Abstract
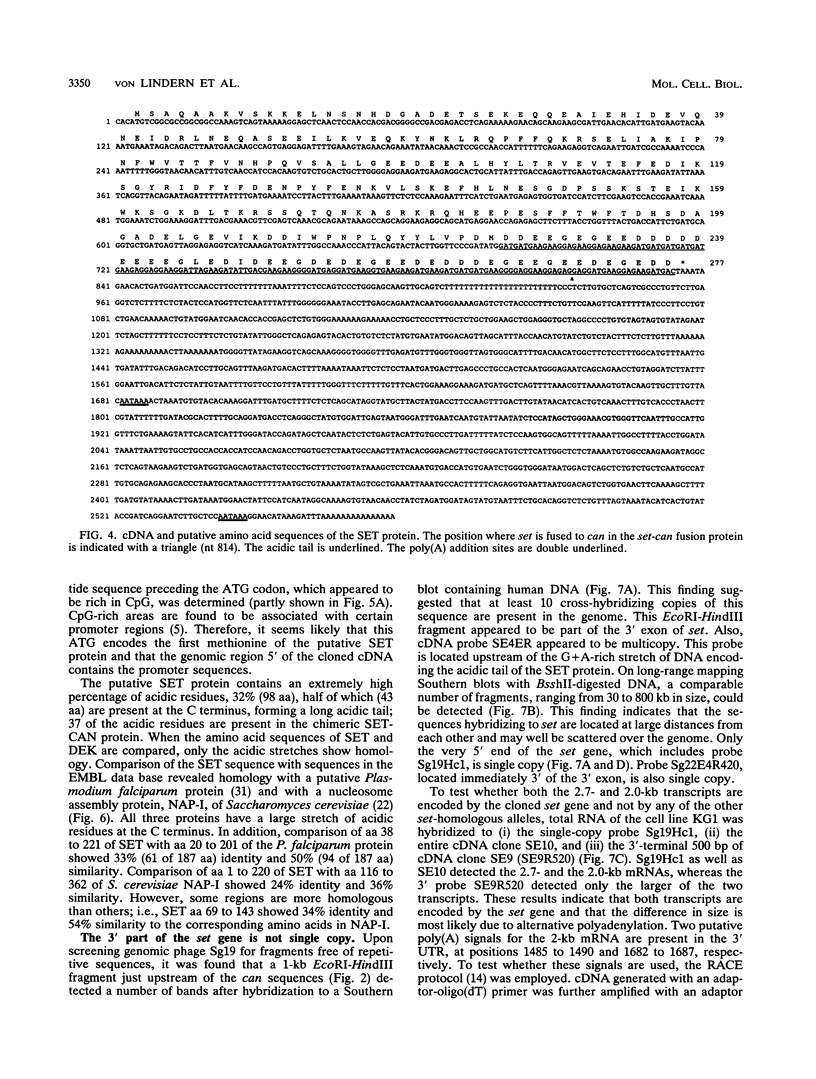
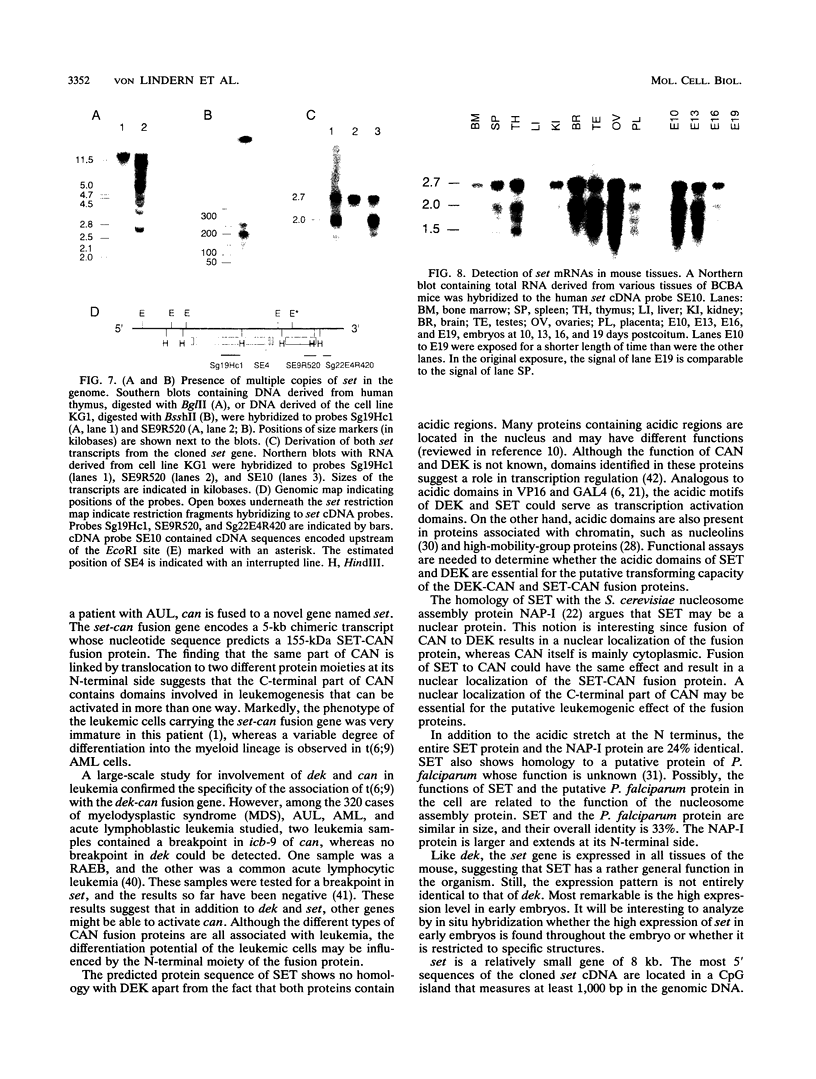
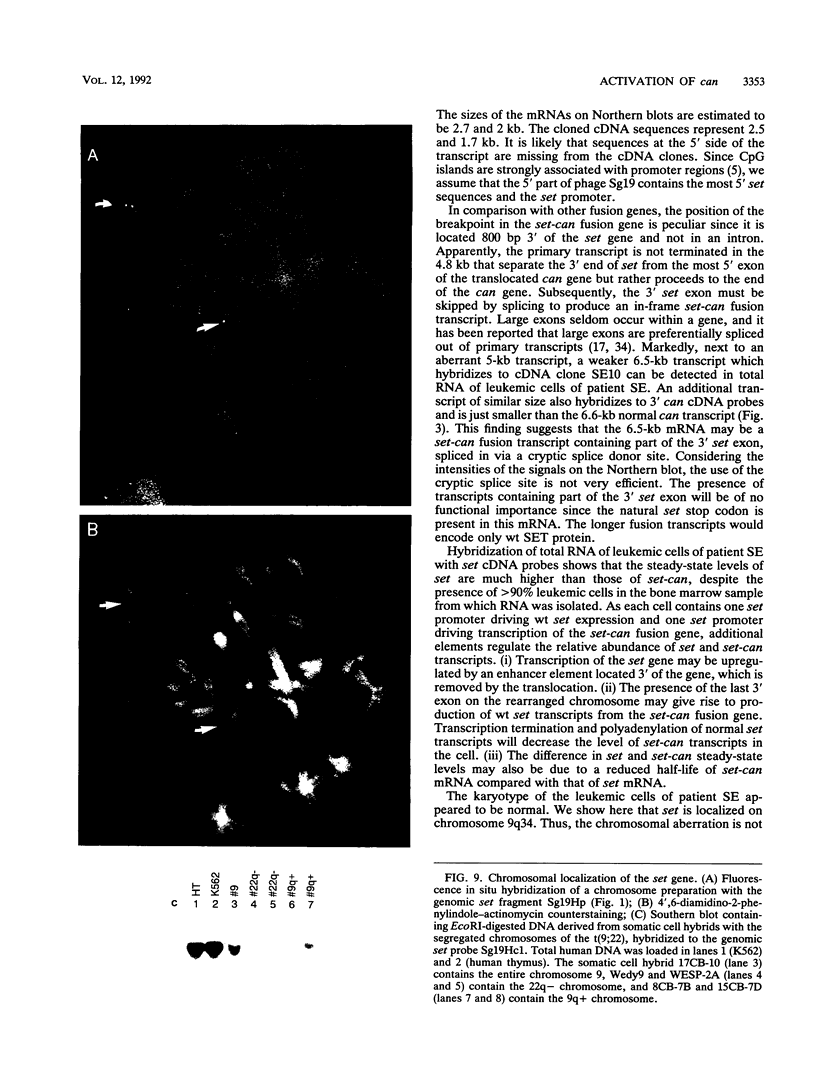
The translocation (6;9)(p23;q34) in acute nonlymphocytic leukemia results in the formation of a highly consistent dek-can fusion gene. Translocation breakpoints invariably occur in single introns of dek and can, which were named icb-6 and icb-9, respectively. In a case of acute undifferentiated leukemia, a breakpoint was detected in icb-9 of can, whereas no breakpoint could be detected in dek. Genomic and cDNA cloning showed that instead of dek, a different gene was fused to can, which was named set. set encodes transcripts of 2.0 and 2.7 kb that result from the use of alternative polyadenylation sites. Both transcripts contain the open reading frame for a putative SET protein with a predicted molecular mass of 32 kDa. The set-can fusion gene is transcribed into a 5-kb transcript that contains a single open reading frame predicting a 155-kDa chimeric SET-CAN protein. The SET sequence shows homology with the yeast nucleosome assembly protein NAP-I. The only common sequence motif of SET and DEK proteins is an acidic region. SET has a long acidic tail, of which a large part is present in the predicted SET-CAN fusion protein. The set gene is located on chromosome 9q34, centromeric of c-abl. Since a dek-can fusion gene is present in t(6;9) acute myeloid leukemia and a set-can fusion gene was found in a case of acute undifferentiated leukemia, we assume that can may function as an oncogene activated by fusion of its 3' part to dek, set, or perhaps other genes.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adriaansen H. J., Soeting P. W., Wolvers-Tettero I. L., van Dongen J. J. Immunoglobulin and T-cell receptor gene rearrangements in acute non-lymphocytic leukemias. Analysis of 54 cases and a review of the literature. Leukemia. 1991 Sep;5(9):744–751. [PubMed] [Google Scholar]
