Summary
DNA double strand breaks (DSBs) in B lymphocytes arise stochastically during replication or as a result of targeted DNA damage by activation induced cytidine deaminase (AID). Here we identify recurrent, early replicating and AID independent DNA lesions, termed early replication fragile sites (ERFS), by genome-wide localization of DNA repair proteins in B cells subjected to replication stress. ERFS colocalize with highly expressed gene clusters and are enriched for repetitive elements and CpG dinucleotides. Although distinct from late-replicating common fragile sites (CFS), the stability of ERFSs and CFSs is similarly dependent on the replication-stress response kinase ATR. ERFSs break spontaneously during replication, but their fragility is increased by hydroxyurea, ATR inhibition or deregulated c-Myc expression. Moreover, greater than 50% of recurrent amplifications/deletions in human diffuse large B cell lymphoma map to ERFSs. In summary, we have identified a source of spontaneous DNA lesions that drives instability at preferred genomic sites.
Introduction
DSBs arise spontaneously during DNA replication, as a result of oncogenic stress, and as a part of the gene diversification programs in lymphocytes (Bartek et al., 2007; Gostissa et al., 2011; Halazonetis et al., 2008). When B lymphocytes are activated, they undergo rapid proliferation and simultaneously initiate two genome remodeling reactions, termed somatic hypermutation (SHM) and class-switch recombination (CSR). The coupling of rapid cycling and programmed DNA damage poses the B cell genome at high risk for destabilization.
SHM introduces point mutations in the variable region of immunoglobulin (Ig) genes, which can increase antibody affinity, whereas CSR is a DNA deletion event that replaces one Ig constant region gene for another. Both of these reactions are initiated by the enzyme AID, which deaminates cytosine residues in single stranded DNA exposed during Ig gene transcription (Chaudhuri and Alt, 2004). In addition to Ig genes, AID causes a considerable amount of collateral genomic damage (Chiarle et al., 2011; Kato et al., 2012; Klein et al., 2011; Liu et al., 2008), including oncogenic targets such as c-Myc (Robbiani et al., 2008). Nevertheless, many recurrent mutations in B cell lymphoma are not associated with AID activity, and the mechanisms of rearrangements at these sites remain unclear.
The DNA damage response (DDR) is activated during programmed rearrangements in lymphocytes to ensure faithful DNA repair and prevent chromosomal translocation (Chen et al., 2000; Petersen et al., 2001). The DDR is also trigged by aberrant oncogene expression which induces precocious entry into S phase, and perturbs replication fork progression (Bartek et al., 2007; Bester et al., 2011; Halazonetis et al., 2008). Replication fork instability can also be triggered by exogenous agents such as hydroxyurea (HU), which depletes deoxynucleotide pools, or by deficiencies in homologous recombination pathways that are needed to complete DNA replication after fork stalling or collapse (Schlacher et al., 2012).
Oncogenic stress has been shown to preferentially target genomic regions called common fragile sites (CFSs) (Bartek et al., 2007; Halazonetis et al., 2008). Historically, CFSs have been mapped in lymphocytes but are induced in all cell types under conditions that obstruct replication, such as treatment with low doses of the DNA polymerase inhibitor aphidicolin. DNA breakage within CFSs spans megabase regions. Nevertheless, CFSs share characteristic features including association with very large genes, enrichment of long stretches of AT dinucleotide-rich repeats, and incomplete DNA replication (Durkin and Glover, 2007).
Replication stress-induced DNA damage is also observed in yeast. Similar to CFSs, sites located in “replication slow zones” (RSZs) are late replicating and breakage-prone (Cha and Kleckner, 2002). In addition to late replicating areas, irreversible replication fork collapse in response to acute doses of hydroxyurea has been observed preferentially around a subset of early firing replication origins in yeast (Raveendranathan et al., 2006), which do not overlap with RSZs (Cha and Kleckner, 2002; Hashash et al., 2011). Although the molecular mechanisms governing replication initiation in yeast and mammalian cells are distinct, we wondered if fragility at early firing origins also exists in mammalian cells. Here we identify highly unstable regions of the B cell genome designated as “Early Replicating Fragile Sites” (ERFSs). We propose that ERFS are a new class of fragile sites in mammalian cells that contribute to recurrent rearrangements during lymphomagenesis.
Results
Genome-wide mapping of replication-induced DNA damage
Single strand DNA (ssDNA) mapping has been used to localize origins of replication in yeast (Feng et al., 2006). To identify potential sites of fork collapse, we first profiled the location and extent of ssDNA genome-wide using chromatin immunoprecipitation (ChIP) with an anti-RPA antibody (Figure 1). RPA associates with ssDNA at stalled forks near early firing origins when fork movement is inhibited by HU (Tanaka and Nasmyth, 1998).
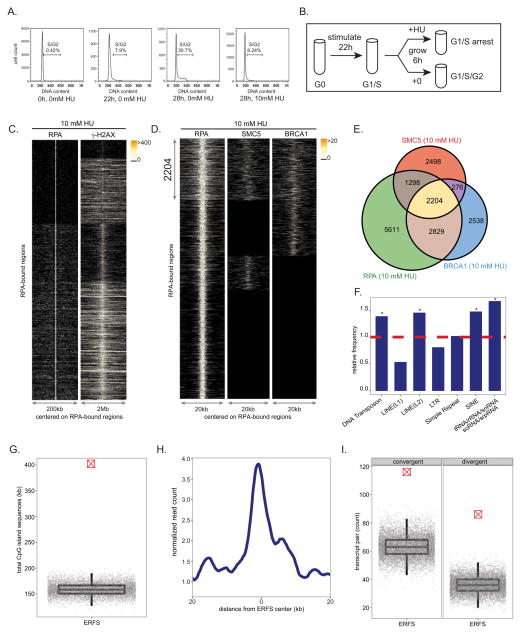
Figure 1. Mapping replication-induced DNA damage in murine B lymphocytes.
(A) FACS analysis showing DNA content of freshly isolated and ex-vivo stimulated splenic murine B lymphocytes in the absence and presence of HU. (B) Experimental plan describing cell synchronization and isolation for samples used in ChIP-Seq and RNA-Seq experiments. (C) For each RPA-bound site in response to 10 mM HU (y axis), each column depicts the presence of RPA (left) and γ-H2AX (right) within a window centered on the RPA-bound sites. Color-map corresponds to binding intensities where “black” represents no binding. K-mean clustering algorithm was used to group the protein-bound sites. (D) RPA, SMC5 and BRCA1 co-occupy 2204 genomic regions in response to 10 mM HU. The plot in each column, from left to right, represents the pattern of RPA, SMC5 and BRCA1 genomic occupancy in response to HU centered on RPA-bound sites. K-mean clustering algorithm is used to group the protein-bound sites. (E) The Venn diagram shows the overlap of sites bound by RPA, SMC5, and BRCA1 in response to 10 mM HU. The total number of bound sites is indicated for each shared and unique area. (F) Relative frequency of ERFSs in classes of repetitive sequences is shown. Dashed line indicates the expected frequency based on the permutation model (*: enriched repetitive element classes, p < 1×10−3). (G) ERFSs are enriched in CpG islands. Total CpG island sequences in all the 2204 ERFSs as indicated by the crossed red point is compared to the permutation model as indicated by the gray points. Each gray point corresponds to the total CpG island sequences covered in an iteration of the permutation model. The box-plot depicts the quantiles of total CpG sequences based on the permutation model (p < 1×10−5). (H) ERFS genomic regions are transcriptionally active. The line-plot represents the average RNA tag count (loess-smoothed) in a genomic window around the center of the ERFSs. (I) ERFSs are enriched in transcriptionally active convergent and divergent gene pairs. Count of divergent/convergent gene pairs coinciding with ERFSs as indicated by the crossed red point is compared to the permutation model as indicated by the gray points. Each gray point corresponds to the total number of divergent/convergent gene pairs observed in an iteration of the permutation model. The box-plot depicts the quantiles of the total convergent/divergent transcript pair count based on the permutation model (p < 1×10−5). For definition of convergent/divergent gene pairs see methods. See also Figures S1, S3, S4.
