Abstract
Remarkable advances in techniques for gene expression profiling have radically changed our knowledge of the transcriptome. Recently, the mammalian brain was reported to express many long intergenic noncoding (lincRNAs) from loci downstream from protein-coding genes. Our experimental tests failed to validate specific accumulation of lincRNA transcripts, and instead revealed strongly distal 3′ UTRs generated by alternative cleavage and polyadenylation (APA). With this perspective in mind, we analyzed deep mammalian RNA-seq data using conservative criteria, and identified 2035 mouse and 1847 human genes that utilize substantially distal novel 3′ UTRs. Each of these extends at least 500 bases past the most distal 3′ termini available in Ensembl v65, and collectively they add 6.6 Mb and 5.1 Mb to the mRNA space of mouse and human, respectively. Extensive Northern analyses validated stable accumulation of distal APA isoforms, including transcripts bearing exceptionally long 3′ UTRs (many >10 kb and some >18 kb in length). The Northern data further illustrate that the extensions we annotated were not due to unprocessed transcriptional run-off events. Global tissue comparisons revealed that APA events yielding these extensions were most prevalent in the mouse and human brain. Finally, these extensions collectively contain thousands of conserved miRNA binding sites, and these are strongly enriched for many well-studied neural miRNAs. Altogether, these new 3′ UTR annotations greatly expand the scope of post-transcriptional regulatory networks in mammals, and have particular impact on the central nervous system.
The 3′ untranslated regions (3′ UTRs) of mRNAs contain cis elements that confer post-transcriptional regulation by RNA-binding proteins (RBPs) and microRNAs (miRNAs) (Licatalosi and Darnell 2010). It is now appreciated that alternative cleavage and polyadenylation (APA) generates tremendous transcript diversity, and the majority of genes have multiple functional polyadenylation (polyA) sites. The dominant class of APA events occurs within terminal exons, causing 3′ UTR shortening or lengthening. Global 3′ UTR shortening is characteristic of proliferating cells and cancer cells (Sandberg et al. 2008; Mayr and Bartel 2009), whereas 3′ UTR lengthening was reported to occur during embryonic development and differentiation (Ji and Tian 2009).
Microarray analysis of assorted tissues indicated that the mammalian brain broadly utilizes distal 3′ UTR species (Sandberg et al. 2008; Wang et al. 2008). Deep sequencing of 3′ ends of polyadenylated transcripts uncovered hundreds of distal APA events in cultured neurons compared with embryonic stem (ES) cells (Shepard et al. 2011), and the picture was broadened by the recognition of more than 1000 3′ UTR extensions in mouse cerebellum (Pal et al. 2011). Most recently, data from Drosophila tiling microarrays (Hilgers et al. 2011) and tissue-specific RNA-seq (Smibert et al. 2012) revealed central nervous system (CNS)-specific 3′ UTR extensions across hundreds of transcripts, indicating a conserved phenomenon for 3′ UTR lengthening in the nervous system.
Diverse regulatory consequences of APA in the nervous system have been described. Variation of 3′ UTR lengths can alter transcript regulation and stability by inclusion or exclusion of miRNA binding sites (Chi et al. 2009) or other RBP sites. Global analysis of protein–RNA interactions by high-throughput sequencing of RNAs isolated by crosslinking immunoprecipitation (HITS-CLIP) revealed that NOVA1, a neural RBP best-characterized as a splicing regulator, also has extensive influence on APA (Licatalosi et al. 2008). As well, some neural 3′ UTR extensions direct localization to dendrites and axons, which can provide spatial specificity and/or facilitate their translation (An et al. 2008; Yudin et al. 2008; Andreassi et al. 2010).
Advances in expression profiling, from tiling microarrays to RNA-seq, promise to reveal a comprehensive view of the transcriptome. This includes a fuller accounting of protein-coding transcript isoforms as well as noncoding transcripts, such as small RNAs and long intergenic noncoding RNAs (lincRNAs). De novo construction of gene models from high-throughput data has identified a plethora of unannotated exons, which have been inferred to include thousands of lincRNAs. However, accurate reconstruction of parent transcripts, especially when alternative products emanate from a given locus, remains challenging. For example, initial tiling microarray studies of the Drosophila transcriptome (Manak et al. 2006) showed that a substantial fraction of novel intergenic transcribed regions actually represented alternative 5′ noncoding exons of downstream protein-coding gene models, sometimes located tens of kilobases away. On the other end, we recognized that APA frequently generates unanticipated Drosophila 3′ UTRs of exceptional length, also ranging up to tens of kilobases (Smibert et al. 2012).
Here, we reassessed previously studied lincRNAs expressed in mammalian brain (Ponjavic et al. 2009; Chodroff et al. 2010), and found that many represent stable 3′ UTR extensions of upstream protein-coding genes. We then used RNA-seq data to uncover thousands of previously unannotated 3′ UTR extensions in mouse and human. The predominant tissue-specific trend was for utilization of 3′ UTR extensions in brain, and this has substantial impact on post-transcriptional regulatory networks, including by thousands of conserved miRNA binding sites. These findings strongly revise the scope of mammalian transcriptomes, and highlight that a full appreciation of even their protein-coding gene models remains to be realized.
Results
Reevaluation of proposed neural mRNA/lincRNA pairs instead reveals distal APA isoforms
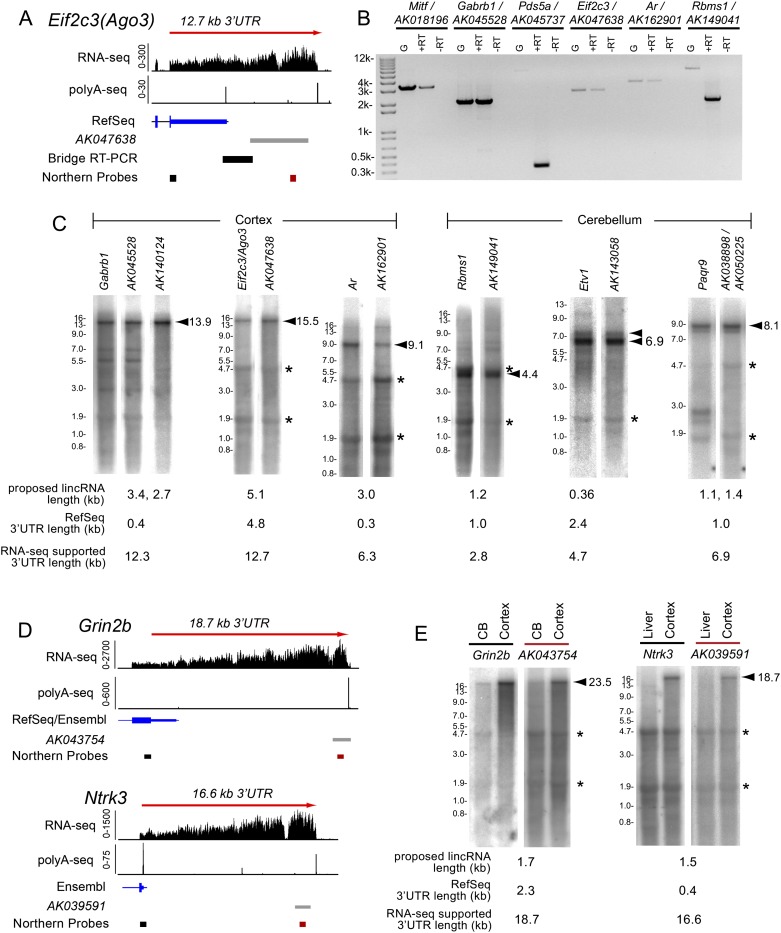
Previous searches for evolutionarily constrained long intergenic noncoding RNAs (lincRNAs) noted more than 200 brain-expressed lincRNAs originating downstream from RefSeq protein-coding genes, and some were spatially coexpressed with their upstream neighbors (Ponjavic et al. 2009). Experimental and bioinformatic tests argued against connectivity of these coding/noncoding pairs. Nevertheless, we observed many lincRNAs resided in regions of RNA-seq coverage continuous with upstream genes (e.g., Ago3 [also known as eIF2C3]) (Fig. 1A). Consequently, we reevaluated mRNA–lincRNA pairs previously reported as experimentally negative for connectivity (Ponjavic et al. 2009).
Figure 1.
