Abstract
Human cytomegalovirus (HCMV) is a member of the Herpesviridae family that manipulates host immune responses and establishes life-long latent infection, in part through mimicry of cytokines, chemokines, and chemokine receptors. The HCMV US27 gene product is a putative chemokine receptor with no known ligands. We generated a stable US27 cell line to screen for chemokine ligands but unexpectedly found that US27 potentiated the activity of an endogenous human chemokine receptor, CXCR4. Cells expressing both US27 and CXCR4 exhibited greater calcium mobilization and enhanced chemotaxis in response to CXCL12/ SDF-1α than controls. Quantitative RT-PCR revealed a significant increase in CXCR4 expression when US27 was present, and elevated CXCR4 receptor levels were detected via flow cytometry, western blot, and immunofluorescence microscopy. Potentiation of CXCR4 signaling by US27 could represent a novel strategy by which HCMV targets virus-infected cells to the bone marrow in order to expand the reservoir of latently infected cells.
Keywords: Cytomegalovirus, chemokines, chemokine receptors, immune modulation
Introduction
Human cytomegalovirus (HCMV) is a member of the β-herpesvirus subgroup and a prevalent pathogen infecting a vast majority of the human population. Although immune-competent hosts are generally asymptomatic upon infection, HCMV poses a serious health risk for immune-compromised hosts (Kesson and Kakakios, 2007). HCMV is the primary infectious cause of birth defects, including deafness and mental retardation (Revello et al., 2008).
Despite a vigorous host immune response to HCMV infection, the virus is able to avoid immune clearance and establish lifelong latency (Jackson et al., 2011). HCMV has evolved tactics to suppress inflammatory cytokines (Spencer et al., 2002), avoid elimination by natural killer cells (Beck and Barrell, 1988; Tomasec et al., 2000), and prevent apoptosis of infected cells (McCormick, 2008). Another immune evasion technique is the codification of proteins with structural similarity to the G-protein coupled receptor (GPCR) superfamily (Chee et al., 1990). Chemokine receptors are a subset of GPCR that play a pivotal role in the immune response by receiving and reacting to chemokine signals, which attract and direct immune cells to sites of injury and infection (Charo and Ransohoff, 2006). Viral mimicry of chemokine receptors provides an effective mechanism for manipulation of human chemokine signaling that could facilitate both virus dissemination and avoidance of harmful host immune cells (Holst and Rosenkilde, 2003; Vischer et al., 2006).
Chemokine receptor homologs encoded by HCMV include US27, US28, UL33, and UL78. Of these, US28 is the most extensively researched and was found to bind to several human CC chemokines (Gao and Murphy, 1994; Neote et al., 1993; Stropes et al., 2009). This binding can lead to calcium mobilization, IP3 production, and activation of transcription factors such as NFAT, CREB, and NF-κB (Billstrom et al., 1998; Casarosa et al., 2001; Vieira et al., 1998). US28 has also been shown to act as a co-receptor for HIV entry (Pleskoff et al., 1997), promote fusion between cells (Ples&kappakoff et al., 1998), trigger migration of smooth muscle cells (Streblow et al., 1999), and contribute to the characteristic angiogenic and invasive phenotype of glioblastoma cells (Soroceanu et al., 2011). Most recently, US28 has been shown to form heteromers with the other HCMV chemokine receptors US27, UL33 and UL78 (Tschische et al., 2011). While no functional changes were observed with the US28:US27 dimer, the US28:UL33 dimer and the US28:UL78 dimer both ablated activation of NF-κB by US28, suggesting a complex level of regulation in which these viral receptors may act in concert to either promote or block signaling through particular pathways during the course of virus infection.
US27 and US28 are adjacent in the HCMV genome and share 31 % sequence identity. Both proteins are expressed in infected cells (Fraile-Ramos et al., 2002), although is US28 expressed throughout lytic and latent infection, while US27 is expressed late in lytic infection only, indicating the two genes are independently regulated and function at different stages of the virus infection cycle. To date, US27 remains an orphan receptor that binds no known cellular ligands (Bodaghi et al., 1998; Stapleton et al., 2012). US27 exhibits many structural features of chemokine receptors, including seven transmembrane domains with conserved cysteine residues in the extracellular loops, and extensive glycosylation of the extracellular domain (Margulies and Gibson, 2007), a common characteristic for receptors with small peptide or chemokine ligands (Katritch et al., 2012). US27 also contains a DRY (aspartic acid, arginine, tyrosine) motif in the second intracellular loop that is critical for activation of associated G proteins following ligand engagement (Flanagan, 2005), and a di-leucine motif in the carboxy-terminal intracellular domain that mediates receptor endocytosis (Stapleton et al., 2012).
US27 is present in the envelope of the virus particle (Margulies and Gibson, 2007), but in virus-infected cells, the receptor is rapidly internalized with the majority of the US27 protein found in endosomes, the Golgi apparatus, and perinuclear compartments (Fraile-Ramos et al., 2002). Intracellular localization was found to be mediated by the carboxy-terminal domain of the US27 protein product (Stapleton et al., 2012). While viral mutants that lack US27 are replication competent (Bodaghi et al., 1998), these viruses are incapable of spreading via the extracellular route (O’Connor and Shenk, 2011), leading to speculation that US27 may play a rolge in virion assembly or egress.
Here, we set out to “de-orphanize” HCMV US27 and instead report the surprising discovery that US27 can directly enhance the calcium signaling activity of a human chemokine receptor, CXCR4. This finding is in direct contrast to the effects of other viral chemokine receptors, which have been found to impair CXCR4 function. Moreover, we found elevated levels of CXCR4 receptor in cells expressing US27, which resulted in increased cell migration in vitro and could have profound implications for immune cell trafficking in HCMV patients. Potentiation of CXCR4 activity by US27 demonstrates yet another highly sophisticated method of immune modulation employed by HCMV.
Results
CXCR4 induces greater calcium mobilization in response to CXCL12/SDF-1α in the presence of HCMV US27
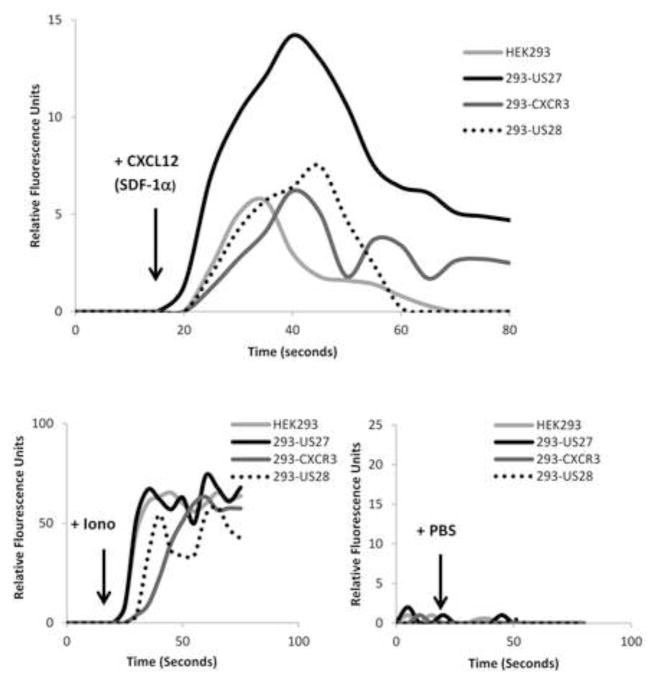
We previously attempted to investigate the functional activity of US27 by performing a chemokine ligand screen (Stapleton et al., 2012). HEK293 cells stably expressing US27 (293-US27) were loaded with a calcium sensitive dye and exposed to more than 100 different individual human chemokines. Only one chemokine elicited a calcium flux response: CXCL12/SDF-1α. The response to CXCL12/SDF-1α was expected due to the presence of CXCR4 endogenously expressed on HEK293 cells (Hoffmann et al., 2012). However, the magnitude of the calcium response to CXCL12/SDF-1α in 293-US27 cells was consistently 2–3 times greater than the response in HEK293 cells, which express only CXCR4 (Figure 1). This difference in the level of calcium mobilization was not attributable to the transfection and selection process, since HEK293 cell lines that express endogenous CXCR4 in combination with either stably transfected HCMV US28 or human chemokine receptor CXCR3 did not exhibit enhanced signaling to CXCL12/SDF-1α. Ionomycin served as a positive control and demonstrated that all cell lines were capable of producing an equivalent calcium flux, while PBS treatment served as a negative control for the addition of stimulus. These results suggested that US27 might potentiate signaling of human CXCR4.