Freshly isolated mouse B cells are arrested in the G0 phase of the cell cycle (Figure 1A). Upon stimulation with LPS/IL4, cells synchronously enter into the cell cycle so that by 22 hours, approximately 8% of cells have entered S phase, while at 28 hours over 30% are in S/G2 phases (Figure 1A). To profile early replication origins, we treated cells at 22 hours with 10 mM HU for 6 hours to fully arrest cells at G1/S (Figures 1A and 1B). We then performed ChIP-Seq of RPA in both untreated and HU-treated cells at 28h (Figures 1A and 1B). Two independent experiments showed reproducibility of genome-wide RPA association in HU-treated cells (Figure S1A). We generated profiles of RPA in untreated and treated samples, centered on individual RPA-bound sites (Figure S1B), and observed a marked increase in the intensity of RPA in HU-treated B cells relative to untreated cells where 5939 out of 11942 genomic regions (49.7%) displayed more than a 4-fold increase in RPA recruitment. In addition to the 53% overlap of RPA-associated regions between HU-untreated vs. treated cells, we also observed that 1441 regions were present only in HU-treated samples (Figure S1B). These HU dependent ssDNA regions may correspond to the firing of new replication origins to compensate for inefficient replication.
To confirm that RPA recruitment maps early replication zones, we used the Repli-Seq approach (Hansen et al., 2010) to identify replication origins in B cells during HU arrest. Approximately 12,000 early activating replication origins across the murine B cell genome were identified (Figure S1C). By comparing the distribution of BrdU incorporation relative to the individual RPA-occupied genomic regions, we observed association of BrdU incorporation with nearly 80% of RPA-bound regions (Figure S1C). Moreover, more than 86% of RPA/BrdU enriched genomic sites coincided with previously mapped early replicating regions in the mouse B cell line CH12 (Stamatoyannopoulos et al., 2012) (p(permutation) < 1×10−5, Figure S1D). Thus, HU-arrested B cells exhibited an enrichment of RPA at early replicating zones, consistent with an early S phase cell cycle arrest (Figure 1A).
Early replicating regions are associated with accessible chromatin configuration (MacAlpine et al., 2004). In agreement with this, we found that more than 67% of RPA-bound regions in HU-arrested cells reside within intragenic sequences (Figure S1E and S1I), a frequency significantly higher than expected (p(permutation) <1×10−5). Moreover, RPA preferentially associated with DNaseI hypersensitive sites (DHS) and euchromatic promoters marked by H3K4me3 (Figure S1F). Finally, we measured transcriptional activity in HU-treated B cells directly by genome-wide RNA sequencing. We observed high transcription activity within the RPA-occupied genomic regions as shown by the aggregated pattern of RNA-Seq centered on those regions (Figure S1G). Moreover, 6100 RPA-bound RefSeq genes exhibited significantly higher average mRNA abundance than those that did not show RPA binding (p< 1×10−16, Figure S1H). Thus, HU-induced RPA recruitment in early S phase maps to actively transcribed genes that show the hallmarks of euchromatin..
Replisome stalling in response to HU triggers the activation of the ATR kinase (Ward and Chen, 2001), which protects forks from collapse (Cimprich and Cortez, 2008). and leads to phosphorylation of H2AX (γ-H2AX) (Ward and Chen, 2001), which colocalizes with RPA (Petermann et al., 2010). To examine the relative distribution of γ-H2AX and RPA genome-wide, we carried out ChIP-Seq with an antibody that recognizes γ-H2AX (Figure S1A) and examined their profiles with respect to the center of RPA-bound sites. γ-H2AX-associated genomic regions were much broader than RPA, but these regions overlapped with 93% of RPA-bound sites marking ssDNA in HU-treated cells (Figure 1C), consistent with the finding that γ-H2AX marks stalled forks even prior to DSB formation (Petermann et al., 2010). γ-H2AX/RPA enriched loci may therefore correspond to a combination of stalled and broken replisomes.
Cells deficient in homologous recombination (HR) pathway components, such as XRCC2, often accumulate spontaneous chromosome breaks and exhibit hypersensitivity to HU (Sonoda et al., 1998). Consistent with increased spontaneous DNA damage at replication forks, untreated XRCC2−/− cells exhibited accumulation of γ-H2AX at similar genomic regions and at almost similar levels observed in HU-treated WT B cells (Figure S2A–C). 90% of γ-H2AX –associated genomic regions in untreated XRCC2−/− cells correlate with the regions enriched for this protein in HU-treated WT B cells (Figure S2B), and nearly 80% of the regions with enriched γ-H2AX observed in HU-treated WT B cells overlapped with those seen in HU-treated XRCC2−/− cells (Figure S2C). These data indicate that XRCC2 deficiency leads to increased endogenous levels of replication stress mostly at the same loci where HU induces replication fork stalling and/or breakage in WT cells.
RPA, BRCA1 and SMC5 colocalization marks the sites of replication stress in early replicating zones
Like XRCC2, BRCA1 and members of the structural maintenance of chromosome (SMC) family have been implicated in promoting replication fork restart (Schlacher et al., 2012; Stephan et al., 2011). To determine whether HR proteins bind to a subset of stalled forks marked by RPA and γ-H2AX, we also defined the genome-wide profile of BRCA1 and SMC5. We confirmed BRCA1 and SMC5 ChIP-Seq efficacy by observing their association at both Sμ and Sγ1 in 53BP1−/− cells, where the breaks in IgH persist unrepaired and undergo extensive resection (Figure S3A) (Bothmer et al., 2010; Bunting et al., 2010; Yamane et al., 2011; Yamane et al., 2013).
We then determined the localization of BRCA1 and SMC5 in HU-arrested B cells. Two independent experiments showed reproducibility of genome-wide BRCA1 and SMC5 association (Figures S3B and S3C). To identify the RPA genomic sites co-occupied by the HR proteins BRCA1 and SMC5, we plotted the distribution of their binding with respect to the center of individual RPA-bound regions. Overall, 2204 regions spanning 10 kbp on average showed RPA/BRCA1/SMC5 triple co-localization (Figures 1D and 1E). We found that RPA was recruited to more than 88% of genomic sites exhibiting BRCA1 and SMC5 association (Figure 1E). Furthermore, genome-wide analysis of RPA/BRCA1/SMC5 profiles in untreated cells revealed more than a 21% increase in the number of genomic regions occupied by these three proteins after HU treatment (Figure S4A). Nevertheless, 48% of RPA/BRCA1/SMC5 triple co-localizations were common between the unperturbed and HU-arrested B cells (Figure S4A). Therefore, we hypothesized that chromatin with concomitant RPA, BRCA1 and SMC5 binding might correspond to regions undergoing replication fork collapse both in response to replication stress and during normal DNA replication. Given that our analysis focused on early replicating sites, which contrasts with late replicating CFSs, we designated these regions as “Early Replication Fragile Sites” (ERFS).
