Summary
The Cajal body (CB) is a domain of concentrated components found within the nucleus of cells in an array of species that is functionally important for the biogenesis of telomerase and small nuclear ribonucleoproteins. The CB is a dynamic structure whose number and size change during the cell cycle and is associated with other nuclear structures and gene loci. Coilin, also known as the marker protein for the CB, is a phosphoprotein widely accepted for its role in maintaining CB integrity. Recent studies have been done to further elucidate functional activities of coilin apart from its structural role in the CB in an attempt to explore the rationale for coilin expression in cells that have few CBs or lack them altogether. Here we show that the RNA association profile of coilin changes in mitosis with respect to that during interphase. We provide evidence of transcriptional and/or processing dysregulation of several CB-related RNA transcripts as a result of ectopic expression of both wild-type and phosphomutant coilin proteins. We also show apparent changes in transcription and/or processing of these transcripts upon coilin knockdown in both transformed and primary cell lines. Additionally, we provide evidence of specific coilin RNase activity regulation, on both U2 and hTR transcripts, by phosphorylation of a single residue, serine 489. Collectively, these results point to additional functions for coilin that are regulated by phosphorylation.
Key words: Cajal body, snRNA, Telomerase
Introduction
The protein coilin was identified in 1991 during a screen of sera from patients with autoantibody diseases (Andrade et al., 1991). Since coilin was the first highly enriched protein identified in the Cajal body (CB), it has become known as the CB marker protein. CBs are nuclear structures that participate in the biogenesis of ribonucleoproteins (RNPs) involved in splicing (small nuclear ribonucleoproteins, snRNPs), telomere maintenance (telomerase) and rRNA processing (small nucleolar ribonucleoproteins, snoRNPs) (Machyna et al., 2013). Although coilin is known as the CB marker protein, it is constitutively expressed in all human cells, including those that lack or have few CBs (Young et al., 2000). Studies have shown that coilin is essential for the formation and composition of CBs in several knockout and knockdown models, such as human cell lines, mouse, zebrafish, Arabidopsis and Drosophila (Collier et al., 2006; Lemm et al., 2006; Liu et al., 2009; Strzelecka et al., 2010b; Tucker et al., 2001). In addition to accumulations in the CB, coilin is also localized in the nucleoplasm. In fact, the majority of coilin is found in the nucleoplasm, not the CB (Lam et al., 2002). The function of this nucleoplasmic pool of coilin remains elusive, as does coilin function in cell types or primary cell lines, such as WI-38, that do not have many robust CBs (Young et al., 2000). Therefore, until recently, coilin could be thought of as a crucial factor necessary to bring together the players required for CB function, but was not implicated directly in any of the activities ascribed to the CB. We have found that coilin can bind both RNA and DNA and has RNase activity (Broome and Hebert, 2012), hinting that coilin takes part directly in aspects of CB function. These findings also indicate that coilin serves a purpose in cell types and lines that lack or have few CBs.
Regardless of its exact function, knockdown and knockout studies in human cell lines and mouse and zebrafish animal models, but not Arabidopsis and Drosophila models, demonstrate that reduced levels of coilin result in decreased viability and proliferation (Collier et al., 2006; Lemm et al., 2006; Liu et al., 2009; Strzelecka et al., 2010b; Tucker et al., 2001; Walker et al., 2009). In the zebrafish model, lethality caused by coilin depletion can be rescued by the addition of snRNPs, highlighting the importance of coilin in CB formation and activity. Other components present in CBs include small Cajal body-specific RNAs (scaRNAs), which help guide modifications that take place on the small nuclear RNA component of snRNPs, and SMN, the survivor of motor neuron protein. Loss of SMN causes most cases of spinal muscular atrophy, the leading genetic cause of infant mortality (Faustino and Cooper, 2003).
In addition to its localization in the CB and the nucleoplasm, certain conditions relocate coilin to the nucleolus or its periphery. These conditions include DNA damage, such as that caused by cisplatin (Gilder et al., 2011), transcription inhibition (Carmo-Fonseca et al., 1992), hypomethylation of coilin (Tapia et al., 2010), coilin overexpression (Hebert and Matera, 2000) or knockdown of SMN (Lemm et al., 2006). Although the reason behind this association of coilin with the nucleolus is not clear, we have found that coilin depletion increases the association of RNA pol I with rDNA (Gilder et al., 2011). We have also found that nucleoli that have coilin accumulations have less RNA pol I activity than other nucleoli within the same cell (Gilder et al., 2011). These findings indicate that coilin may take part in the regulation of rDNA transcription or precursor rRNA processing. In support of this hypothesis, we have observed that coilin immunoprecipitation complexes contain 47/45S pre-processed rRNA (Broome and Hebert, 2013).
