Abstract
Advances in genomics and innovative strategies such as reverse vaccinology have changed the concepts and approaches to vaccine candidate selection and design. Genome mining and blind selection of novel antigens provide a novel route to investigate the mechanisms that underpin pathogenesis. The resulting lists of novel candidates are revealing new aspects of pathogenesis of target organisms, which in turn drives the rational design of optimal vaccine antigens. Here we use the discovery, characterization, and exploitation of fHbp, a vaccine candidate and key virulence factor of meningococcus, as an illustrative case in point. Applying genomic approaches to study both the pathogen and host will ultimately increase our fundamental understanding of pathogen biology, mechanisms responsible for the development of protective immunity, and guide next-generation vaccine design.
Genome-based approaches have led to the discovery of vaccine candidates and mechanisms of bacterial pathogenesis. For example, reverse vaccinology led to discovery of fHbp, a key virulence factor of meningococcus.
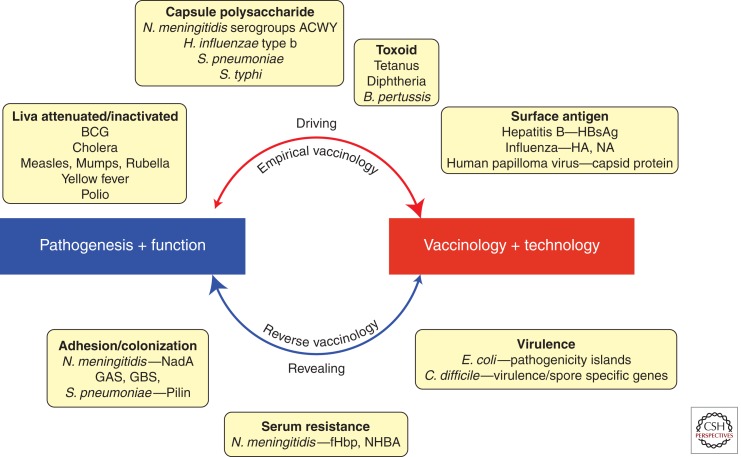
Along with improved sanitation and the discovery and use of antibiotics, vaccination is the intervention that has had the greatest impact on human health and the standard of living in recent history. Vaccines also represent the most cost-effective way of improving health and saving lives (Levine and Lagos 2004). In the past, the development of new vaccines and therapeutics was aided, and largely driven, by increased understanding of the pathogenesis of infectious agents. The antigens used in vaccines do not necessarily have to be virulence factors; however, understanding and interrupting the cycle of infection by directing the immune responses toward key virulence determinants has historically been a successful rationale in eliciting protective immunity. However, the development of vaccines for many pathogens remains elusive, and there is a growing requirement for the fast development of effective vaccines for emerging diseases (Morens et al. 2008). During the last three decades, the vaccine field has been transformed by new technologies such as recombinant DNA and chemical conjugation. More recently, new methods of antigen discovery and design as well as investigation of vaccine responses have been applied, including reverse vaccinology, structural biology, and systems biology (Rinaudo et al. 2009). Genome-based technologies have enabled functionally blind selection of vaccine candidates, and have not only led to the discovery of novel protective antigens but have also revealed new virulence factors of several pathogens. Consequently, the pathogenesis-to-vaccine paradigm has been reversed in several situations, and vaccine development frequently leads to a better understanding of pathogenesis, which has in turn led to novel approaches in studying not only the organism itself but also the strategies for the design of more successful vaccines (Fig. 1). The continuing advancement of genome-based technologies will hopefully lead to new vaccines for unmet diseases, as well as increasingly efficient development pathways for these vaccines (Rappuoli et al. 2011).
Figure 1.
Interactions between microbial pathogenesis and vaccine development. Historically, the understanding of microbial mechanisms of pathogenesis have driven vaccine development, with empirical vaccinology approaches being based on inactivating, attenuating, and/or targeting and interrupting the function of virulence factors, such as capsule polysaccharides, toxins, and other surface proteins. However, with new technologies and vaccine approaches, the focus has shifted to functionally blind antigen discovery based on high-throughput screening of the pathogen’s genome and proteome. These approaches have identified many promising vaccine candidates, which have subsequently been found to be involved in the pathogenic process, such as colonization of the host or serum resistance.
HOW OUR UNDERSTANDING OF BACTERIAL PATHOGENESIS HAS LED TO NEW VACCINES
Since the first observations of Jenner at the turn of the nineteenth century, that milkmaids exposed to cowpox appeared to be protected from the more severe human smallpox disease, the understanding of pathogenesis has been fundamental to the development of vaccines. Once it became evident in the early 1800s that diseases were caused by microbes, Louis Pasteur started the rational development of vaccines and established the basic rules of vaccinology. Most of the vaccines licensed to date have been developed based on Pasteur’s principles of “isolate, inactivate and inject” the causative agent of disease (Plotkin 2009). One could say that they have been developed empirically, with little or no understanding of the complex immunological mechanisms by which they induce protective immunity. However, the strategy was based on a unifying rationale, that by reducing the virulence of the disease-causing organism or by inhibiting its ability to replicate, the consequent attenuation or inactivation robs infectious organisms of their pathogenicity while preserving their immunogenicity. The mechanism of action of vaccines relies entirely on activation of the human body’s own protective immunological mechanisms, and live-attenuated or inactivated vaccines basically mimic the kind of protective immunity induced in people who survive a normal infection.
Pasteur applied the principles of “isolate, inactivate and inject” in the late 1800s to both viruses (dried rabies virus isolated from rabbits) and bacteria (heat-inactivated anthrax bacilli). These strategies have been successful for those diseases that result in natural immunity to reinfection, in particular for many viral diseases that have invariant antigens (e.g., smallpox, yellow fever, polio, rabies, mumps, measles, rubella, varicella, herpes zoster, hepatitis A, and Japanese encephalitis) (Rappuoli 2007). Furthermore, inactivated or live-attenuated vaccines against some bacterial infectious diseases have also been developed with varying degrees of success; for instance, Bacille Calmette-Guerin (BCG; live-attenuated Mycobacterium bovis) is possibly the most widely used vaccine worldwide and currently the only vaccine available against tuberculosis (Parida and Kaufmann 2010). Whole-cell vaccines against enteric diseases such as typhoid and cholera, and a whole-cell pertussis vaccine are currently licensed and in use, although there is a general need and trend to replace these with newer improved vaccines.
A further development to Pasteur’s strategy was undertaken when essential components of the organism, usually virulence determinants, were isolated and inactivated, for use as “subunit” vaccines. This approach was first used for diphtheria and tetanus in which the pathology of the disease is largely owing to the toxins produced by the bacterial pathogen, and initial toxoid vaccines were chemically inactivated toxins isolated from the bacteria (Glenny and Hopkins 1923; Ramon 1924). Similarly, capsule polysaccharides of Gram-negative bacteria are common virulence factors and subunit vaccines based on purified polysaccharides were initially developed for Streptococcus pneumoniae in the 1940s (MacLeod et al. 1945), Neisseria meningitidis in the 1960s (Gotschlich et al. 1969), Haemophilus influenzae in the 1970s (Peltola et al. 1977), and Salmonella typhi Vi in the 1980s (Acharya et al. 1987). With advances in new technologies, such as recombinant DNA and chemical conjugation of proteins to polysaccharides, subunit vaccines have advanced notably and are able to provide improved memory responses and increased protection in infants. These vaccines are safe and efficacious, and have also been developed in multivalent forms to provide protection against numerous pneumococcal serotypes and meningococcal serogroups. For the three major causes of bacteremic disease—H. influenzae type b (Hib), pneumococci, and meningococci—the use of polysaccharide conjugate vaccines has led to successful control of invasive diseases caused by many serogroups in various countries worldwide (Kelly et al. 2004; Gill et al. 2010; Jefferies et al. 2011).
Genetic inactivation by targeted mutagenesis as well as modern recombinant techniques for the expression and purification of protein-based subunit vaccines have led to the development of safer live-attenuated vaccines and subunit vaccines through rational design. For example, recombinant DNA technology enabled the generation of the detoxified Bordetella pertussis toxin (Pizza et al. 1989), and the recombinant hepatitis B surface antigen (HBsAg) (Buynak et al. 1976; Ellis 1990). Ultimately, recombinant DNA technology has formed the basis of a wide range of new platforms for vaccine design and delivery.