Evidence that annotated lincRNAs downstream from protein-coding gene pairs are 3′ UTR extensions. (A) Experimental strategy to test connectivity between a protein-coding gene and a downstream lincRNA. RNA-seq and polyA-seq evidence in the vicinity of eIF2C3 (also known as Ago3) and the proposed lincRNA AK047638. We designed primers to amplify bridge rt-PCR products and Northern probes, as shown. (B) Bridge rt-PCR using adult cerebral cortex RNA connects many protein-coding genes with their proposed downstream neighboring lincRNAs (Ponjavic et al. 2009); note Ar was previously referred to as Adr, and Ube2k was termed Hip2. Note that AK045737 was proposed to be a pair with Ube2k (Ponjavic et al. 2009); however, stranded RNA-seq data revealed that AK045737 is continuous with a spliced exon of Pds5a transcribed from the other strand (see Supplemental Figure S1A–C). (C) Northern analysis demonstrates that the predominant transcripts detected by probes for the protein-coding loci assayed in B are codetected by probes against their neighboring downstream lincRNAs. Conversely, we did not detect stable transcripts corresponding to the sizes of the annotated lincRNAs. Northern blots are also shown for ncRNAs described by Mattick and colleagues (Clark et al. 2012) and their protein-coding pairs Etv1 and Paqr9. (D) RNA-seq and PolyA-Seq tracks for cases of annotated lincRNAs that appear to be contained with exceptionally long, continuous 3′ UTRs of stable mRNAs. (E) Northern blots for proposed lincRNAs show a band of exceptional length that is of the same molecular weight as the bands identified by probes corresponding to the upstream protein-coding transcripts. Arrowheads identify dominant bands that correspond to size estimates based on RNA-seq data. Note that the sizes of the bands on the Northern blot are consistent with the RNA-seq evidence–based size estimates. Asterisks denote 28S and 18S ribosomal bands corresponding to 4.7 kb and 1.9 kb, respectively. Ladder information can be found in Supplemental Figure S4. For RNA-seq tracks, probe locations, and gene annotations, see Supplemental Figure S1D.
Interestingly, we observed reverse transcriptase–dependent PCR products from post-natal brain that join the annotated mRNAs of Mitf, Gabrb1, Ago3/eIF2C3, Ar, and Rbms1 with their reported downstream lincRNAs (Fig. 1B; Supplemental Fig. S1). The only pair not validated was Ube2k–AK045737. However, stranded RNA-seq data revealed transcription of AK045737 exclusively on the opposite strand (Supplemental Fig. S1). This places AK045737 downstream from Pds5a, with intervening spliced reads, and rt-PCR products joined these loci (Fig. 1B).
More definitive information on transcript connectivity and alternative isoforms is provided by Northern analysis. We designed paired probes targeting the terminal coding region/proximal 3′ UTR of the annotated mRNAs and their proposed downstream noncoding RNAs (Ponjavic et al. 2009). We did not detect Mitf1 (data not shown), we but observed robust signals for the rest in cortex or cerebellum (Fig. 1C). Notably, the dominant bands detected by mRNA probes were always substantially larger than the RefSeq-annotated transcripts, and these always cohybridized with their downstream lincRNA probes (Fig. 1C). Moreover, no distal probes identified shorter bands of lengths predicted for the reported lincRNAs. While these data do not rule out the possibility of distinct lowly expressed lincRNAs, they demonstrate the bulk of stable transcripts bearing these noncoding sequences to be 3′ UTR extensions of mRNAs.
Some lincRNAs are substantially separated from known mRNA termini. The lincRNA AK043754 was recently studied in detail (Chodroff et al. 2010), and its locus resides 14.9 kb downstream from Grin2b. Examination of hippocampal RNA-seq data (Keane et al. 2011) revealed continuous coverage from the annotated Grin2b 3′ UTR to AK043754 (Fig. 1D). Our Northern analysis for AK043754 revealed a single, strong ∼23.5-kb band in cerebral cortex. A probe against Grin2b coding sequence identified the same band; thus, Grin2b expresses an ∼19-kb 3′ UTR in brain (Fig. 1E). We obtained similar results by Northern analysis of the proposed lincRNA AK039591 (Ponjavic et al. 2009), which is contained within the 16.6-kb 3′ UTR of Ntrk3 (Fig. 1D,E). That the same large bands were specifically detected by probes against very proximal and very distal portions of these long 3′ UTRs rules out potential alternative events 5′ of the stop codon (i.e., retained internal introns, alternative splicing, or alternative promoters). Grin2b and Ntrk3 contain some of the longest stable 3′ UTRs ever demonstrated in mammals, highlighting that some “very downstream” lincRNAs can be reinterpreted as 3′ UTR extensions.
A bioinformatic pipeline to call 3′ UTR extensions using RNA-seq data
None of the 3′ UTR extensions analyzed in Figure 1 are present in RefSeq, although many are well-studied genes. We therefore sought to annotate 3′ UTR extensions more comprehensively. Searching other databases, we observed that Ensembl currently annotates Ago3/eIF2C3, Ar, and Rbms1 3′ UTR extensions; however, neither database includes the distal 3′ UTRs for Gabrb1, Pds5a, Ntrk3, or Grin2b. We consequently used the latest Ensembl version 65 (v65) (Flicek et al. 2012) as a conservative reference for annotating novel 3′ UTRs.
We took advantage of deep RNA-seq data from six mouse tissues (Keane et al. 2011) and reprocessed the raw data to map ∼1.7 billion reads (Supplemental Table S1). We initially generated transcript models using Cufflinks (Trapnell et al. 2012), but we noticed from browsing its outputs that 3′ UTR extents were frequently truncated due to variable read depth, discontinuous coverage, and/or multimapper reads. We therefore developed an alternate approach to identify 3′ UTR extensions.
The key features included a sliding window that identified continuously transcribed genomic segments ≥1.0 fragments per kilobase of transcript per million mapped fragments (FPKM) (empirically determined from recall of known, internal, nonalternative exons), followed by judicious merging of adjacent contigs split by lower coverage regions or repeats. To ensure that merging was conservative, we demanded that gaps were bridged by paired-end reads, limited nonrepetitive gaps to <150 nucleotides (nt), and restricted novel extensions from containing >20% of repetitive sequence. We then made extensive efforts to cull potentially ambiguous 3′ UTR extensions using many additional filtering steps. We grouped together novel 3′ ends from different tissues that were within 30 nt of each other, and extensively confirmed the final calls by visual inspection. Our stringent filtering steps removed some genuine 3′ UTR extensions (e.g., Supplemental Fig. S3), but we preferred this conservative approach to focus on 3′ UTRs of high confidence. The pipeline is described in detail in the Methods, Supplemental Text, and Supplemental Figures S2 and S3.
Analysis of mouse and human RNA-seq data using stringent criteria reveals more than 3850 novel 3′ UTR extensions
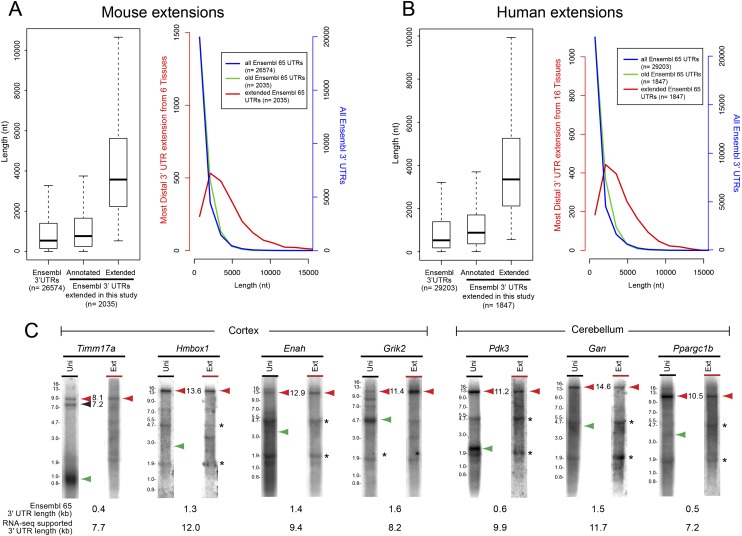
We compared our 3′ UTR calls from six mouse tissues to Ensembl v65 annotations to identify 3′ UTR extensions. To focus on substantially novel isoforms, we required Ensembl gene models be extended >500 nt. Although some stable 3′ UTR extensions that we validated by Northern failed our conservative annotation pipeline (e.g., Hmbox1, Ntrk3, and Etv1) (see Supplemental Fig. S3), this analysis strikingly identified 2035 confident 3′ UTR extensions over Ensembl v65 mouse gene models (Supplemental Table S2). These 3′ UTRs comprise ∼6.6 Mb of unannotated sequence and average 4347 nt in length, far above the Ensembl v65 average of 989 nt (Fig. 2A; Supplemental Table S2). A recent analysis of mouse cerebellum identified many 3′ UTR extensions (Pal et al. 2011), of which 600 remain exclusive of Ensembl v65. Still, our annotations comprised 6.2 Mb sequence beyond these models (Supplemental Table S3; Supplemental Text).
Figure 2.