We have also observed that coilin immunoprecipitation complexes contain U2 snRNA (Broome and Hebert, 2013). CBs are known to associate with certain gene loci, including those that give rise to U1 and U2 snRNA (Frey and Matera, 1995; Smith et al., 1995) and this association is dependent upon U snRNA gene transcription (Frey et al., 1999; Frey and Matera, 2001). U1 snRNA, U2 snRNA and the RNA component of telomerase (hTR) are initially produced as longer transcripts that require 3′ end processing (Jády et al., 2003; Nesic et al., 2004; Theimer et al., 2007; Zhu et al., 2004). U1 and U2 snRNA 3′ end processing is facilitated by the Integrator complex in order to generate the appropriately sized mature RNA that is incorporated into the U1 or U2 snRNP (Baillat et al., 2005; Egloff et al., 2007), but it is not known how hTR processing takes place in mammalian cells. WRAP53 (also known as WDR79 or TCAB), which interacts with coilin and is required for CB formation, is required for hTR and scaRNA localization to the CB (Mahmoudi et al., 2010; Stern et al., 2012; Tycowski et al., 2009; Venteicher et al., 2009). We have also found that hTR is highly enriched in coilin immunoprecipitation complexes (Broome and Hebert, 2013). Based on our observations that coilin binds DNA, RNA and has RNase activity, we have hypothesized that coilin is important for the association of CBs with U snRNA gene loci and the 3′ end processing of U1 snRNA, U2 snRNA and hTR (Broome and Hebert, 2012; Broome and Hebert, 2013).
Since it is known that coilin is a phosphoprotein, and the amount of this phosphorylation increases during mitosis (Carmo-Fonseca et al., 1993), it is logical to examine how this post-translational modification regulates coilin activity. In particular, we are interested in investigating how phosphorylation impacts coilin association with RNAs known to accumulate in the CB or associate with coilin. We are also interested in determining if phosphorylation modulates coilin RNA degradation activity. Towards this end, we have examined the association of hyperphosphorylated coilin with RNAs and monitored the level of these RNAs in the presence of overexpressed wild-type (WT) or phosphomutant coilin proteins. Additionally, we have examined the relative levels of precursor and total RNAs upon coilin knockdown in both primary and transformed cell lines. We have also conducted in vitro degradation assays with purified, nucleic acid free WT, phosphomimic, or deletion coilin proteins using U2 snRNA or hTR substrates. The results of these experiments demonstrate that phosphorylation regulates coilin activity and association with RNA. The results presented here also indicate that coilin levels may influence the transcription of certain genes and or processing of specific RNAs.
Results
Coilin hyperphosphorylation correlates with a decrease in its association with U2 snRNA and hTR
We have found (Broome and Hebert, 2013) that coilin associates with specific RNA and DNA. In particular, we have observed that coilin immunoprecipitation complexes contain a 3.6-fold enrichment of U2 snRNA and a 17.5-fold enrichment of hTR relative to control IgG reactions in untreated HeLa cells (Broome and Hebert, 2013). These untreated data are also shown in Fig. 1 for reference. To determine if the phosphorylation of coilin alters the amount of RNA found in the coilin immunoprecipitation complex, nocodazole was used to enrich for cells in mitosis. Coilin is a phosphoprotein, and the amount of this phosphorylation increases during mitosis (Carmo-Fonseca et al., 1993). Evidence from MS/MS analysis indicates that at least four residues, in addition to those that are phosphorylated during interphase, are phosphorylated specifically during mitosis (Beausoleil et al., 2004; Dephoure et al., 2008; Toyota et al., 2010). Relative to untreated cells, therefore, coilin is hyperphosphorylated in nocodazole treated cells. For these experiments, lysate was immunoprecipitated with control (purified rabbit IgG) or coilin antibodies, followed by extensive washing of the beads and RNA isolation. The isolated RNA was then subjected to reverse transcriptase real-time PCR using primers specific to RNAs known to accumulate in the CB (U1 snRNA, U2 snRNA and hTR). Given that certain conditions, such as DNA damage (Gilder et al., 2011), result in the relocalization of coilin to the nucleolus, we also examined the association of coilin with the RNA pol I derived pre-processed (47/45S) and processed 5.8S rRNAs. Associated GAPDH message level was used as a control. Immunoprecipitation complexes containing hyperphosphorylated coilin contain significantly less U2 snRNA and hTR than that recovered from untreated cells (Fig. 1, nocodazole), demonstrating that increased coilin phosphorylation correlates with a reduction in the association of coilin with these RNAs. In contrast, the amount of pre-rRNA (47/45S) recovered in the coilin complex does not change after nocodazole treatment.
Fig. 1. Mitotic arrest results in differential RNA association with coilin.