Subunit vaccine antigens are usually better tolerated than inactivated or live-attenuated pathogens, and this has led to a push for the development of new and improved vaccines using recombinant techniques. Acellular pertussis combination vaccines, containing between one and five pertussis virulence factors are substantially less reactogenic than the original whole-cell vaccines, and have increasingly replaced whole-cell-containing vaccines in developed countries (Kitchin 2011). However, a limitation of subunit vaccines is that they are generally less immunogenic and often require the addition of an adjuvant to elicit protective immune responses (Mbow et al. 2010). Unfortunately, few adjuvants are currently licensed and widely used (e.g., aluminum salts, Alum, that have been used for more than 80 years). However, newer adjuvants (e.g., MF59 [O’Hagan et al. 2011] and AS04 [Garcon et al. 2011]) have been developed, each of which have specific properties designed to enhance the magnitude and quality of the immune response to target specific diseases.
Although the virtual disappearance of diseases like diphtheria, tetanus, pertussis, and invasive H. influenzae B are landmarks to the success of vaccines, there still remain many bacterial diseases for which vaccination strategies have so far been unsuccessful. Often this is owing to the hypervariability of their antigens, an incomplete understanding of pathogenic mechanisms, and/or a need for cell-mediated immune responses (Rappuoli 2007). For instance, for those diseases that do not induce immunity after natural infection (e.g., malaria, or respiratory syncytial virus [RSV]) or those that cause persistent or latent infection (e.g., HIV and hepatitis C virus), a vaccine-induced protective immune response must go one step beyond the mechanisms that nature has evolved and vaccine development must go beyond the traditional empirical methodologies.
REVERSING THE PARADIGM: REVERSE VACCINOLOGY APPROACHES LEAD TO INCREASED UNDERSTANDING OF BACTERIAL PATHOGENESIS
With the arrival of whole-genome sequencing, genome-based antigen selection has played a major role in antigen discovery and vaccine design. One approach that has been used to mine pathogenic bacterial genomes has been coined “Reverse vaccinology,” and allows the investigation of the complete potential antigenic repertoire of an organism from its genome sequence. Reverse vaccinology involves the cloning and expression of all the proteins in an organism’s genome sequence that are predicted in silico to be surface exposed or secreted. Then each protein is screened, through high-throughput immunization, for their ability to elicit antibodies in mice that can kill or neutralize the target organism. The first pathogen addressed using reverse vaccinology was N. meningitidis serogroup B (MenB) (Pizza et al. 2000), for which no broadly protective vaccine exists owing to the similarity of its capsular polysaccharide to a self-antigen and the hypervariability of its major outer membrane protein antigens. This approach led to the identification of 29 novel antigens that can elicit bactericidal antibodies against the pathogen in vitro. After successful preclinical studies, the vaccine deriving from the genome approach entered phase I testing in 2002 (Giuliani et al. 2006). In November 2012, the European Medicinal Agency recommended the granting of a marketing authorization for Bexsero, the first vaccine to provide broad coverage against meningococcal serogroup B. This was followed by the approval of the European Commission in January 2013. The three main antigens identified by reverse vaccinology that are formulated as components of the MenB vaccine, Neisserial heparin-binding antigen (NHBA), factor H-binding protein (fHbp), and N. meningitidis adhesin A (NadA), have all subsequently been implicated as playing important roles in meningococcal virulence (Capecchi et al. 2005; Madico et al. 2006; Schneider et al. 2006; Serruto et al. 2010) (see below for more details on fHbp).
Since the first application of reverse vaccinology, based on the single genome of a MenB strain (Pizza et al. 2000), various advancements have been made to this genomic approach. A multigenome or pan-genome reverse vaccinology approach was applied to Group B streptococcus (GBS) to identify antigens from the extended gene repertoire of the species rather than from a single organism (Maione et al. 2005), and the subtractive reverse vaccinology approach has been used to identify antigens present in pathogenic but not commensal strains of Escherichia coli (Rasko et al. 2008; Moriel et al. 2010). Reverse vaccinology has been applied to a wide range of bacterial pathogens and has provided a long list of promising antigens from functionally blind interrogation of their genomes, and the subsequent studies on antigen function are leading to increased understanding of the biology of the pathogens (Fig. 1; Table 1).
Table 1.
Microbial virulence factors identified during vaccine development
| Organism | Disease | Vaccine approach/genomic-based technology | Antigen/virulence factor | Role | References |
|---|---|---|---|---|---|
| Neisseria meningitidis | Meningococcal meningitis and sepsis | Reverse Vaccinology: genome analysis of serogroup B strain Structural Vaccinology: chimeric fHbp engineered based on variant 1 backbone-containing epitopes from variants 2 and 3 |
Factor H-binding protein (fHbp) | Binding of complement factor H (fH), serum resistance | Pizza et al. 2000, 2008; Giuliani et al. 2006; Madico et al. 2006; Scarselli et al. 2011 |
| Reverse Vaccinology: genome analysis of serogroup B strain | Neisserial adhesin A (NadA) | Adhesion, colonization | Comanducci et al. 2002; Capecchi et al. 2005 | ||
| Reverse Vaccinology: genome analysis of serogroup B strain | Neisserial heparin-binding antigen (NHBA) | Binding of heparin, serum resistance | Serruto et al. 2010 | ||
| Group B Streptococcus (Streptococcus agalactiae) | Sepsis, pneumonia, and neonatal meningitis | Pan-Genome Reverse Vaccinology: genome analysis of 8 GBS strains Comparative Genomics and Genome Hybridization: Streptococcus pneumoniae, GAS genomes, 19 GBS strains |
Pili (BP-1, BP-2a, AP-1, AP-2a, GBS67) | Adhesion, colonization, biofilm formation, translocation through epithelial cells | Lauer et al. 2005; Maione et al. 2005; Tettelin et al. 2005; Dramsi et al. 2006; Rosini et al. 2006; Pezzicoli et al. 2008; Margarit et al. 2009; Rinaudo et al. 2010; Nuccitelli et al. 2011 |
| Group A Streptococcus (Streptococcus pyogenes) | Pharyngitis, necrotizing faciitis, pneumonia, sepsis, arthritis, myositis | Pan-Genome Reverse Vaccinology/Comparative Genomics: analysis of five GAS strains for pilus encoding (LPXTG motif) genes | Pili (Lancefield antigens) | Adhesion, colonization, biofilm formation | Mora et al. 2005; Abbot et al. 2007; Manetti et al. 2007 |
| Streptococcus pneumoniae | Pneumonia, meningitis, otitis media, sepsis | Reverse Vaccinology/Comparative Genomics: analysis of T4 and INV104 genomes for putative virulence factors and pilus genes | Pili (RrgB, RrgA, RrgC encoded by rlrA pathogenicity Islet; PitB, pilus islet 2) | Adhesion, colonization, biofilm formation | Hava and Camilli 2002; Barocchi et al. 2006; LeMieux et al. 2006; Bagnoli et al. 2008; Hilleringmann et al. 2009; Donati et al. 2010 |
| Escherichia coli | Intestinal (diarrhea) and extraintestinal (urinary tract infection, sepsis, meningitis, hemolytic-uremic syndrome) disease | Comparative/Subtractive Reverse Vaccinology: comparison of genomes of commensal and pathogenic strains Proteomics: analysis of OMV proteins |
Pathogenicity islands, pathogen-specific OMPs (e.g., ECOK1_0290) | Adhesion, colonization | Berlanda Scorza et al. 2008; Moriel et al. 2010; Nesta et al. 2012 |
| Staphylococcus aureus | Nosocomial, skin, wound infections, endocarditis, pneumonia, bacteremia, osteomyelitis, toxic shock syndrome | Reverse Vaccinology: genome analysis of eight strains for the presence of sortase substrate genes (based on loss of pathogenesis of sortase mutant) Comparative Genomics: genome analysis of 58 strains for variation of putative surface proteins |
Conserved virulence-associated surface proteins (e.g., IsdA, IsdB, SdrD, SdrE) | Pathogenesis in murine renal abscess model | Mazmanian et al. 2000; Stranger-Jones et al. 2006; McCarthy and Lindsay 2010 |
| Clostridium difficile | Nosocomial diarrhea | Comparative Genomics: genome analysis of historic nonepidemic and newly emerged hypervirulent epidemic strains, and evolutionary dynamics of 30 isolates Proteomics: investigation of core spore gene, spore–host interactions |
Genetic regions specific for epidemic strain (e.g., flagella biosynthesis and glycosylation proteins) 29 C. difficile spore specific proteins | Motility, autoagglutination, sporulation | Stabler et al. 2006, 2009; Lawley et al. 2009; He et al. 2010 |
| Chlamydia trachomatis | Most prevalent sexually transmitted infection worldwide (causes ectopic pregnancy, infertility), trachoma | Reverse Vaccinology: genome analysis Comparative Genomics: genome analysis of five chlamydial species Proteomics: investigation of elementary bodies |
Outer membrane complex and elementary body proteins | Host–pathogen interactions | Montigiani et al. 2002; Thorpe et al. 2007; Heinz et al. 2009; Liu et al. 2010 |
OMV, outer membrane vesicle; OMPs, outer membrane proteins.
