0.001 | 0.002 | 0.878 |
| miR-29a | 6.30 | 6.40 | 6.81 | 0.003 | 0.055 | 0.328 |
| miR-92a | 3.08 | 3.01 | 3.29 | 0.058 | 0.013 | 0.597 |
| miR-106b | 7.33 | 7.36 | 7.62 | 0.088 | 0.128 | 0.984 |
| miR-133a | 11.44 | 11.46 | 12.31 | 0.003 | 0.012 | 0.935 |
| miR-143 | 8.63 | 8.93 | 9.61 | <0.001 | 0.007 | 0.210 |
| miR-145 | 8.53 | 8.65 | 9.26 | 0.003 | 0.029 | 0.711 |
| miR-181b | 10.36 | 9.80 | 10.16 | 0.932 | 0.035 | 0.024 |
| miR-342-3p | 7.53 | 7.23 | 7.24 | 0.052 | 0.777 | 0.174 |
| miR-532-3p | 11.76 | 11.62 | 11.92 | 0.445 | 0.084 | 0.405 |
Data were normalized to miR-16.
P values are based on Wilcoxon tests.
Abbreviations: CRC, colorectal cancer.
(iii) Multivariate analysis
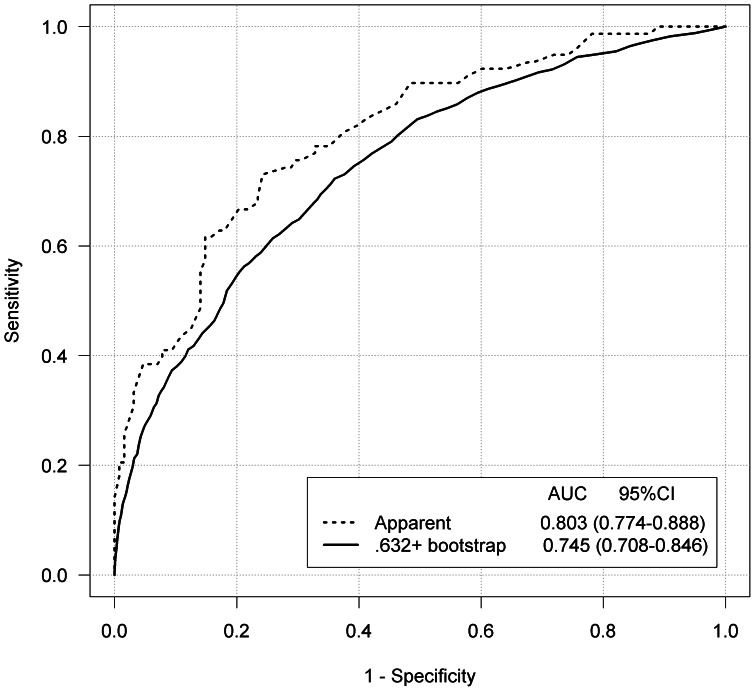
With the panel of 12 miRNAs (miR-18a, -20a, -21, -29a, -92a, -106b, -133a, -143, -145, -181b, -342-3p, -532-3p), the ROC analyses yielded an unadjusted AUC of 0.803 (95% CI: 0.774–0.888) and an adjusted, i.e. optimism-corrected, AUC of 0.745 (95% CI: 0.708–0.846). The ROC curves are depicted in Figure 2. For each miRNA, the ROC curves are depicted in Figure S1.
Figure 2. Receiver operating characteristic curves using the panel of 12 selected microRNAs for discrimination of 80 colorectal cancer patients and 144 neoplasm-free controls.
Adjustment for over-optimism was done by the.632+ bootstrap method. Abbreviations: AUC, area under receiver operating characteristic curve.
Expression of miRNAs in Advanced Adenoma Patients and Neoplasm-free Controls
Advanced colorectal adenomas represent a precursor stage of CRC. To assess potential use for early detection even of advanced adenomas, the identified 12 miRNAs were determined by qRT-PCR in plasma samples from 100 study participants (50 with advanced adenoma and 50 neoplasm-free controls). No statistically significant differences in miRNA expression levels were observed in the plasma of advanced adenoma patients and neoplasm-free controls.