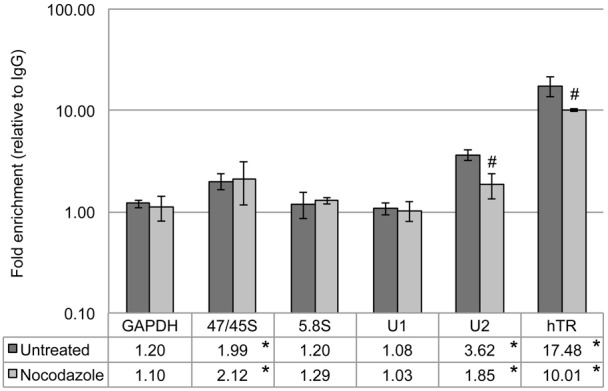
Untreated or nocodazole treated HeLa cell lysate was used for RNA immunoprecipitations with rabbit IgG and α-coilin antibodies. RNA isolated from the IP complexes was analyzed by qRT-PCR with specific primers. Histogram of qRT-PCR data from RNA IP with IgG or α-coilin in untreated or nocodazole treated HeLa cells; fold enrichment of α-coilin IP over IgG control is shown on a log10 scale; specific primer target RNA is noted beneath each column pair; error bars represent 1 s.d. (data derived from at least 3 independent experiments with at least 2 technical repeats per experiment); *P<0.02 relative to GAPDH within a treatment; #P<4E−4 relative to untreated. The data for the untreated samples have been reported elsewhere (Broome and Hebert, 2013).
Transient expression of coilin wild type or S489 phosphomutants differentially impact the level of specific RNAs
Transient expression of coilin wild type (WT) and various phosphomutants demonstrate that mutation of the S489 phosphorylation site to aspartic acid (S489D), which mimics constitutive phosphorylation, results in the reduction of cell proliferation compared to cells with ectopically expressed WT coilin (Carrero et al., 2011). The phosphorylation of coilin S489 is enriched during mitosis (Beausoleil et al., 2004), indicating that this may be a mechanism for regulation of coilin activity. Since we have found that coilin directly binds RNA and DNA (Broome and Hebert, 2012), and can associate with U2 gene loci, as assessed by chromatin immunoprecipitation (ChIP) (Broome and Hebert, 2013), we decided to test if the transient expression of GFP-coilin (S489D) alters the message levels of specific RNAs. Towards this end, cells were transfected with GFP-tagged WT, phosphomimic (S489D) or phosphonull (S489A) coilin constructs, followed by RNA isolation and detection of messages by reverse transcriptase real-time PCR (qRT-PCR). The level of RNAs known to localize to the CB, such as U1 and U2 snRNA and hTR, as well as the pre-processed forms of these transcripts, were examined. As mentioned above, RNA pol I derived transcripts (the pre-processed 47/45S and the processed 5.8S rRNAs) were also examined since coilin nucleolar relocalization is observed in certain stress conditions. The levels of these various RNAs were normalized to the amount of GAPDH in cells expressing GFP. To our surprise, ectopic expression of GFP-coilin WT significantly decreased the levels of all RNAs tested except for U2 snRNA, relative to GAPDH and normalized to cells expressing GFP alone (Fig. 2). We have shown previously that GFP-coilin WT overexpression results in the decrease of both hTR and pre-hTR levels (Broome and Hebert, 2013), and this result was also found in this study (Fig. 2). We then tested if the transient expression of S489 phosphomutants would likewise decrease specific RNA messages as found for WT coilin expression. For the most part, message levels in cells expressing GFP-tagged S489D or S489A were reduced in a similar trend as observed for cells expressing GFP-coilin (Fig. 2). However, there are a few notable differences. The most striking of which is the decrease observed in the level of mature U2 snRNA in cells expressing GFP-coilin (S489A) compared to the relative increase of this snRNA in GFP-coilin or GFP-coilin (S489D) expressing cells. Regarding S489D expression, the amount of pre-U1 is not reduced by this protein to the same extent as that found upon the expression of WT or S489A coilin. When comparing RNA levels from cells expressing GFP-coilin to that found in cells expressing S489D or S489A, WT coilin is most divergent from the phosphomutant proteins in the level of U1 snRNA and pre-hTR. In summary, these results demonstrate that coilin overexpression reduces the level of several RNAs known to accumulate in the CB or associate with coilin. The extent of this change varies in cells expressing S489D or S489A phosphomutant proteins. Taken together, these findings suggest that coilin overexpression decreases the transcription of these genes and/or increases the degradation or processing of these specific RNAs, and that mimicking phosphorylation of serine 489 alters this profile for certain transcripts.
Fig. 2. Coilin phosphomutants alter non-coding RNA levels.