We then characterized ERFSs to determine if they share common underlying primary sequence characteristics. Indeed, these loci were enriched at known repetitive elements, including LINE L2, SINE, DNA transposons and tRNA elements (p(permutation) < 1×10−3, Figure 1F) which are known replication fork barriers (Mirkin and Mirkin, 2007). Furthermore, ERFSs showed significantly higher G and C nucleotide content compared to the whole mouse genome, in contrast to CFSs that are enriched in A+T sequences (p(Wilcoxon) < 1×10−16, Figure S4B). 26% of the ERFSs regions overlapped with CpG islands, which are highly enriched at translocation breakpoints in B cell lymphoma (Tsai et al., 2008). Conversely, CpG islands covered approximately 400,000 nucleotides within these regions (p(permutation) < 1×10−5, Figure 1G). As anticipated, ERFSs clustered at early replication origins (Figure S4C), and over 66% of the loci overlapped with intragenic or promoter sequences of RefSeq annotated protein coding genes (p(permutation) < 1×10−3, Figures S4D–E). Moreover, ERFSs are more transcriptionally active relative to flanking genomic regions shown by relative mRNA enrichment by RNA-Seq (Figure 1H). Indeed, more than 86% of the RefSeq annotated genes with ERFSs are among the highest transcribed genes (p(binomial) < 1×10−16, Figure S4F). Finally, ERFSs were significantly enriched in gene pairs that are transcribed in converging or diverging directions (see methods), such as the convergent transcription pair of IKZF1 and FIGNL1 shown in Figure 2A. Compared to expected values, ERFSs were at least 2 times more likely to localize in regions containing gene pairs exhibiting convergent and/or divergent gene pairs ((p(permutation) < 1×10−5, Figure 1I).
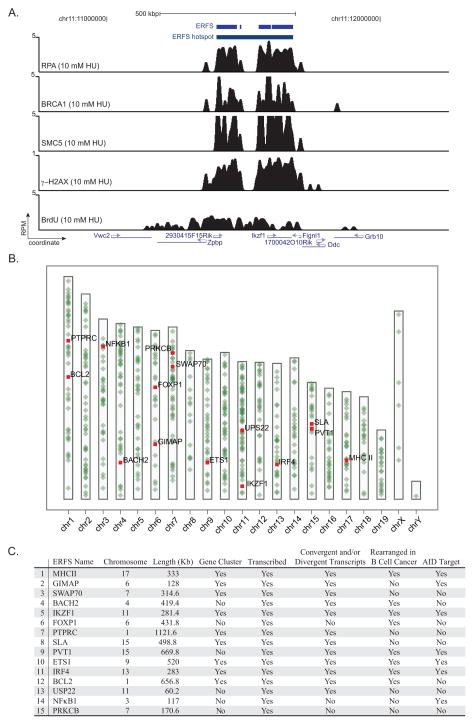
Figure 2. ERFS “hotspots” associate with highly transcribed gene clusters.
(A) Gene tracks represent, from the top, ERFS and ERFS hotspot demarcations, bindings of RPA, BRCA1, SMC5, γH2AX occupancy, and BrdU incorporation near the IKZF1 locus. The y-axis represents the total number of mapped reads per million of mapped reads (RPM) in 200 nucleotide windows (sliding-window smoothed). (B) Genome-wide map of 619 ERFS hotspots. Each hotspot is represented by a green dot on the ideograms. The top fifteen hotspots are color-coded in red. (C) Table of the top fifteen ERFS hotspots. ERFS hotspots are ordered based on a ranked statistics of RPA/SMC5/BRCA1 binding strength (see methods). The first column depicts a representative gene within the hotspot. A hotspot containing at least three genes is designated as a “gene-cluster.” A hotspot with a gene transcript value greater than 1 RPKM (reads per kilobase exon model per million mapped reads) is designated as transcribed. ERFS rearrangements in B cell cancers are listed in Table S2. ERFS is designated as “AID-target” according to (Chiarle et al., 2011; Klein et al., 2011). For complete definition of columns see methods. See also Tables S1 and S2.
Replication is organized into discrete zones (30–450 kbp in size) containing multiple replication origins that exhibit similar replication timing (Costa and Blow, 2007). Similarly, approximately 80% of the ERFSs are within 300 kbp of one another (Figure 2A; Figure S4G). We therefore integrated these neighboring clustered ERFSs and removed those with footprints less than 10 kbp to define 619 triple co-localized hotspot regions (Figure 2B; Table S1). Interestingly, whereas these hotspots were distributed throughout the genome, the density of hotspots on autosomes was higher than on the sex chromosomes, which have a lower gene density (Figure 2B). An examination of the top 15 hotspots based on a ranked statistics of RPA/BRCA/SMC5 binding strength showed that 9 out of the 15 regions contained gene clusters with at least 3 genes, and 12 out of 15 exhibited divergent/convergent gene pairs (Figure 2A and 2C, Table S1). Of note, 8 out of 15 hotspots are also rearranged in B cell lymphomas (Figure 2C; Table S2), suggesting a possible link between ERFSs, genome rearrangements and cancer (see below).
Early S phase arrest by HU induces DNA damage at ERFSs but not at CFSs
DNA damage at CFSs is visualized by conventional cytogenetic analysis of metaphase chromosomes (Durkin and Glover, 2007). To investigate whether the ERFSs defined by RPA/BRCA1/SMC5 binding are prone to actual breakage, we again treated cells with 10 mM HU, released them into fresh medium overnight, and examined metaphase spreads. Chromatid breaks, chromosome breaks and rearrangements could be discerned in 20–60% of WT cells after HU treatment (Figure S2D). To determine whether ERFSs are more sensitive to breakage under replication stress than regions lacking RPA/BRCA1/SMC5 binding (i.e. coldspots), we hybridized metaphases with BAC probes corresponding to six ERFS hotspots (MHCII, GIMAP, SWAP70, BACH2, IKZF1 and FOXP1) (Figures 3A and 3B), two coldspots (CNTNAP4 and SLITRK6) and two CFSs (FRA8E1 and FRA14A2). For each of the six ERFS hotspots a total of at least forty chromosome aberrations were counted (Table S3). Notably, all 6 ERFS hotspots displayed chromosome aberrations in metaphases from HU-treated samples (Figure 3B). In contrast, neither of the cold regions or CFSs was broken under the same conditions (Figure 3B). Overall, 8–15% of the total damage localized to individual ERFS hotspots, representing a significant fraction of the total damage (Figure 3B). DNA lesions were observed on either the centromeric or telomeric sides of ERFS-specific hybridized BAC (Figure S2E), suggesting that an ERFS represents a potentially large fragile genomic region.
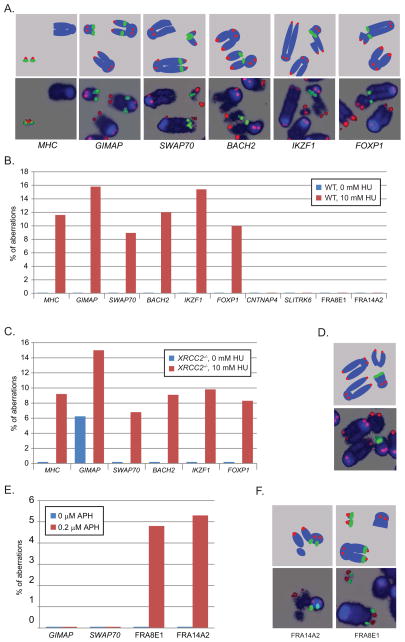
Figure 3. ERFS break in response to HU.
(A) Upper panel, diagram of FISH probes. Lower panel, representative DNA aberrations identified by FISH. Blue is DAPI-stained DNA, green represents the BAC probe (MHC, GIMAP, SWAP70, BACH2, IKZF1 or FOXP1) and red marks telomeric DNA. (B) HU induced aberrations were found at ERFSs but not at “cold sites” (CNTNAP4, SLITRK6) or CFSs (FRA16D, FRA3B). Quantitation of abnormalities from FISH analysis of untreated cells (blue bars) or cells treated with 10 mM HU (red bars). The percent aberrations specifically at the BAC probes relative to the total damage is plotted. (C) Abnormalities detected by FISH in untreated (blue bars) and 10 mM HU-treated (red bars) XRCC2−/− cells. (D) Upper panel, diagram of FISH probes. Lower panel: representative metaphase showing a spontaneous break at the GIMAP locus in an XRCC2−/− cell. (E) Quantitation of abnormalities detected by FISH in untreated (blue bars) and 0.2 μM aphidicolin-treated (red bars) WT cells. (F) Upper panel, diagram of FISH probes. Lower panel: representative metaphases showing aphidicolin-induced breaks at the FRA14A2 and FRA8E1 loci in WT cells. See also Figure S2 and Table S3.