Although a vaccine antigen does not have to be a virulence factor, focusing on proteins with a crucial function in bacterial pathogenesis or survival is a useful selection criterion, because antibodies elicited to these antigens may block their function and result in neutralization. For example, the use of protective antigens involved in adhesion is particularly attractive, as there may be important implications for providing herd immunity through reduction of the pathogen’s colonization and circulation. Indeed, functional studies of many protective antigens have subsequently revealed them to be novel adhesins involved in colonization of the pathogen (Table 1) (Capecchi et al. 2005; Rosini et al. 2006). The pili of gram-negative bacteria are well-described virulence factors, however, little was known about pili in gram-positive bacteria before their discovery as a result of reverse vaccinology on GBS (Lauer et al. 2005; Maione et al. 2005), and subsequent studies on Streptococcus pyogenes, and S. pneumoniae (reviewed in Telford et al. 2006). The presence of pili that contain protective antigens in all three principal streptococcal pathogens indicates that these structures play an important role in virulence. Genome-based comparison of pathogenic and nonpathogenic strains has also been applied to enable vaccine targets to be identified on the basis of proteins that are specifically present in pathogenic strains and therefore implicated in virulence. Recently, subtractive reverse vaccinology has been applied to E. coli and five out of the nine protective antigens identified are located on putative pathogenicity islands specific for pathogenic E. coli strains (Moriel et al. 2010).
A CASE STUDY OF HOW VACCINE DEVELOPMENT AND UNDERSTANDING OF BACTERIAL PATHOGENESIS ARE LINKED: DISCOVERY, CHARACTERIZATION, AND EXPLOITATION OF fHbp
Reverse vaccinology, based on functionally blind selection of candidate antigens from genome screening, led to the discovery and characterization of factor H-binding protein (fHbp), which is a novel protective antigen and virulence determinant of meningococcal infection. The continuing study of fHbp has led to significant advances in the understanding of meningococcal pathogenesis, contributed to the development of improved animal models for N. meningitidis, and provided greater clarity for host susceptibility studies, which has in turn led to novel approaches in studying the organism itself as well as the design of more successful vaccines (Fig. 2).
Figure 2.
The meningococcal fHbp antigen as an example of how vaccine development has led to an increased understanding of pathogenesis. Genome-derived Neisseria antigen 1870 (GNA1870) was identified as a novel vaccine antigen from the genome sequence of N. meningitidis by reverse vaccinology. Since then it has been studied extensively for vaccine development (top panel) in terms of its distribution and sequence variation, as well as its ability to induce cross-protective bactericidal antibodies. This information has also been used to engineer meningococcal strains overexpressing fHbp for outer membrane vesicle (OMV) vaccines. In addition, structural studies have been used for epitope mapping and the generation of a chimeric fHbp antigen that is able to induce broad cross protection. Parallel to this work, the understanding of meningococcal pathogenesis has greatly advanced by studying this antigen (bottom panel). GNA1870 was renamed as factor H-binding protein (fHbp) owing to the discovery of its functional role in binding human factor H (fH), which increases serum resistance. This role has also led to increased understanding of meningococcal host specificity and the development of transgenic mice models expressing human fH. Furthermore, human fH alleles have been identified that increase host susceptibility to meningococcal disease.
Identification of fHbp as a Vaccine Antigen
fHbp was initially identified as the genome-derived Neisseria antigen 1870 (GNA1870), and was described as a Neisseria-specific putative surface lipoprotein of unknown function (Pizza et al. 2000). An unrelated vaccine discovery project also identified this antigen (referred to as lipoprotein [LP] 2086) using a proteomic approach to analyze soluble outer membrane proteins (Fletcher et al. 2004). fHbp induces high levels of bactericidal antibodies in mice (Fletcher et al. 2004; Giuliani et al. 2006) and humans (Jacobsson et al. 2009; Plested et al. 2009; Findlow et al. 2010; Giuliani et al. 2010; Halperin et al. 2010; Toneatto et al. 2011a,b). Protection against meningococcal disease correlates with the level of bactericidal antibodies in the serum bactericidal activity (SBA) assay (Borrow et al. 2006; Frasch et al. 2009), indicating that fHbp is an important vaccine antigen. Antibodies to fHbp also confer passive protection in the infant rat model of meningococcal bacteremia (Masignani et al. 2003). Development of fHbp as a vaccine antigen has greatly progressed and it is currently a component of the Novartis multivalent 4CMenB vaccine (Giuliani et al. 2006) that was submitted for licensure to the European Medicines Agency in 2010. Another vaccine contains two alleles of the recombinant fHbp protein (rLP2086) and is in late stages of development by Pfizer (Fletcher et al. 2004). In another strategy for the production of outer membrane vesicle (OMV)-based vaccines, meningococcal strains have been engineered to overexpress variants of fHbp, which has shown to provide broad protection against several MenB strains (Keiser et al. 2011; Koeberling et al. 2011a,b).
fHbp Function and Role in Pathogenesis
Before its identification as a vaccine candidate the function of fHbp was unknown. Two independent laboratories subsequently found that this antigen binds human complement factor H (fH) (Madico et al. 2006; Schneider et al. 2006), which is a key regulatory protein that acts as an inhibitor of the alternative complement pathway (reviewed in Meri et al. 2008). fHbp is an important meningococcal survival factor in human serum and blood (Madico et al. 2006; Welsch et al. 2008; Seib et al. 2009), because recruitment of fH to the bacterial surface via fHbp binding enables the pathogen to evade alternative complement-mediated killing by the host innate immune system. This has implications for fHbp not only as a virulence factor in vivo but also as a target antigen for vaccination as interestingly, anti-fHbp antibodies can also increase the susceptibility of the bacterium to killing by the alternative complement pathway by blocking fH binding to the bacteria (Madico et al. 2006). Additional functions for fHbp have also been proposed, including resistance to the antimicrobial LL-37 (Seib et al. 2009) and binding of the siderophore enterobactin (Veggi et al. 2012). Investigation of fHbp transcription and regulation revealed that fHbp expression is induced under oxygen limitation and implicates an important role in oxygen-poor microenvironments, such as the submucosa or intracellular locations (Bartolini et al. 2006; Oriente et al. 2010).
Meningococcal Epidemiology and Typing
fHbp is expressed by almost all N. meningitidis strains studied to date, although levels of expression vary between isolates (Masignani et al. 2003; Fletcher et al. 2004; McNeil et al. 2009; Murphy et al. 2009). Furthermore, a significant degree of fHbp sequence variability exists in MenB strains circulating globally, and sequencing of fHbp in ∼2000 MenB strains led to the identification of ∼300 subvariant polypeptides (Bambini et al. 2009; Murphy et al. 2009). Different sequence variants can be classified into three groups (variants 1, 2, and 3) (Masignani et al. 2003; Brehony et al. 2009) or two subfamilies (A and B: corresponding to variants 2/3 and 1, respectively) (Fletcher et al. 2004; McNeil et al. 2009; Murphy et al. 2009; Jiang et al. 2010) that do not induce cross-protective immunity. However, all variants studied to date are able to bind fH, mediate serum resistance, and induce bactericidal antibodies (Dunphy et al. 2011; Seib et al. 2011). Members of the variant 1, 2, and 3 families are present in 65%, 25%, and 10% of the MenB global population, respectively (Beernink et al. 2007; Bambini et al. 2009; Murphy et al. 2009).