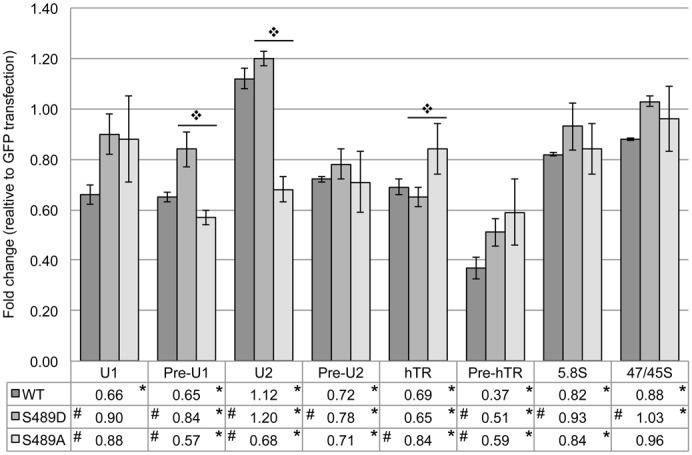
HeLa cells were transfected with various GFP expression vectors, the RNA isolated and then analyzed by qRT-PCR with specific primers. Histogram of qRT-PCR data from HeLa cells transfected with GFP, GFP-coilin wild type (WT), GFP-coilin S489D (S489D) or GFP-coilin S489A (S489A); fold change relative to both GFP transfection and GAPDH amplification is shown; specific primer target RNA is noted beneath each column group; error bars represent 1 s.d. (data derived from at least 3 independent experiments with at least 2 technical repeats for each experiment, although one set has an n of 5); *P<0.05 relative to GAPDH; #P<0.05 relative to WT within a primer set;  P<0.05 between S489D and S489A.
P<0.05 between S489D and S489A.
Coilin knockdown results in transcription and/or processing consequences in both transformed and primary cell lines
In addition to exploring how the presence of ectopic coilin affects RNA expression and processing, we examined what effect the transient knockdown of endogenous protein would have on these transcripts. We chose to examine total U2 snRNA, pre-U2 snRNA, 47/45S rRNA and 5.8S rRNA given their association with coilin and/or altered levels upon coilin overexpression. We utilized both HeLa and WI-38 cell lines for these experiments due to the fact that they differ not only in terms of transformed/non-transformed status but also in the number and prominence of CBs. While HeLa cells contain 2–6 CBs per cell (Leeflang et al., 1992), only 2–3% of WI-38 cells have a single CB (Spector et al., 1992). Cells were transfected with either a control non-targeting siRNA or one targeting coilin mRNA for 48 hours, and then the RNA was analyzed by qRT-PCR using specific primers. We have previously shown that overexpression of coilin results in a significant decrease in relative 47/45S pre-rRNA levels in HeLa cells (Gilder et al., 2011), and this result was also observed in this study (Fig. 2). In Fig. 3A, we show that following a 48-hour knockdown of coilin in HeLa cells, there are no significant changes in the levels of either 5.8S rRNA or 47/45S pre-processed rRNA. We have previously published that coilin knockdown in HeLa cells results in a significant decrease in total U2 snRNA and an increase in the pre-processed U2 transcript (Broome and Hebert, 2012). Also, we have found that coilin knockdown in HeLa cells results in a significant decrease in pre-hTR, and that coilin overexpression results in a significant decrease in both total and pre-processed hTR (Broome and Hebert, 2013). In Fig. 3B, we show the effect of coilin knockdown on U2 snRNA, hTR and rRNA levels in WI-38 cells. While coilin knockdown resulted in a nearly 1.6 fold increase in total U2 levels, there was essentially no change in the levels of the pre-processed U2 transcript. This is an interesting finding due to the fact that an opposite trend is seen in HeLa cells following coilin knockdown (Broome and Hebert, 2012), in which there is an increase of pre-processed and a small decrease of total U2 snRNA. In agreement with what we find in HeLa cells (Broome and Hebert, 2013), we find a slight decrease in total hTR with a 57% decrease in pre-hTR upon coilin knockdown in WI-38 cells. In addition, we find a 1.9 fold increase in the 47/45S pre-rRNA with no change in 5.8S rRNA levels. Interestingly, the relative amount of coilin message is 2.5 fold greater in WI-38 versus HeLa (Fig. 3C). Collectively, these data suggest a role for coilin in the transcription or processing of specific RNAs in cell types with or without CBs.
Fig. 3. Coilin levels correlate with altered nascent U2 snRNA, hTR and rRNA levels.
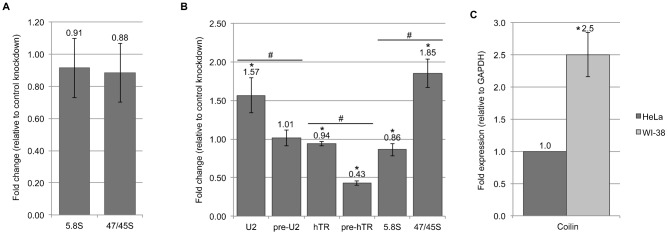
RNA isolated from HeLa or WI-38 cells following RNAi targeting coilin was analyzed by qRT-PCR using primers specific to U2 snRNA, telomerase RNA and rRNA. (A) Histogram of relative 47/45S precursor rRNA to 5.8S rRNA following 48-hour control or coilin knockdown in HeLa cells resulting in 87% reduction of coilin mRNA. (B) Histogram of the fold change of specific RNA transcripts following 48-hour coilin knockdown in WI-38 cells resulting in an average 96% reduction of coilin mRNA; *P<0.05 relative to control knockdown; #P<0.05 between data pairs. (C) Histogram of coilin expression, relative to GAPDH, in HeLa and WI-38 cells treated with control siRNA for 48 hours; *P<0.05 relative to HeLa. Error bars represent 1 s.d. of 3 experimental repeats (2 for WI-38 hTR) and 3 technical repeats within each experiment. All message levels were normalized to GAPDH prior to comparison with either control knockdown or between cell lines.