Correlations of Expression Levels of Plasma miRNAs
Among participants of the validation phase, strong correlations were observed between expression levels of miR-18a and miR-20a (r = 0.800, p<0.001), which are both encoded in the miR-17-92 cluster; miR-143 and miR-145 (r = 0.762, p<0.001) which are both encoded in the miR-143/145 cluster; and miR-29a and miR-106b (r = 0.694, p<0.001), two closely residing miRNAs on chromosome 7q. The expression levels of some other miRNAs which are not colocated in the genome were also highly correlated. On the other hand, miR-92a which is also encoded in miR-17-92 cluster was not highly correlated with miR-18a or miR-20a (r = 0.340 and 0.251 respectively) (Table S3).
Discussion
In this study, we compared the expression levels of twelve miRNAs in plasma samples of CRC patients and neoplasm-free controls. To our knowledge, this is the first report on circulating miRNA for CRC detection using TaqMan MicroRNA Array for miRNA profiling. Five miRNAs (miR-29a, -106b, -133a, -342-3p, -532-3p) were identified from the microarray analyses and seven were selected from the literature (miR-18a, -20a, -21, -92a, -143, -145, -181b) for further investigation [17]. In our validation study, the expression levels of miR-18a, -20a, -21, -29a, -92a, -106b, -133a, -143, and -145 were found to be significantly higher in CRC patients than in neoplasm-free controls. ROC analysis of the identified miRNAs adjusted for over-optimism by bootstrap yielded an AUC of 0.745 (95% CI: 0.708–0.846). This result compares favorably with results of other blood-based tests for CRC detection [5].
Diagnostic characteristics of circulating miRNAs for CRC were evaluated previously in two recent studies. Ng et al. reported 0.89 sensitivity and 0.70 specificity at a certain cut-off point of miR-92a, and an AUC of 0.885 (95% CI: 0.83–0.94) based on 90 CRC patients (TNM stage I/II/III/IV: 6/34/23/27) and 50 controls [10]. Huang et al. reported 0.830 sensitivity and 0.847 specificity, and an AUC of 0.883 (95% CI: 0.830–0.937) obtained in a multivariate analysis, including miR-29a and miR-92a based on 100 CRC patients (TNM stage I/II/III/IV: 27/25/38/10) and 59 controls [9]. Thus, both studies found higher AUC estimates than obtained in the present study. These studies had not adjusted for over-optimism. While the 95% confidence interval (0.774–0.888) of the unadjusted AUC in our study encompassed the AUCs reported by Ng et al. and Huang et al., adjustment for over-optimism attenuated the AUC by approximately 6 percent units. Nevertheless, even our unadjusted AUC estimate was slightly lower than the previous reported ones, despite the larger number of included miRNAs. Factors that could partly account for these differences include stage distribution of CRCs and chance. The percentage of early stage CRC patients (AJCC stage I and II) was higher in our study (59%) than in the previously reported studies (44% and 52%, respectively) and probably closer to the stage distribution expected in a screening setting [5]. However, no major difference of expression levels by stage was observed in our study.
Colorectal adenomas represent a precursor stage of adenocarinoma. A previous study had reported that the differential expression of plasma miR-29a and miR-92a could also discriminate advanced adenoma patients from neoplasm-free controls even though discrimination was less pronounced than for CRC patients [9]. In our study, none of the investigated miRNAs was found significantly differentially expressed in the advanced adenoma patients compared to the neoplasm-free controls. These results suggest that the investigated miRNAs may not be useful for the diagnosis of advanced adenomas. However, lack of significant differences may also be due to limited power to detect moderate differences with the the sample size of our substudy comparing carriers of advanced adenomas and neoplasm-free subjects Given the observed lack of differential expression of individual miRNAs, a multivariate model for prediction of advanced adenomas was not applied.
Although the differential expression of plasma miR-18a, -20a, -21, -106b, -133a, -143, -145 and -181b in CRC patients was examined in some tissue sample based studies [24]–[27], this is, to our knowledge, the first study reporting on the expression of these miRNAs in plasma samples of a large number of patients with CRC and neoplasm-free controls. Statistically significant differences in the expression of all analyzed miRNAs except miR-181b, miR-342-3p and miR-532-3p were observed. Intriguingly, our data showed that the expression levels of miR-133a -143, and -145 were significantly over-expressed in the plasma of CRC patients compared to the neoplasm-free controls, which seems to contradict data consistently showing less expression of these three miRNAs in cancer in tissue sample based studies [28]–[34]. Further studies with larger numbers of patients and simultaneous measurements of miRNA expression in plasma and tissue may be warranted to clarify this issue.