Figure 1. Increased calcium mobilization in cells expressing CXCR4 and HCMV US27.
HEK293 cells and stable 293-US27, 293-US28, and 293-CXCR3 cell lines were loaded with Fluo-4 dye, treated with 100 μg/ml CXCL12/SDF-1α in a volume of 10 μl, and fluorescence intensity monitored over time. An equal volume of PBS was added as a negative control and 1 mg/ml ionomycin in a volume of 5 μl served as a positive control. These results are representative of four independent experiments.
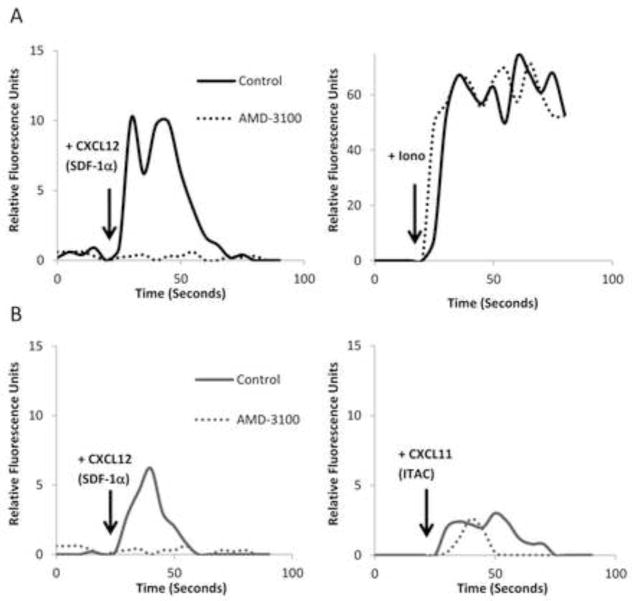
One other explanation for the increased calcium mobilization in 293-US27 cells was that CXCL12/SDF-1α was actually a ligand for US27. To examine this possibility, 293-US27 cells were incubated with 100uM AMD-3100 (plerixafor) for 10 minutes prior to stimulation with CXCL12/SDF-1α. AMD-3100 is a highly selective antagonist that blocks signaling through the CXCR4 receptor (Hendrix et al., 2004). As shown in Figure 2A, the calcium response to CXCL12/SDF-1α in 293-US27 cells was completely ablated in the presence of the inhibitor. Treatment with ionomycin demonstrated that the cells were still capable of eliciting calcium flux in the presence of the CXCR4 antagonist. As shown in Figure 2B, AMD-3100 is highly selective for CXCR4, and treatment with the antagonist had no impact on the ability of CXCR3 to induce calcium mobilization in response to its natural ligand, CXCL11/ITAC, in 293-CXCR3 cells. These results confirm that CXCL12/SDF-1α is not a ligand for US27 since there is no calcium flux when CXCR4 is blocked, further supporting the notion that the presence of US27 enhances the signaling activity of CXCR4.
Figure 2. Treatment with AMD-3100 completely inhibits CXCL12/SDF-1α-induced calcium mobilization.
Cells were loaded with Fluo-4 dye, and then relative fluorescence intensity was monitored over time. A) 293-US27 cells were pre treated with either 100 μM AMD-3100 or an equal volume of PBS for 10 minutes before treatment with 100 μg/ml CXCL12/SDF-1α or 1 μg/ml ionomycin. B) 293-CXCR3 cells were pre-treated with AMD-3100 or PBS before treatment with 100 μg/ml CXCL12/SDF-1α or CXCL11/ITAC. These results are representative of three independent experiments.
Enhanced CXCR4 calcium signaling requires the DRY box and C-tail of US27
To investigate which domains of US27 might be required for the potentiation of CXCR4 signaling, we made use of two stable cell lines expressing US27 mutants. US27/CXCR3-CT is a chimeric receptor that lacks the C-terminal intracellular domain of US27 (Stapleton et al., 2012). Instead, the receptor contains the extracellular domain of US27 through the seventh transmembrane α-helix fused to the C-terminal intracellular domain of human CXCR3. The US27-DAY mutant contains a substitution in the DRY box motif, with arginine 128 replaced with an alanine. These mutant receptors were stably expressed in the HEK293 cell line with endogenous CXCR4 also present. When the cells were treated with CXCL12/SDF-1α, calcium mobilization was observed; however, the magnitude of the response was comparable to the parent HEK293 and control 293-CXCR3 cell lines (Figure 3). Again, ionomycin served as the positive control to demonstrate all cell lines were capable of producing an equivalent calcium flux. Only cells expressing wild type US27 along with endogenous CXCR4 exhibited an enhanced response to CXCL12/SDF-1α, which suggests that both the C-terminal intracellular domain and the DRY box of US27 may play important roles in potentiating CXCR4 signaling.
Figure 3. CXCR4 calcium responses in cells expressing US27 mutants are not enhanced.
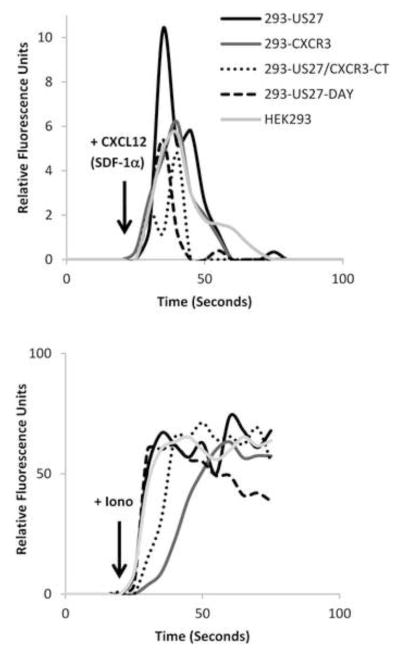
Cells were loaded with Fluo-4 dye, and then relative fluorescence intensity was monitored over time. HEK293, 293-US27, 293-CXCR3, 293-US27/CXCR3-CT, and 293-US27-DAY cells were treated with 10 μl of CXCL12/SDF-1α (100ug/ml) or 5 μl ionomycin (1 mg/ml). These results are representative of two independent experiments.
US27 demonstrates specificity for CXCR4 and fails to enhances CXCR3 signaling
We also wondered whether the effect of US27 on CXCR4 was specific or if US27 could enhance the signaling of any other cellular chemokine receptors. Thus, we examined signaling from CXCR3 in response to its ligand CXCL11/ITAC in the presence of US27. HEK293 cells and 293-US27 cells were transiently transfected with p3XFLAG-CXCR3, loaded with Fluo-4 dye, and then stimulated with CXCL11/ITAC. As shown in Figure 4, the magnitude of the calcium response induced by CXCL11/ITAC in these cells was comparable whether US27 was present or not. With all three chemokine receptors (CXCR3, US27, and endogenous CXCR4) present in the same cell, the response to CXCL12/SDF-1α was still higher (lower panel), indicating that US27 preferentially affected the signaling of CXCR4 and not CXCR3. While not every human chemokine receptor was tested here, these results strongly suggest that the signal potentiation mediated by US27 has some specificity for CXCR4.
