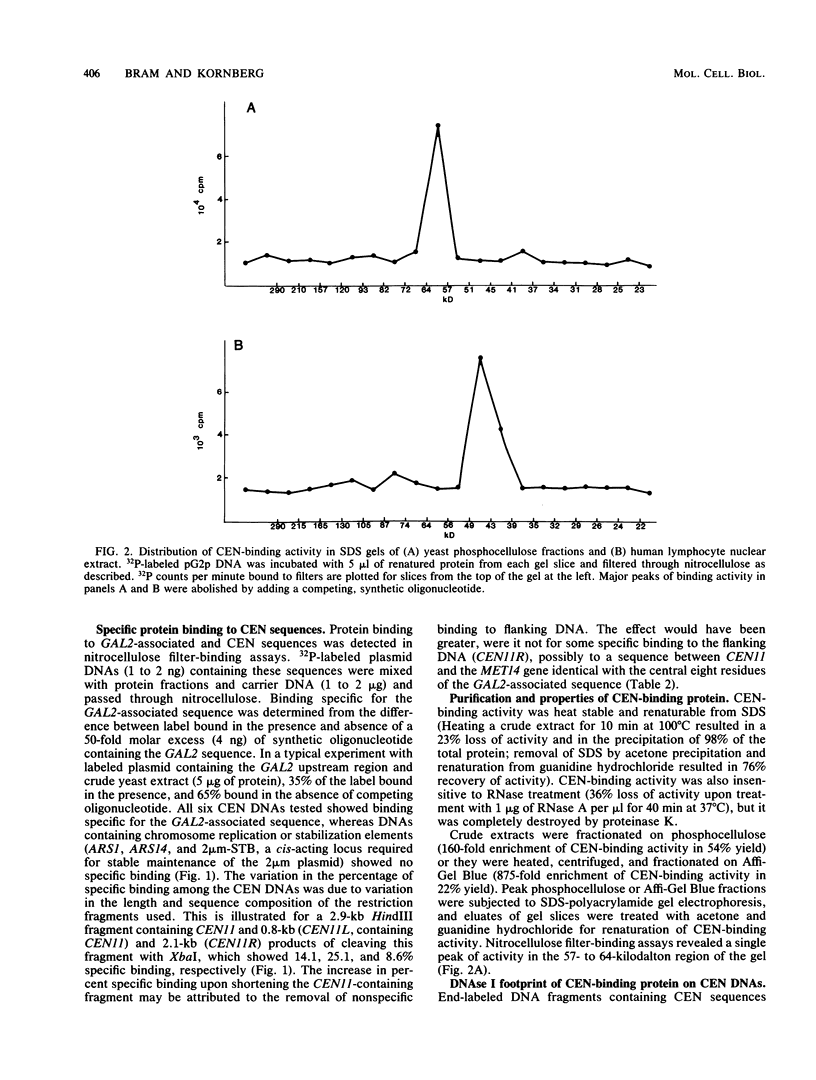
Abstract
A protein that binds specifically to Saccharomyces cerevisiae centromere DNA element I was purified on the basis of a nitrocellulose filter-binding assay. This protein, termed centromere-binding protein 1 (CP1), was heat stable and renaturable from sodium dodecyl sulfate (SDS), and assays of eluates from SDS gels indicated a molecular weight of 57,000 to 64,000. An activity with similar specificity and stability was detected in human lymphocyte extracts, and analysis in SDS gels revealed a molecular weight of 39,000 to 49,000. CP1-binding sites occurred not only at centromeres but also near many transcription units, for example, adjacent to binding sites for the GAL4-positive regulatory protein upstream of the GAL2 gene in S. cerevisiae and adjacent to the TATA element of the adenovirus major late promoter. A factor (termed USF) that binds to the latter site and stimulates transcription has been isolated from HeLa cells by others.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baralle F. E., Shoulders C. C., Goodbourn S., Jeffreys A., Proudfoot N. J. The 5' flanking region of human epsilon-globin gene. Nucleic Acids Res. 1980 Oct 10;8(19):4393–4404. doi: 10.1093/nar/8.19.4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrois M., Riou G., Galibert F. Complete nucleotide sequence of minicircle kinetoplast DNA from Trypanosoma equiperdum. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3323–3327. doi: 10.1073/pnas.78.6.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayev A. A., Jr, Krayev A. S., Lyubomirskaya N. V., Ilyin Y. V., Skryabin K. G., Georgiev G. P. The transposable element Mdg3 in Drosophila melanogaster is flanked with the perfect direct and mismatched inverted repeats. Nucleic Acids Res. 1980 Aug 11;8(15):3263–3273. doi: 10.1093/nar/8.15.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom K. S., Carbon J. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 1982 Jun;29(2):305–317. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- Bloom K. S., Fitzgerald-Hayes M., Carbon J. Structural analysis and sequence organization of yeast centromeres. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1175–1185. doi: 10.1101/sqb.1983.047.01.133. [DOI] [PubMed] [Google Scholar]
- Bram R. J., Kornberg R. D. Specific protein binding to far upstream activating sequences in polymerase II promoters. Proc Natl Acad Sci U S A. 1985 Jan;82(1):43–47. doi: 10.1073/pnas.82.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram R. J., Lue N. F., Kornberg R. D. A GAL family of upstream activating sequences in yeast: roles in both induction and repression of transcription. EMBO J. 1986 Mar;5(3):603–608. doi: 10.1002/j.1460-2075.1986.tb04253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew R. W., Chodosh L. A., Sharp P. A. An RNA polymerase II transcription factor binds to an upstream element in the adenovirus major late promoter. Cell. 1985 Dec;43(2 Pt 1):439–448. doi: 10.1016/0092-8674(85)90174-6. [DOI] [PubMed] [Google Scholar]
- Celniker S. E., Campbell J. L. Yeast DNA replication in vitro: initiation and elongation events mimic in vivo processes. Cell. 1982 Nov;31(1):201–213. doi: 10.1016/0092-8674(82)90420-2. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Howley P. M., Levinson A. D., Seeburg P. H. The primary structure and genetic organization of the bovine papillomavirus type 1 genome. Nature. 1982 Oct 7;299(5883):529–534. doi: 10.1038/299529a0. [DOI] [PubMed] [Google Scholar]
- Clarke L., Carbon J. Genomic substitutions of centromeres in Saccharomyces cerevisiae. Nature. 1983 Sep 1;305(5929):23–28. doi: 10.1038/305023a0. [DOI] [PubMed] [Google Scholar]