Analysis of the 4CMenB vaccine antigens fHbp, NadA, and NHBA variant distribution in meningococcal population (Bambini et al. 2009) indicated that vaccine coverage could not be predicted on the basis of multilocus sequence typing (MLST), the gold standard for meningococcal classification (Brehony et al. 2007). As a result of this, a high-throughput meningococcal antigen typing system (MATS) was developed based on a unique vaccine antigen-specific enzyme-linked immunosorbent assay (ELISA), which measures both immunologic cross reactivity and quantity of fHbp, NHBA, and NadA, and correlates with bactericidal activity (Donnelly et al. 2010). MATS analysis of large panels of strains has predicted that adults and infants immunized with the 4CMenB vaccine would be protected from disease caused by 86% and 77% of the diverse MenB strains circulating globally, respectively. This typing approach could also be applied for vaccines against other variable pathogens, including nontypable H. influenzae, Streptococcus groups A and B, and S. pneumoniae.
fHbp and Structural Vaccinology
Analysis of the three-dimensional structure of fHbp, by nuclear magnetic resonance (NMR) of the carboxy-terminal portion (Cantini et al. 2009) and the crystal structure of the entire fHbp protein in complex with a fragment of fH (Schneider et al. 2009) has led to identification of residues important for binding of bactericidal antibodies and fH, respectively. Using this data a structural vaccinology approach has enabled the rational design of a chimeric fHbp antigen, which combines the antigenic repertoire of the three major fHbp variant groups into a single molecule, using the variant 1 scaffold carrying patches of amino acids from the surfaces of variants 2 and 3. The chimeric fHbp protein G1 is capable of inducing antibodies that are bactericidal against a diverse panel of strains of meningococcus B, suggesting that it could be used to produce a broadly protective vaccine (Scarselli et al. 2011).
Understanding fHbp-fH Binding Leads to Better Animal Models and Improved Vaccine Candidates
Meningococcus is an obligate human pathogen that has no known reservoir outside the human host, and one of the biggest limitations in studying the pathogenesis of the disease is the lack of good animal models of infection. The finding that fHbp specifically binds human fH, but not rat or mouse fH, provided a key element to understanding meningococcal species specificity (Granoff et al. 2009). Indeed, the administration of human fH to infant rats challenged with MenB greatly increased meningococcal survival (>10-fold) (Granoff et al. 2009). Recently, a human fH transgenic mouse model has been developed by microinjection of mouse embryos with human fH cDNA, leading to concentrations of human fH in the sera of the transgenic mice comparable to that seen in human sera (Beernink et al. 2011). This model was used to test the immunogenicity of an fHbp mutant with a single amino acid substitution, R41S, that is no longer able to bind human fH, to investigate whether epitopes may be obscured when human fH is bound to the wild-type fHbp vaccine given to humans. The fHbp R41S mutant induced higher serum bactericidal antibody responses than the wild-type fHbp and these antibodies had increased ability to block fH binding to wild-type fHbp (Beernink et al. 2011).
Host SNPs and fH Levels Are Involved in Susceptibility to Meningococcal Diseases
The increased incidence and recurrence of infection and disease in people with deficiencies (Nielsen et al. 1989) or overproduction of fH (Haralambous et al. 2006) indicated the importance of host fH in controlling meningococcal disease. A single-nucleotide polymorphism (SNP; C-496T C/C) in the promoter region of the gene encoding fH (CFH) is associated with slightly increased fH concentrations, reduced bactericidal activity against meningococci, and an increased risk of disease (Haralambous et al. 2006). Genome-wide association studies have also revealed a set of strongly correlated SNPs in CFH and the gene encoding CFH-related protein 3 (CFHR3) that are associated with decreased susceptibility to meningococcal disease for minor allele carriers. Interestingly, the protective alleles of the identified SNPs are rare in Africa, suggesting that these variants may play a role in the increased disease rate seen in the meningitis belt (Davila et al. 2010). However, it has been highlighted that association studies typically do not provide sufficient coverage of African populations, although new tools such as the “The 1000 Genomes Project” may help gain a better understanding of the link between host fH polymorphisms and meningococcal disease in high-risk African populations (de Bakker and Telenti 2010).
In conclusion, the discovery of fHbp through the reverse vaccinology approach and its characterization and study as a vaccine antigen has considerably advanced the understanding of meningococcal pathogenesis, and furthermore, this enhanced knowledge has led to advancements in the use of fHbp as a vaccine candidate (Fig. 2).
THE WAY AHEAD FOR VACCINOLOGY
The use of reverse vaccinology has enabled identification of numerous promising vaccine candidates against meningococcus, GBS, group A streptococcus, pneumococcus, pathogenic E. coli, and also for antibiotic-resistant bacteria such as Staphylococcus aureus (Table 1). The subsequent challenge is the refinement of these long lists of new antigens, enabling detection of the best candidate antigens and formulations to take forward for clinical development. This selection of the best candidates will largely depend on the use of novel approaches, such as structural vaccinology to engineer optimal antigens, and systems biology to gain a better understanding of signatures of immunogenicity and correlates of protection.
STRUCTURAL VACCINOLOGY
Although the mining of bacterial genome sequences has identified excellent targets that could be used in a vaccine, the sequence variability of antigens is often a major challenge to their use and development as broadly protective vaccine components. Traditionally, vaccines have been developed using natural antigens, however, rational optimization of antigens could improve their properties as immunogens by combining, exposing, and/or improving the immunogenicity of epitopes. After all, native surface proteins of pathogens have adapted to evade, not induce, immunity (Dormitzer et al. 2008).
Structural biological studies enable the atomic resolution of an antigen, and structure-based design allows the engineering of multiple immunodominant epitopes in one molecule to induce broad immune responses against different protein variants. Analogous to three musketeers, who together make a formidable team (all for one), one can combine all immunogenic epitopes of non-cross-reacting variants of an antigen into a rationally designed individual molecular champion (one for all), as achieved for the meningococcal fHbp (Scarselli et al. 2011) and a group B streptococcal pilus antigen (Nuccitelli et al. 2011). Structure-based antigen design has also been used in HIV vaccine development to render conserved epitopes immunogenic (Tobin et al. 2008; Burton 2010). Furthermore, stabilizing the structure of difficult antigens can improve their value as vaccine components, by optimizing their efficient production and storage stability.
SYSTEMS VACCINOLOGY
Systems biology is an interdisciplinary approach that systematically investigates the structure and complex interactions between all parts of a biological system with the ultimate goal of predicting the behavior of the system (Kitano 2002). A systems-based approach is increasingly being applied to vaccinology with two main goals in mind, first to understand the mechanisms by which vaccines stimulate protective immunity, and secondly to identify markers or molecular signatures that can be used to predict the immunogenicity or efficacy of vaccines (Pulendran et al. 2010; Nakaya et al. 2011).
Systems vaccinology, which relies on next-generation sequencing and postgenomic technologies for the high-throughput acquisition and analysis of data, can be used to define and monitor the global architecture of the human immune response and the changes that occur following vaccination. For example, analysis of vaccine-induced changes in blood, including patterns of global gene expression or alterations in immune cells, with respect to several immunological parameters, has successfully been used to identify molecular signatures induced early after vaccination with the live-attenuated yellow fever vaccine (YF-17D) that could be used to predict immunogenicity of the vaccine (Gaucher et al. 2008; Querec et al. 2009). Such signatures of immunogenicity, when identified, would also allow early identification of unsafe vaccine candidates and guide selection of the most effective vaccine composition, in terms of optimal candidate combinations, formulations, delivery systems, and immunization schemes, thus bridging the gap between antigen discovery and clinical research. Furthermore, such signatures would enable monitoring of suboptimal immune responses in certain individuals (e.g., elderly, infants, or immunocompromised populations) in whom vaccination may not confer protective immunity and may remain at risk of infection (Nakaya et al. 2011).
Other relevant data that can be gained from this field of study is the characterization of human genetic variation, at the individual or population level, and how this impacts on vaccine success. The application of state-of-the-art next-generation sequencing technology and tools that allow high-throughput detection of gene variations can provide genome-wide information, which when integrated with immune-response signature profiles will further fill the gap in our understanding of the relationship between the phenotype–genotype of an immune response. A large and growing family of polymorphisms in genes of the immune system have been identified that lead to significant variations in immune responses and that are critical to vaccine efficacy (Poland et al. 2007, 2009; Haralambieva and Poland 2010). For example, many studies have revealed associations with human leukocyte antigen (HLA) haplotypes and immune outcomes after vaccination with the MMR (measles, mumps, and rubella) vaccine (Ovsyannikova et al. 2007) hepatitis B vaccine (Kruskall et al. 1992) and the rubella vaccine (Dhiman et al. 2010). This raises the notion of “personalized vaccinology” in which a prevaccination genome-wide screen would allow individualized vaccination approaches that can be tailored and guided by the genetic uniqueness of each patient, maximizing immunogenicity and minimizing the risk of either vaccine failure or adverse events (Poland et al. 2008; Dhiman et al. 2009).