Reevaluation of mouse and human RNA-seq data reveals abundant 3′ UTR extensions. (A, left) Box plot comparing aggregate Ensembl v65 3′ UTR lengths (longest annotation per terminal exon) and those Ensembl v65 3′ UTRs that were specifically extended in this study. (Right) Histogram of the same data to highlight the abundance of newly annotated long 3′ UTRs. (B) Analyses similar to A, except plotting known and novel Ensembl v65 human 3′ UTRs. (C) Northern analysis validates the stable accumulation of many transcripts utilizing very distal polyadenylation signals in cerebellum or cortex, in several cases yielding 3′ UTRs >10 kb in length. Green arrowheads indicate predicted mRNA length of Ensembl v65 gene model. Red arrowheads indicate inferred mRNA lengths of novel 3′ UTR extension isoforms. Asterisks denote background hybridization to ribosomal RNAs.
We performed similar analysis of the human transcriptome using the Illumina Body Map 2.0 of 16 tissues, including a pooled stranded data set that confirmed expression directionality. We reprocessed these from the raw data and mapped more than 4 billion reads (Supplemental Table S1). We used the same pipeline to annotate 3′ UTR extensions, except that we increased expression cutoffs to 1.5 FPKM (based on recall of known nonalternative exons). This analysis identified 1847 confident 3′ UTR extensions over Ensembl v65 (Fig. 2B; Supplemental Table S4), comprising 5.1 Mb of unannotated sequence (Fig. 2B).
Altogether, these thousands of confident 3′ UTR extensions add ∼11.1 Mb to mouse and human protein-coding gene models. Moreover, we further designated thousands of candidate loci in mouse (Supplemental Table S2) and human (Supplemental Table S4) with compelling evidence for extension but failed to meet our full criteria. These included loci bearing 300- to 499-nt extensions relative to Ensembl v65, that were expressed at 0.5–0.99 FPKM, or that had small gaps not bridged by paired-end reads. Thus, the scope of mammalian 3′ UTR extensions is likely even larger than we currently annotate.
Experimental support for extended 3′ UTR isoforms
We vetted the veracity of 3′ UTR extensions by systematic visual inspection, coupled with extensive “gold-standard” Northern evidence. Because commercial RNA ladders only size to 9 kb, we generated a “virtual ladder” composed of endogenous transcripts from 0.8–16 kb (Supplemental Fig. S4). We implemented this by hybridizing mixed probe to stripped blots, a strategy that furthermore reported the integrity of long transcripts.
We focused on genes expressed in the cortex and/or cerebellum. We did not detect specific Kcna4 or Nxph1 transcripts (data not shown), reflecting technical failures or low abundance. However, all proximal 3′ UTR probes that detected specific transcripts (e.g., Timm17a, Grik2, Enah, Pdk3, Gan, and Ppargc1b) revealed long species corresponding in length to distal APA isoforms inferred from RNA-seq (Fig. 2C). We re-probed for their distal extensions using amplicons separated from the proximal probes by the length of the 3′ UTR extension. We consistently detected the same large transcripts with paired proximal and distal probes (Figs. 1C,E, 2C), constituting unambiguous evidence for stable mRNAs bearing long 3′ UTRs. Moreover, these data comprise strong evidence against possibilities that the underlying RNA-seq data reflect heterogeneous runaway transcription products, pre-mRNA intermediates, or unstable transcripts in the process of being degraded.
Several of these genes exhibit >10-kb 3′ UTRs, adding to other long extensions validated earlier (Fig. 1D). Overall, our high validation rate, using the modestly sensitive Northern technique, reflects the stringency of the annotation pipeline. We also note validation of some loci that did not meet our full bioinformatic criteria. For example, we clearly observed the strongly extended 12-kb 3′ UTR of Hmbox1 by Northern, even though it overlaps an annotated alternative last exon and was therefore culled from our pipeline (Fig. 2C; Supplemental Fig. S3). This highlights the conservative nature of our annotations, and that our candidate lists undoubtedly contain additional genuine 3′ UTR extensions.
Analysis of polyA signals and conservation among novel distal 3′ termini
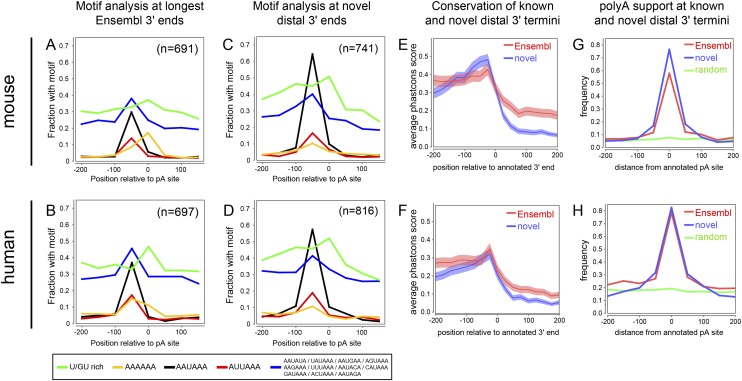
We investigated the characteristics of novel distal 3′ termini. Although our pipeline assesses confident 3′ UTR extensions, it does not necessarily pinpoint precise 3′ ends, especially as RNA-seq protocols undersample near transcript termini. We further noted many instances of likely extensions whose most distal regions did not satisfy our expression cutoff and are thus truncated by our pipeline. We therefore implemented a “dropoff” filter to identify extension calls that coincide with a sharp drop in RNA-seq coverage. We required that two consecutive 100-nt windows downstream from the 3′ end call exhibit greater than eightfold reduction in reads, relative to the final 100-nt window of the 3′ extension (Supplemental Text; Supplemental Fig. S5). Since some extensions terminated in repetitive regions, and thus lacked precise 3′ end calls, we also culled these from motif analysis. This yielded 691 extended mouse loci comprising 741 distinct 3′ ends (Supplemental Table S2; Supplemental Fig. S6) and 697 extended human loci totaling 816 distinct 3′ ends (Supplemental Table S4; Supplemental Fig. S7).
We compared the properties of our novel 3′ ends with their annotated Ensembl v65 counterparts. Known 3′ termini usually bear canonical polyadenylation signals (PASs) AAUAAA or AUUAAA ∼35 nt upstream of transcript ends, with lower and less specific enrichment for various noncanonical PAS (Tian et al. 2005). In addition, U/GU motifs are enriched downstream from known PAS (for motif definitions, see Methods). We observed all of these features in the vicinity of annotated mouse (Fig. 3A) and human (Fig. 3B) 3′ termini, in proportions consistent with previous global analyses of mammalian 3′ ends. Curiously, we also observed ∼15% genomically encoded AAAAAA among both mouse and human Ensembl v65 termini, suggesting that some of these annotations may potentially derive from internally primed cDNAs.
Figure 3.
Sequence and conservation features of known and novel 3′ termini. All analyses in this figure concern those mouse (top graphs) and human (bottom graphs) genes whose 3′ UTRs were confidently extended in this study. These comprise 691 Ensembl65 mouse gene models for which we precisely annotate 741 novel 3′ termini in one or more tissues, and 697 Ensembl65 human genes for which we precisely annotate 816 novel 3′ termini in one or more tissues. (A,B) Motif frequency in 50-nt bins in the vicinity of annotated 3′ termini. Motifs are listed at bottom, and include the downstream U/GU-rich region that promotes 3′ cleavage, the canonical PAS AAUAAA and its most common variant AUUAAA, a panel of low-frequency PAS variants, and genomically encoded hexa-A tracts. As expected for annotated mouse (A) and human (B) 3′ termini, there is strong positional enrichment of functional PAS upstream of the polyadenylation site and U/GU downstream. The collection of low-frequency PAS variants exhibits a broad background frequency, with mild enrichment at the normal location of canonical PAS. Unexpectedly, we observed enrichment of A6 at annotated 3′ termini, potentially reflecting internal priming events in this collection of curated 3′ termini. (C,D) The frequency and positional specificity of PAS and U/GU motifs in our novel mouse (C) and human (D) 3′ termini are relatively similar to known termini but lack substantial A6 enrichment at transcript ends. (E,F) Analysis of average phastCons scores in the vicinity of known and newly annotated 3′ termini in mouse (E) and human (F) shows that both populations of termini exhibit selective constraint that rises to a peak in the local sequence upstream of 3′ termini, and drops sharply in the downstream sequence. Note also that the aggregate conservation of the last ∼500 nt of proximal 3′ UTR sequences is higher than that of the distal novel 3′ UTR sequences, but the overall level of conservation 3′ of our mouse and human extensions drops to background. (G,H) Analysis of location of polyA-seq tags relative to known and newly annotated 3′ termini shows a similar positional enrichment at transcript 3′ termini. Comparison with a randomly selected set of 3′ ends from these transcripts shows no positional enrichment of polyA-seq tags, indicating that our novel annotations include genuine 3′ ends.