A common problem in research concerning circulating miRNAs is that no consensus internal control has been established. We evaluated miR-16, miR-188-5p and RNU6B, and due to the consistent, stable and high expression across all plasma samples, miR-16 was selected as internal control in the plasma sample based tests, but more empirical validations are still needed for a consensus on robust and accurate internal controls.
The nine miRNAs found to be over-expressed in plasma of CRC patients in our study have also been investigated with respect to the other diseases in previous research [9], [10], [33], [35]–[48]. (Table 3) For example, plasma miR-21 was found to be over-expressed in a wide variety of malignancies including hepatocellular carcinoma, prostate cancer, and gastric cancer [35], [36], [42], [46]–[49]. These commonalities in miR-21 expression pattern in different cancers suggestes that miR-21 might function as an oncogene. Functional investigations showed that miR-21 targets tumor suppressor genes such as phosphatase and tensin homolog (PTEN) and inversely regulates its expression [50]. PTEN is ranked the second most commonly mutated tumor suppressor gene after p53. It can be inactivated by mutation with loss of heterozygosity, promoter methylation, miRNA interference and some other mechanisms in a number of cancers including brain, prostate, uterine cancer [51]. Differential expression of some miRNAs in multiple cancers support suggestions that they should typically be combined with additional miRNAs when used for detection of specific cancers.
Table 3. Circulating microRNAs involved in cancer and other diseases.
| MiRNAs | Chromosomal location | Number of studies (references) | Expression in patients | Samples sizes | Expression of potential target gene |
| miR-18a | 13q31.3 | 1 [42] | Up in pancreatic cancer | 76 | |
| miR-20a | 13q31.3 | 2 [53] | Up in multiple myeloma | 40 | |
| miR-21 | 17q23.1 | 8 [35], [36], [42], [46]–[49] | Up in HCC | 407 | |
| Up in ESCC (the miR-21/miR-375 ratio) | 70 | ||||
| Up in NSCLC | 93 | ||||
| Up in prostate cancer | 71 | ||||
| Up in pancreatic cancer | 60 | PTEN, PDCD4, Maspin and TPM1 reduced | |||
| Up in GC | 96 | ||||
| Up in pancreatic cancer | 85 | ||||
| Up in CHC | 81 | ||||
| miR-29a | 7q32.3 | 2 [9], [40] | Up in CRC | 159 | |
| Up in active pulmonary tuberculosis | 127 | ||||
| miR-92a | 13q31.3 | 6 [9], [10], [37], [39], [43], [44], [54] | Up in CRC | 159 | |
| Up in CRC | 140 | ||||
| Down in HCC | 20 | ||||
| Down in acute leukemia | 77 | ||||
| Down in CRC | 102 | ||||
| Down in CAD | 53 | ||||
| miR-106b | 7q22.1 | 1 [42] | Up in GC | 96 | |
| miR-133a | 18q11.2 | 2 [39], [45] | Up in CAD | 53 | |
| Up in AMI | 63 | ||||
| miR-143 | 5q32 | 1 [38] | Up in enterovirus infection | 40 | |
| miR-145 | 5q32 | 1 [39] | Down in CAD | 53 |
Abbreviations: CRC, colorectal cancer; HCC, hepatocellular carcinoma; ESCC, oesophageal squamous cell carcinoma; NSCLC, non-small cell lung cancer; GC, gastric cancer; CAD, coronary artery disease; CHC, chronic hepatitis C virus infection; AMI, acute myocardial infarction; PTEN, phosphatase and tensin homolog; PDCD4, programmed cell death 4; TPM1, tropomyosin.
There are some limitations that need to be taken into account when interpreting the results of this study. First, the sample size is still small, especially in the marker selection phase. Second, a lot of miRNAs’ abundances in plasma are too low to be accurately quantified by qRT-PCR, therefore, some potential relevant markers could not be considered. Third, it remains to be determined to what extent modified expression levels of miRNAs found among CRC patients in this study are CRC specific.