Figure 4. US27 preferentially increases CXCR4, but not CXCR3, calcium signaling.
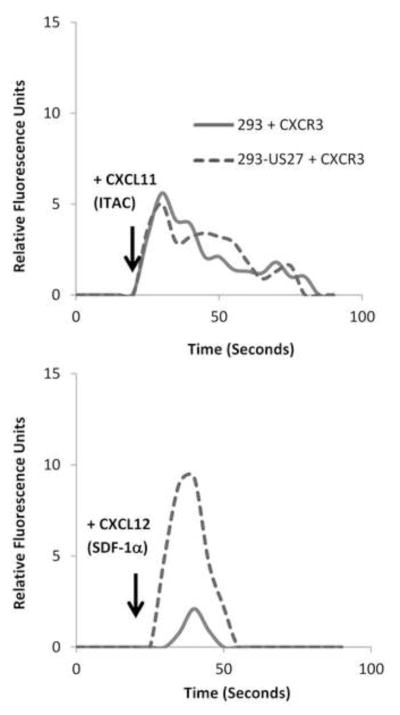
HEK293 and 293-US27 cells were transfected with p3XFLAG-CXCR3, loaded with Fluo-4 dye, and then relative fluorescence intensity monitored over time. Cells were stimulated with 10 μl of CXCL11/ITAC or CXCL12/SDF-1α (100ug/ml) as indicated. These results are representative of two independent experiments.
Chemotaxis toward CXCL12/SDF-1α is also enhanced in the presence of US27
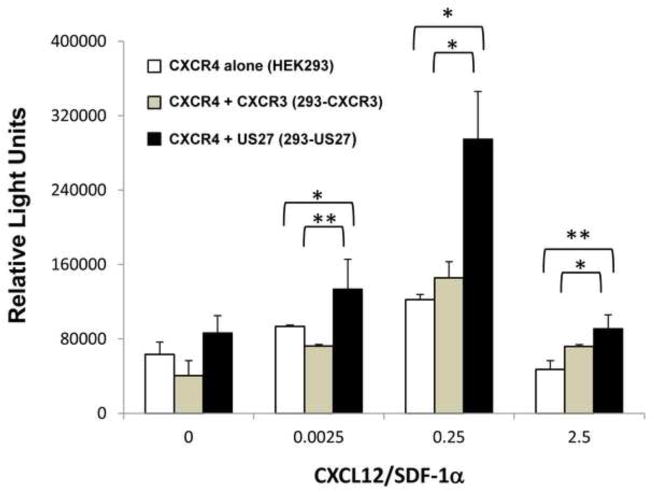
Next we investigated whether the presence of US27 also enhanced other downstream signaling outcomes of the CXCR4 receptor, such as chemotaxis. Using a modified Boyden chamber assay, cell migration was examined. Briefly, cells were placed in the upper chamber of an 8 um trans-well filter, while migration media containing varying doses of CXCL12/SDF-1α was placed in the lower chamber. After four hours, cells that traversed the filter to the lower chamber were harvested and quantified. While each cell type exhibited some chemotaxis toward CXCL12/SDF-1α, 293-US27 cells showed significantly more migration than either HEK293 cells expressing CXCR4 alone or 293-CXCR3 controls (Figure 5). Basal movement toward migration media without CXCL12/SDF-1α was comparable between cell lines, and the migration response toward the chemokine exhibited the classic bell-shaped curve for chemotaxis, with optimal movement to a median dose of 0.25 ng/ml CXCL12/SDF-1α and less migration to higher and lower doses. These findings show that the presence of US27 also results in increased CXCR4 directed cell migration toward CXCL12/SDF-1α.
Figure 5. Increased migration of 293-US27 cells toward CXCL12/SDF-1α.
HEK293, 293-US27 or 293-CXCR3 cells were seeded at a density of 2 x 105 cells in a total volume of 0.1 ml in the upper chamber of an 8 μm trans-well filter. Migration media containing the indicated concentrations of CXCL12/SDF-1α was placed in the lower chamber. After four hours, cells in the lower chamber were harvested and quantified by the addition of CellTiter-Glo reagent and measurement of luminescence. Error bars represent standard error. Statistical analysis was performed using the two-tailed student’s t-test and * indicates p < 0.05; ** indicates p < 0.01. Results are representative of three independent experiments.
CXCR4 protein and mRNA levels are increased in cells expressing US27
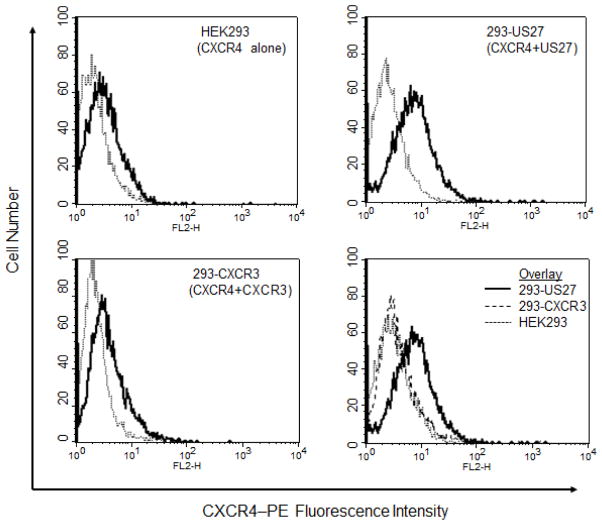
Given the enhanced chemotaxis and calcium flux responses to CXCL12/SDF-1α in cells expressing US27, we wondered whether there might be increased levels of CXCR4 on the surface of these 293-US27 cells. To address this question, cells were stained with an anti-CXCR4-PE antibody and examined via flow cytometry. As shown in Figure 6, HEK293 cells naturally express CXCR4, as indicated by the right-shifted fluorescence histogram compared to isotype control antibody. The level of CXCR4 present on 293-CXCR3 cells was also comparable to the HEK293 parent cells, whereas 293-US27 cells exhibited greater fluorescence intensity, indicating that more CXCR4 receptors were present on the cell surface.
Figure 6. Surface expression of CXCR4 is higher in 293-US27 cells.
Cells were stained with phycoerythrin-conjugated anti-CXCR4 antibody (solid line) or IgG2b-PE isotype control (dotted line) and fluorescence intensity measured via flow cytometry. The lower right panel is an overlay of all three cell types stained with anti-CXCR4 antibody.
Increased levels of CXCR4 on the cell surface could be the result of several events, such as increased gene transcription, decreased receptor recycling, or a redistribution of receptors in the cell. Reverse transcriptase-PCR was employed to compare CXCR4 gene transcription between HEK293, 293-US27, and 293-CXCR3 cell lines. RNA was extracted from each cell line, reverse transcribed into cDNA, and CXCR4 specific primers were used to carry out PCR. The resulting bands were visualized using agarose gel electrophoresis, as shown in Figure 7A. β–actin served as a positive control, and the intensity of these bands was comparable between cell lines. The intensity of the CXCR4 band was significantly greater for the 293-US27 cells than for either HEK293 or 293-CXCR3 lanes, indicating that US27 confers an increase in CXCR4 mRNA levels. To determine more precisely how much greater CXCR4 gene expression was in 293-US27 cells, quantitative real time PCR (qPCR) was used and when compared to the HEK293 cell line, there was a 2.2-fold increase in CXCR4 expression (Figure 7B). In contrast, the level of CXCR4 expression in the 293-CXCR3 cell line was virtually identical to that of the parent HEK293 cells. These results indicate that CXCR4 mRNA levels are significantly increased in the presence of US27.
Figure 7. 293-US27 cells have elevated CXCR4 expression.