- Adriaansen H. J., van Dongen J. J., Hooijkaas H., Hählen K., van 't Veer M. B., Löwenberg B., Hagemeijer A. Translocation (6;9) may be associated with a specific TdT-positive immunological phenotype in ANLL. Leukemia. 1988 Mar;2(3):136–140. [PubMed] [Google Scholar]
- Arnoldus E. P., Wiegant J., Noordermeer I. A., Wessels J. W., Beverstock G. C., Grosveld G. C., van der Ploeg M., Raap A. K. Detection of the Philadelphia chromosome in interphase nuclei. Cytogenet Cell Genet. 1990;54(3-4):108–111. doi: 10.1159/000132972. [DOI] [PubMed] [Google Scholar]
- Auffray C., Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980 Jun;107(2):303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986 May 15;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Brent R., Ptashne M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell. 1985 Dec;43(3 Pt 2):729–736. doi: 10.1016/0092-8674(85)90246-6. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Earnshaw W. C. Anionic regions in nuclear proteins. J Cell Biol. 1987 Oct;105(4):1479–1482. doi: 10.1083/jcb.105.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Frischauf A. M., Lehrach H., Poustka A., Murray N. Lambda replacement vectors carrying polylinker sequences. J Mol Biol. 1983 Nov 15;170(4):827–842. doi: 10.1016/s0022-2836(83)80190-9. [DOI] [PubMed] [Google Scholar]
- Geurts van Kessel A. H., Tetteroo P. A., von dem Borne A. E., Hagemeijer A., Bootsma D. Expression of human myeloid-associated surface antigens in human-mouse myeloid cell hybrids. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3748–3752. doi: 10.1073/pnas.80.12.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosveld G., Verwoerd T., van Agthoven T., de Klein A., Ramachandran K. L., Heisterkamp N., Stam K., Groffen J. The chronic myelocytic cell line K562 contains a breakpoint in bcr and produces a chimeric bcr/c-abl transcript. Mol Cell Biol. 1986 Feb;6(2):607–616. doi: 10.1128/mcb.6.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins J. D. A survey on intron and exon lengths. Nucleic Acids Res. 1988 Nov 11;16(21):9893–9908. doi: 10.1093/nar/16.21.