Our novel distal 3′ UTR extensions exhibited strong positional enrichments of upstream PAS and downstream U/GU motifs, in both mouse (Fig. 3C) and human (Fig. 3D). In fact, the enrichment of canonical AAUAAA PAS was greater among our novel 3′ termini, compared with their Ensembl termini. Moreover, we did not observe enrichment of AAAAAA polymers at our newly defined termini. These observations provided strong support for the quality of our extension annotations. Therefore, while we do not presume that every novel terminus was defined precisely, this set of more than 1500 novel distal termini exhibits motif properties of genuine transcript ends that meet or exceed those of well-annotated Ensembl transcript ends.
We next analyzed the conservation of proximal and distal termini using phastCons values. For both mouse (Fig. 3E) and human (Fig. 3F), we observed a local spike in conservation 5′ to the proximal polyadenylation sites, followed by a drop in conservation. The local conservation surrounding aggregate distal PAS was comparable to corresponding proximal PAS, indicating their similarly strong evolutionary selection. We also note that the overall conservation 100–500 nt downstream from proximal PAS was higher than downstream from our novel distal PAS. These trends applied to both mouse (Fig. 3E) and human (Fig. 3F) and are compatible with the scenario that our thousands of novel 3′ UTR extensions contain functional cis-regulatory information that distinguishes them from background intergenic sequence.
Finally, we assessed our novel 3′ termini for overlap with recent 3′-sequencing of mouse and human transcripts (Derti et al. 2012). These data are a valuable resource for the discovery of novel mRNA ends, although the initial study did not systematically annotate these. We reprocessed the polyA-seq data (Supplemental Table S1) to precisely annotate 3′ ends that correspond with the end of RNA-seq coverage. For comparison, we analyzed the extent of polyA-seq support for Ensembl v65 ends of all loci whose models we extended in this study. Nearly 80% of mouse (Fig. 3G) and 85% of human (Fig. 3H) annotated termini were supported by polyA-seq tags. We did not necessarily expect such a high validation rate a priori, since RNA-seq data do not demarcate transcript ends precisely, and distal 3′ UTR extension transcripts often accumulate to lower levels and thus contribute fewer polyA-seq tags than shorter isoforms.
Altogether, these bioinformatic analyses demonstrate that we annotated a large population of functional mammalian transcript termini, adding large expanses of currently unannotated mouse and human genomic sequence to expressed 3′ UTR space. Strikingly, these thousands of novel 3′ termini exhibit motif and conservation properties that are comparable to known mRNA termini in these well-annotated genomes.
Tissue-specific 3′ UTR lengthening is strongly biased toward neural tissue
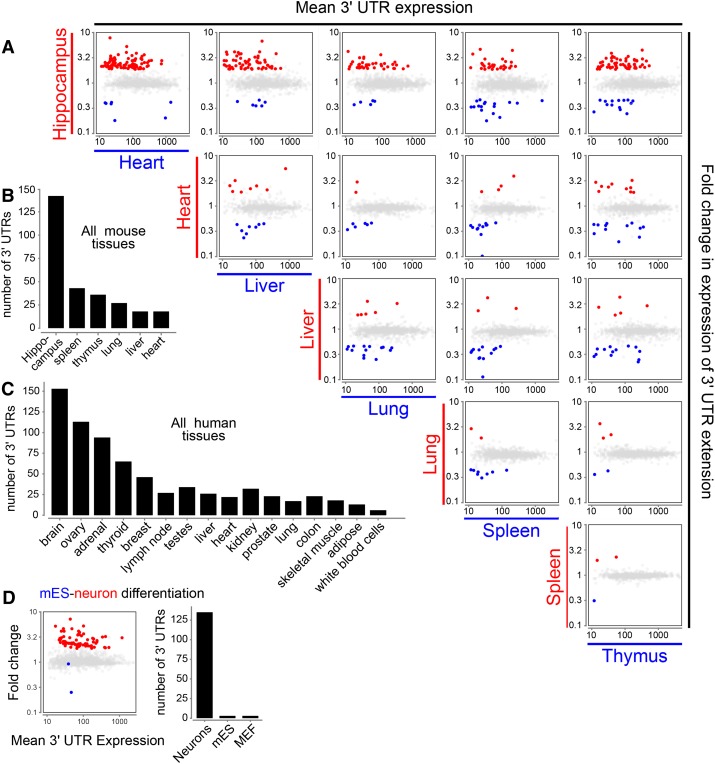
We reannotated 3′ UTRs from RNA-seq data across a variety of tissues, but until this point, our experimental validation focused on brain. We utilized DEXSeq (Anders et al. 2012), a statistical approach to detect differential exon usage, to identify tissue-biased APA events. Analysis of 15 pairwise combinations of mouse tissues yielded at least some genes with significant differential expression of 3′ UTR extensions between each tissue pair. In addition, many of the novel 3′ UTR extensions we annotated were not differentially expressed across tissues. However, we consistently observed that the hippocampus exhibited the highest number and expression of 3′ UTR extensions, relative to all other tissues (Fig. 4A). We repeated this analysis for 16 human tissues in the Illumina BodyMap 2.0. We show representative comparisons in Figure 4C and provide all 120 pairwise comparisons in Supplemental Figure S8. These tests recapitulate the pattern observed in mouse, in that the absolute number and relative abundance of the novel 3′ UTR extensions are highest in the human brain.
Figure 4.
Systematic tissue comparisons show that 3′ UTR lengthening occurs preferentially in the brain. (A) Pairwise analysis of tissue-specific preferences of novel mouse 3′ UTR extensions using DEXSeq. Each gene is represented as a single point, such that the relative expression of the 3′ UTR extension between the pair of tissues (indicated at the left of each row and the bottom of each column) is plotted as the Y-coordinate, and the average expression of the 3′ UTR in that pair of tissues is plotted as the X-coordinate. For genes exhibiting a significant (greater than twofold, FDR < 0.01) difference between the two tissues the point is colored red if the relative usage is higher in the tissue indicated at the left of the row and blue if it was higher in the tissue indicated at the bottom of the column; all other 3′ UTRs are shown in gray. We observed a broad tissue-wide trend toward increased expression of lengthened 3′ UTRs in hippocampus, seen as a substantial excess of red points across the top row of tissue comparisons against hippocampus. No particular trend is observed among the other pairwise tissue comparisons. (B) Summary of the pairwise analysis of novel 3′ UTR extensions annotated in mouse. For each tissue, the set of genes that are detected by DEXSeq to have a higher fold expression of an extended 3′ UTR extension compared to at least one other tissue were counted. (C) Summary of DEXSeq tissue comparisons of novel 3′ UTR extensions in human (for all pairwise scatterplots, see also Supplemental Fig. 8). (D) DEXSeq analysis of our novel mouse 3′ UTR extensions, assessed in RNA-seq data from mES/neuron/MEF cells. In the scatterplot, mES data are in blue and differentiated neuron data are in red.
To strengthen the conjecture that preferential usage of unannotated distal polyadenylation sites is a property of neuronal cells, we examined RNA-seq data from mouse ES cells and differentiated neurons derived from these cells (Lienert et al. 2011). By using the coordinates from DEXSeq analysis of mouse tissues (Fig. 4A, top row), we performed DEXSeq expression analysis for ES cells and derived neurons, as well as for mouse embryonic fibroblasts (MEFs). These tests robustly reproduced the pattern of preferential neural expression of novel extensions (Fig. 4D).
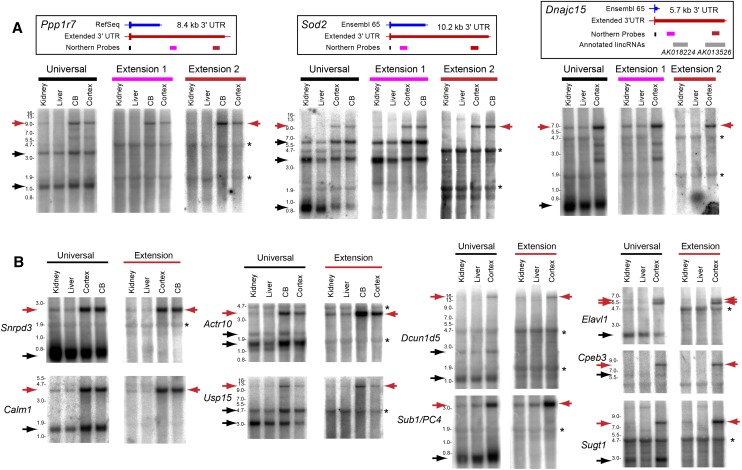
We confirmed these bioinformatic trends using Northern analysis of mouse kidney, liver, cerebellum, and cortex. As many transcripts whose 3′ UTR extensions we validated earlier (Figs. 1, 2) were not necessarily subject to APA, we focused on genes with tissue-specific 3′ UTR extensions. Figure 5A illustrates stringent APA analysis using proximal probes and two different extension probes for Ppp1r7, Sod2, and Dnajc15. Their universal probes detected shorter transcripts across the tissue panel and longer transcripts that were brain-specific. In all cases, both sets of extension probes detected exclusively the longer isoforms and only in brain. This was particularly notable for Sod2 and Dnajc15, whose intermediate extension probes detected APA isoforms of intermediate length, which were not detected by the most distal extension probes.