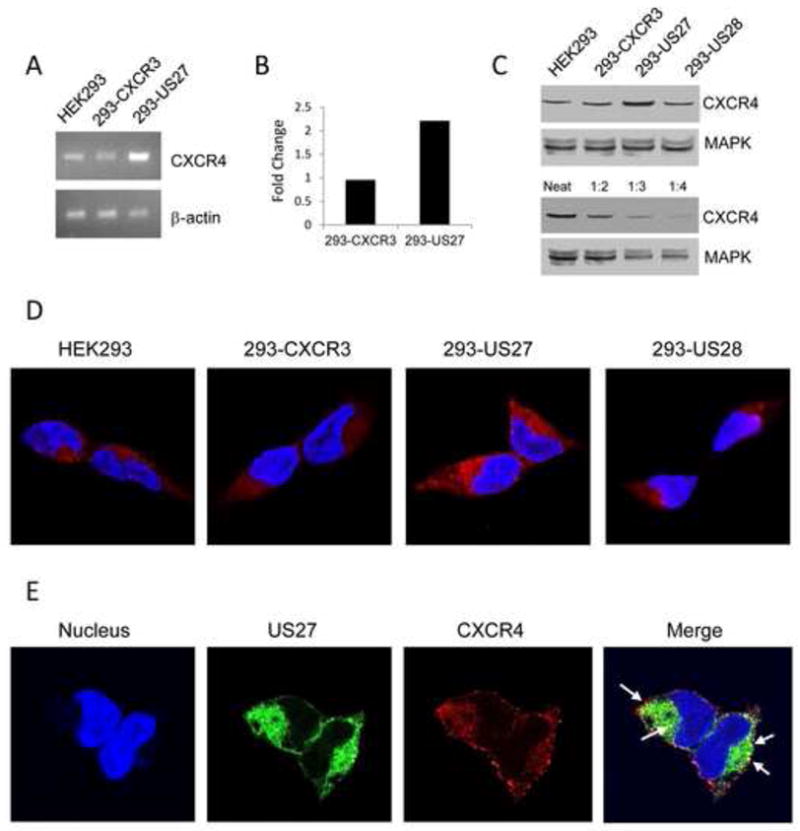
RNA was isolated from each cell type, reverse transcribed, then gene specific primers were used to amplify either CXCR4 or β-actin (A). For qPCR (B), data was normalized to the β-actin control and expressed as fold change relative to CXCR4 levels from HEK293 cells. For Western blotting (C), cell lysates were probed with either anti-CXCR4 antibody or anti-MAPK antibody as indicated. In the lower panel, 293-US27 cell lysates were analyzed either undiluted (neat) or diluted as indicated. For immunofluorescence microscopy (D), cells were seeded onto glass coverslips in 6-well plates at a density of 2 x 105 cells/well, and then stained with an anti-CXCR4 antibody followed by TRITC-conjugated secondary antibody (red). Nuclei appear blue due to DAPI staining. Co-localization of US27 and CXCR4 (E) is shown in cells that were fixed, permeabilized and stained with anti-FLAG (US27) and anti-CXCR4 antibodies followed by appropriate FITC- or TRITC-conjugated secondary antibodies. The merged image results from a compilation of the three colored images into one image where areas of red (CXCR4) and green (US27) overlap appear as discrete yellow spots indicated by white arrows.
To examine CXCR4 protein levels, Western blotting was performed. Lysates from each of the indicated cell types were solubilized in 4 M urea, separated by SDS-PAGE, transferred to a nitrocellulose membrane and probed with an antibody to CXCR4. As shown in Figure 7C, a band of 45 kD, the expected size for CXCR4, was detected in each cell type. However, the CXCR4 band present in 293-US27 cells was approximately double size of the bands detected for each of the other cell types, which supports the notion that increased expression of CXCR4 mRNA also correlates with higher protein levels in the cell. Identical membranes were probed with an antibody to MAPK, which appeared as the expected doublet at 42/44 kD and served as a control to ensure there was equal loading of samples. In the lower panel, the US27 cell lysate was diluted in an effort to quantitate the level of CXCR4 present in these cells compared to the HEK293, 293-CXCR3, and 293-US28 cell lines. The intensity of the band for the 1:2 dilution of the 293-US27 was most comparable to the band for CXCR4 in the other cell lines (upper blot), again indicating that there is a nearly two-fold increase in CXCR4 protein expression in the 293-US27 cells.
US27 and CXCR4 co-localize to discrete locations in the cell
Immunofluorescence staining and confocal microscopy were also used to confirm that the elevated CXCR4 gene expression correlated with an increase in CXCR4 protein levels throughout the cell. Each cell line was cultured on glass coverslips and then fixed, permeabilized, and stained with anti-CXCR4 antibody followed by TRITC-conjugated secondary antibody. As expected, 293-US27 cells showed a dramatic increase in CXCR4 expression, both at the cell surface and throughout the entire cell, as compared to control cell lines (Figure 7D). Notably, there was no difference in distribution of the CXCR4 receptor in 293-US27 cells compared to control cells, only a change in the total amount of CXCR4 protein present. When 293-US27 cells were co-stained with an anti-FLAG antibody for US27 (green) and an anti-CXCR4 antibody (red), as shown in Figure 7E, a typical US27 distribution pattern and the increased red fluorescence for CXCR4 were seen. In addition, there appeared to be a number of yellow punctae (indicated with white arrows in the Merge panel), suggesting that there is either co-localization of US27 and CXCR4 in the cell or internalization of both receptors in the same endocytic vesicles.
Discussion
The interaction between virus and host can be intricate and convoluted. HCMV has evolved many unique adaptations to evade and manipulate the human immune system, including the codification of four chemokine-like receptors. In this study, we made the unexpected observation that US27 potentiates the signaling of human CXCR4. This is the first report of functional activity for HCMV US27, as well as the first evidence of a viral chemokine receptor enhancing calcium mobilization and chemotactic activity of CXCR4 in response to its cognate ligand CXCL12/SDF-1α. Given the importance of the CXCR4-CXCL12/SDF-1α signaling axis in hematopoiesis, manipulation of CXCR4 activity could represent a mechanism for controlling migration of infected cells to the bone marrow to expand the reservoir of latently infected cells, as well as the release of latently infected cells back into the periphery for lytic infection.
Enhanced signaling of CXCR4 in cells expressing HCMV US27 was first observed during a ligand screen aimed at identifying chemokines that bind and signal through US27 (Stapleton et al., 2012). This effect was highly specific to US27, as control cell lines stably expressing either HCMV US28 or human CXCR3 exhibited no enhancement of CXCL12/SDF1α-induced calcium mobilization. In addition, this potentiation effect on CXCR4 was also seen in cells transiently transfected with either p3X-FLAG-US27 or pEGFP-US27 (data not shown), ruling out the possibility that enhanced CXCR4 signaling was due to a random event in the selection of the clonal stable cell line.