Systems vaccinology can be used to define the signatures of protection that are required to elicit a protective immune response, which will increase our knowledge of the mechanisms of action of currently successful vaccines, as well as enable the improved identification, rational design, and testing of novel vaccine antigens (Pulendran et al. 2010; Bernstein et al. 2011).
CONCLUSIONS
With the growing problem of antimicrobial resistance, and newly emerging or reemerging pathogens (for review see Morens et al. 2008), we are increasingly looking to the development and use of vaccines to control infectious diseases. Furthermore, new and improved vaccines are needed to replace several vaccines that are suboptimal in terms of efficacy or safety. Advancing technologies continue to transform the field of vaccinology, and we are now able to use genomic-based approaches to aid selection of vaccine candidates and structure-based design to optimize the chosen immunogens. In parallel, the increasing use of systems biology will provide essential insights into the immune response elicited by vaccines, and should help identify signatures of immunogenicity and correlates of protection. This improved understanding of both the host and the pathogen is aiding the development of new vaccine technologies, including the use of small molecule adjuvants to target specific immune responses, as well as new delivery systems and immunization schemes to optimize vaccine efficacy, which are essential components for the next generation of vaccine development (Fig. 3). The knowledge gained through systems biology will also increase fundamental understanding of microbial pathogenesis, enabling continued advancements in vaccine development.
Figure 3.
Key areas of next-generation vaccine development. High-throughput (HTP) DNA sequencing and screening technologies are leading to a better understanding of the genetic variation of both pathogens and the human host, which in turn is improving the fundamental understanding of the pathogen and the immune response, respectively. This information can be exploited to enable identification of protective antigens from the pathogen as well as the signatures in the host immune response that lead to protection. These two parallel fields of vaccinology, combined with improving vaccine formulation and delivery technologies, are key components of next-generation vaccine development.
ACKNOWLEDGMENTS
We thank Giorgio Corsi for artwork.
Footnotes
Editors: Pascale Cossart and Stanley Maloy
Additional Perspectives on Bacterial Pathogenesis available at www.perspectivesinmedicine.org
REFERENCES
- Abbot EL, Smith WD, Siou GP, Chiriboga C, Smith RJ, Wilson JA, Hirst BH, Kehoe MA 2007. Pili mediate specific adhesion of Streptococcus pyogenes to human tonsil and skin. Cell Microbiol 9: 1822–1833 [DOI] [PubMed] [Google Scholar]
- Acharya IL, Lowe CU, Thapa R, Gurubacharya VL, Shrestha MB, Cadoz M, Schulz D, Armand J, Bryla DA, Trollfors B, et al. 1987. Prevention of typhoid fever in Nepal with the Vi capsular polysaccharide of Salmonella typhi. A preliminary report. N Engl J Med 317: 1101–1104 [DOI] [PubMed] [Google Scholar]
- Bagnoli F, Moschioni M, Donati C, Dimitrovska V, Ferlenghi I, Facciotti C, Muzzi A, Giusti F, Emolo C, Sinisi A, et al. 2008. A second pilus type in Streptococcus pneumoniae is prevalent in emerging serotypes and mediates adhesion to host cells. J Bacteriol 190: 5480–5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bambini S, Muzzi A, Olcen P, Rappuoli R, Pizza M, Comanducci M 2009. Distribution and genetic variability of three vaccine components in a panel of strains representative of the diversity of serogroup B meningococcus. Vaccine 27: 2794–2803 [DOI] [PubMed] [Google Scholar]
- Barocchi MA, Ries J, Zogaj X, Hemsley C, Albiger B, Kanth A, Dahlberg S, Fernebro J, Moschioni M, Masignani V, et al. 2006. A pneumococcal pilus influences virulence and host inflammatory responses. Proc Natl Acad Sci 103: 2857–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolini E, Frigimelica E, Giovinazzi S, Galli G, Shaik Y, Genco C, Welsch JA, Granoff DM, Grandi G, Grifantini R 2006. Role of FNR and FNR-regulated, sugar fermentation genes in Neisseria meningitidis infection. Mol Microbiol 60: 963–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beernink PT, Welsch JA, Harrison LH, Leipus A, Kaplan SL, Granoff DM 2007. Prevalence of factor H-binding protein variants and NadA among meningococcal group B isolates from the United States: Implications for the development of a multicomponent group B vaccine. J Infect Dis 195: 1472–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beernink PT, Shaughnessy J, Braga EM, Liu Q, Rice PA, Ram S, Granoff DM 2011. A meningococcal factor H binding protein mutant that eliminates factor H binding enhances protective antibody responses to vaccination. J Immunol 186: 3606–3614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlanda Scorza F, Doro F, Rodriguez-Ortega MJ, Stella M, Liberatori S, Taddei AR, Serino L, Gomes Moriel D, Nesta B, Fontana MR, et al. 2008. Proteomics characterization of outer membrane vesicles from the extraintestinal pathogenic Escherichia coli ΔtolR IHE3034 mutant. Mol Cell Proteomics 7: 473–485 [DOI] [PubMed] [Google Scholar]
- Bernstein A, Pulendran B, Rappuoli R 2011. Systems vaccinomics: The road ahead for vaccinology. OMICS 15: 529–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrow R, Carlone GM, Rosenstein N, Blake M, Feavers I, Martin D, Zollinger W, Robbins J, Aaberge I, Granoff DM, et al. 2006. Neisseria meningitidis group B correlates of protection and assay standardization—International meeting report Emory University, Atlanta, Georgia, United States, 16–17 March 2005. Vaccine 24: 5093–5107 [DOI] [PubMed] [Google Scholar]
- Brehony C, Jolley KA, Maiden MC 2007. Multilocus sequence typing for global surveillance of meningococcal disease. FEMS Microbiol Rev 31: 15–26 [DOI] [PubMed] [Google Scholar]
- Brehony C, Wilson DJ, Maiden MC 2009. Variation of the factor H-binding protein of Neisseria meningitidis. Microbiology 155: 4155–4169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR 2010. Scaffolding to build a rational vaccine design strategy. Proc Natl Acad Sci 107: 17859–17860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buynak EB, Roehm RR, Tytell AA, Bertland AU II, Lampson GP, Hilleman MR 1976. Vaccine against human hepatitis B. JAMA 235: 2832–2834 [PubMed] [Google Scholar]
- Cantini F, Veggi D, Dragonetti S, Savino S, Scarselli M, Romagnoli G, Pizza M, Banci L, Rappuoli R 2009. Solution structure of the factor H binding protein, a survival factor and protective antigen of Neisseria meningitidis. J Biol Chem 284: 9022–9026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi B, Adu-Bobie J, Di Marcello F, Ciucchi L, Masignani V, Taddei A, Rappuoli R, Pizza M, Arico B 2005. Neisseria meningitidis NadA is a new invasin which promotes bacterial adhesion to and penetration into human epithelial cells. Mol Microbiol 55: 687–698 [DOI] [PubMed] [Google Scholar]
- Comanducci M, Bambini S, Brunelli B, Adu-Bobie J, Arico B, Capecchi B, Giuliani MM, Masignani V, Santini L, Savino S, et al. 2002. NadA, a novel vaccine candidate of Neisseria meningitidis. J Exp Med 195: 1445–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila S, Wright VJ, Khor CC, Sim KS, Binder A, Breunis WB, Inwald D, Nadel S, Betts H, Carrol ED, et al. 2010. Genome-wide association study identifies variants in the CFH region associated with host susceptibility to meningococcal disease. Nat Genet 42: 772–776 [DOI] [PubMed] [Google Scholar]
- de Bakker PI, Telenti A 2010. Infectious diseases not immune to genome-wide association. Nat Genet 42: 731–732 [DOI] [PubMed] [Google Scholar]