Figure 5.
Northern analysis validates brain-specific 3′ UTR extensions. (A) Northern analyses that compare universal (proximal) probes with two probes directed against an intermediate and a very distal portion of a 3′ UTR extension. The gene models above show the known and newly recognized 3′ UTR extensions and locations of Northern probes. In all cases, the universal probes detect broadly expressed transcripts bearing short 3′ UTRs as well as longer 3′ UTR isoforms that are specific to cerebellum (CB) and/or cortex, while the extension probes detect exclusively the longer 3′ UTR isoforms in brain. Note that the intermediate probes (extension 1) for Sod2 and Dnajc15 detect intermediate 3′ UTR isoforms that are codetected by their respective universal probes but not by their most distal 3′ UTR probes. Asterisks denote cross-hybridization to abundant rRNA bands. (B) Additional examples of brain-specific distal APA events validated by Northern blots. Northern analysis using universal Northern probes (black bars) designed to detect all 3′ UTR isoforms reveal dominant isoforms used by all tissues examined along with brain-specific long 3′ UTR isoforms. Extension probes (red bars) designed to detect the 3′ UTR extensions reveal expression only in the brain and not in other tissues. Asterisks denote background hybridization to ribosomal RNAs; (CB) cerebellum.
We extended this analysis using paired universal and extension probes for nine other genes exhibiting brain-specific 3′ UTR lengthening (Fig. 5B; Supplemental Fig. S9). These data broadly support the bioinformatic inference of brain-specific distal APA usage (Fig. 4) and further validate the existence of exceptionally long 3′ UTRs on stable neural transcript isoforms (e.g., 10.2-kb Sod2 3′ UTR and 14-kb Dcun1d5 3′ UTR). Altogether, a process of tissue-biased transcript lengthening that generates hundreds of novel, distal 3′ UTRs in the nervous system occurs on a global scale in Drosophila, mouse, and human.
In situ hybridization validates expression of neural-specific 3′ UTR extensions
We used in situ hybridization of E13.5 mice to assess the spatial expression patterns of several genes undergoing neural 3′ UTR extension. We were particularly interested if paired proximal and distal probes ever detected differential patterns, as observed in Drosophila (Hilgers et al. 2011; Smibert et al. 2012).
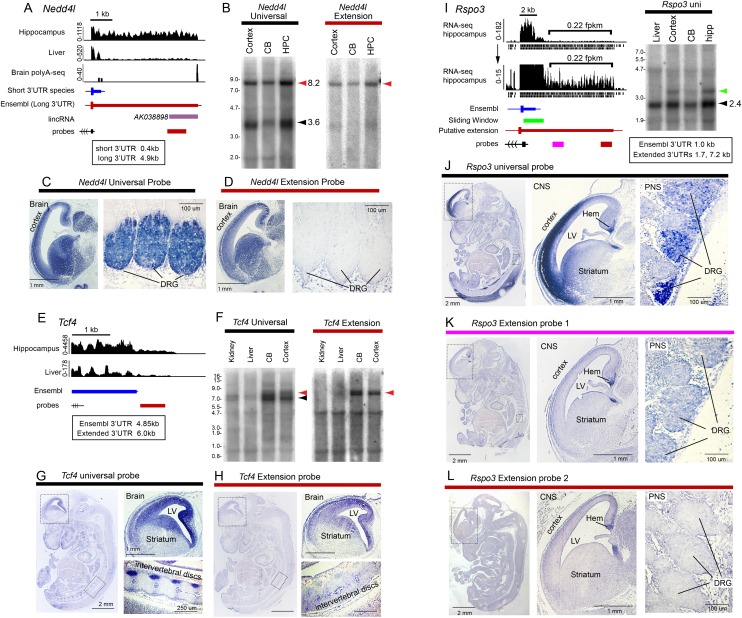
Nedd4l encodes an ubiquitin ligase whose substrates include receptors and kinases in several signaling pathways. By using paired universal and extension probes, we observed expression of both short and long 3′ UTR isoforms in brain Northerns (Fig. 6A,B). In situ analysis using a Nedd4l universal probe detected expression in brain and dorsal root ganglion (DRG) (Fig. 6C). The extended 3′ UTR isoform probe similarly detected expression in brain; however, no signals were obtained in PNS (Fig. 6D).
Figure 6.
In situ hybridization of mouse embryos reveals localization of extended 3′ UTR isoforms in specific brain regions. (A) RNA-seq data for Nedd4l indicate an alternative 4.9-kb-long 3′ UTR isoform that includes a proposed lincRNA AK038898. (B) Northern blotting demonstrates that an AK038898 probe detects the long 3′ UTR isoform of Nedd4l. (C) A Nedd4l universal probe detects expression in both brain and dorsal root ganglia (DRG). (D) A probe directed against the very distal portion of the Nedd4l 3′ UTR extension detects only brain expression. (E) RNA-seq data for Tcf4 indicate the existence of a 3′ UTR extension of the annotated gene model, with preferential expression in hippocampal data. (F) Tissue Northern blot using a distal probe confirms the existence of a discrete band expressed in brain that corresponds to a 3′ UTR extension isoform. (G) In situ hybridization to a probe in the common 3′ coding exon detects Tcf4 predominantly in the CNS and the intervertebral discs; (LV) lateral ventricle. A whole-embryo cross-section is shown at left, and the regions boxed are enlarged at right. (H) The Tcf4 3′ UTR extension probe only detects expression in the brain. (I) RNA-seq data for Rspo3 indicate a candidate 3′ UTR extension, although this level of expression (0.22 FPKM) was below our cutoff for genome-wide calls of 3′ UTR extensions. (J) A universal Rspo3 probe predominantly detects CNS expression in the cortex and hem, as well as PNS expression in the spinal cord, mainly in dorsal root ganglia (DRG). (K) The intermediate Rspo3 extension probe hybridizes specifically to the cortical hem. (L) A probe directed against the very low abundance Rspo3 extension region similarly detects expression in cortical hem.
We next analyzed Tcf4, a transcription factor in the Wnt pathway. Recent studies proposed differential stability of TCF4 and its downstream lincRNAs (Clark et al. 2012). However, RNA-seq data indicate continuous transcription downstream from Tcf4 (Fig. 6E). Northern analysis using a probe to the downstream unannotated region supported the presence of a 3′ UTR extension of Tcf4 and did not detect shorter ncRNAs (Fig. 6F). The extended APA isoform was spatially restricted, since a Tcf4 universal probe detected expression in forebrain and intervertebral discs (Fig. 6G), whereas the extended isoform was only found in forebrain (Fig. 6H).
Finally, we analyzed Rspo3, which encodes a thrombospondin type 1 repeat family protein, members of which are also involved in Wnt signaling. Our pipeline predicted a ∼700-nt extension of its annotated 3′ UTR. However, visual inspection of RNA-seq data revealed a potential 3′ UTR extension of ∼7.2 kb that was below our expression cutoff (0.22 FPKM) (Fig. 6I) and was therefore excluded from our confident bioinformatic list. Northern analysis validated the short 3′ UTR extension, but not its substantially longer counterpart. Nevertheless, in situ hybridization revealed discrete and distinct expression of the extended isoform. The universal probe detected expression broadly in the pallium, cortical hem, spinal cord, and DRG (Fig. 6J). In contrast, the Rspo3 3′ UTR extension probe hybridized exclusively to the cortical hem and did not reveal PNS expression (Fig. 6K). A probe against the distal unique sequence of the 3′ UTR extension revealed the same patterns (Fig. 6L), suggesting the existence of a stable isoform not detected by Northern. The restricted spatial pattern of Rspo3 extensions explains its seemingly low expression in total hippocampal RNA and emphasizes that our computational identification of thousands of 3′ UTR extensions still underestimate the magnitude of this phenomenon.