While we have found that US27 potentiates host CXCR4-mediated calcium mobilization and cell migration, the specific mechanism responsible for these effects remains to be determined. One possibility is that US27 forms heteromers with CXCR4. CXCR4 has previously been shown to form homodimers both in the absence of ligand (Babcock et al., 2003) and upon CXCL12/SDF-1α binding (Vila-Coro et al., 1999). In addition, CXCR4 is known to form heterodimers with other chemokine receptors, including CCR2 (See et al., 2011; Sohy et al., 2007) and CXCR7 (Levoye et al., 2009). CXCR7 was found to interfere with the ability of CXCR4 to activate G proteins and trigger calcium responses in response to CXCL12/SDF-1α suggesting that there are numerous levels of regulation of CXCR4 signaling activity. Several viral chemokine receptors have been shown to form heterodimers with CXCR4, including the BILF1 receptor of Epstein-Barr virus (EBV) and the UL33 and UL78 gene products of HCMV. In transfected cells, expression of UL33 or UL78 with CXCR4 was found to significantly reduce signaling and migration responses to CXCL12/SDF-1α, as well as inhibit the ability of CXCR4 to function as a co-factor for HIV entry (Tadagaki et al., 2012). For EBV BILF1, formation of heterodimers with CXCR4 was shown to impair binding of CXCL12/SDF-1α and thus reduce G protein and CREB activation by CXCR4 (Nijmeijer et al., 2010). In contrast, we found that the presence of HCMV US27 increased the signaling activity of CXCR4, making it the first viral receptor documented to enhance the signaling activity of CXCR4. The observation that US27 and CXCR4 co-localized to discrete sites in the cell could support the idea that US27 forms heteromers with CXCR4 that result in increased signaling.
While both this study and the Tadagaki et al. work (Tadagaki et al., 2012) were performed in transfected cells, the notion that HCMV would express multiple receptors during infection that have the potential to either enhance or reduce CXCR4 signaling is compelling because bone marrow progenitor cells are a primary site of HCMV latency. Differential interaction of CXCR4 with UL33 and UL78 could lead to retention of infected cells in the bone marrow, whereas enhanced CXCR4 activity resulting from interaction with US27 could lead to migration of productively infected cells into the bone marrow, enabling the virus to orchestrate an exquisite level of control for the movement of infected cells throughout the host. Impaired migration of macrophages upon HCMV infection has been reported, but no change in CXCR4 levels or activity was observed (Frascaroli et al., 2009). Additional studies are necessary to examine the complex interactions of each of the HCMV encoded chemokine receptors with each other and CXCR4 during infection in a variety of cell types.
Another possible mechanism for the enhanced CXCR4 signaling observed here is that US27 might function as a sponge to absorb negative regulators of signaling or endocytic sorting proteins that mediate receptor internalization, leaving more CXCR4 receptors on the cell surface to engage ligand and transduce signaling. CXCR4 receptor trafficking is highly complex and variable; constitutive recycling back to the cell surface occurs in the absence of ligand (Zhang et al., 2004), while ligand engagement leads to lysosomal targeting (Kumar et al., 2011) mediated by a di-leucine motif and phosphorylation of key serine residues in the C-terminal domain (Orsini et al., 1999). US27 is found predominantly in intracellular compartments (Fraile-Ramos et al., 2002) and also contains a di-leucine motif in the C-terminal intracellular domain, removal of which results in greatly increased cell surface expression (Stapleton et al., 2012). The shared di-leucine motif would account for the co-localization patterns in instances where the two proteins were internalized into the same endocytic vesicles. If US27 is binding to intracellular proteins, thus allowing CXCR4 to remain on the surface longer, it is unclear at this point which accessory proteins might be involved. Numerous proteins have been found to function in CXCR4 internalization, including β-arrestin (Busillo et al., 2010), Hip (Fan et al., 2002), myosin II (Rey et al., 2007), cortactin (Luo et al., 2006), and the ESCRT machinery (Sierra et al., 2010). To date, US27 has been shown to interact directly with only one endocytic sorting protein, N-ethylmaleimide sensitive factor (NSF), which typically mediates recycling back to the cell surface rather than targeting the receptor for degradation (Heydorn et al., 2004). Further examination of the endocytic sorting pathways for US27 and CXCR4 will be required to in order to determine whether US27 might be competing with CXCR4 for binding of intracellular proteins.
Neither of the two previous models accounts for the observation that CXCR4 mRNA transcript levels were elevated in cells expressing US27. This apparent upregulation of gene expression might be the result of an actual increase in transcriptional activity at the CXCR4 promoter, or it could be due to increased mRNA transcript stability. Nuclear Respiratory Factor-1 (NRF-1) is the primary transcription factor regulating CXCR4 gene expression (Moriuchi et al., 1997; Wegner et al., 1998), and positive and negative regulatory roles have been identified for SP-1 and YY1 (Moriuchi et al., 1999), respectively. It may be that the US27 protein perturbs a signaling pathway ultimately leading to enhanced CXCR4 transcription. Alternatively, US27 could function to stabilize CXCR4 mRNA transcripts, which are normally rapidly turned over, with a half-life of two hours (Gupta et al., 1998; Lam et al., 2001).
While follow-up studies are needed to fully understand the effect of US27 on host CXCR4 during virus infection, the results presented here are highly significant because we demonstrate for the first time functional activity for the US27 gene product. This activity, namely, modulation of signaling of host chemokine receptor CXCR4 could have a dramatic impact on cell trafficking during infection. The many immune modulation mechanisms of HCMV are dynamic, inter-related, and complex, and additional studies will be necessary to clarify how US27 contributes to these processes.
Materials and Methods
Cells, Viruses, and Reagents
HCMV strain AD169 was used to infect monolayer cultures of WI-38 human lung fibroblasts (American Type Culture Collection, Manassas, VA), and DNA from the infected cells was harvested and used as a template for PCR with US27- and US28-specific primers. The resulting PCR products were cloned into the p3X-FLAG vector (Sigma-Aldrich, St. Louis, MO) as previously described (Stapleton et al., 2012). The human CXCR3 gene was cloned from RNA isolated from peripheral blood mononuclear cells from the buffy coat of a healthy human donor purchased from the Blood Centers of the Pacific (San Francisco, CA) and cloned into the p3X-FLAG vector. The US27-DAY mutant was prepared from wild-type US27 via site-directed mutagenesis, and the US27/CXCR3-CT chimera was constructed as described (Stapleton et al., 2012). Human embryonic kidney (HEK) 293 cells were grown in Eagles minimal essential media (MEM) with 10 % fetal bovine serum (FBS) (Cellgro, Herndon, VA) in a humidified incubator at 37 ° C and 5 % CO2 atmosphere. The cells were transfected with p3X-FLAG plasmid DNA encoding target genes (US27, US28, CXCR3, or US27 mutants) using Fugene reagent (Promega, Madison, WI). Stable cell lines were developed via selection and cultivation in 1 mg/ml geneticin (Invitrogen, Carlsbad, CA). Purified recombinant chemokines CXCL11/ITAC and CXCL12/SDF-1α were purchased from Peprotech (Rocky Hill, NJ) and AMD-3100 and ionomycin were from Sigma-Aldrich (St. Louis, MO).
Calcium Mobilization Assays
HEK293 and stable 293 cell lines were harvested and resuspended in calcium loading buffer (RPMI + 25mM HEPES) at a density of 1 x 106 cells per ml. Aliquots of 2 ml cell suspension were treated with 1 nM Fluo-4AM (Invitrogen) for 30 minutes at 37 °C with agitation every 10 minutes. Following incubation, samples were washed with calcium loading buffer, centrifuged for 5 minutes at 1000 rpm, and then re-suspended in 0.5 ml calcium loading buffer. Calcium mobilization was monitored using a BD FACSCalibur flow cytometer (BD Biosciences, San Jose, CA) as described by Burns and Lewis (Burns and Lewis, 1997). Briefly, the sample was placed on the flow cytometer sample injection port with a small micro-stir bar containing a volume of 10μl chemokine (100 μg/ml) or 5 μl ionomycin (1mg/ml) suspended above the sample. Unstimulated cells were collected for 25 seconds, then the stir bar with the stimulus was dropped into the sample and collection continued for a total of 3 minutes per sample. Where indicated, calcium mobilization was also performed on cells pretreated with 100uM AMD-3100 in calcium loading buffer for 10 minutes prior to stimulation. Data was collected using CellQuestPro software and analyzed using FlowJo software (FlowJo, Ashland, Oregon).