- Dhiman N, Smith DI, Poland GA 2009. Next-generation sequencing: A transformative tool for vaccinology. Expert Rev Vaccines 8: 963–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhiman N, Haralambieva IH, Kennedy RB, Vierkant RA, O’Byrne MM, Ovsyannikova IG, Jacobson RM, Poland GA 2010. SNP/haplotype associations in cytokine and cytokine receptor genes and immunity to rubella vaccine. Immunogenetics 62: 197–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati C, Hiller NL, Tettelin H, Muzzi A, Croucher NJ, Angiuoli SV, Oggioni M, Dunning Hotopp JC, Hu FZ, Riley DR, et al. 2010. Structure and dynamics of the pan-genome of Streptococcus pneumoniae and closely related species. Genome Biol 11: R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly J, Medini D, Boccadifuoco G, Biolchi A, Ward J, Frasch C, Moxon ER, Stella M, Comanducci M, Bambini S, et al. 2010. Qualitative and quantitative assessment of meningococcal antigens to evaluate the potential strain coverage of protein-based vaccines. Proc Natl Acad Sci 107: 19490–19495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormitzer PR, Ulmer JB, Rappuoli R 2008. Structure-based antigen design: A strategy for next generation vaccines. Trends Biotechnol 26: 659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dramsi S, Caliot E, Bonne I, Guadagnini S, Prevost MC, Kojadinovic M, Lalioui L, Poyart C, Trieu-Cuot P 2006. Assembly and role of pili in group B streptococci. Mol Microbiol 60: 1401–1413 [DOI] [PubMed] [Google Scholar]
- Dunphy KY, Beernink PT, Brogioni B, Granoff DM 2011. Effect of factor H-binding protein sequence variation on factor H binding and survival of Neisseria meningitidis in human blood. Infect Immun 79: 353–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RW 1990. New and improved vaccines against hepatitis B. In New generation vaccines (ed. Woodrow G, Levine M), pp. 439–447 Marcel Dekker, New York [Google Scholar]
- Findlow J, Borrow R, Snape MD, Dawson T, Holland A, John TM, Evans A, Telford KL, Ypma E, Toneatto D, et al. 2010. Multicenter, open-label, randomized phase II controlled trial of an investigational recombinant Meningococcal serogroup B vaccine with and without outer membrane vesicles, administered in infancy. Clin Infect Dis 51: 1127–1137 [DOI] [PubMed] [Google Scholar]
- Fletcher LD, Bernfield L, Barniak V, Farley JE, Howell A, Knauf M, Ooi P, Smith RP, Weise P, Wetherell M, et al. 2004. Vaccine potential of the Neisseria meningitidis 2086 lipoprotein. Infect Immun 72: 2088–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch CE, Borrow R, Donnelly J 2009. Bactericidal antibody is the immunologic surrogate of protection against meningococcal disease. Vaccine 27 (Suppl 2): B112–B116 [DOI] [PubMed] [Google Scholar]
- Garcon N, Wettendorff M, Van Mechelen M 2011. Role of AS04 in human papillomavirus vaccine: Mode of action and clinical profile. Expert Opin Biol Ther 11: 667–677 [DOI] [PubMed] [Google Scholar]
- Gaucher D, Therrien R, Kettaf N, Angermann BR, Boucher G, Filali-Mouhim A, Moser JM, Mehta RS, Drake DR III, Castro E, et al. 2008. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med 205: 3119–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill CJ, Baxter R, Anemona A, Ciavarro G, Dull P 2010. Persistence of immune responses after a single dose of Novartis meningococcal serogroup A, C, W-135 and Y CRM-197 conjugate vaccine (Menveo) or Menactra among healthy adolescents. Hum Vaccin 6: 881–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliani MM, Adu-Bobie J, Comanducci M, Arico B, Savino S, Santini L, Brunelli B, Bambini S, Biolchi A, Capecchi B, et al. 2006. A universal vaccine for serogroup B meningococcus. Proc Natl Acad Sci 103: 10834–10839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliani MM, Biolchi A, Serruto D, Ferlicca F, Vienken K, Oster P, Rappuoli R, Pizza M, Donnelly J 2010. Measuring antigen-specific bactericidal responses to a multicomponent vaccine against serogroup B meningococcus. Vaccine 28: 5023–5030 [DOI] [PubMed] [Google Scholar]
- Glenny AT, Hopkins BE 1923. Diphtheria toxoid as an immunizing agent. Br J Exp Pathol 4: 283–288 [Google Scholar]
- Gotschlich EC, Goldschneider I, Artenstein MS 1969. Human immunity to the meningococcus. IV. Immunogenicity of group A and group C meningococcal polysaccharides in human volunteers. J Exp Med 129: 1367–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granoff DM, Welsch JA, Ram S 2009. Binding of complement factor H (fH) to Neisseria meningitidis is specific for human fH and inhibits complement activation by rat and rabbit sera. Infect Immun 77: 764–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halperin SA, Diaz-Mitoma F, Dull P, Anemona A, Ceddia F 2010. Safety and immunogenicity of an investigational quadrivalent meningococcal conjugate vaccine after one or two doses given to infants and toddlers. Eur J Clin Microbiol Infect Dis 29: 259–267 [DOI] [PubMed] [Google Scholar]
- Haralambieva IH, Poland GA 2010. Vaccinomics, predictive vaccinology and the future of vaccine development. Future Microbiol 5: 1757–1760 [DOI] [PubMed] [Google Scholar]
- Haralambous E, Dolly SO, Hibberd ML, Litt DJ, Udalova IA, O’Dwyer C, Langford PR, Simon Kroll J, Levin M 2006. Factor H, a regulator of complement activity, is a major determinant of meningococcal disease susceptibility in UK Caucasian patients. Scand J Infect Dis 38: 764–771 [DOI] [PubMed] [Google Scholar]
- Hava DL, Camilli A 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol Microbiol 45: 1389–1406 [PMC free article] [PubMed] [Google Scholar]
- He M, Sebaihia M, Lawley TD, Stabler RA, Dawson LF, Martin MJ, Holt KE, Seth-Smith HM, Quail MA, Rance R, et al. 2010. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc Natl Acad Sci 107: 7527–7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz E, Tischler P, Rattei T, Myers G, Wagner M, Horn M 2009. Comprehensive in silico prediction and analysis of chlamydial outer membrane proteins reflects evolution and life style of the Chlamydiae. BMC Genomics 10: 634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleringmann M, Ringler P, Muller SA, De Angelis G, Rappuoli R, Ferlenghi I, Engel A 2009. Molecular architecture of Streptococcus pneumoniae TIGR4 pili. EMBO J 28: 3921–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsson S, Molling P, Olcen P 2009. Seroprevalence of antibodies against fHbp and NadA, two potential vaccine antigens for Neisseria meningitidis. Vaccine 27: 5755–5759 [DOI] [PubMed] [Google Scholar]
- Jefferies JM, Macdonald E, Faust SN, Clarke SC 2011. 13-valent pneumococcal conjugate vaccine (PCV13). Hum Vaccin 7: 1012–1018 [DOI] [PubMed] [Google Scholar]
- Jiang HQ, Hoiseth SK, Harris SL, McNeil LK, Zhu D, Tan C, Scott AA, Alexander K, Mason K, Miller L, et al. 2010. Broad vaccine coverage predicted for a bivalent recombinant factor H binding protein based vaccine to prevent serogroup B meningococcal disease. Vaccine 28: 6086–6093 [DOI] [PubMed] [Google Scholar]
- Keiser PB, Biggs-Cicatelli S, Moran EE, Schmiel DH, Pinto VB, Burden RE, Miller LB, Moon JE, Bowden RA, Cummings JF, et al. 2011. A phase 1 study of a meningococcal native outer membrane vesicle vaccine made from a group B strain with deleted lpxL1 and synX, over-expressed factor H binding protein, two PorAs and stabilized OpcA expression. Vaccine 29: 1413–1420 [DOI] [PubMed] [Google Scholar]
- Kelly DF, Moxon ER, Pollard AJ 2004. Haemophilus influenzae type b conjugate vaccines. Immunology 113: 163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H 2002. Systems biology: A brief overview. Science 295: 1662–1664 [DOI] [PubMed] [Google Scholar]
- Kitchin NR 2011. Review of diphtheria, tetanus and pertussis vaccines in clinical development. Expert Rev Vaccines 10: 605–615 [DOI] [PubMed] [Google Scholar]