Aberrations at ERFS hotspots were also detected in XRCC2−/− cells treated with HU (Figure 3C). XRCC2−/− cells are more sensitive to HU than WT, as evidenced by the higher level of total damage in these cells (Figure S2D). Breaks at MHCII, GIMAP, SWAP70, BACH2, IKZF1 and FOXP1 were found in 5–10% of HU-treated XRCC2−/− cells compared with 1–6% of WT cells damaged in these regions (Table S3). Nevertheless, the frequency of ERFS-specific instability relative to the total damage was similar in XRCC2−/− and WT cells (Figures 3B and 3C). Interestingly, spontaneous breaks in the vicinity of the GIMAP hotspot were detectable spontaneously in XRCC2−/− cells (Figures 3C and 3D; Table S3), which is consistent with increased γ-H2AX observed in unchallenged XRCC2-mutant cells (Figure S2A).
None of the eight CFSs defined in mouse (Helmrich et al., 2006) were among our 619 ERFS hotspots (Table S1). Consistent with this, DNA aberrations at two of the most expressed CFSs in mouse lymphocytes, FRA14A2 and FRA8E1 (Helmrich et al., 2006) were undetectable in HU-treated WT samples (Figure 3B). Absence of CFS expression could be explained by the fact that high concentrations of HU stall replication forks in early S phase (Figure 1A), whereas CFSs replicate late (Durkin and Glover, 2007). Conversely, we found that overnight treatment with low doses of aphidicolin (0.2 μM for 20 h) induced damage at the CFSs FRA14A2 and FRA8E1, while the ERFSs GIMAP and SWAP70 were largely insensitive (Figures 3E and 3F). These data are consistent with the idea that ERFS arise from fork collapse during early replication, while breakage at CFSs arises from a failure to replicate (Debatisse et al., 2012), and the two forms of replication stress induce distinct types of recurrent DNA lesions.
ATR inhibition promotes ERFS and CFS expression
The ATR kinase protects the genome from chromosomal aberrations at late replicating CFSs, (Durkin and Glover, 2007) and is essential for stabilizing stalled forks and facilitates fork restart in early S phase (Cimprich and Cortez, 2008). To confirm that ATR inactivation induces CFSs and determine whether it similarly leads to damage at ERFSs, we treated asynchronous B cells on day 2 with 1 μM of a recently described ATR inhibitor (ATRi) (Toledo et al., 2011). We found that approximately 2.5% and 7.0% of the total chromosomal aberrations localized to the two CFSs, FRA8E1 and FRA14A2, respectively (Figure 4A). ATR deficiency also led to chromosomal aberrations at ERFSs at a similar frequency (Figure 4A; Table S3). Moreover, ERFSs and CFSs were both damaged in XRCC2−/− cells treated with ATRi (Figure 4A). Thus, the rupture of un-replicated regions at CFSs and fork collapse at ERFSs are similarly sensitive to ATR inhibition.
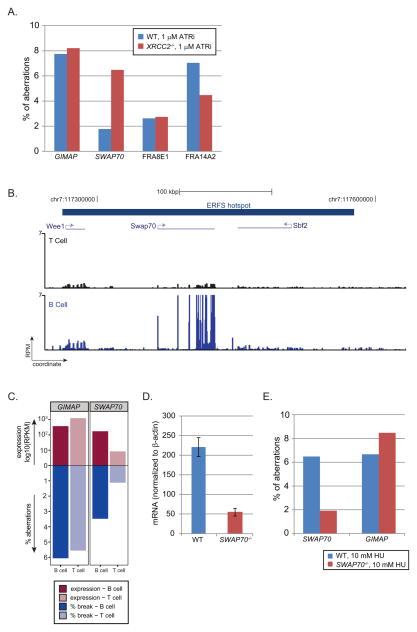
Figure 4. ERFS break in response to ATR inhibition and high transcription.
(A) Quantitation of aberrations observed by FISH in response to overnight exposure to 1 μM ATRi in WT (blue bars) and XRCC2−/− cells (red bars). (B) Gene tracks represent, from the top, ERFS demarcation, and transcription measured by RNA-Seq in T and B cells at the region flanking SWAP70 locus. (C) Relative transcriptional activities of GIMAP and SWAP70 loci in B and T cells and their relation to the ERFS fragility. GIMAP and SWAP70 hotspots are shown in separate facets. The x-axis shows the cell lineage. The y-axis upward depicts the log10(RPKM) in B and T cells by dark and light reds, respectively; the y-axis downward depicts the quantitation of aberrations observed by FISH in response to overnight exposure to 1 μM ATRi in B and T cells in dark and light blue, respectively. (D) Relative SWAP70 mRNA abundance (measured across exon 4) normalized to β-actin in WT and SWAP70−/− B cells. (E) Quantitation of aberrations in WT and SWAP70−/− cells at the GIMAP and SWAP70 regions in response to 10 mM HU. See also Figure S5 and Table S3.
Transcriptional activity can increase ERFS fragility
As described above, ERFSs are enriched in regions with high transcriptional activity (Figure 1H; Figure 2C; Figure S4F; Table S1). To determine the contribution of transcriptional activity to individual ERFSs, we focused on loci with tissue-specific transcription patterns. SWAP70 is a B cell specific developmental regulator whereas genes within the GIMAP cluster are expressed both in B and in T cells (Figure 4B; Figure S5A). Treatment with ATRi led to a similar frequency of damage at GIMAP in B and T cells, consistent with insignificant changes in gene expression between the two cell types (Figure 4C). In contrast, damage near SWAP70 was 3-fold lower in T than in B cells (Figure 4C; Table S3), which correlated with the decreased transcription of SWAP70 in T cells (Figure 4B). Nevertheless, the replication timing near SWAP70 was similar in both cell types (Figure S5B). To further delineate the role of transcription on ERFS breakage, we used SWAP70−/− mice in which 2.7 kbp including the first exon and part of the 5′ untranslated region is removed (Borggrefe et al., 2001), allowing us to compare the fragility of ERFSs in the same genomic region in knockout B cells. We determined that SWAP70 mRNA in SWAP70−/− B cells was reduced by approximately 4-fold relative to WT (Figure 4D). Moreover, DNA damage near SWAP70 was approximately 2.5 fold lower in SWAP70−/− relative to WT B cells (Figure 4E). In contrast, DNA damage near GIMAP remained at a similar level both in WT and SWAP70−/− cells (Figure 4E). While our data indicates that high level of transcription contributes to the breakage of some ERFSs, other molecular features including repetitive elements (Figure 1F), covalently bound protein complexes and RNA:DNA hybrids might also be sources of ERFS fragility.