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisterkamp N., Groffen J. Duplication of the bcr and gamma-glutamyl transpeptidase genes. Nucleic Acids Res. 1988 Aug 25;16(16):8045–8056. doi: 10.1093/nar/16.16.8045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans A., Heisterkamp N., von Linden M., van Baal S., Meijer D., van der Plas D., Wiedemann L. M., Groffen J., Bootsma D., Grosveld G. Unique fusion of bcr and c-abl genes in Philadelphia chromosome positive acute lymphoblastic leukemia. Cell. 1987 Oct 9;51(1):33–40. doi: 10.1016/0092-8674(87)90007-9. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Ishimi Y., Kikuchi A. Identification and molecular cloning of yeast homolog of nucleosome assembly protein I which facilitates nucleosome assembly in vitro. J Biol Chem. 1991 Apr 15;266(11):7025–7029. [PubMed] [Google Scholar]
- Jeffreys A. J., Flavell R. A. A physical map of the DNA regions flanking the rabbit beta-globin gene. Cell. 1977 Oct;12(2):429–439. doi: 10.1016/0092-8674(77)90119-2. [DOI] [PubMed] [Google Scholar]
- Kadowaki T., Kadowaki H., Taylor S. I. A nonsense mutation causing decreased levels of insulin receptor mRNA: detection by a simplified technique for direct sequencing of genomic DNA amplified by the polymerase chain reaction. Proc Natl Acad Sci U S A. 1990 Jan;87(2):658–662. doi: 10.1073/pnas.87.2.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakizuka A., Miller W. H., Jr, Umesono K., Warrell R. P., Jr, Frankel S. R., Murty V. V., Dmitrovsky E., Evans R. M. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991 Aug 23;66(4):663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- Kamps M. P., Murre C., Sun X. H., Baltimore D. A new homeobox gene contributes the DNA binding domain of the t(1;19) translocation protein in pre-B ALL. Cell. 1990 Feb 23;60(4):547–555. doi: 10.1016/0092-8674(90)90658-2. [DOI] [PubMed] [Google Scholar]
- Konopka J. B., Watanabe S. M., Witte O. N. An alteration of the human c-abl protein in K562 leukemia cells unmasks associated tyrosine kinase activity. Cell. 1984 Jul;37(3):1035–1042. doi: 10.1016/0092-8674(84)90438-0. [DOI] [PubMed] [Google Scholar]