- Koeberling O, Delany I, Granoff DM 2011a. A critical threshold of meningococcal factor H binding protein expression is required for increased breadth of protective antibodies elicited by native outer membrane vesicle vaccines. Clin Vaccine Immunol 18: 736–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeberling O, Seubert A, Santos G, Colaprico A, Ugozzoli M, Donnelly J, Granoff DM 2011b. Immunogenicity of a meningococcal native outer membrane vesicle vaccine with attenuated endotoxin and over-expressed factor H binding protein in infant rhesus monkeys. Vaccine 29: 4728–4734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruskall MS, Alper CA, Awdeh Z, Yunis EJ, Marcus-Bagley D 1992. The immune response to hepatitis B vaccine in humans: Inheritance patterns in families. J Exp Med 175: 495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer P, Rinaudo CD, Soriani M, Margarit I, Maione D, Rosini R, Taddei AR, Mora M, Rappuoli R, Grandi G, et al. 2005. Genome analysis reveals pili in Group B Streptococcus. Science 309: 105. [DOI] [PubMed] [Google Scholar]
- Lawley TD, Croucher NJ, Yu L, Clare S, Sebaihia M, Goulding D, Pickard DJ, Parkhill J, Choudhary J, Dougan G 2009. Proteomic and genomic characterization of highly infectious Clostridium difficile 630 spores. J Bacteriol 191: 5377–5386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeMieux J, Hava DL, Basset A, Camilli A 2006. RrgA and RrgB are components of a multisubunit pilus encoded by the Streptococcus pneumoniae rlrA pathogenicity islet. Infect Immun 74: 2453–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine MM, Lagos R 2004. Vaccines and vaccination in historical perspective. In New generation vaccines (ed. Levine MM, Kaper JB, Rappouli R, et al. ), pp. 1–10 Marcel Dekker, New York [Google Scholar]
- Liu X, Afrane M, Clemmer DE, Zhong G, Nelson DE 2010. Identification of Chlamydia trachomatis outer membrane complex proteins by differential proteomics. J Bacteriol 192: 2852–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod CM, Hodges GR, Heidelberger M, Bernhard WG 1945. Prevention of pneumococcal pneumonia by immunization with specific capsular polysaccharides. J Exp Med 82: 445–465 [PMC free article] [PubMed] [Google Scholar]
- Madico G, Welsch JA, Lewis LA, McNaughton A, Perlman DH, Costello CE, Ngampasutadol J, Vogel U, Granoff DM, Ram S 2006. The meningococcal vaccine candidate GNA1870 binds the complement regulatory protein factor H and enhances serum resistance. J Immunol 177: 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maione D, Margarit I, Rinaudo CD, Masignani V, Mora M, Scarselli M, Tettelin H, Brettoni C, Iacobini ET, Rosini R, et al. 2005. Identification of a universal Group B streptococcus vaccine by multiple genome screen. Science 309: 148–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manetti AG, Zingaretti C, Falugi F, Capo S, Bombaci M, Bagnoli F, Gambellini G, Bensi G, Mora M, Edwards AM, et al. 2007. Streptococcus pyogenes pili promote pharyngeal cell adhesion and biofilm formation. Mol Microbiol 64: 968–983 [DOI] [PubMed] [Google Scholar]
- Margarit I, Rinaudo CD, Galeotti CL, Maione D, Ghezzo C, Buttazzoni E, Rosini R, Runci Y, Mora M, Buccato S, et al. 2009. Preventing bacterial infections with pilus-based vaccines: The group B streptococcus paradigm. J Infect Dis 199: 108–115 [DOI] [PubMed] [Google Scholar]
- Masignani V, Comanducci M, Giuliani MM, Bambini S, Adu-Bobie J, Arico B, Brunelli B, Pieri A, Santini L, Savino S, et al. 2003. Vaccination against Neisseria meningitidis using three variants of the lipoprotein GNA1870. J Exp Med 197: 789–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Liu G, Jensen ER, Lenoy E, Schneewind O 2000. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc Natl Acad Sci 97: 5510–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbow ML, De Gregorio E, Valiante NM, Rappuoli R 2010. New adjuvants for human vaccines. Curr Opin Immunol 22: 411–416 [DOI] [PubMed] [Google Scholar]
- McCarthy AJ, Lindsay JA 2010. Genetic variation in Staphylococcus aureus surface and immune evasion genes is lineage associated: Implications for vaccine design and host-pathogen interactions. BMC Microbiol 10: 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil LK, Murphy E, Zhao XJ, Guttmann S, Harris SL, Scott AA, Tan C, Mack M, DaSilva I, Alexander K, et al. 2009. Detection of LP2086 on the cell surface of Neisseria meningitidis and its accessibility in the presence of serogroup B capsular polysaccharide. Vaccine 27: 3417–3421 [DOI] [PubMed] [Google Scholar]
- Meri S, Jordens M, Jarva H 2008. Microbial complement inhibitors as vaccines. Vaccine 26: I113–I117 [DOI] [PubMed] [Google Scholar]
- Montigiani S, Falugi F, Scarselli M, Finco O, Petracca R, Galli G, Mariani M, Manetti R, Agnusdei M, Cevenini R, et al. 2002. Genomic approach for analysis of surface proteins in Chlamydia pneumoniae. Infect Immun 70: 368–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora M, Bensi G, Capo S, Falugi F, Zingaretti C, Manetti AG, Maggi T, Taddei AR, Grandi G, Telford JL 2005. Group A streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc Natl Acad Sci 102: 15641–15646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens DM, Folkers GK, Fauci AS 2008. Emerging infections: A perpetual challenge. Lancet Infect Dis 8: 710–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriel DG, Bertoldi I, Spagnuolo A, Marchi S, Rosini R, Nesta B, Pastorello I, Corea VA, Torricelli G, Cartocci E, et al. 2010. Identification of protective and broadly conserved vaccine antigens from the genome of extraintestinal pathogenic Escherichia coli. Proc Natl Acad Sci 107: 9072–9077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E, Andrew L, Lee KL, Dilts DA, Nunez L, Fink PS, Ambrose K, Borrow R, Findlow J, Taha MK, et al. 2009. Sequence diversity of the factor H binding protein vaccine candidate in epidemiologically relevant strains of serogroup B Neisseria meningitidis. J Infect Dis 200: 379–389 [DOI] [PubMed] [Google Scholar]
- Nakaya HI, Li S, Pulendran B 2011. Systems vaccinology: Learning to compute the behavior of vaccine induced immunity. Wiley Interdiscip Rev Syst Biol Med 4: 193–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesta B, Spraggon G, Alteri C, Moriel DG, Rosini R, Veggi D, Smith S, Bertoldi I, Pastorello I, Ferlenghi I, et al. 2012. FdeC, a novel broadly conserved Escherichia coli adhesin eliciting protection against urinary tract infections. MBio 3: e00010–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen HE, Christensen KC, Koch C, Thomsen BS, Heegaard NH, Tranum-Jensen J 1989. Hereditary, complete deficiency of complement factor H associated with recurrent meningococcal disease. Scand J Immunol 30: 711–718 [DOI] [PubMed] [Google Scholar]
- Nuccitelli A, Cozzi R, Gourlay LJ, Donnarumma D, Necchi F, Norais N, Telford JL, Rappuoli R, Bolognesi M, Maione D, et al. 2011. Structure-based approach to rationally design a chimeric protein for an effective vaccine against Group B Streptococcus infections. Proc Natl Acad Sci 108: 10278–10283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hagan DT, Rappuoli R, De Gregorio E, Tsai T, Del Giudice G 2011. MF59 adjuvant: The best insurance against influenza strain diversity. Expert Rev Vaccines 10: 447–462 [DOI] [PubMed] [Google Scholar]
- Oriente F, Scarlato V, Delany I 2010. Expression of factor H binding protein of meningococcus responds to oxygen limitation through a dedicated FNR-regulated promoter. J Bacteriol 192: 691–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova IG, Jacobson RM, Vierkant RA, Pankratz VS, Poland GA 2007. HLA supertypes and immune responses to measles-mumps-rubella viral vaccine: Findings and implications for vaccine design. Vaccine 25: 3090–3100 [DOI] [PubMed] [Google Scholar]
- Parida SK, Kaufmann SH 2010. Novel tuberculosis vaccines on the horizon. Curr Opin Immunol 22: 374–384 [DOI] [PubMed] [Google Scholar]