Novel 3′ UTR extensions harbor thousands of conserved miRNA target sites
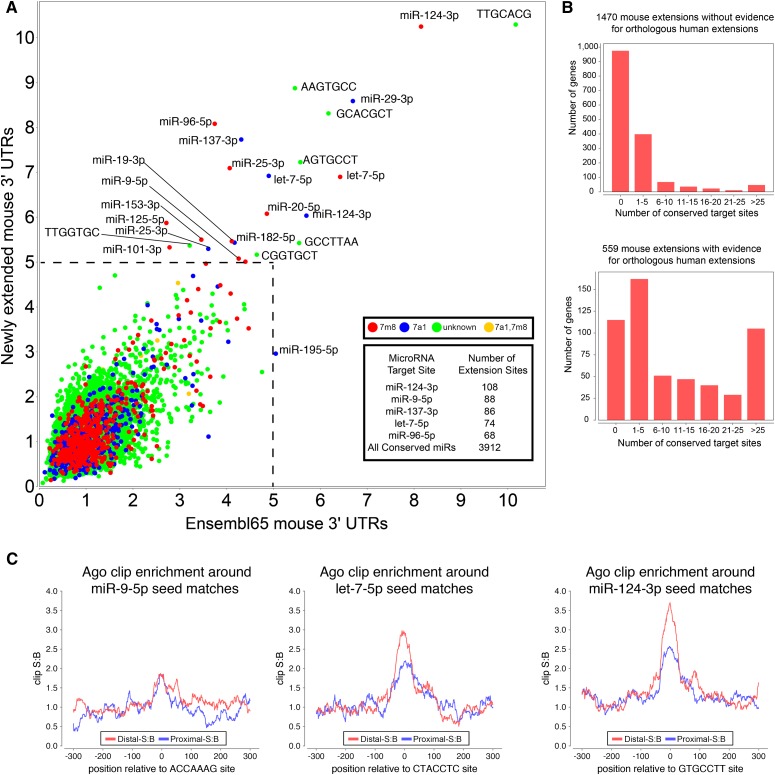
We sought regulatory implications of this large network of 3′ UTR extensions. We assessed all 7-mers for evidence of conservation above background among our 2035 mouse 3′ UTR extensions. Notably, the motifs with highest signals and highest numbers of conserved instances corresponded to seeds for miRNAs with well-described neural functions (Sun et al. 2013), including let-7, miR-124, miR-9, miR-96, miR-125, and miR-137 (Fig. 7A). We asked whether the enrichment for neural miRNA seeds was a property of coherent targeting or mutual exclusion. We segregated the 2035 extensions for those detected in hippocampus, and compared their site properties to the rest. This analysis showed that extensions expressed in hippocampus bore the majority of neural miRNA target sites (Supplemental Fig. S10), indicating that neural 3′ UTR extensions are utilized to confer regulation by neural-specific miRNAs.
Figure 7.
Novel 3′ UTR extensions harbor thousands of functional miRNA target sites. (A) Signal-to-background ratio (S:B) of 7-mers found in the proximal 3′ UTR annotations compared with the novel extended 3′ UTR region annotated in mouse from all tissues analyzed. Note that target sites for several well-characterized neural miRNAs are found among the most well-conserved 7mers in both proximal and novel extended 3′ UTR regions, including miR-124, miR-137, miR-9, let-7, miR-96, and miR-125. Supplemental Figure S10 demonstrates that the signal for neural miRNA seed matches is driven by genes with neural-expressed 3′ UTR extensions. (B) Analysis of seed matches to mammalian-conserved miRNAs, that are present among mouse 3′ UTR extensions that lack companion expression evidence for an orthologous 3′ UTR extension in human (top graph) or that do have such experimental evidence for a human extension (bottom graph). The proportion of conserved miRNA binding sites is much higher among genes with evidence for a conserved 3′ UTR extension. (C) Regions surrounding miRNA target sites located in proximal (in blue) and novel distal 3′ UTR mouse extensions (in red) show enrichment of Ago HITS-CLIP tags over background. The signal:background (S:B) of clip tags at let-7 and miR-124 seed matches is actually higher in the novel 3′ UTR extension regions.
We subsequently performed a directed search of the unannotated mouse extensions for seed matches conserved between rodents and primates, restricting this to mammalian-conserved miRNAs. We identified nearly 4000 conserved miRNA target sites, which substantially extend the scope of post-transcriptional regulatory networks in mammals (Fig. 7A, inset; Supplemental Table S5). Even though the list of human genes with novel extensions overlapped only partially with mouse, de novo analysis revealed mostly the same neural miRNA seeds among their best-conserved 7-mers (Supplemental Fig. S11; Supplemental Table S6).
The presence of conserved miRNA binding sites on sense strands downstream from annotated gene models serves as corroborating evidence for 3′ UTR extensions. For the 2035 novel mouse extensions, 559 have an orthologous extension in human (469 of which are annotated in Ensembl v65, and 96 of which are newly identified here). The remaining 1470 loci are tentatively supported only in mouse, although at least some are associated with “downstream” human RNA-seq evidence that did not meet the 1.5 FPKM threshold (Supplemental Table S2). We asked whether the conserved miRNA sites preferentially partitioned into mouse 3′ UTR extensions that did, or did not, have an orthologous human extension. We observed highly significant (P < 1 × 10−15, binomial test) enrichment of conserved miRNA binding sites among genes that currently share experimental evidence for neural 3′ UTR extensions in both mouse and human (Fig. 7B; Supplemental Table S7). Nevertheless, a population of “mouse-only” extensions harbor miRNA binding sites conserved in human (in some cases more than 25 sites), indicating that the catalog of neural 3′ UTR APA events is still not complete.
To further address the functionality of miRNA binding sites in neural 3′ UTR extensions, we queried Ago binding sites in mouse brain using published HITS-CLIP data (Chi et al. 2009). In this study, some Ago-bound tags were noted downstream from certain gene models, and inferred to represent potentially unannotated 3′ UTR sequences. We surveyed our 3′ UTR extensions and observed robust signals for Ago binding at miRNA seeds in both proximal and extended 3′ UTRs (Supplemental Table 8). Exemplar seeds enriched in Ago-CLIP tags in extended 3′ UTRs included miR-124-3p, let-7-5p, and miR-9-5p (Fig. 7C). While a lower fraction of 7-mers overlapped Ago-CLIP tags in the extended portion of UTR, the ratio of Ago-IP frequency at these sites to background UTR Ago-IP was actually higher in extended regions. The lower CLIP frequency may be due to differential isoform abundance, since CLIP tags are sampled more frequently from abundant mRNAs, and shorter isoforms tend to accumulate to higher levels than the distal extension isoforms. Nevertheless, the Ago HITS-CLIP data strongly support that these novel neural 3′ UTR extensions confer substantial regulation by many mammalian neural miRNAs.
Discussion
Extensive usage of highly distal APA is a well-conserved feature of the nervous system
Genes expressed in the nervous system contain longer 3′ UTRs, on average, compared with other tissues (Stark et al. 2005; Wang et al. 2008; Ramskold et al. 2009). Moreover, a number of transcripts have been noted to undergo alternative polyadenylation (APA) in the nervous system, yielding longer 3′ UTRs in their neural isoforms. Very recently, the Drosophila central nervous system was recognized to utilize novel distal APA sites across hundreds of transcripts, often generating 3′ UTRs of exceptional length (Hilgers et al. 2011; Smibert et al. 2012). Analysis of mammalian brain and cultured neurons similarly provides evidence of distal APA events (Pal et al. 2011; Shepard et al. 2011).
In this study, we substantially increase the number and magnitude of neural distal APA events in the mammalian brain, including the accumulation of a multitude of stable mRNAs bearing exceptionally long 3′ UTRs (nearly 20 kb in length). In a day and age where the amount of “extragenic” transcription that contributes to discrete, stable transcripts remains hotly contested (Ponting and Belgard 2010; van Bakel et al. 2010; Clark et al. 2011), it is striking that we can add 6.6 Mb and 5.1 Mb of currently unannotated sequence to the confident mRNA space of the mouse and human transcriptomes, respectively. These transcript extensions have substantial impact on post-transcriptional networks, especially those mediated by the particularly extensive collection of neural 3′ UTR extensions.
We find that short and long tissue-specific 3′ UTRs exhibit marked usage of canonical PAS hexamers (AAUAAA or AUUAAA) and downstream U/GU-rich elements, although it is striking that our plethora of novel, distal 3′ ends exhibit motif properties that are slightly stronger than their Ensembl-annotated counterparts (Fig. 3A–D). Still, Northern analysis of mouse brain tissue revealed that although a longer 3′ UTR is used, the shorter 3′ UTR continues to be expressed, often at relatively high levels. This contrasts with the “switch-like” behavior of many Drosophila genes to dominantly express the distal 3′ UTR in heads and dissected CNS (Smibert et al. 2012). At present, it is unclear if this reflects that the mammalian brain does not bypass proximal PAS as efficiently, or whether this reflects differences in cellular composition in samples analyzed. The mammalian brain may contain a higher proportion of glial cells and lower proportion of neurons, compared with Drosophila heads, which may potentially comprise a higher proportion of neurons. Our analysis of RNA-seq data from ES cells differentiated into neurons showed more robust switching to distal 3′ UTR isoforms (Fig. 4D) than observed in the tissue comparisons (Fig. 4B, C).