Oncogenic stress can trigger ERFS and CFS fragility
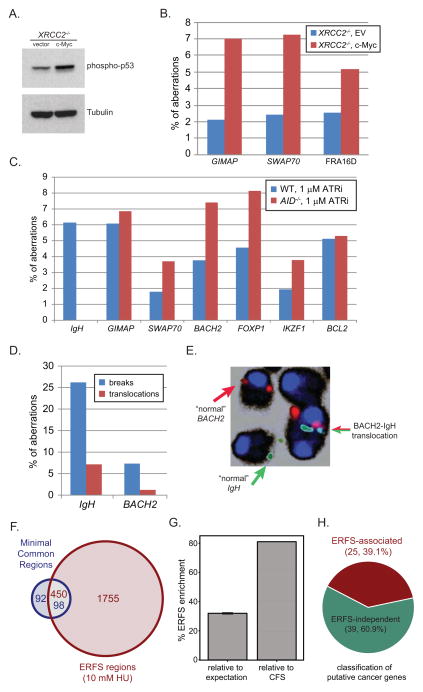
Oncogene deregulation is thought to compromise genome integrity preferentially at CFSs (Bartek et al., 2007; Halazonetis et al., 2008), and CFS deletion has been associated with various cancers (Bignell et al., 2010). To determine whether oncogenic stress similarly induces DNA damage at ERFSs, we overexpressed c-myc in B cells, as it has been implicated in regulating replication initiation and origin firing (Dominguez-Sola et al., 2007). XRCC2−/− cells were utilized to increase the amount of replicative stress and DNA damage as a result of decreased HR (Figure S2D). c-myc overexpression lead to induction of p53 (Figure 5A), which correlated with an approximately 1.6-fold increase in overall DNA damage in XRCC2−/− cells overexpressing c-myc compared to empty vector (EV) infected cells (Table S3). Moreover, 7.3% of the total breaks generated in c-myc overexpressing cells were found near SWAP70, compared to 2.4% of total breaks at this ERFS in EV-infected B cells (Figure 5B). Similarly, out of 43 breaks observed in c-myc-infected cells, 3 (7%) were found at the GIMAP cluster and 3(6.7%) were found near BACH2. c-myc overexpression also induced breaks at FRA8E1, showing a 2-fold relative increase in breaks relative to EV-infected cells (Figure 5B). Thus, DNA damage induced by c-myc overexpression can occur at ERFSs and CFSs.
Figure 5. ERFS fragility is observed in response to oncogenic stress and in human cancer.
(A) Western blot for phosphorylated p53 in c-myc and EV-infected XRCC2−/− B cells. (B) Aberrations in c-myc-infected and EV-infected XRCC2−/− B cells. (C) Aberrations in WT (blue bars) and AID−/− B cells (red bars) treated with 1 μM ATRi. (D) Spontaneous chromosome breaks (blue bars) and translocations (red bars) at the IgH and BACH2 locus in IgκAID/53BP1−/− B cells. (E) Normal chromosomes and a translocation of BACH2 ERFS (red) to the IgH locus (green) is shown. (F, G) ERFSs significantly overlap with MCRs detected in DLBCL. The Venn diagram shows the overlap of ERFSs with MCR found in DLBCL. The total number of regions is indicated for each shared and unique area and color-coded based on the region’s title. (G) Significance of correlation between the ERFSs and MCRs is evaluated relative to the permutation model and CFS. The percent increase in the overlap between the ERFSs and MCRs relative to the permutation model’s expectation (p< 1×10−4) and CFSs are shown in the left and right bar-graphs, respectively. (H) ERFSs are enriched for known cancer genes. The pie-chart shows the fraction of putative cancer genes (Bignell et al., 2010) associated with ERFSs (p<6×10−20). See also Figure S6 and Tables S3 and S4.
ERFS fragility is AID independent
Mutations and DSBs at various oncogenes including c-myc are due to AID off-target activity (Robbiani et al., 2008). Recently, a number of genome-wide studies in primary B cells mapped AID-induced DNA translocation events, and identified several novel hotspots for AID-dependent translocations at non-Ig genes (Chiarle et al., 2011; Kato et al., 2012; Klein et al., 2011). Among these translocation hotspots, MHCII, GIMAP, IKZF1, PVT1, ETS1, IRF4 and NFkB1 were located within top 15 ERFS hotspots in this study, whereas the IgH locus (the physiologic target of AID) was not ranked high among the list (Figure 2C; Table S1). To determine if AID contributes to ERFS fragility, we stimulated WT and AID knockout B cells with LPS/IL4 for two days, and then treated them with ATRi overnight. These conditions induce robust AID-dependent DNA damage simultaneously with replication stress. We probed metaphases with BACs spanning the IgH locus, the GIMAP cluster, and IKZF1- all AID translocation hotspots- as well as BACH2, SWAP70, FOXP1 and BCL2 (Figure S2E)- ERFSs that are frequently rearranged in B cell lymphoma (Figure 2C; Table S1; Table S2). In WT, the IgH locus was damaged in 3.8% of cells, but the frequency of IgH specific instability did not increase with ATRi (Figure S2F), despite the fact that ATRi greatly increased overall damage (Table S3). Upon ATRi treatment, the frequency of breaks at the ERFSs GIMAP, IKZF1, BACH2, SWAP70, FOXP1 and BCL2 were elevated to the levels similar to those observed at the IgH in activated B cells (Figure S2F). Breaks at some ERFSs were even spontaneously detected (FOXP1 and GIMAP, Figure S2F).
To determine if AID expression contributes to aberrations observed at ERFSs, we next analyzed their breakage frequency in AID−/− cells. Unlike WT cells, IgH breaks were absent in AID−/− cells. In contrast, all ERFSs exhibited similar levels of breakage both in WT and AID−/− cells (Figure 5C; Table S3). Therefore, whereas IgH breaks in B cells are entirely AID-dependent, the breakage of ERFSs is AID-independent. Altogether, these data suggest that some recurrent rearrangements in B cell lymphoma are due to AID-independent replicative stress at ERFSs.
Genome instability at ERFSs is observed in mouse models and human cancer
Among the top 15 ERFS hotspot that break in response to AID-independent replication stress, we have identified three partners that recurrently translocate to IgH in lymphomas: BACH2, FOXP1, and BCL2 (Table S2). We hypothesized that if AID-dependent DSBs in G1 persisted into early S phase, translocations between AID-dependent breaks and ERFS might be detectable. To test this, we examined cells transgenically overexpressing AID and are simultaneously deficient for 53BP1 (IgκAID/53BP1−/−), thus allowing the persistence of G1 IgH breaks into S phase where they could be joined to ERFSs. Indeed, 26% and 7% of IgκAID/53BP1−/− B cells carried IgH locus and BACH2 breaks, respectively (Figure 5D). These breaks are fusogenic, since IgH- and BACH2- associated translocations to unidentified partner chromosomes were found in 7.3% and 1.2% of the metaphases respectively (Figure 5D). Importantly, we also detected one IgH/BACH2 translocation among 750 cells (Figure 5E), reminiscent of the translocations observed in human B cell lymphoma (Kobayashi et al., 2011). Thus, AID-dependent breaks generated in G1 (Petersen et al., 2001) can join to ERFS breaks triggered in early S phase.
A hallmark of cancer genomes is widespread copy number changes, insertions and deletions. To determine whether deletions and/or amplifications at ERFSs are a general feature of the B cell lymphoma genome, we compared our ERFSs with high resolution copy number changes detected in biopsies of patients with diffuse large B cell lymphoma (DLBCL), the most common type of non-Hodgkins lymphoma (Lenz et al., 2008). A total of 190 “minimal common regions” (MCRs) were found among 203 biopsies, carrying a gain or a loss of a chromosomal region ranging in size from 5 kbp to 21Mbp (Lenz et al., 2008). Mouse ERFS coordinates were overlaid onto the human genome using two methods, yielding 2205 syntenic regions (Figure S6B–D). Notably, 51.6% of the MCRs observed in primary DLBCL overlapped with syntenic ERFS regions (p(permutation)<1×10−4, Figure 5F,). Moreover, 20.4% of ERFSs overlapped with MCRs, 32% higher than expectation (p(permutation)< 1×10−6, Figure 5G). Surprisingly, ERFS were deleted or amplified in DLBCL at least 81% more frequently as compared to CFSs, despite their cancer-specific propensity for breakage (Figure 5G). Moreover, our analysis indicated that the DLBCL copy number alterations exhibited 2-fold higher correlation with B-cell ERFSs compared to deletions and/or amplifications in T-lineage acute lymphoblastic leukemia (Figure S6A) (Zhang et al., 2012a). Finally, by examining homozygous deletions in cancer genomes (Bignell et al., 2010), we found that 25 out of 64 genes known to contribute to oncogenesis coincide with ERFSs (p(hypergeometric) < 6×10−20, Figure 5H; Table S4). Based on these findings, we conclude that ERFSs are a significant feature of the mutational landscape of diffuse large B cell lymphomas, and potentially other cancers.