- Peltola H, Kayhty H, Sivonen A, Makela H 1977. Haemophilus influenzae type b capsular polysaccharide vaccine in children: A double-blind field study of 100,000 vaccinees 3 months to 5 years of age in Finland. Pediatrics 60: 730–737 [PubMed] [Google Scholar]
- Pezzicoli A, Santi I, Lauer P, Rosini R, Rinaudo D, Grandi G, Telford JL, Soriani M 2008. Pilus backbone contributes to group B Streptococcus paracellular translocation through epithelial cells. J Infect Dis 198: 890–898 [DOI] [PubMed] [Google Scholar]
- Pizza M, Covacci A, Bartoloni A, Perugini M, Nencioni L, De Magistris MT, Villa L, Nucci D, Manetti R, Bugnoli M, et al. 1989. Mutants of pertussis toxin suitable for vaccine development. Science 246: 497–500 [DOI] [PubMed] [Google Scholar]
- Pizza M, Scarlato V, Masignani V, Giuliani MM, Arico B, Comanducci M, Jennings GT, Baldi L, Bartolini E, Capecchi B, et al. 2000. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287: 1816–1820 [DOI] [PubMed] [Google Scholar]
- Pizza M, Donnelly J, Rappuoli R 2008. Factor H-binding protein, a unique meningococcal vaccine antigen. Vaccine 26 (Suppl 8): 146–148 [DOI] [PubMed] [Google Scholar]
- Plested JS, Welsch JA, Granoff DM 2009. Ex vivo model of meningococcal bacteremia using human blood for measuring vaccine-induced serum passive protective activity. Clin Vaccine Immunol 16: 785–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin SA 2009. Vaccines: The fourth century. Clin Vaccine Immunol 16: 1709–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland GA, Ovsyannikova IG, Jacobson RM, Smith DI 2007. Heterogeneity in vaccine immune response: The role of immunogenetics and the emerging field of vaccinomics. Clin Pharmacol Ther 82: 653–664 [DOI] [PubMed] [Google Scholar]
- Poland GA, Ovsyannikova IG, Jacobson RM 2008. Personalized vaccines: The emerging field of vaccinomics. Expert Opin Biol Ther 8: 1659–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland GA, Ovsyannikova IG, Jacobson RM 2009. Application of pharmacogenomics to vaccines. Pharmacogenomics 10: 837–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B, Li S, Nakaya HI 2010. Systems vaccinology. Immunity 33: 516–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, Pirani A, Gernert K, Deng J, Marzolf B, et al. 2009. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol 10: 116–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon G 1924. Sur la toxine et sur l’anatoxine diphtériques. Pouvoir floculant et propriétés immunisantes. Ann Inst Pasteur 38: 1–106 [Google Scholar]
- Rappuoli R 2007. Bridging the knowledge gaps in vaccine design. Nat Biotechnol 25: 1361–1366 [DOI] [PubMed] [Google Scholar]
- Rappuoli R, Black S, Lambert PH 2011. Vaccine discovery and translation of new vaccine technology. Lancet 378: 360–368 [DOI] [PubMed] [Google Scholar]
- Rasko DA, Rosovitz MJ, Myers GS, Mongodin EF, Fricke WF, Gajer P, Crabtree J, Sebaihia M, Thomson NR, Chaudhuri R, et al. 2008. The pangenome structure of Escherichia coli: Comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol 190: 6881–6893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaudo CD, Telford JL, Rappuoli R, Seib KL 2009. Vaccinology in the genome era. J Clin Invest 119: 2515–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaudo CD, Rosini R, Galeotti CL, Berti F, Necchi F, Reguzzi V, Ghezzo C, Telford JL, Grandi G, Maione D 2010. Specific involvement of pilus type 2a in biofilm formation in group B Streptococcus. PLoS ONE 5: e9216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosini R, Rinaudo CD, Soriani M, Lauer P, Mora M, Maione D, Taddei A, Santi I, Ghezzo C, Brettoni C, et al. 2006. Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol Microbiol 61: 126–141 [DOI] [PubMed] [Google Scholar]
- Scarselli M, Arico B, Brunelli B, Savino S, Di Marcello F, Palumbo E, Veggi D, Ciucchi L, Cartocci E, Bottomley MJ, et al. 2011. Rational design of a meningococcal antigen inducing broad protective immunity. Sci Transl Med 3: 91ra62. [DOI] [PubMed] [Google Scholar]
- Schneider MC, Exley RM, Chan H, Feavers I, Kang YH, Sim RB, Tang CM 2006. Functional significance of factor H binding to Neisseria meningitidis. J Immunol 176: 7566–7575 [DOI] [PubMed] [Google Scholar]
- Schneider MC, Prosser BE, Caesar JJ, Kugelberg E, Li S, Zhang Q, Quoraishi S, Lovett JE, Deane JE, Sim RB, et al. 2009. Neisseria meningitidis recruits factor H using protein mimicry of host carbohydrates. Nature 458: 890–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seib KL, Serruto D, Oriente F, Delany I, Adu-Bobie J, Veggi D, Arico B, Rappuoli R, Pizza M 2009. Factor H-binding protein is important for meningococcal survival in human whole blood and serum and in the presence of the antimicrobial peptide LL-37. Infect Immun 77: 292–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seib KL, Brunelli B, Brogioni B, Palumbo E, Bambini S, Muzzi A, Dimarcello F, Marchi S, van der Ende A, Arico B, et al. 2011. Characterisation of diverse sub-variants of the meningococcal factor H binding protein (fHbp) for their ability to bind fH, mediate serum resistance and induce bactericidal antibodies. Infect Immun 79: 970–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serruto D, Spadafina T, Ciucchi L, Lewis LA, Ram S, Tontini M, Santini L, Biolchi A, Seib KL, Giuliani MM, et al. 2010. Neisseria meningitidis GNA2132, a heparin-binding protein that induces protective immunity in humans. Proc Natl Acad Sci 107: 3770–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabler RA, Gerding DN, Songer JG, Drudy D, Brazier JS, Trinh HT, Witney AA, Hinds J, Wren BW 2006. Comparative phylogenomics of Clostridium difficile reveals clade specificity and microevolution of hypervirulent strains. J Bacteriol 188: 7297–7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, Lawley TD, Sebaihia M, Quail MA, Rose G, et al. 2009. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol 10: R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger-Jones YK, Bae T, Schneewind O 2006. Vaccine assembly from surface proteins of Staphylococcus aureus. Proc Natl Acad Sci 103: 16942–16947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G 2006. Pili in gram-positive pathogens. Nat Rev Microbiol 4: 509–519 [DOI] [PubMed] [Google Scholar]
- Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, et al. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome.” Proc Natl Acad Sci 102: 13950–13955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe C, Edwards L, Snelgrove R, Finco O, Rae A, Grandi G, Guilio R, Hussell T 2007. Discovery of a vaccine antigen that protects mice from Chlamydia pneumoniae infection. Vaccine 25: 2252–2260 [DOI] [PubMed] [Google Scholar]
- Tobin GJ, Trujillo JD, Bushnell RV, Lin G, Chaudhuri AR, Long J, Barrera J, Pena L, Grubman MJ, Nara PL 2008. Deceptive imprinting and immune refocusing in vaccine design. Vaccine 26: 6189–6199 [DOI] [PubMed] [Google Scholar]
- Toneatto D, Ismaili S, Ypma E, Vienken K, Oster P, Dull P 2011a. The first use of an investigational multicomponent meningococcal serogroup B vaccine (4CMenB) in humans. Hum Vaccin 7: 646–653 [DOI] [PubMed] [Google Scholar]
- Toneatto D, Oster P, Deboer AC, Emerson A, Santos GF, Ypma E, Detora L, Pizza M, Kimura A, Dull P 2011b. Early clinical experience with a candidate meningococcal B recombinant vaccine (rMenB) in healthy adults. Hum Vaccin 7: 781–791 [DOI] [PubMed] [Google Scholar]
- Veggi D, Gentile MA, Cantini F, Lo Surdo P, Nardi-Dei V, Seib KL, Pizza M, Rappuoli R, Banci L, Savino S, et al. 2012. The factor H binding protein of Neisseria meningitidis interacts with xenosiderophores in vitro. Biochemistry 51: 9384–9393 [DOI] [PubMed] [Google Scholar]
- Welsch JA, Ram S, Koeberling O, Granoff DM 2008. Complement-dependent synergistic bactericidal activity of antibodies against factor H-binding protein, a sparsely distributed meningococcal vaccine antigen. J Infect Dis 197: 1053–1061 [DOI] [PubMed] [Google Scholar]