Challenges for accurate transcript assembly from RNA-seq data
The exponential rise in the throughput of next-generation sequencing has outpaced many aspects of its analysis and interpretation. With respect to the task of assembling transcripts from shards of RNA sequence, solutions such as ERANGE, Cufflinks, and Trinity have proven effective and are widely used. Nevertheless, even current ENCODE project efforts to annotate the human transcriptome, undertaken by the HAVANA and GENCODE subgroups (http://www.sanger.ac.uk/research/projects/vertebrategenome/havana/), acknowledge that substantial manual curation is required to provide confident gene models from RNA-seq data.
Our study highlights the substantial extent to which bioinformatics must advance to fully exploit the power of RNA-seq. Although Cufflinks was invaluable for initial processing of RNA-seq data, we found numerous truncations of clear 3′ UTR extensions evident from visual inspection. A major issue is that current conceptions of the transcriptome do not generally include the broad possibility for large processed exons, yet we show that hundreds of continuous 3′ UTRs ranging from 5–25 kb are encoded by the Drosophila, mouse and human genomes. In general, understanding the connectivity of transcripts from RNA-seq data remains challenging. Major impediments include the highly nonlinear representation of reads across individual transcripts, the difficulty of deconvolving overlapping and/or alternative transcripts of unequal abundance, and the existence of intervening multi-mapping sequences, which can be common in 3′ UTRs that include fragments of repetitive elements. It is hoped that some of these issues may be ameliorated as technologies for direct and/or long read sequencing improve.
We supported the veracity of our computational 3′ UTR inferences with extensive experimental analysis. While >10-kb 3′ UTRs are rare in current annotations and only a handful have been experimentally validated, we provide Northern support for many 3′ UTRs in this size range in this study alone. Notably, in all cases where purported lincRNAs reside in our 3′ UTR extensions, our extensive Northern studies failed to reveal evidence for lincRNA transcripts of annotated lengths. We do not formally exclude that some of these 3′ UTR extensions might be processed into shorter RNAs (Mercer et al. 2010); for example, multiple pathways digest 3′ UTR segments into endo-siRNAs or even piRNAs (Okamura 2012). Nevertheless, the parsimonious interpretation of our studies is that alternative 3′ UTR extensions of stable mRNAs remain a strongly under-appreciated aspect of the mammalian protein-coding transcriptome. This has consequences for interpreting lincRNAs, which are well-documented to exhibit tissue-specific expression, to be conserved, and to be associated with phenotypes when depleted. In fact, all of these are also characteristics of 3′ UTRs, and our studies provide extensive evidence that simply being distant from an annotated gene model is not a reliable predictor of being transcribed independently (Figs. 1, 2, 5). Our studies suggest that additional loci currently annotated as lincRNAs may actually correspond to unannotated 3′ UTR extensions. For example, we provide Northern evidence that lincRNAs described by Mattick and colleagues (Clark et al. 2012) can be detected as stable 3′ UTR extensions of Etv1, Paqr9, and Tcf4 mRNAs (Figs. 1C, 6F).
We are confident that the myriad and unexpected roles for long noncoding RNAs are just beginning to be unraveled (Rinn et al. 2007; Khalil et al. 2009; Gendrel and Heard 2011; Guttman et al. 2011). At the same time, our findings serve as a reminder that establishing transcript connectivity and full-length structures from tiling array and RNA-seq data are not trivial operations, but present and ongoing challenges for transcriptome studies.
Methods
RNA preparation
Adult male ICR (CD-1) mice (Taconic) were euthanized by CO2 overdose. Total RNA from dissected brain samples was extracted using RNeasy lipid tissue kit (Qiagen). Other tissues were extracted using TRIzol (Invitrogen). Poly(A)+ RNA was prepared from total RNA using Oligotex mRNA kit.
Northern analysis and RT-qPCR
For Northern analysis, 1.5–2 μg of poly(A)+ RNA was denatured using glyoxal, and electrophoresis was performed using 1% agarose BPTE gels and blotted and probed as described. Internal size standards were prepared using a mix of probes against several highly expressed genes with known sizes (Supplemental Fig. S4). Note that the migration of the lower bands at 0.8, 1, and 3 kb differ by ∼200 nt from commercially single-stranded RNA ladders (New England Biolabs) due to the presence of endogenous polyA tails.
For cDNA preparation, reverse transcription was performed using superscript III reverse transcriptase (Invitrogen) on DNase I (Ambion) treated total RNA. End-point PCR was performed using Taq DNA polymerase (New England Biolabs) with 55°C–62°C annealing temperatures and 28–35 cycles. To rule out amplification of genomic DNA, first-strand synthesis was performed using control reactions lacking reverse transcriptase (−RT). qPCR was performed using SYBR green PCR mastermix (Qiagen).
Next-generation sequencing data sets analyzed
Mouse RNA-seq data (GSE30617) generated by the Wellcome Trust Sanger Institute included six pooled libraries from the hippocampus, spleen, heart, lung, thymus, and liver (Keane et al. 2011). We also analyzed data from mouse ES cells differentiated into neurons (GSE27866) (Lienert et al. 2011). Wiggle tracks from the mouse ENCODE project were downloaded from the ENCODE DCC (http://hgdownload.cse.ucsc.edu/goldenPath/mm9/encodeDCC/) and merged into a single track (Rosenbloom et al. 2012). Human RNA-seq data were from the Illumina Human Body Map 2.0 Project (GSE30611), which includes nonstranded RNA-seq libraries from 16 tissues, as well as stranded RNA-seq data for a mixture of these tissues. We used 3′-seq data (SRP007359/GSE3019) from the brain, kidney, liver, muscle, and testis for both mouse and human (Derti et al. 2012). All RNA-Seq data were mapped to the human (hg19) and mouse (mm9) genomes using TopHat with the default parameters (Trapnell et al. 2012). The mapping statistics are shown in Supplemental Table S1.
Identification of 3′ UTR extensions
As a first step to identifying candidate 3′ UTR extensions, we ran a sliding window across the genome to identify all continuously transcribed regions. We defined a 100-nt window to be continuously transcribed if more than 80/100 positions were covered by more than 10 reads. This criterion was applied at single-nucleotide increments across the genome, and the overlapping segments were merged. We provisionally made further refinement by merging neighboring regions separated by <150 nt of nonrepetitive sequence. We also merged expressed windows separated by repetitive elements (as identified by RepeatMasker). The resulting segmentation was subjected to extensive filtering that aggressively culled potentially ambiguous extension cases, which might not be distinguished from intragenic transcription, overlapping transcripts, retained introns, etc.
In brief, we identified continuously transcribed regions that overlap annotated stop codons, and subjected them to the following criteria: (1) The identified segment overlaps only a single gene. (2) The extended region does not overlap any exons, only either intronic or intergenic space. (3) The called 3′ UTR overlaps only a single stop codon. (4) There is at least 500 nt of nontranscribed space before the next gene or exon. (5) The gaps bridged between neighboring segments in an extension accounts for <20% of the extension, and these gaps are spanned by at least one paired-end read. (6) The extension is expressed above 1.0 FPKM in mouse and 1.5 FPKM in human. The requirement of 1 FPKM (1.5 FPKM Illumina bodymap data) was selected by comparing the accuracy of the sliding window for annotating known exon boundaries at different FPKM cutoffs and selecting a cutoff with >90% sensitivity within 100 nt. (7) The extension increases current Ensembl v65 gene models by >500 nt. (8) Less than 20% of reads from available stranded libraries derive from the opposite strand. (9) For any extension containing spliced reads, the percentage of reads supporting splicing across either junction account for <20% of reads mapped across that genomic location. For further details, see the Supplemental Methods.
Analysis of polyadenylation site features
Although gene models might bear confidently extended 3′ UTRs, the genomic regions that satisfied expression FPKM cutoffs did not always correspond to a clean 3′ terminus. In cases where lower-level transcription continued for some distance past the called end, our sliding window truncated the 3′ terminus. Alternatively, a called end might terminate in a repetitive element, and thus no specific 3′ end was identified. Such loci, although confidently extended, are not germane for the analysis of 3′ terminal motifs.
Visual inspection of RNA-seq data on the IGV Browser indicated that bioinformatically called 3′ ends were likely to be precise when there was a substantial dropoff in expression emanating from the downstream genomic sequence. We selected those 3′ termini that did not overlap an annotated repetitive element, and exhibited greater than eightfold coverage dropoff between the 100 nt upstream of the annotated 3′ end and both of two subsequent 100-nt windows downstream from the 3′ end.