- Krohne G., Franke W. W. A major soluble acidic protein located in nuclei of diverse vertebrate species. Exp Cell Res. 1980 Sep;129(1):167–189. doi: 10.1016/0014-4827(80)90341-9. [DOI] [PubMed] [Google Scholar]
- Kurzrock R., Gutterman J. U., Talpaz M. The molecular genetics of Philadelphia chromosome-positive leukemias. N Engl J Med. 1988 Oct 13;319(15):990–998. doi: 10.1056/NEJM198810133191506. [DOI] [PubMed] [Google Scholar]
- Lapeyre B., Bourbon H., Amalric F. Nucleolin, the major nucleolar protein of growing eukaryotic cells: an unusual protein structure revealed by the nucleotide sequence. Proc Natl Acad Sci U S A. 1987 Mar;84(6):1472–1476. doi: 10.1073/pnas.84.6.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenstra R., d'Auriol L., Andrieu B., Le Bras J., Galibert F. Cloning and sequencing of Plasmodium falciparum DNA fragments containing repetitive regions potentially coding for histidine-rich proteins: identification of two overlapping reading frames. Biochem Biophys Res Commun. 1987 Jul 15;146(1):368–377. doi: 10.1016/0006-291x(87)90734-0. [DOI] [PubMed] [Google Scholar]
- Nourse J., Mellentin J. D., Galili N., Wilkinson J., Stanbridge E., Smith S. D., Cleary M. L. Chromosomal translocation t(1;19) results in synthesis of a homeobox fusion mRNA that codes for a potential chimeric transcription factor. Cell. 1990 Feb 23;60(4):535–545. doi: 10.1016/0092-8674(90)90657-z. [DOI] [PubMed] [Google Scholar]
- Pendergast A. M., Muller A. J., Havlik M. H., Maru Y., Witte O. N. BCR sequences essential for transformation by the BCR-ABL oncogene bind to the ABL SH2 regulatory domain in a non-phosphotyrosine-dependent manner. Cell. 1991 Jul 12;66(1):161–171. doi: 10.1016/0092-8674(91)90148-r. [DOI] [PubMed] [Google Scholar]