- Clarke L., Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980 Oct 9;287(5782):504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- Comings D. E., Okada T. A. Fine structure of kinetochore in Indian muntjac. Exp Cell Res. 1971 Jul;67(1):97–110. doi: 10.1016/0014-4827(71)90625-2. [DOI] [PubMed] [Google Scholar]
- DesGroseillers L., Villemur R., Jolicoeur P. The high leukemogenic potential of Gross passage A murine leukemia virus maps in the region of the genome corresponding to the long terminal repeat and to the 3' end of env. J Virol. 1983 Jul;47(1):24–32. doi: 10.1128/jvi.47.1.24-32.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiovanni L., Haynes S. R., Misra R., Jelinek W. R. Kpn I family of long-dispersed repeated DNA sequences of man: evidence for entry into genomic DNA of DNA copies of poly(A)-terminated Kpn I RNAs. Proc Natl Acad Sci U S A. 1983 Nov;80(21):6533–6537. doi: 10.1073/pnas.80.21.6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan C. H., Jagadeeswaran P., Wang R. R., Weissman S. M. Structural analysis of templates and RNA polymerase III transcripts of Alu family sequences interspersed among the human beta-like globin genes. Gene. 1981 Mar;13(2):185–196. doi: 10.1016/0378-1119(81)90007-x. [DOI] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M., Buhler J. M., Cooper T. G., Carbon J. Isolation and subcloning analysis of functional centromere DNA (CEN11) from Saccharomyces cerevisiae chromosome XI. Mol Cell Biol. 1982 Jan;2(1):82–87. doi: 10.1128/mcb.2.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M., Clarke L., Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982 May;29(1):235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- Hager D. A., Burgess R. R. Elution of proteins from sodium dodecyl sulfate-polyacrylamide gels, removal of sodium dodecyl sulfate, and renaturation of enzymatic activity: results with sigma subunit of Escherichia coli RNA polymerase, wheat germ DNA topoisomerase, and other enzymes. Anal Biochem. 1980 Nov 15;109(1):76–86. doi: 10.1016/0003-2697(80)90013-5. [DOI] [PubMed] [Google Scholar]
- Hieter P., Pridmore D., Hegemann J. H., Thomas M., Davis R. W., Philippsen P. Functional selection and analysis of yeast centromeric DNA. Cell. 1985 Oct;42(3):913–921. doi: 10.1016/0092-8674(85)90287-9. [DOI] [PubMed] [Google Scholar]
- Hsiao C. L., Carbon J. Direct selection procedure for the isolation of functional centromeric DNA. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3760–3764. doi: 10.1073/pnas.78.6.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B., Miklos G. L. Functional aspects of satellite DNA and heterochromatin. Int Rev Cytol. 1979;58:1–114. doi: 10.1016/s0074-7696(08)61473-4. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Maine G. T., Surosky R. T., Tye B. K. Isolation and characterization of the centromere from chromosome V (CEN5) of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Jan;4(1):86–91. doi: 10.1128/mcb.4.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw P., Tzagoloff A. Assembly of the mitochondrial membrane system. Characterization of a yeast nuclear gene involved in the processing of the cytochrome b pre-mRNA. J Biol Chem. 1983 Aug 10;258(15):9459–9468. [PubMed] [Google Scholar]
- Miyamoto N. G., Moncollin V., Egly J. M., Chambon P. Specific interaction between a transcription factor and the upstream element of the adenovirus-2 major late promoter. EMBO J. 1985 Dec 16;4(13A):3563–3570. doi: 10.1002/j.1460-2075.1985.tb04118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman A. F., De Boer P. A., De Laaf R. T., Destrée O. H. Primary structure of the histone H2A and H2B genes and their flanking sequences in a minor histone gene cluster of Xenopus laevis. FEBS Lett. 1982 Aug 2;144(2):235–241. doi: 10.1016/0014-5793(82)80645-5. [DOI] [PubMed] [Google Scholar]