Conclusions
Plasma miRNAs appear to be potentially useful biomarkers for early detection and diagnosis of CRC. While research on plasma based miRNA profiling is still at very early stage compared to research on tissue based miRNA profiling, it might have the potential to contribute to development of new approaches of non-invasive, blood based CRC screening. Nevertheless, the diagnostic performance of the identified panel of miRNAs might not yet be sufficient to compete with performance of some other non-invasive tests, in particular immunochemical faecal occult blood tests [52]. Further research on a multi-marker blood based test, potentially including the panel of miRNAs identified in this study might be a promising approach to enhance the repertoire for non-invasive cancer screening.
Supporting Information
Receiver operating characteristic curves using 12 selected microRNAs (miR-18a, -20a, -21, -29a, -92a, -106b, -133a, -143, -145, -181b, -342-3p and miR-532-3p) for discrimination of 80 colorectal cancer patients and 144 neoplasm-free controls. Abbreviations: FPR, false positive rate. TPR, true positive rate. AUC, area under receiver operating characteristic curve.
(DOC)
667 human microRNAs tested using TaqMan MicroRNA Array.
(DOC)
RT-primer pools used for multiplex Real-Time quantitative PCR.
(DOC)
Spearman correlation coefficients of microRNAs expression levels among 324 participants of the validation (p<0.001 if not annotated).
(DOC)
Funding Statement
The authors have no support or funding to report.
References
- 1. Jemal A, Bray F (2011) Center MM, Ferlay J, Ward E, et al (2011) Global cancer statistics. CA Cancer J Clin 61: 69–90. [DOI] [PubMed] [Google Scholar]
- 2. Gupta AK, Brenner DE, Turgeon DK (2008) Early detection of colon cancer: new tests on the horizon. Mol Diagn Ther 12: 77–85. [DOI] [PubMed] [Google Scholar]
- 3. Pawa N, Arulampalam T, Norton JD (2011) Screening for colorectal cancer: established and emerging modalities. Nat Rev Gastroenterol Hepatol 8: 711–22. [DOI] [PubMed] [Google Scholar]
- 4. Hundt S, Haug U, Brenner H (2007) Blood markers for early detection of colorectal cancer: a systematic review. Cancer Epidemiol Biomarkers Prev 16: 1935–53. [DOI] [PubMed] [Google Scholar]
- 5. Tao S, Hundt S, Haug U, Brenner H (2011) Sensitivity estimates of blood-based tests for colorectal cancer detection: impact of overrepresentation of advanced stage disease. Am J Gastroenterol 106: 242–53. [DOI] [PubMed] [Google Scholar]
- 6. Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–97. [DOI] [PubMed] [Google Scholar]
- 7. Garzon R, Marcucci G, Croce CM (2010) Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov 9: 775–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, et al. (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 105: 10513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang Z, Huang D, Ni S, Peng Z, Sheng W, et al. (2010) Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer 127: 118–26. [DOI] [PubMed] [Google Scholar]
- 10. Ng EK, Chong WW, Jin H, Lam EK, Shin VY, et al. (2009) Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut 58: 1375–81. [DOI] [PubMed] [Google Scholar]
- 11. Pu XX, Huang GL, Guo HQ, Guo CC, Li H, et al. (2010) Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J Gastroenterol Hepatol 25: 1674–80. [DOI] [PubMed] [Google Scholar]
- 12. Brenner H, Chang-Claude J, Seiler CM, Rickert A, Hoffmeister M (2011) Protection from colorectal cancer after colonoscopy: a population-based, case-control study. Ann Intern Med 154: 22–30. [DOI] [PubMed] [Google Scholar]
- 13. Brenner H, Chang-Claude J, Seiler CM, Hoffmeister M (2011) Long-term risk of colorectal cancer after negative colonoscopy. J Clin Oncol 29: 3761–7. [DOI] [PubMed] [Google Scholar]
- 14. Hundt S, Haug U, Brenner H (2009) Comparative evaluation of immunochemical fecal occult blood tests for colorectal adenoma detection. Ann Intern Med 150: 162–9. [DOI] [PubMed] [Google Scholar]
- 15. Brenner H, Tao S, Haug U (2010) Low-dose aspirin use and performance of immunochemical fecal occult blood tests. JAMA 304: 2513–20. [DOI] [PubMed] [Google Scholar]