We centered on each of these novel 3′ termini and analyzed their genomic vicinity for canonical polyadenylation signals (PAS) AAUAAA or its closest variant AUUAAA, as well as noncanonical polyadenylation motifs defined previously (Tian et al. 2005). The U/GU-rich motif was defined as six consecutive U and/or G, with at least three Us (corresponding to 22 distinct 6-mers).
We assessed the positional enrichment of published polyA-seq tags (Derti et al. 2012) around our novel extended 3′ UTRs. We counted the fraction of ends that have at least one 3′ sequencing read in 50-nt bins 200 nt upstream of and downstream from the annotated polyadenylation site. We estimated the background frequency of 3′ sequencing tag enrichment by sampling random points distributed uniformly between the longest Ensembl v65 annotation and the location of our confident 3′ end annotation.
Post-transcriptional regulatory motif analysis
MULTIZ multiple species alignments projected onto the human (hg19) and mouse (mm9) genomes were downloaded from the UCSC Genome Browser. Signal-to-noise ratios of all 7-mers were calculated according to the method previously described (Wang et al. 2008). We considered a 7-mer to be conserved if it was aligned without mismatches or gaps between human, mouse, rat, dog. The conservation frequency was calculated by dividing the number of conserved instances of a 7-mer by the total number of occurrences of that sequence. The signal to background ratio was calculated for each 7-mer by dividing the conservation frequency by the average conservation frequency of a set of at least 10 control sequences. Control sequences exhibited matched GC content and occurred within twofold frequency of the query 7-mer in the selected regions. The 7-mers were cross-referenced with mature miRNA seed sequences from miRBase.
We also downloaded FASTQ files of 130-kD AGO-CLIP data (Chi et al. 2009) from http://ago.rockefeller.edu/rawdata.php, and mapped them to mm9 using Bowtie with default parameters. For each 7-mer matching a miRNA 7m8 target site, the 300 nt upstream of and downstream from that sequence were queried for overlapping Ago-CLIP reads. The fraction of sequences that overlapped a CLIP tag at each nucleotide position relative to the seed match were counted and compared with the background rate of clip frequency around 7-mers with similar GC content. The signal:background ratio was calculated at each nucleotide relative to the target site by dividing the fraction of sequences overlapped by an Ago-CLIP read at each position by the average fraction of Ago-CLIP reads overlapping GC-matched control 7-mers in the same genomic regions.
Third party software used
We used picard liftOver (http://picard.sourceforge.net/) to identify orthologous mouse and human positions, SAM-JDK to query BAM files, apache-commons and R to perform statistical calculations, BigWig (http://code.google.com/p/bigwig/) to parse phastCons conservation scores, and JFreeChart and R for plotting.
Mouse in situ hybridization
E13.5 embryos were fixed in 4% paraformaldehyde overnight. The following day, embryos were washed with PBS, dehydrated, and paraffin embedded. Sagittal 7-μm embryo sections were processed using a Leica microtome. We prepared DIG-labeled antisense RNA probes according to the method previously described (Smibert et al. 2012) and performed in situ hybridization according to the method previously described (Blaess et al. 2011). Probe primer sets are listed in Supplemental Table S9.
Acknowledgments
We thank the many researchers who made their deep sequencing data available for this study. P.M. was supported by a fellowship from the Canadian Institutes of Health Research. S.S. was supported by the Tri-Institutional Training Program in Computational Biology and Medicine. C.A-A. was supported by a NYSTEM post-doctoral fellowship. Work in E.C.L.'s group was supported by the Burroughs Wellcome Fund, the Starr Cancer Consortium (I3-A139), and the NIH (R01-GM083300 and RC2-HG005639).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.146886.112.
References
- An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, Torre ER, Jones KR, Feng Y, Lu B, et al. 2008. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell 134: 175–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Reyes A, Huber W 2012. Detecting differential usage of exons from RNA-seq data. Genome Res 22: 2008–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassi C, Zimmermann C, Mitter R, Fusco S, De Vita S, Saiardi A, Riccio A 2010. An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nat Neurosci 13: 291–301 [DOI] [PubMed] [Google Scholar]
- Blaess S, Bodea GO, Kabanova A, Chanet S, Mugniery E, Derouiche A, Stephen D, Joyner AL 2011. Temporal-spatial changes in Sonic Hedgehog expression and signaling reveal different potentials of ventral mesencephalic progenitors to populate distinct ventral midbrain nuclei. Neural Dev 6: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB 2009. Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps. Nature 460: 479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodroff RA, Goodstadt L, Sirey TM, Oliver PL, Davies KE, Green ED, Molnar Z, Ponting CP 2010. Long noncoding RNA genes: Conservation of sequence and brain expression among diverse amniotes. Genome Biol 11: R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MB, Amaral PP, Schlesinger FJ, Dinger ME, Taft RJ, Rinn JL, Ponting CP, Stadler PF, Morris KV, Morillon A et al. 2011. The reality of pervasive transcription. PLoS Biol 9: e1000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, Dinger ME, Mattick JS 2012. Genome-wide analysis of long noncoding RNA stability. Genome Res 22: 885–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, Babak T 2012. A quantitative atlas of polyadenylation in five mammals. Genome Res 22: 1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. 2012. Ensembl 2012. Nucleic Acids Res 40: D84–D90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel AV, Heard E 2011. Fifty years of X-inactivation research. Development 138: 5049–5055 [DOI] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. 2011. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V, Perry MW, Hendrix D, Stark A, Levine M, Haley B 2011. Neural-specific elongation of 3′ UTRs during Drosophila development. Proc Natl Acad Sci 108: 15864–15869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Tian B 2009. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS ONE 4: e8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, et al. 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477: 289–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. 2009. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci 106: 11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB 2010. RNA processing and its regulation: Global insights into biological networks. Nat Rev Genet 11: 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. 2008. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456: 464–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienert F, Mohn F, Tiwari VK, Baubec T, Roloff TC, Gaidatzis D, Stadler MB, Schubeler D 2011. Genomic prevalence of heterochromatic H3K9me2 and transcription do not discriminate pluripotent from terminally differentiated cells. PLoS Genet 7: e1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manak JR, Dike S, Sementchenko V, Kapranov P, Biemar F, Long J, Cheng J, Bell I, Ghosh S, Piccolboni A, et al. 2006. Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat Genet 38: 1151–1158 [DOI] [PubMed] [Google Scholar]
- Mayr C, Bartel DP 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Bracken CP, Kolle G, Szubert JM, Korbie DJ, Askarian-Amiri ME, Gardiner BB, Goodall GJ, Grimmond SM, et al. 2010. Regulated post-transcriptional RNA cleavage diversifies the eukaryotic transcriptome. Genome Res 20: 1639–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K 2012. Diversity of animal small RNA pathways and their biological utility. RNA 3: 351–368 [DOI] [PubMed] [Google Scholar]
- Pal S, Gupta R, Kim H, Wickramasinghe P, Baubet V, Showe LC, Dahmane N, Davuluri RV 2011. Alternative transcription exceeds alternative splicing in generating the transcriptome diversity of cerebellar development. Genome Res 21: 1260–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponjavic J, Oliver PL, Lunter G, Ponting CP 2009. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet 5: e1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Belgard TG 2010. Transcribed dark matter: Meaning or myth? Hum Mol Genet 19: R162–R168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramskold D, Wang ET, Burge CB, Sandberg R 2009. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol 5: e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, Wong MC, Maddren M, Fang R, Heitner SG, et al. 2012. ENCODE data in the UCSC Genome Browser: Year 5 update. Nucleic Acids Res 41: D56–D63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y 2011. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA 17: 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P, Miura P, Westholm JO, Shenker S, May G, Duff MO, Zhang D, Eads B, Carlson J, Brown JB, et al. 2012. Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Reports 1: 277–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM 2005. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell 123: 1133–1146 [DOI] [PubMed] [Google Scholar]
- Sun AX, Crabtree GR, Yoo AS 2013. MicroRNAs: Regulators of neuronal fate. Curr Opin Cell Biol 25: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS 2005. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res 33: 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H, Nislow C, Blencowe BJ, Hughes TR 2010. Most “dark matter” transcripts are associated with known genes. PLoS Biol 8: e1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudin D, Hanz S, Yoo S, Iavnilovitch E, Willis D, Gradus T, Vuppalanchi D, Segal-Ruder Y, Ben-Yaakov K, Hieda M, et al. 2008. Localized regulation of axonal RanGTPase controls retrograde injury signaling in peripheral nerve. Neuron 59: 241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]