Bacterially purified coilin (S489D) co-isolates with more nucleic acid than WT coilin
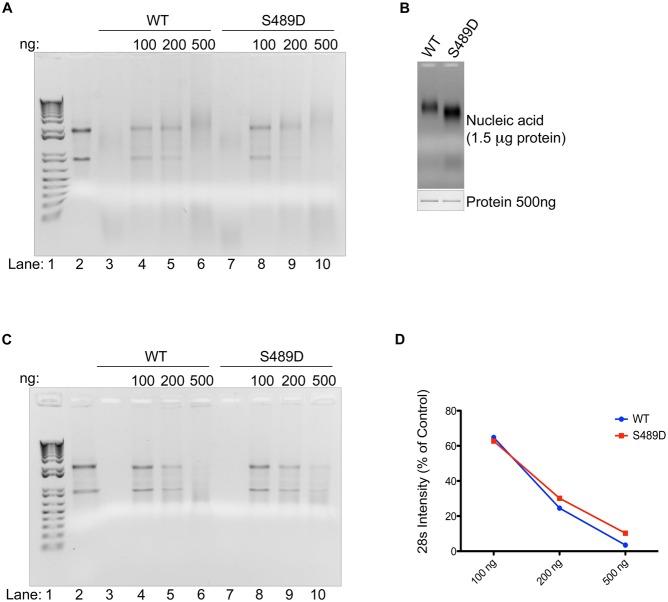
In addition to nucleic acid binding, we have found that coilin has RNase activity (Broome and Hebert, 2012). To further explore the role of S489 phosphorylation on coilin function, with particular emphasis on how this modification may impact the RNase activity of coilin, we purified GST-coilin (S489D) from bacteria using a stringent purification protocol involving removal of the GST-tag and excision of the full-length protein from a SDS-PAGE gel followed by electro-elution (Broome and Hebert, 2012). The RNase activity of S489D when compared to WT coilin was initially found to be equivalent when using total RNA as the substrate (Fig. 4A). Interestingly, the amount of co-purified nucleic acid, which we have previously shown to be mostly RNA (Broome and Hebert, 2012), is greater in S489D compared to WT (Fig. 4B). In order to more definitively characterize the RNase activity of the S489D phosphomutant compared to WT coilin, we utilized a modified coilin purification protocol (Broome and Hebert, 2013) in which nucleic acid free protein is isolated. In brief, protein samples are treated with DNase I and RNase A/T1 before SDS-PAGE and band excision. After electro-elution, the samples are treated with α-cyclodextrin (to strip SDS from the protein) followed by extensive dialysis in PBS containing 250 mM NaCl. The end result of this modified protocol is soluble purified coilin lacking any nucleic acid. Both WT and S489D proteins purified in this manner were subjected to RNA degradation assays using total RNA (Fig. 4C). Both proteins appear to degrade RNA approximately equally, although quantification of the 28S rRNA band demonstrates that there is a very slight reduction of RNase activity in the S489D phosphomutant compared to WT (Fig. 4D).
Fig. 4. Coilin phosphomutant displays differential RNA binding and degradation activities.
Purified proteins were analyzed for RNase activity with total RNA and RNA co-isolation amount. (A) Agarose gel with RNase reactions containing 500 ng HeLa RNA and the indicated amount of nucleic acid-bound coilin wild type (WT) or coilin S489D; lane 1 contains 1 kb+DNA ladder, lane 2 contains RNA alone, lanes 3 and 7 contain 500 ng of protein alone. (B) Agarose gel (top) showing co-purified nucleic acid of 1.5 µg nucleic acid-bound proteins and Coomassie stained SDS-PAGE gel (bottom) of 500 ng nucleic acid bound proteins. (C) Agarose gel with RNase reactions containing 500 ng HeLa RNA and the indicated amount of nucleic acid-free proteins; lane 1 contains 1 kb+DNA ladder, lane 2 contains RNA alone, lanes 3 and 7 contain 500 ng of protein alone. (D) Graph showing densitometric analysis of the 28S bands from C as a percent of the lane 2 control.