Discussion
Replicative stress at ERFSs contributes to genome instability in B cells
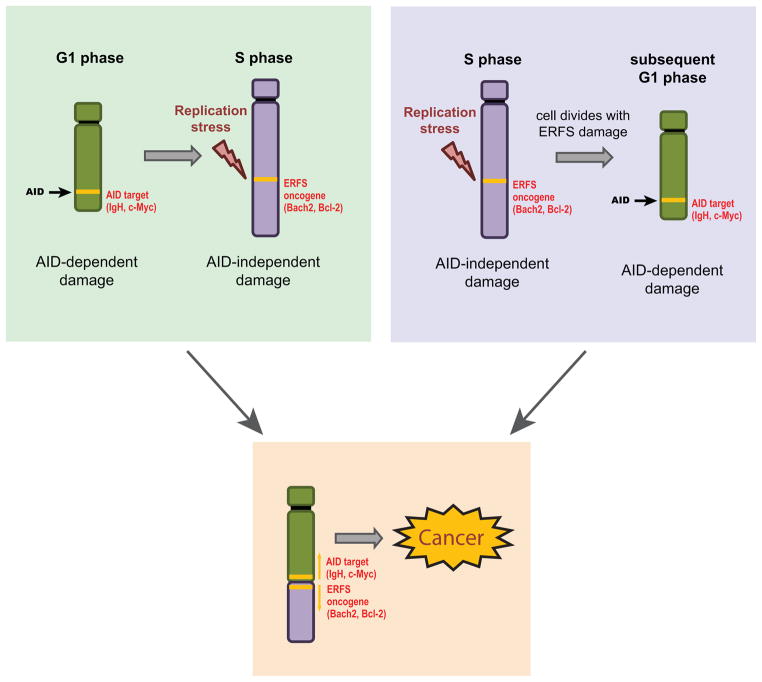
While AID has been implicated in B cell translocations (Gostissa et al., 2011), very little is known about the mechanisms of chromosomal breakage at several IgH-partner loci including BCL2, BACH2 and FOXP1. Besides programmed DNA damage, replication-based mechanisms are a major contributor to chromosomal instability in cancer (Liu et al., 2012). Activated B cells are among the most rapidly dividing mammalian cells (Zhang et al., 1988), which potentially exposes them to high endogenous levels of replicative stress. Here, we have used a genome-wide approach to identify a subset of early replicating regions in the B cell genome that are particularly vulnerable to fork collapse and contribute to rearrangements in B cell malignancies. In our model, ERFS breaks can occur after the generation of unrepaired AID-induced breaks in G1, and the two breaks could recombine during S or G2. Alternatively, ERFS damage might persist through mitosis resulting in DNA breaks in the subsequent G1 phase when AID is predominantly active. In either case, we suggest that AID-mediated DSBs in G1 (Petersen et al., 2001), together with replication stress-induced damage at recurrent loci, can coordinately drive B cell lymphoma initiation and progression (Figure 6).
Figure 6.
Model for recurrent rearrangements in B cell lymphomas. AID is active in G1 (Petersen et al., 2001) and targets IgH and various oncogenes (eg. c-myc). Replication fork collapse at ERFSs in S phase occurs at preferential sites including various cancer-associated genes (eg. BCL2, BACH2). An AID-generated break might be passed from G1 to early S, where it meets an ERFS, which may eventually result in a translocation (left panel). Alternatively, an ERFS (bearing unresolved a replication intermediate of under-replicated DNA) might break in mitosis and then become permissive to translocate to an AID-induced DSB in the next G1 phase of the cell cycle (right).
ERFS versus CFS
CFSs are considered to be the most replication stress-sensitive sites in the genome (Durkin and Glover, 2007). Although no single mechanism accounts for CFS instability, it is hypothesized that a number of different characteristics may contribute to their fragility including co-occurrence with very large genes, late replication, low density of replication origins, high A–T content and sequences prone to form secondary structures, histone hypoacetylation and a condensed chromatin structure (Helmrich et al., 2011; Jiang et al., 2009; Letessier et al., 2011; Ozeri-Galai et al., 2011). In stark contrast to CFSs, our identified ERFSs replicate early, have an open chromatin configuration, and are origin-, gene-, and G-C-rich.
Despite these diametrically opposite properties, both CFS and ERFS fragility are increased by ATR inhibition (Figure 4A), oncogenic stress (Figure 5B) and deficiencies in HR (Figure 3C) (Bartek et al., 2007; Durkin and Glover, 2007; Halazonetis et al., 2008). These conditions decrease the rate of fork progression but concomitantly increase the density of replication initiating events (Bester et al., 2011; Daboussi et al., 2008; Dominguez-Sola et al., 2007; Shechter et al., 2004) which might contribute to the damage at both CFSs and ERFSs respectively. The decrease in fork speed hinders the completion of replication at CFSs, either because of the scarcity of origins near CFSs (Letessier et al., 2011), the heterochromatic nature of the regions that would limit accessibility of DNA replication and/or DSB repair machineries (Jiang et al., 2009), or because of the interference between transcription and replication at very large genes (Helmrich et al., 2011). While additional origins are not activated near CFSs upon replication stress (Letessier et al., 2011), an increase in origin activity at early replicons might paradoxically contribute to genome instability at ERFSs. For example, increasing the replication initiation events near highly transcribed gene clusters with divergent and/or convergent gene pairs could increase conflicts between DNA replication and transcription machineries. The higher density of activated origins at ERFSs would also be expected to prematurely deplete nucleotide pools (Bester et al., 2011), thereby increasing the probability of subsequent fork stalling and collapse. These two outcomes of replication stress are likely to be linked since increased replication initiation and depletion of nucleotide supplies slows replication (Bester et al., 2011; Jones et al., 2012), while slow fork progression causes activation of dormant origins (Ge et al., 2007), and both incomplete replication and increased origin firing are monitored by ATR activity (Shechter et al., 2004). In conclusion, increased initiating events at ERFSs and a paucity of replication initiation at CFSs could both challenge replication fidelity.
ERFSs and cancer
Oncogenic stress is a major driving force in the early stages of cancer development (Halazonetis et al., 2008); nevertheless, the factors that trigger replicative stress in vivo remain unclear. In the case of B cell lymphomas, oncogenic stress can be initiated by the activity of AID, which by targeting non-Ig genes such as c-myc (Robbiani et al., 2008), leads to c-myc/IgH translocations and consequent aberrant c-myc expression. This form of AID induced oncogenic stress or high levels of proliferative activity in activated B cells could generate DNA damage at ERFSs (Figure 6).
Altogether 103 AID hotspots (Chiarle et al., 2011; Klein et al., 2011) —including the GIMAP cluster, MHCII locus, and IKZF1—were also identified as ERFS hotspots in this study (Table S1). It is possible that the overlap observed between a subset of off-target AID sites and ERFSs is due to common underlying features of these loci. For example, AID is recruited to ssDNA regions (Chaudhuri and Alt, 2004), which are also generated during replicative stress; AID-dependent DSBs and ERFSs are also both enriched in repeat elements (Staszewski et al., 2011). In addition, chromosomal regions with the highest transcriptional activity have the highest AID-dependent translocation density (Chiarle et al., 2011; Klein et al., 2011), and early origins and translocations frequently reside near transcription start sites and RNA polymerase II binding sites (Chiarle et al., 2011; Klein et al., 2011). Thus, these euchromatic regions could serve both as AID targets in G1 and also be susceptible to fork collapse during early S phase.