Chemotaxis Assays
HEK293 and stable cell lines were harvested and re-suspended at a density of 2 x 106 cells per ml in migration media (MEM containing 1 % FBS). A total volume of 0.1 ml cell suspension (2 x 105 cells) was placed in the upper chamber of a ThinCert filter with 8 um pores in a 24 well plate (E&K, Santa Clara, CA). A total volume of 0.6 ml of migration media plus the indicated concentrations of human CXCL12/SDF-1α (Peprotech, Rocky Hill, New Jersey) was added to the lower chamber of each well, and plates were incubated for 4 hours at 37 °C. Medium from the lower chamber was collected, used to rinse the bottom of the filter twice, and then centrifuged at 1000 rpm for 10 minutes. The cell pellet was resuspended in 0.1 ml media and transferred to a white 96-well plate. Viable cell number was quantified using the CellTiter-Glo Assay kit (Promega, Madison, WI) according to the manufacturer’s protocol and relative light units were determined using the Glomax 96 Microplate Luminometer (Promega).
Flow Cytometry
HEK293 and stable cells were washed with PBS, harvested, and re-suspended in 100 ul FACS buffer (PBS + 1 % BSA and 0.1 % sodium azide) at a density of 1 x 106 cells per sample. The cells were stained with anti-CXCR4-PE-conjugated antibody (R&D Systems, Minneapolis, MN) or IgG2B-PE isotype control at a dilution of 1:100 for 1 hour at 4 °C protected from light. Cells were washed three times, fixed with FACS buffer containing 1 % paraformaldehyde (PFA), and collected using the BD FACSCalibur flow cytometer (BD Biosciences) and CellQuestPro software.
Gene Expression
RNA was harvested from HEK293 and stable cell lines using the RNeasy Mini Kit (Qiagen, Valencia, CA) according to manufacturer’s instructions, and then cDNA was synthesized from the RNA template using iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA). For standard PCR, each reaction contained cDNA template, primers, dNTP mix, Ex-Taq buffer, and Ex-Taq polymerase (Clontech, Mountain View, California). The gene specific primers for CXCR4 were 5′-TCCTTATGCAAGGCAGTCCATGTCA-3′ (forward) and 5′-ACAATACCAGGCAGGATAAGGCCA-3′ (reverse) and for β-actin 5′-AAGAGAGGCATCCTCACC-3′ (forward) and 5′-TACATGGCTGGGGTGTTG-3′ (reverse). The reaction underwent the following protocol on a MyCycler Thermal Cycler (Bio-Rad): 95 °C for 1 minute followed by 35 cycles of 94 °C for 1 minute, 55 °C for 1 minute, 72 °C for 1 minute, followed by 1 cycle of 72 °C for 10 minutes, and a final hold at 4 °C. The PCR products were visualized on a 2 % agarose gel. For quantitative real time PCR each reaction contained cDNA template, SYBR green mastermix (SABiosciences, Frederick, Maryland) and PrimeTime qPCR primers (IDT, San Diego, California) using a Bio-Rad CFX96 Real Time PCR Detection System. The gene specific primers for CXCR4 were 5′-CCTCGGTGTAGTTATCTGAAGTG-3′ (forward) and 5′-AGCAGGTAGCAAAAGTGACG-3′ (reverse) and for β-actin 5′-CCTTGCACATGCCGGAG-3′ (forward) and 5′-ACAGAGCCTCGCCTTTG-3′ (reverse). The comparative threshold cycle method was used to calculate relative gene expression and measure fold differences in CXCR4 expression between samples.
Western Blotting
Cells were seeded into 60 mm dishes at a density of 5 x 105 cells and lysed in the dish after 48 hours via the addition of lysis buffer (150 mM NaCl, 20 mM HEPES, 0.5 % Cymal-5, 1 mM NaOV4, 1 mM EDTA, 0.1 % NaN3 and 4 M urea). The lysate was sonicated, clarified via centrifugation at 14,000 rpm for 15 minutes at 4°C, and then sonicated again prior to the addition of loading dye. Samples were separated using SDS-PAGE, and then proteins were transferred to a nitrocellulose filter which was blocked in TBS-T + 5 % milk for one hour. The membranes were incubated with either anti-CXCR4 (Santa Cruz Biotechnology, Santa Cruz, CA) or anti-MAPK (Cell Signaling Technology, Danvers, MA) rabbit polyclonal antiserum according to the manufacturer’s instructions, followed by incubation with alkaline phosphatase-conjugated secondary antibody. Bands were visualized via the addition of Western Blue Substrate reagent (Promega).
Immunofluorescence Microscopy
HEK293 and stable cells were seeded in six-well dishes containing FBS coated glass coverslips at a density of 2 x 105 cells per well and then incubated for 48 hours at 37 °C. Cell monolayers were washed with PBS, fixed with 4 % paraformaldehyde, then permeabilized with 0.2 % (w/v) Triton X-100 followed by treatment with ice cold 50 % methanol-50 % acetone for 30 minutes. Cells were then blocked with PBS + 10 % FBS for one hour at 37 °C and stained with either anti-CXCR4 goat polyclonal primary antiserum (Santa Cruz Biotechnology) alone or in combination with anti-FLAG M2 antibody (Sigma-Aldrich) at a 1:100 (CXCR4) and 1:1000 (M2) dilution for one hour at 37 °C. Following three PBS washes, the coverslips were incubated with appropriate fluorochrome-conjugated secondary antibody (donkey-anti-goat-TRITC and goat-anti-mouse-FITC) for one hour, washed again, and then mounted on a glass slide using Prolong Gold anti-fade reagent with DAPI (Invitrogen). Images were acquired using a Zeiss LSM700 laser scanning confocal fluorescence microscope equipped with an AxioCAM imaging system and Zen software (Carl Zeiss, Inc., Oberkochen, Germany).
Highlights.
We show that an HCMV protein functions to manipulate host chemokine responses.
CXCR4 calcium signaling activity is enhanced in the presence of HCMV US27.
US27 triggers increased CXCR4 expression and heightens cell migration toward CXCL12/SDF-1α
US27 and CXCR4 co-localize in the cell and may work in concert to alter cell migration patterns.