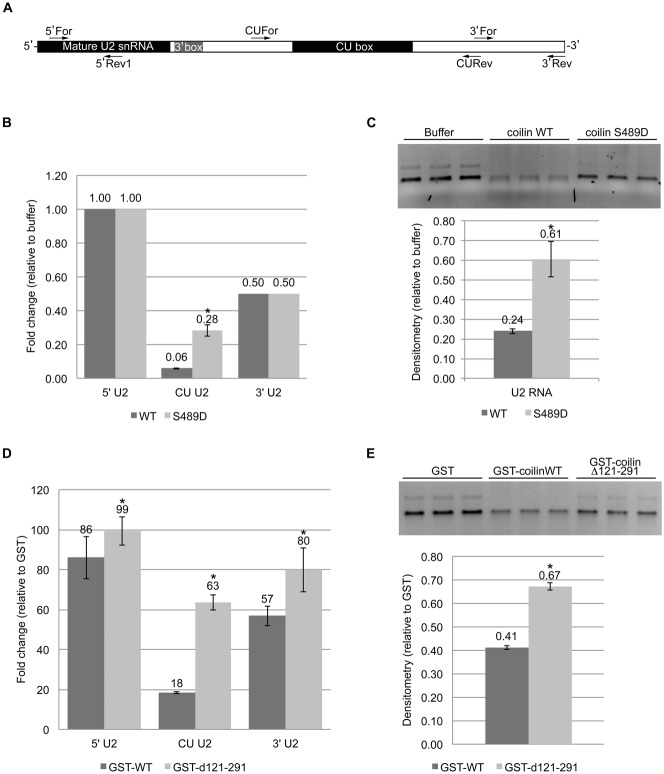
Differential processing of U2 snRNA by coilin WT, S489D and the deletion mutant d121–291
Our previous work demonstrates that in vitro transcribed U2 is selectively degraded by purified coilin, suggesting that coilin may take part in the 3′ end processing of this snRNA (Broome and Hebert, 2012). Specifically, coilin preferentially degraded the CU region of the extended U2 snRNA transcript. Since the coilin protein used in the previous study also contained co-purifying nucleic acid, we wanted to test if purified coilin lacking nucleic acid also shows a similar pattern of specificity. We also tested the activity of the S489D phosphomutant (likewise purified in the absence of nucleic acid) on the processing of an U2 snRNA substrate. For these studies, an 820 nt in vitro transcribed U2 snRNA, containing an extended 3′ that encompasses the CU (CT) box, was used as a substrate for reactions containing buffer or purified nucleic-acid free WT or S489D coilin proteins. After incubation, the reaction mix was DNase treated followed by qRT-PCR and agarose gel electrophoresis. The primers used for qRT-PCR amplify the 5′ end containing the mature region, the middle CU region, and the extreme 3′ end (Fig. 5A). When compared to buffer alone, a 2:1 (nM) ratio of protein to U2 snRNA revealed little degradation of the 5′ end as assessed via reverse transcriptase qRT-PCR (Fig. 5B). In sharp contrast, the amount of the U2 snRNA CU region is reduced 94% in reactions containing WT and 70% for reactions with S489D, relative to control. The U2 snRNA 3′ end is reduced by both WT and S489D proteins to 50% of that found in control reactions. A portion of the same RNase reactions used for the qRT-PCR set-up were also run on an agarose gel, and it is clear that the WT protein shows more degradation of the U2 snRNA substrate while the RNA degradation activity of the S489D protein is relatively blunted (Fig. 5C).
Fig. 5. Coilin mutants display differential U2 processing activity with specificity toward the CU region.
Purified proteins were incubated with in vitro transcribed U2 snRNA and the reactions analyzed by both qRT-PCR and agarose gel electrophoresis. (A) Illustration of U2 snRNA pre-processed transcript with relative primer binding locations for the 5′, CU and 3′ regions; relative length of the mature snRNA is denoted. (B) Histogram of qRT-PCR data following incubation of 100 nM U2 RNA transcript with buffer alone, coilin wild type (WT) or coilin S489D at 200 nM protein. (C) Agarose gel (top) with a portion of the same reactions used as a template for qRT-PCR in B and densitometry graph (bottom) of the major U2 RNA band from gel. (D) Histogram of qRT-PCR data following incubation of 100 nM U2 RNA transcript with GST, GST-coilin WT or GST-coilin d121–291 at 300 ng protein. (E) Agarose gel (top) with a portion of the same reactions used as a template for qRT-PCR in D and densitometry graph (bottom) of the major U2 RNA band from gel. *P<0.05 relative to the wild-type protein in each pair. Three independent incubations of protein (or buffer) with U2 snRNA substrate were conducted. These incubations were used for qRT-PCR with three technical repeats per incubation.
We have previously identified a deletion mutant of coilin, lacking amino acids 121–291 (d121–291), that binds less RNA and has less total RNA degradation activity compared to WT coilin (Broome and Hebert, 2013). Since the S489D mutant has reduced degradation of the U2 snRNA CU region compared to WT, we next wanted to test if the coilin (d121–291) mutant would also show similar impaired U2 snRNA degradation. For these studies, GST, GST-coilin and GST-coilin (d121–291) were purified using the modified protocol (Broome and Hebert, 2013) such that nucleic acid free, dialyzed protein was isolated. Note that for these studies the GST tag was left fused to the WT and d121–291 coilin proteins. The same 820 nt U2 snRNA substrate used above was incubated with GST, GST-coilin or GST-coilin d121–291, followed by DNase treatment, qRT-PCR and agarose gel electrophoresis. Relative to the reactions containing GST, WT coilin only modestly reduces the U2 5′ end, while the d121–291 protein has no effect (Fig. 5D). However, the CU region was reduced 80% in WT reactions, and 37% in d121–291 reactions. The 3′ end was reduced by both proteins, but more so for WT compared to d121–291. The same reactions used as the template for qRT-PCR were also run on an agarose gel for visualization (Fig. 5E), and it is clear that relative to GST, WT coilin is more effective at degrading the U2 snRNA substrate. In contrast, and in agreement with that observed with the S489D mutation, the RNA degradation activity of the d121–291 protein for the U2 snRNA substrate is reduced compared to WT.