A number of different hypotheses have been put forward about the mechanisms that promote recurrent translocations in mature B cell lymphomas. These include recurrent genomic damage by AID, random DNA damage followed by selection, and a non-random three-dimensional organization of the genome (Chiarle et al., 2011; Hakim et al., 2012; Klein et al., 2011; Zhang et al., 2012b). To date, replication stress-induced DNA damage has been associated with late-replicating CFS. By using an alternative experimental approach for the discovery of fragile site expression during early replication, we have identified a novel source of recurrent AID-independent DNA breaks that may play a mechanistic role in some of the most common genome rearrangements during B cell lymphomagenesis. Because transcriptional activity and replication timing of a genomic region vary among different cell lineages (Hansen et al., 2010), different sets of ERFSs might also account for recurrent chromosomal rearrangements in cancers of distinct cellular origins.
Experimental Procedures
Mice
XRCC2−/−(Frappart et al., 2009), 53BP1−/− (Ward et al., 2004), Igκ AID (Robbiani et al., 2009), AID−/− (Muramatsu et al., 2000) and SWAP70−/− (Borggrefe et al., 2001) mice have been described. SWAP70−/− and WT control mice used in Figure 4D are C57BL/6 background, all other mice are 129/Sv x C57BL/6 background.
ChIP-Seq, Repli-Seq, RNA-Seq, DHS I Mapping, and FISH analysis
ChIP-Seq and RNA-Seq procedures were performed as in (Yamane et al., 2011), Repli-Seq was performed as described in (Hansen et al., 2010). DHSI mapping was performed as described (Sekimata et al., 2009), and FISH analysis is described in (Callen et al., 2007). For detailed methods, see Extended Experimental Procedures.
BACs
Individual BACs to ERFSs were identified using NCBI clone finder, and purchased from BACPAC For complete list of BAC probes used in FISH experiments see Extended Experimental Procedures.
Retroviral infection
Cells were infected with pMX-c-Myc-IRES-GFP or empty vector and GFP-positive cells were sorted as described (Robbiani et al., 2008).
Statistical and computational analyses
Detailed description is available in Extended Experimental Procedures.
Supplementary Material
Early replicating fragile sites (ERFS) break spontaneously during replication
ERFS translocate to AID-induced breaks
Recurrent genomic aberrations in B cell lymphomas map to ERFS
ERFS are distinct from late replicating common fragile sites
Acknowledgments
We thank Rolf Jessberger and Alessandra Pernis for SWAP70−/− mice, Mila Jankovic for pMX-c-Myc-IRES-GFP, Jean Gautier, Mirit Aladjem and Toren Finkel for discussions and Lars Grontved for help with DHS mapping. This work was supported by the Intramural Research Program of the NIH, the National Cancer Institute, and the Center for Cancer Research, and by a Department of Defense grant to A.N. (BC102335).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bartek J, Bartkova J, Lukas J. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene. 2007;26:7773–7779. doi: 10.1038/sj.onc.1210881. [DOI] [PubMed] [Google Scholar]
- Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, Bensimon A, Zamir G, Shewach DS, Kerem B. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell GR, Greenman CD, Davies H, Butler AP, Edkins S, Andrews JM, Buck G, Chen L, Beare D, Latimer C, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–898. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borggrefe T, Keshavarzi S, Gross B, Wabl M, Jessberger R. Impaired IgE response in SWAP-70-deficient mice. European journal of immunology. 2001;31:2467–2475. doi: 10.1002/1521-4141(200108)31:8<2467::aid-immu2467>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Bothmer A, Robbiani DF, Feldhahn N, Gazumyan A, Nussenzweig A, Nussenzweig MC. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. The Journal of experimental medicine. 2010;207:855–865. doi: 10.1084/jem.20100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen E, Jankovic M, Difilippantonio S, Daniel JA, Chen HT, Celeste A, Pellegrini M, McBride K, Wangsa D, Bredemeyer AL, et al. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 2007;130:63–75. doi: 10.1016/j.cell.2007.06.016. [DOI] [PubMed] [Google Scholar]
- Cha RS, Kleckner N. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science. 2002;297:602–606. doi: 10.1126/science.1071398. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nature reviews Immunology. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- Chen HT, Bhandoola A, Difilippantonio MJ, Zhu J, Brown MJ, Tai X, Rogakou EP, Brotz TM, Bonner WM, Ried T, et al. Response to RAG-mediated VDJ cleavage by NBS1 and gamma-H2AX. Science. 2000;290:1962–1965. doi: 10.1126/science.290.5498.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarle R, Zhang Y, Frock RL, Lewis SM, Molinie B, Ho YJ, Myers DR, Choi VW, Compagno M, Malkin DJ, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nature reviews Molecular cell biology. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa S, Blow JJ. The elusive determinants of replication origins. EMBO reports. 2007;8:332–334. doi: 10.1038/sj.embor.7400954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daboussi F, Courbet S, Benhamou S, Kannouche P, Zdzienicka MZ, Debatisse M, Lopez BS. A homologous recombination defect affects replication-fork progression in mammalian cells. Journal of cell science. 2008;121:162–166. doi: 10.1242/jcs.010330. [DOI] [PubMed] [Google Scholar]
- Debatisse M, Le Tallec B, Letessier A, Dutrillaux B, Brison O. Common fragile sites: mechanisms of instability revisited. Trends in genetics: TIG. 2012;28:22–32. doi: 10.1016/j.tig.2011.10.003. [DOI] [PubMed] [Google Scholar]
- Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448:445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- Durkin SG, Glover TW. Chromosome fragile sites. Annual review of genetics. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- Feng W, Collingwood D, Boeck ME, Fox LA, Alvino GM, Fangman WL, Raghuraman MK, Brewer BJ. Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nature cell biology. 2006;8:148–155. doi: 10.1038/ncb1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frappart PO, Lee Y, Russell HR, Chalhoub N, Wang YD, Orii KE, Zhao J, Kondo N, Baker SJ, McKinnon PJ. Recurrent genomic alterations characterize medulloblastoma arising from DNA double-strand break repair deficiency. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1880–1885. doi: 10.1073/pnas.0806882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge XQ, Jackson DA, Blow JJ. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes & development. 2007;21:3331–3341. doi: 10.1101/gad.457807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostissa M, Alt FW, Chiarle R. Mechanisms that promote and suppress chromosomal translocations in lymphocytes. Annual review of immunology. 2011;29:319–350. doi: 10.1146/annurev-immunol-031210-101329. [DOI] [PubMed] [Google Scholar]
- Hakim O, Resch W, Yamane A, Klein I, Kieffer-Kwon KR, Jankovic M, Oliveira T, Bothmer A, Voss TC, Ansarah-Sobrinho C, et al. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature. 2012;484:69–74. doi: 10.1038/nature10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- Hansen RS, Thomas S, Sandstrom R, Canfield TK, Thurman RE, Weaver M, Dorschner MO, Gartler SM, Stamatoyannopoulos JA. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:139–144. doi: 10.1073/pnas.0912402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashash N, Johnson AL, Cha RS. Regulation of fragile sites expression in budding yeast by MEC1, RRM3 and hydroxyurea. Journal of cell science. 2011;124:181–185. doi: 10.1242/jcs.077313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmrich A, Ballarino M, Tora L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Molecular cell. 2011;44:966–977. doi: 10.1016/j.molcel.2011.10.013. [DOI] [PubMed] [Google Scholar]