Acknowledgments
The authors thank Carolyn Tu, Annette Chan, and Geoff Lambright for excellent technical assistance and helpful discussions. This work was supported by funding from the National Institutes of Health (AI074029 to JVS), the Lily Drake Cancer Fund, and the USF Faculty Development Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Babcock GJ, Farzan M, Sodroski J. Ligand-independent dimerization of CXCR4, a principal HIV-1 coreceptor. J Biol Chem. 2003;278:3378–3385. doi: 10.1074/jbc.M210140200. [DOI] [PubMed] [Google Scholar]
- Beck S, Barrell BG. Human cytomegalovirus encodes a glycoprotein homologous to MHC class-I antigens. Nature. 1988;331:269–272. doi: 10.1038/331269a0. [DOI] [PubMed] [Google Scholar]
- Billstrom MA, Johnson GL, Avdi NJ, Worthen GS. Intracellular signaling by the chemokine receptor US28 during human cytomegalovirus infection. J Virol. 1998;72:5535–5544. doi: 10.1128/jvi.72.7.5535-5544.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodaghi B, Jones TR, Zipeto D, Vita C, Sun L, Laurent L, Arenzana-Seisdedos F, Virelizier JL, Michelson S. Chemokine sequestration by viral chemoreceptors as a novel viral escape strategy: withdrawal of chemokines from the environment of cytomegalovirus infected cells. J Exp Med. 1998;188:855–866. doi: 10.1084/jem.188.5.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JM, Lewis GK. Improved measurement of calcium mobilization by flow cytometry. Biotechniques. 1997;23:1022–1024. 1026. doi: 10.2144/97236bm11. [DOI] [PubMed] [Google Scholar]
- Busillo JM, Armando S, Sengupta R, Meucci O, Bouvier M, Benovic JL. Site-specific phosphorylation of CXCR4 is dynamically regulated by multiple kinases and results in differential modulation of CXCR4 signaling. J Biol Chem. 2010;285:7805–7817. doi: 10.1074/jbc.M109.091173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casarosa P, Bakker RA, Verzijl D, Navis M, Timmerman H, Leurs R, Smit MJ. Constitutive signaling of the human cytomegalovirus-encoded chemokine receptor US28. J Biol Chem. 2001;276:1133–1137. doi: 10.1074/jbc.M008965200. [DOI] [PubMed] [Google Scholar]
- Charo IF, Ransohoff RM. The many roles of chemokines and chemokine receptors in inflammation. N Engl J Med. 2006;354:610–621. doi: 10.1056/NEJMra052723. [DOI] [PubMed] [Google Scholar]
- Chee MS, Satchwell SC, Preddie E, Weston KM, Barrell BG. Human cytomegalovirus encodes three G protein-coupled receptor homologues. Nature. 1990;344:774–777. doi: 10.1038/344774a0. [DOI] [PubMed] [Google Scholar]
- Fan GH, Yang W, Sai J, Richmond A. Hsc/Hsp70 interacting protein (hip) associates with CXCR2 and regulates the receptor signaling and trafficking. J Biol Chem. 2002;277:6590–6597. doi: 10.1074/jbc.M110588200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan CA. A GPCR that is not “DRY”. Mol Pharmacol. 2005;68:1–3. doi: 10.1124/mol.105.014183. [DOI] [PubMed] [Google Scholar]
- Fraile-Ramos A, Pelchen-Matthews A, Kledal TN, Browne H, Schwartz TW, Marsh M. Localization of HCMV UL33 and US27 in endocytic compartments and viral membranes. Traffic. 2002;3:218–232. doi: 10.1034/j.1600-0854.2002.030307.x. [DOI] [PubMed] [Google Scholar]
- Frascaroli G, Varani S, Blankenhorn N, Pretsch R, Bacher M, Leng L, Bucala R, Landini MP, Mertens T. Human cytomegalovirus paralyzes macrophage motility through down-regulation of chemokine receptors, reorganization of the cytoskeleton, and release of macrophage migration inhibitory factor. J Immunol. 2009;182:477–488. doi: 10.4049/jimmunol.182.1.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao JL, Murphy PM. Human cytomegalovirus open reading frame US28 encodes a functional beta chemokine receptor. J Biol Chem. 1994;269:28539–28542. [PubMed] [Google Scholar]
- Gupta SK, Lysko PG, Pillarisetti K, Ohlstein E, Stadel JM. Chemokine receptors in human endothelial cells. Functional expression of CXCR4 and its transcriptional regulation by inflammatory cytokines. J Biol Chem. 1998;273:4282–4287. doi: 10.1074/jbc.273.7.4282. [DOI] [PubMed] [Google Scholar]
- Hendrix CW, Collier AC, Lederman MM, Schols D, Pollard RB, Brown S, Jackson JB, Coombs RW, Glesby MJ, Flexner CW, Bridger GJ, Badel K, MacFarland RT, Henson GW, Calandra G. Safety, pharmacokinetics, and antiviral activity of AMD3100, a selective CXCR4 receptor inhibitor, in HIV-1 infection. J Acquir Immune Defic Syndr. 2004;37:1253–1262. doi: 10.1097/01.qai.0000137371.80695.ef. [DOI] [PubMed] [Google Scholar]
- Heydorn A, Sondergaard BP, Ersboll B, Holst B, Nielsen FC, Haft CR, Whistler J, Schwartz TW. A library of 7TM receptor C-terminal tails. Interactions with the proposed post-endocytic sorting proteins ERM-binding phosphoprotein 50 (EBP50), N-ethylmaleimide-sensitive factor (NSF), sorting nexin 1 (SNX1), and G protein-coupled receptor-associated sorting protein (GASP) J Biol Chem. 2004;279:54291–54303. doi: 10.1074/jbc.M406169200. [DOI] [PubMed] [Google Scholar]
- Hoffmann F, Muller W, Schutz D, Penfold ME, Wong YH, Schulz S, Stumm R. Rapid Uptake and Degradation of CXCL12 Depend on CXCR7 Carboxyl-terminal Serine/Threonine Residues. J Biol Chem. 2012;287:28362–28377. doi: 10.1074/jbc.M111.335679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst PJ, Rosenkilde MM. Microbiological exploitation of the chemokine system. Microbes Infect. 2003;5:179–187. doi: 10.1016/s1286-4579(02)00081-3. [DOI] [PubMed] [Google Scholar]
- Jackson SE, Mason GM, Wills MR. Human cytomegalovirus immunity and immune evasion. Virus Res. 2011;157:151–160. doi: 10.1016/j.virusres.2010.10.031. [DOI] [PubMed] [Google Scholar]
- Katritch V, Cherezov V, Stevens RC. Diversity and modularity of G protein-coupled receptor structures. Trends Pharmacol Sci. 2012;33:17–27. doi: 10.1016/j.tips.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesson AM, Kakakios A. Immunocompromised children: conditions and infectious agents. Paediatr Respir Rev. 2007;8:231–239. doi: 10.1016/j.prrv.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Kumar A, Kremer KN, Dominguez D, Tadi M, Hedin KE. Galpha13 and Rho mediate endosomal trafficking of CXCR4 into Rab11+ vesicles upon stromal cell-derived factor-1 stimulation. J Immunol. 2011;186:951–958. doi: 10.4049/jimmunol.1002019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam LT, Pickeral OK, Peng AC, Rosenwald A, Hurt EM, Giltnane JM, Averett LM, Zhao H, Davis RE, Sathyamoorthy M, Wahl LM, Harris ED, Mikovits JA, Monks AP, Hollingshead MG, Sausville EA, Staudt LM. Genomic-scale measurement of mRNA turnover and the mechanisms of action of the anti-cancer drug flavopiridol. Genome Biol. 2001;2:RESEARCH0041. doi: 10.1186/gb-2001-2-10-research0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levoye A, Balabanian K, Baleux F, Bachelerie F, Lagane B. CXCR7 heterodimerizes with CXCR4 and regulates CXCL12-mediated G protein signaling. Blood. 2009;113:6085–6093. doi: 10.1182/blood-2008-12-196618. [DOI] [PubMed] [Google Scholar]
- Luo C, Pan H, Mines M, Watson K, Zhang J, Fan GH. CXCL12 induces tyrosine phosphorylation of cortactin, which plays a role in CXC chemokine receptor 4-mediated extracellular signal-regulated kinase activation and chemotaxis. J Biol Chem. 2006;281:30081–30093. doi: 10.1074/jbc.M605837200. [DOI] [PubMed] [Google Scholar]