- 16. Brenner H, Haug U, Hundt S (2010) Inter-test agreement and quantitative cross-validation of immunochromatographical fecal occult blood tests. Int J Cancer 127: 1643–9. [DOI] [PubMed] [Google Scholar]
- 17. Luo X, Burwinkel B, Tao S, Brenner H (2011) MicroRNA signatures: novel biomarker for colorectal cancer? Cancer Epidemiol Biomarkers Prev 20: 1272–86. [DOI] [PubMed] [Google Scholar]
- 18. Turchinovich A, Weiz L, Langheinz A, Burwinkel B (2011) Characterization of extracellular circulating microRNA. Nucleic Acids Res 39: 7223–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kroh EM, Parkin RK, Mitchell PS, Tewari M (2010) Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods 50: 298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Efron B, Tibshirani R (1997) Improvements on cross-validation: The.632+ bootstrap method. J Am Stat Assoc. 92: 548–60. [Google Scholar]
- 21. Adler W, Lausen B (2009) Bootstrap estimated true and false positive rates and ROC curve. Comput Stat Data An 53: 718–29. [Google Scholar]
- 22.Canty A, Ripley B (2012) boot: bootstrap R (S-Plus) functions. R package version 1.3–4. Available: http://CRAN.R-project.org/package=boot. Accessed 27 Jul 2012.
- 23.Potapov S, Adler W, Lausen B (2012) Daim: Diagnostic accuracy of classification models. R package version 1.0.0. Available: http://CRAN.R-project.org/package=Daim. Accessed 24 Jun 2012.
- 24. Arndt GM, Dossey L, Cullen LM, Lai A, Druker R, et al. (2009) Characterization of global microRNA expression reveals oncogenic potential of miR-145 in metastatic colorectal cancer. BMC Cancer 2009 9: 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Diosdado B, van de Wiel MA, Terhaar Sive Droste JS, Mongera S, Postma C, et al. (2009) MiR-17–92 cluster is associated with 13q gain and c-myc expression during colorectal adenoma to adenocarcinoma progression. Br J Cancer 101: 707–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schmitz KJ, Hey S, Schinwald A, Wohlschlaeger J, Baba HA, et al. (2009) Differential expression of microRNA 181b and microRNA 21 in hyperplastic polyps and sessile serrated adenomas of the colon. Virchows Arch 455: 49–54. [DOI] [PubMed] [Google Scholar]
- 27. Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, et al. (2007) Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology 72: 397–402. [DOI] [PubMed] [Google Scholar]
- 28. Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, et al. (2008) MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 299: 425–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen X, Guo X, Zhang H, Xiang Y, Chen J, et al. (2009) Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene 28: 1385–92. [DOI] [PubMed] [Google Scholar]
- 30. Earle JS, Luthra R, Romans A, Abraham R, Ensor J, et al. (2010) Association of microRNA expression with microsatellite instability status in colorectal adenocarcinoma. J Mol Diagn 12: 433–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kulda V, Pesta M, Topolcan O, Liska V, Treska V, et al. (2010) Relevance of miR-21 and miR-143 expression in tissue samples of colorectal carcinoma and its liver metastases. Cancer Genet Cytogenet 200: 154–60. [DOI] [PubMed] [Google Scholar]
- 32. Motoyama K, Inoue H, Takatsuno Y, Tanaka F, Mimori K, et al. (2009) Over- and under-expressed microRNAs in human colorectal cancer. Int J Oncol 34: 1069–75. [DOI] [PubMed] [Google Scholar]
- 33. Wang J, Chen J, Chang P, LeBlanc A, Li D, et al. (2009) MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res 2: 807–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ng EK, Tsang WP, Ng SS, Jin HC, Yu J, et al. (2009) MicroRNA-143 targets DNA methyltransferases 3A in colorectal cancer. Br J Cancer 101: 699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ali S, Almhanna K, Chen W, Philip PA, Sarkar FH (2010) Differentially expressed miRNAs in the plasma may provide a molecular signature for aggressive pancreatic cancer. Am J Transl Res 3: 28–47. [PMC free article] [PubMed] [Google Scholar]