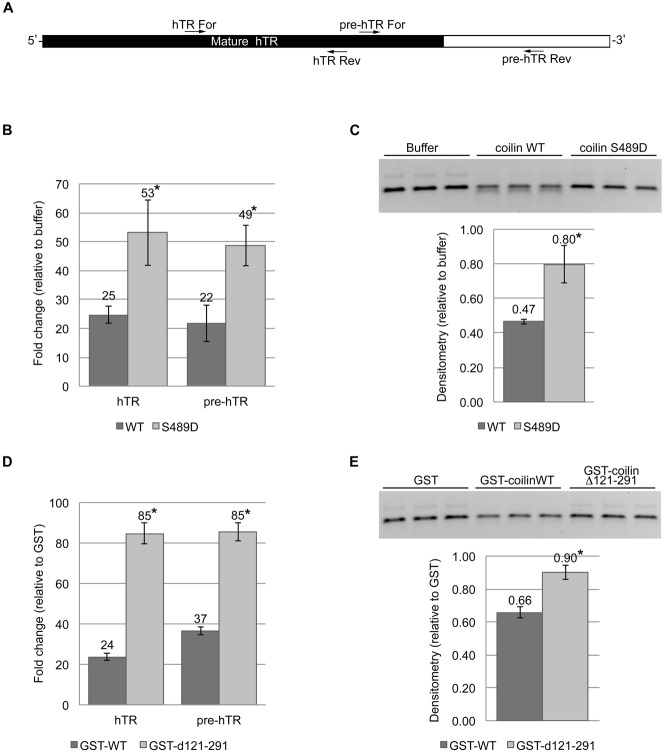
Mutation of S489 and deletion of amino acids 121–291 impair coilin degradation of hTR
In the context of total RNA, incubation with purified WT coilin specifically reduces the amount of the 3′ end of hTR (Broome and Hebert, 2013), which is removed during pre-hTR processing in order to become the mature hTR that is incorporated into telomerase. In contrast, incubation of total RNA with purified coilin d121–291 does not result in reduced hTR 3′ end levels (Broome and Hebert, 2013). To examine if the coilin S489D and d121–291 proteins have reduced degradation activity using hTR as a substrate, as observed with the U2 snRNA substrate as described above, we conducted experiments with hTR that contains an extended 3′ end. Primers were used that amplify the mature region and the 3′ end spanning the processing site (Fig. 6A). For the first set of experiments, substrate was incubated with buffer or nucleic acid free WT or S489D coilin proteins, followed by DNase treatment. The reactions were then used as a template for reverse transcriptase qRT-PCR or were subject to agarose gel electrophoresis. As seen in Fig. 6B, the amount of both the mature hTR and 3′ end was reduced 75% in reactions containing WT coilin compared to control. As found previously for the CU region of the U2 snRNA substrate, this reduction was not as drastic in reactions containing coilin S489D (Fig. 6B). Visualization of the reactions by agarose gel confirms this observation (Fig. 6C).
Fig. 6. Coilin mutants display differential hTR processing activity but lack specificity toward the 5′ or 3′ regions.
(A) Illustration of human telomerase RNA (hTR) pre-processed transcript with relative primer binding locations for 5′ (for amplification of all hTR transcripts) and 3′ primers (which span the cleavage position); black region indicates the relative length of mature hTR. (B) Histogram of qRT-PCR data following incubation of 100 nM hTR transcript with buffer alone, coilin wild type (WT) or coilin S489D at 200 nM protein. (C) Agarose gel (top) with a portion of the same reactions used as a template for qRT-PCR in B and densitometry graph (bottom) of the hTR RNA band from gel. (D) Histogram of qRT-PCR data following incubation of 100 nM hTR transcript with GST, GST-coilin WT or GST-coilin d121–291 at 300 ng protein. (E) Agarose gel (top) with a portion of the same reactions used as a template for qRT-PCR in D and densitometry graph (bottom) of the hTR band from gel. *P<0.05 relative to the wild-type protein in each pair. Three independent incubations of protein with hTR substrate were conducted. These incubations were used for qRT-PCR with three technical repeats per incubation.