- Margulies BJ, Gibson W. The chemokine receptor homologue encoded by US27 of human cytomegalovirus is heavily glycosylated and is present in infected human foreskin fibroblasts and enveloped virus particles. Virus Res. 2007;123:57–71. doi: 10.1016/j.virusres.2006.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick AL. Control of apoptosis by human cytomegalovirus. Curr Top Microbiol Immunol. 2008;325:281–295. doi: 10.1007/978-3-540-77349-8_16. [DOI] [PubMed] [Google Scholar]
- Moriuchi H, Moriuchi M, Fauci AS. Cloning and analysis of the promoter region of CCR5, a coreceptor for HIV-1 entry. J Immunol. 1997;159:5441–5449. [PubMed] [Google Scholar]
- Moriuchi M, Moriuchi H, Margolis DM, Fauci AS. USF/c-Myc enhances, while Yin-Yang 1 suppresses, the promoter activity of CXCR4, a coreceptor for HIV-1 entry. J Immunol. 1999;162:5986–5992. [PubMed] [Google Scholar]
- Neote K, DiGregorio D, Mak JY, Horuk R, Schall TJ. Molecular cloning, functional expression, and signaling characteristics of a C-C chemokine receptor. Cell. 1993;72:415–425. doi: 10.1016/0092-8674(93)90118-a. [DOI] [PubMed] [Google Scholar]
- Nijmeijer S, Leurs R, Smit MJ, Vischer HF. The Epstein-Barr virus-encoded G protein-coupled receptor BILF1 hetero-oligomerizes with human CXCR4, scavenges Galphai proteins, and constitutively impairs CXCR4 functioning. J Biol Chem. 2010;285:29632–29641. doi: 10.1074/jbc.M110.115618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor CM, Shenk T. Human cytomegalovirus pUS27 G protein-coupled receptor homologue is required for efficient spread by the extracellular route but not for direct cell-to-cell spread. J Virol. 2011;85:3700–3707. doi: 10.1128/JVI.02442-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsini MJ, Parent JL, Mundell SJ, Marchese A, Benovic JL. Trafficking of the HIV coreceptor CXCR4. Role of arrestins and identification of residues in the c-terminal tail that mediate receptor internalization. J Biol Chem. 1999;274:31076–31086. doi: 10.1074/jbc.274.43.31076. [DOI] [PubMed] [Google Scholar]
- Pleskoff O, Treboute C, Alizon M. The cytomegalovirus-encoded chemokine receptor US28 can enhance cell-cell fusion mediated by different viral proteins. J Virol. 1998;72:6389–6397. doi: 10.1128/jvi.72.8.6389-6397.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleskoff O, Treboute C, Brelot A, Heveker N, Seman M, Alizon M. Identification of a chemokine receptor encoded by human cytomegalovirus as a cofactor for HIV-1 entry. Science. 1997;276:1874–1878. doi: 10.1126/science.276.5320.1874. [DOI] [PubMed] [Google Scholar]
- Revello MG, Campanini G, Piralla A, Furione M, Percivalle E, Zavattoni M, Gerna G. Molecular epidemiology of primary human cytomegalovirus infection in pregnant women and their families. J Med Virol. 2008;80:1415–1425. doi: 10.1002/jmv.21243. [DOI] [PubMed] [Google Scholar]
- Rey M, Valenzuela-Fernandez A, Urzainqui A, Yanez-Mo M, Perez Martinez M, Penela P, Mayor F, Jr, Sanchez-Madrid F. Myosin IIA is involved in the endocytosis of CXCR4 induced by SDF-1alpha. J Cell Sci. 2007;120:1126–1133. doi: 10.1242/jcs.03415. [DOI] [PubMed] [Google Scholar]
- See HB, Seeber RM, Kocan M, Eidne KA, Pfleger KD. Application of G protein-coupled receptor-heteromer identification technology to monitor beta-arrestin recruitment to G protein-coupled receptor heteromers. Assay and drug development technologies. 2011;9:21–30. doi: 10.1089/adt.2010.0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra MI, Wright MH, Nash PD. AMSH interacts with ESCRT-0 to regulate the stability and trafficking of CXCR4. J Biol Chem. 2010;285:13990–14004. doi: 10.1074/jbc.M109.061309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohy D, Parmentier M, Springael JY. Allosteric transinhibition by specific antagonists in CCR2/CXCR4 heterodimers. J Biol Chem. 2007;282:30062–30069. doi: 10.1074/jbc.M705302200. [DOI] [PubMed] [Google Scholar]
- Soroceanu L, Matlaf L, Bezrookove V, Harkins L, Martinez R, Greene M, Soteropoulos P, Cobbs CS. Human cytomegalovirus US28 found in glioblastoma promotes an invasive and angiogenic phenotype. Cancer Res. 2011;71:6643–6653. doi: 10.1158/0008-5472.CAN-11-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer JV, Lockridge KM, Barry PA, Lin G, Tsang M, Penfold ME, Schall TJ. Potent immunosuppressive activities of cytomegalovirus-encoded interleukin-10. J Virol. 2002;76:1285–1292. doi: 10.1128/JVI.76.3.1285-1292.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton LK, Arnolds KL, Lares AP, Devito TM, Spencer JV. Receptor chimeras demonstrate that the C-terminal domain of the human cytomegalovirus US27 gene product is necessary and sufficient for intracellular receptor localization. Virology journal. 2012;9:42. doi: 10.1186/1743-422X-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streblow DN, Soderberg-Naucler C, Vieira J, Smith P, Wakabayashi E, Ruchti F, Mattison K, Altschuler Y, Nelson JA. The human cytomegalovirus chemokine receptor US28 mediates vascular smooth muscle cell migration. Cell. 1999;99:511–520. doi: 10.1016/s0092-8674(00)81539-1. [DOI] [PubMed] [Google Scholar]
- Stropes MP, Schneider OD, Zagorski WA, Miller JL, Miller WE. The carboxy-terminal tail of human cytomegalovirus (HCMV) US28 regulates both chemokine independent and chemokine dependent signaling in HCMV-infected cells. J Virol. 2009;83:10016–10027. doi: 10.1128/JVI.00354-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadagaki K, Tudor D, Gbahou F, Tschische P, Waldhoer M, Bomsel M, Jockers R, Kamal M. Human cytomegalovirus-encoded UL33 and UL78 heteromerize with host CCR5 and CXCR4 impairing their HIV coreceptor activity. Blood. 2012;119:4908–4918. doi: 10.1182/blood-2011-08-372516. [DOI] [PubMed] [Google Scholar]
- Tomasec P, Braud VM, Rickards C, Powell MB, McSharry BP, Gadola S, Cerundolo V, Borysiewicz LK, McMichael AJ, Wilkinson GW. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science. 2000;287:1031. doi: 10.1126/science.287.5455.1031. [DOI] [PubMed] [Google Scholar]
- Tschische P, Tadagaki K, Kamal M, Jockers R, Waldhoer M. Heteromerization of human cytomegalovirus encoded chemokine receptors. Biochem Pharmacol. 2011;82:610–619. doi: 10.1016/j.bcp.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J, Schall TJ, Corey L, Geballe AP. Functional analysis of the human cytomegalovirus US28 gene by insertion mutagenesis with the green fluorescent protein gene. J Virol. 1998;72:8158–8165. doi: 10.1128/jvi.72.10.8158-8165.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila-Coro AJ, Rodriguez-Frade JM, Martin De Ana A, Moreno-Ortiz MC, Martinez AC, Mellado M. The chemokine SDF-1alpha triggers CXCR4 receptor dimerization and activates the JAK/STAT pathway. Faseb J. 1999;13:1699–1710. [PubMed] [Google Scholar]
- Vischer HF, Vink C, Smit MJ. A viral conspiracy: hijacking the chemokine system through virally encoded pirated chemokine receptors. Curr Top Microbiol Immunol. 2006;303:121–154. doi: 10.1007/978-3-540-33397-5_6. [DOI] [PubMed] [Google Scholar]
- Wegner SA, Ehrenberg PK, Chang G, Dayhoff DE, Sleeker AL, Michael NL. Genomic organization and functional characterization of the chemokine receptor CXCR4, a major entry co-receptor for human immunodeficiency virus type 1. J Biol Chem. 1998;273:4754–4760. doi: 10.1074/jbc.273.8.4754. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Foudi A, Geay JF, Berthebaud M, Buet D, Jarrier P, Jalil A, Vainchenker W, Louache F. Intracellular localization and constitutive endocytosis of CXCR4 in human CD34+ hematopoietic progenitor cells. Stem Cells. 2004;22:1015–1029. doi: 10.1634/stemcells.22-6-1015. [DOI] [PubMed] [Google Scholar]