- 36. Bihrer V, Waidmann O, Friedrich-Rust M, Forestier N, Susser S, et al. (2011) Serum microRNA-21 as marker for necroinflammation in hepatitis C patients with and without hepatocellular carcinoma. PloS One 6: e26971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cheng H, Zhang L, Cogdell DE, Zheng H, Schetter AJ, et al. (2011) Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PloS One 6: e17745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cui L, Qi Y, Li H, Ge Y, Zhao K, et al. (2011) Serum microRNA expression profile distinguishes enterovirus 71 and coxsackievirus 16 infections in patients with hand-foot-and-mouth disease. PloS One 6: e27071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fichtlscherer S, De Rosa S, Fox H, Schwietz T, Fischer A, et al. (2010) Circulating microRNAs in patients with coronary artery disease. Circ Res 107: 677–84. [DOI] [PubMed] [Google Scholar]
- 40. Fu Y, Yi Z, Wu X, Li J, Xu F (2011) Circulating microRNAs in patients with active pulmonary tuberculosis. J Clin Microbiol 49: 4246–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huang JJ, Yu J, Li JY, Liu YT, Zhong RQ (2012) Circulating microRNA expression is associated with genetic subtype and survival of multiple myeloma. Med Oncol. 29: 2402–8. [DOI] [PubMed] [Google Scholar]
- 42. Komatsu S, Ichikawa D, Takeshita H, Tsujiura M, Morimura R, et al. (2011) Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer 105: 104–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Morimura R, Komatsu S, Ichikawa D, Takeshita H, Tsujiura M, et al. (2011) Novel diagnostic value of circulating miR-18a in plasma of patients with pancreatic cancer. Br J Cancer 105: 1733–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shigoka M, Tsuchida A, Matsudo T, Nagakawa Y, Saito H, et al. (2010) Deregulation of miR-92a expression is implicated in hepatocellular carcinoma development. Pathol Int 60: 351–7. [DOI] [PubMed] [Google Scholar]
- 45. Tanaka M, Oikawa K, Takanashi M, Kudo M, Ohyashiki J, et al. (2009) Down-regulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PloS One 4: e5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, et al. (2010) Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer 102: 1174–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, et al. (2010) Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J 31: 659–66. [DOI] [PubMed] [Google Scholar]
- 48. Wei J, Gao W, Zhu CJ, Liu YQ, Mei Z, et al. (2011) Identification of plasma microRNA-21 as a biomarker for early detection and chemosensitivity of non-small cell lung cancer. Chin J Cancer 30: 407–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yaman Agaoglu F, Kovancilar M, Dizdar Y, Darendeliler E, Holdenrieder S, et al. (2011) Investigation of miR-21, miR-141, and miR-221 in blood circulation of patients with prostate cancer. Tumour Biol 32: 583–8. [DOI] [PubMed] [Google Scholar]
- 50. Zhou J, Yu L, Gao X, Hu J, Wang J, et al. (2011) Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol 29: 4781–8. [DOI] [PubMed] [Google Scholar]
- 51. Wang J, Chen J, Chang P, LeBlanc A, Li D, et al. (2009) MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Cancer Prev Res 2: 807–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, et al. (2007) MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133: 647–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Georgescu MM (2010) PTEN Tumor Suppressor Network in PI3K-Akt Pathway Control. Genes Cancer 1: 1170–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tao S, Haug U, Kuhn K, Brenner H (2012) Comparison and combination of blood-based inflammatory markers with faecal occult blood tests for non-invasive colorectal cancer screening. Br J Cancer 106: 1424–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Receiver operating characteristic curves using 12 selected microRNAs (miR-18a, -20a, -21, -29a, -92a, -106b, -133a, -143, -145, -181b, -342-3p and miR-532-3p) for discrimination of 80 colorectal cancer patients and 144 neoplasm-free controls. Abbreviations: FPR, false positive rate. TPR, true positive rate. AUC, area under receiver operating characteristic curve.
(DOC)
667 human microRNAs tested using TaqMan MicroRNA Array.
(DOC)
RT-primer pools used for multiplex Real-Time quantitative PCR.
(DOC)
Spearman correlation coefficients of microRNAs expression levels among 324 participants of the validation (p<0.001 if not annotated).
(DOC)