- Muesing M. A., Smith D. H., Cabradilla C. D., Benton C. V., Lasky L. A., Capon D. J. Nucleic acid structure and expression of the human AIDS/lymphadenopathy retrovirus. Nature. 1985 Feb 7;313(6002):450–458. doi: 10.1038/313450a0. [DOI] [PubMed] [Google Scholar]
- Neitz M., Carbon J. Identification and characterization of the centromere from chromosome XIV in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Nov;5(11):2887–2893. doi: 10.1128/mcb.5.11.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikai M., Okano Y., Yamashita H., Watanabe M. Characterisation of centromere (kinetochore) antigen reactive with sera of patients with a scleroderma variant (CREST syndrome). Ann Rheum Dis. 1984 Dec;43(6):819–824. doi: 10.1136/ard.43.6.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hare K., Rubin G. M. Structures of P transposable elements and their sites of insertion and excision in the Drosophila melanogaster genome. Cell. 1983 Aug;34(1):25–35. doi: 10.1016/0092-8674(83)90133-2. [DOI] [PubMed] [Google Scholar]
- Panzeri L., Landonio L., Stotz A., Philippsen P. Role of conserved sequence elements in yeast centromere DNA. EMBO J. 1985 Jul;4(7):1867–1874. doi: 10.1002/j.1460-2075.1985.tb03862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzeri L., Philippsen P. Centromeric DNA from chromosome VI in Saccharomyces cerevisiae strains. EMBO J. 1982;1(12):1605–1611. doi: 10.1002/j.1460-2075.1982.tb01362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pech M., Streeck R. E., Zachau H. G. Patchwork structure of a bovine satellite DNA. Cell. 1979 Nov;18(3):883–893. doi: 10.1016/0092-8674(79)90140-5. [DOI] [PubMed] [Google Scholar]
- Ris H., Witt P. L. Structure of the mammalian kinetochore. Chromosoma. 1981;82(2):153–170. doi: 10.1007/BF00286101. [DOI] [PubMed] [Google Scholar]
- Roos U. P. Light and electron microscopy of rat kangaroo cells in mitosis. II. Kinetochore structure and function. Chromosoma. 1973;41(2):195–220. doi: 10.1007/BF00319696. [DOI] [PubMed] [Google Scholar]
- Sawadogo M., Roeder R. G. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell. 1985 Nov;43(1):165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- Singer M. F. Highly repeated sequences in mammalian genomes. Int Rev Cytol. 1982;76:67–112. doi: 10.1016/s0074-7696(08)61789-1. [DOI] [PubMed] [Google Scholar]
- Stinchcomb D. T., Mann C., Davis R. W. Centromeric DNA from Saccharomyces cerevisiae. J Mol Biol. 1982 Jun 25;158(2):157–190. doi: 10.1016/0022-2836(82)90427-2. [DOI] [PubMed] [Google Scholar]
- Temple M., Antoine G., Delius H., Stahl S., Winnacker E. L. Replication of mouse adenovirus strain FL DNA. Virology. 1981 Feb;109(1):1–12. doi: 10.1016/0042-6822(81)90466-9. [DOI] [PubMed] [Google Scholar]
- Tschumper G., Carbon J. Sequence of a yeast DNA fragment containing a chromosomal replicator and the TRP1 gene. Gene. 1980 Jul;10(2):157–166. doi: 10.1016/0378-1119(80)90133-x. [DOI] [PubMed] [Google Scholar]
- Villanueva J., Bull P., Valenzuela P., Venegas A. Nucleotide sequence of a yeast tRNAArg3A gene and its transcription in a homologous in vitro system. FEBS Lett. 1984 Feb 13;167(1):165–169. doi: 10.1016/0014-5793(84)80854-6. [DOI] [PubMed] [Google Scholar]
- Williamson V. M., Cox D., Young E. T., Russell D. W., Smith M. Characterization of transposable element-associated mutations that alter yeast alcohol dehydrogenase II expression. Mol Cell Biol. 1983 Jan;3(1):20–31. doi: 10.1128/mcb.3.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth T., Glöggler K., Baumruker T., Schmidt M., Horak I. Family of middle repetitive DNA sequences in the mouse genome with structural features of solitary retroviral long terminal repeats. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3327–3330. doi: 10.1073/pnas.80.11.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]