- Helmrich A, Stout-Weider K, Hermann K, Schrock E, Heiden T. Common fragile sites are conserved features of human and mouse chromosomes and relate to large active genes. Genome research. 2006;16:1222–1230. doi: 10.1101/gr.5335506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Lucas I, Young DJ, Davis EM, Karrison T, Rest JS, Le Beau MM. Common fragile sites are characterized by histone hypoacetylation. Human molecular genetics. 2009;18:4501–4512. doi: 10.1093/hmg/ddp410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RM, Mortusewicz O, Afzal I, Lorvellec M, Garcia P, Helleday T, Petermann E. Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress. Oncogene. 2012 doi: 10.1038/onc.2012.387. [DOI] [PubMed] [Google Scholar]
- Kato L, Begum NA, Burroughs AM, Doi T, Kawai J, Daub CO, Kawaguchi T, Matsuda F, Hayashizaki Y, Honjo T. Nonimmunoglobulin target loci of activation-induced cytidine deaminase (AID) share unique features with immunoglobulin genes. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2479–2484. doi: 10.1073/pnas.1120791109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein IA, Resch W, Jankovic M, Oliveira T, Yamane A, Nakahashi H, Di Virgilio M, Bothmer A, Nussenzweig A, Robbiani DF, et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell. 2011;147:95–106. doi: 10.1016/j.cell.2011.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Taki T, Chinen Y, Tsutsumi Y, Ohshiro M, Kobayashi T, Matsumoto Y, Kuroda J, Horiike S, Nishida K, et al. Identification of IGHCdelta-BACH2 fusion transcripts resulting from cryptic chromosomal rearrangements of 14q32 with 6q15 in aggressive B-cell lymphoma/leukemia. Genes, chromosomes & cancer. 2011;50:207–216. doi: 10.1002/gcc.20845. [DOI] [PubMed] [Google Scholar]
- Lenz G, Wright GW, Emre NC, Kohlhammer H, Dave SS, Davis RE, Carty S, Lam LT, Shaffer AL, Xiao W, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13520–13525. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letessier A, Millot GA, Koundrioukoff S, Lachages AM, Vogt N, Hansen RS, Malfoy B, Brison O, Debatisse M. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011;470:120–123. doi: 10.1038/nature09745. [DOI] [PubMed] [Google Scholar]
- Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- Liu P, Carvalho CM, Hastings PJ, Lupski JR. Mechanisms for recurrent and complex human genomic rearrangements. Current opinion in genetics & development. 2012;22:211–220. doi: 10.1016/j.gde.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine DM, Rodriguez HK, Bell SP. Coordination of replication and transcription along a Drosophila chromosome. Genes & development. 2004;18:3094–3105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiology and molecular biology reviews: MMBR. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Ozeri-Galai E, Lebofsky R, Rahat A, Bester AC, Bensimon A, Kerem B. Failure of origin activation in response to fork stalling leads to chromosomal instability at fragile sites. Molecular cell. 2011;43:122–131. doi: 10.1016/j.molcel.2011.05.019. [DOI] [PubMed] [Google Scholar]
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Molecular cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen S, Casellas R, Reina-San-Martin B, Chen HT, Difilippantonio MJ, Wilson PC, Hanitsch L, Celeste A, Muramatsu M, Pilch DR, et al. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 2001;414:660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveendranathan M, Chattopadhyay S, Bolon YT, Haworth J, Clarke DJ, Bielinsky AK. Genome-wide replication profiles of S-phase checkpoint mutants reveal fragile sites in yeast. The EMBO journal. 2006;25:3627–3639. doi: 10.1038/sj.emboj.7601251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, Bolland DJ, Chen HT, Corcoran AE, Nussenzweig A, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani DF, Bunting S, Feldhahn N, Bothmer A, Camps J, Deroubaix S, McBride KM, Klein IA, Stone G, Eisenreich TR, et al. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Molecular cell. 2009;36:631–641. doi: 10.1016/j.molcel.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Wu H, Jasin M. A Distinct Replication Fork Protection Pathway Connects Fanconi Anemia Tumor Suppressors to RAD51-BRCA1/2. Cancer cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimata M, Perez-Melgosa M, Miller SA, Weinmann AS, Sabo PJ, Sandstrom R, Dorschner MO, Stamatoyannopoulos JA, Wilson CB. CCCTC-binding factor and the transcription factor T-bet orchestrate T helper 1 cell-specific structure and function at the interferon-gamma locus. Immunity. 2009;31:551–564. doi: 10.1016/j.immuni.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechter D, Costanzo V, Gautier J. ATR and ATM regulate the timing of DNA replication origin firing. Nature cell biology. 2004;6:648–655. doi: 10.1038/ncb1145. [DOI] [PubMed] [Google Scholar]
- Sonoda E, Sasaki MS, Buerstedde JM, Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y, Takeda S. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. The EMBO journal. 1998;17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, Gingeras T, Gilbert DM, Groudine M, Bender M, Kaul R, Canfield T, et al. An encyclopedia of mouse DNA elements (Mouse ENCODE) Genome biology. 2012;13:418. doi: 10.1186/gb-2012-13-8-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staszewski O, Baker RE, Ucher AJ, Martier R, Stavnezer J, Guikema JE. Activation-induced cytidine deaminase induces reproducible DNA breaks at many non-Ig Loci in activated B cells. Molecular cell. 2011;41:232–242. doi: 10.1016/j.molcel.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan AK, Kliszczak M, Morrison CG. The Nse2/Mms21 SUMO ligase of the Smc5/6 complex in the maintenance of genome stability. FEBS letters. 2011;585:2907–2913. doi: 10.1016/j.febslet.2011.04.067. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Nasmyth K. Association of RPA with chromosomal replication origins requires an Mcm protein, and is regulated by Rad53, and cyclin- and Dbf4-dependent kinases. The EMBO journal. 1998;17:5182–5191. doi: 10.1093/emboj/17.17.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo LI, Murga M, Zur R, Soria R, Rodriguez A, Martinez S, Oyarzabal J, Pastor J, Bischoff JR, Fernandez-Capetillo O. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nature structural & molecular biology. 2011;18:721–727. doi: 10.1038/nsmb.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai AG, Lu H, Raghavan SC, Muschen M, Hsieh CL, Lieber MR. Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell. 2008;135:1130–1142. doi: 10.1016/j.cell.2008.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward IM, Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. The Journal of biological chemistry. 2001;276:47759–47762. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- Ward IM, Reina-San-Martin B, Olaru A, Minn K, Tamada K, Lau JS, Cascalho M, Chen L, Nussenzweig A, Livak F, et al. 53BP1 is required for class switch recombination. The Journal of cell biology. 2004;165:459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane A, Resch W, Kuo N, Kuchen S, Li Z, Sun HW, Robbiani DF, McBride K, Nussenzweig MC, Casellas R. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nature immunology. 2011;12:62–69. doi: 10.1038/ni.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane A, Robbiani DF, Resch W, Bothmer A, Nakahashi H, Oliveira T, Rommel PC, Brown EJ, Nussenzweig A, Nussenzweig MC, et al. RPA accumulation during class switch recombination represents 5′–3′ DNA end resection during S-G2/M phase of the cell cycle. Cell reports. 2013 doi: 10.1016/j.celrep.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012a;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, MacLennan IC, Liu YJ, Lane PJ. Is rapid proliferation in B centroblasts linked to somatic mutation in memory B cell clones? Immunology letters. 1988;18:297–299. doi: 10.1016/0165-2478(88)90178-2. [DOI] [PubMed] [Google Scholar]
- Zhang Y, McCord RP, Ho YJ, Lajoie BR, Hildebrand DG, Simon AC, Becker MS, Alt FW, Dekker J. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012b;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.