- Robberson B. L., Cote G. J., Berget S. M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990 Jan;10(1):84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg A. A., Morgan R., McCallister J. A., Kaiser-McCaw B., Hecht F. Acute myeloblastic leukemia (AML) with t(6;9) (p23;q34): a specific subgroup of AML? Cancer Genet Cytogenet. 1983 Oct;10(2):139–142. doi: 10.1016/0165-4608(83)90117-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtivelman E., Lifshitz B., Gale R. P., Canaani E. Fused transcript of abl and bcr genes in chronic myelogenous leukaemia. Nature. 1985 Jun 13;315(6020):550–554. doi: 10.1038/315550a0. [DOI] [PubMed] [Google Scholar]
- Walker L. C., Ganesan T. S., Dhut S., Gibbons B., Lister T. A., Rothbard J., Young B. D. Novel chimaeric protein expressed in Philadelphia positive acute lymphoblastic leukaemia. 1987 Oct 29-Nov 4Nature. 329(6142):851–853. doi: 10.1038/329851a0. [DOI] [PubMed] [Google Scholar]
- de Klein A., van Kessel A. G., Grosveld G., Bartram C. R., Hagemeijer A., Bootsma D., Spurr N. K., Heisterkamp N., Groffen J., Stephenson J. R. A cellular oncogene is translocated to the Philadelphia chromosome in chronic myelocytic leukaemia. Nature. 1982 Dec 23;300(5894):765–767. doi: 10.1038/300765a0. [DOI] [PubMed] [Google Scholar]
- de Thé H., Lavau C., Marchio A., Chomienne C., Degos L., Dejean A. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991 Aug 23;66(4):675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- von Lindern M., Fornerod M., van Baal S., Jaegle M., de Wit T., Buijs A., Grosveld G. The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can, and the expression of a chimeric, leukemia-specific dek-can mRNA. Mol Cell Biol. 1992 Apr;12(4):1687–1697. doi: 10.1128/mcb.12.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Lindern M., Poustka A., Lerach H., Grosveld G. The (6;9) chromosome translocation, associated with a specific subtype of acute nonlymphocytic leukemia, leads to aberrant transcription of a target gene on 9q34. Mol Cell Biol. 1990 Aug;10(8):4016–4026. doi: 10.1128/mcb.10.8.4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Lindern M., van Agthoven T., Hagemeijer A., Adriaansen H., Grosveld G. The human pim-1 gene is not directly activated by the translocation (6;9) in acute nonlymphocytic leukemia. Oncogene. 1989 Jan;4(1):75–79. [PubMed] [Google Scholar]