To test if the d121–291 mutant will again share a reduced degradation phenotype with S489D using the hTR substrate, reactions were conducted using purified GST, GST-coilin or GST-coilin d121–291. While the amount of both the mature hTR and 3′ end were reduced upon incubation with WT coilin compared to GST, the level of reduction was not as severe in reactions containing d121–291 (Fig. 6D). Agarose gel electrophoresis of the same samples that were used as the template for the qRT-PCR reactions corroborates this observation (Fig. 6E). In summary, therefore, differential processing is observed for hTR upon coilin incubation in the context of total RNA (Broome and Hebert, 2013). However, when using only hTR RNA in a reaction, coilin appears to degrade both the 5′ and 3′ ends without preference. Interestingly, as observed for the U2 snRNA substrate, the S489D and d121–291 coilin mutants both display reduced RNase activity for the hTR substrate compared to WT, with the d121–291 protein being the least active.
Discussion
Numerous publications have clearly shown the importance of coilin in the formation and composition of CBs, justifying its unofficial title as the CB marker protein. In particular, knockout and knockdown studies in human, mouse, zebrafish, Arabidopsis and Drosophila all demonstrate that canonical CB structures are not formed in the absence of coilin (Collier et al., 2006; Lemm et al., 2006; Liu et al., 2009; Strzelecka et al., 2010a; Tucker et al., 2001). Therefore, in an array of species, coilin is known to serve as an important factor necessary for CB formation but is not thought to play a role in the mechanisms that occur within the CB. Our recently published work (Broome and Hebert, 2012) showing that coilin has RNase activity and can bind DNA as well as different types of RNA (Broome and Hebert, 2013) strongly suggests that coilin may take part more directly in CB activity than was previously appreciated. Additionally, the description of these coilin activities will shed light on what coilin may be doing in primary cells, some of which lack CBs. In cell types/lines with CBs, it is known that CBs associate with certain gene loci, for example U2 gene loci (Frey and Matera, 1995; Smith et al., 1995). It is also known that some snRNAs, such as U1 and U2 snRNA, as well as hTR, are generated as longer transcripts that require 3′ end processing and trimming in order to produce the mature snRNA or hTR that is incorporated into snRNPs or telomerase. The Integrator complex plays a role in the 3′ end processing of U2 snRNA (Baillat et al., 2005), and we hypothesize that coilin is also involved in these events and those required for U1 snRNA and hTR processing. In support of this hypothesis, we have shown that coilin preferentially associates with the 3′ end of the U2 gene as determined by ChIP (Broome and Hebert, 2013), suggesting that the DNA binding activity of coilin may in part underlie the association of CBs with U2 (and other) gene loci. Further support for this hypothesis comes from data showing that U2 snRNA can be found in a complex with coilin (H.J.B. and M.D.H., unpublished; this study) and purified coilin, lacking any co-purifying nucleic acid, preferentially cleaves the CU region and 3′ end of pre-processed U2 snRNA (Fig. 5). Since U1 snRNA and hTR are also known to undergo 3′ end processing, we suspect that coilin directly impacts the processing of these transcripts as well, and provide evidence for coilin hTR processing in this work (Fig. 6). Our data here demonstrate that in cells that contain many CBs as well as in those that contain few, there is an apparent dysregulation of U2 snRNA and rRNA transcription and/or processing upon coilin knockdown (Fig. 2). Interestingly, while the trend of a decrease in pre-hTR is consistent for both cell lines, there is an effect on pre-rRNA only in WI-38 cells and opposite effects with the relative levels of pre-processed U2 snRNA (Broome and Hebert, 2012) (Fig. 3). We also show that there is significantly more coilin mRNA in WI-38 cells relative to HeLa cells (Fig. 3). It is a logical assumption that cell transformation and/or CB number would result in these relative U2 snRNA abundance differences between cell lines, but this significant difference in coilin expression could also play a role.
A major focus of the experiments presented here was to determine if phosphorylation impacts coilin RNase activity. Coilin is phosphorylated on at least 11 residues, and this number increases during mitosis (Carmo-Fonseca et al. 1993; Toyota et al., 2010). Since S489 of coilin is preferentially phosphorylated during mitosis (Dephoure et al., 2008), and transient transfection of GFP-coilin S489D decreases cell proliferation (Carrero et al., 2011), we wanted to test if the RNA degradation activity of a purified S489D phosphomimic would differ compared to WT coilin. WT and S489D protein purified with co-isolated nucleic acid did not show any obvious differences in their capacity to degrade total RNA (Fig. 4A), but slightly more nucleic acid, presumably RNA, was co-isolated with S489D protein preparations compared to WT (Fig. 4B). When protein was isolated in the absence of co-purifying nucleic acid, a slight decrease in the RNA degradation activity of S489D compared to WT was observed (Fig. 4C,D), and this difference was more obvious when U2 snRNA and hTR substrates were used (Figs 5, 6). These findings demonstrate that phosphorylation of coilin S489 reduces its RNA degradation activity and suggest that mitotic coilin will have less RNase activity compared to interphase coilin. Thus S489 phosphorylation may be a key event that regulates coilin activity. Interestingly, coilin association with both U2 snRNA and hTR is reduced in mitotic (nocodazole treated) cells (Fig. 1), further supporting the idea that coilin RNA degradation activity is reduced during mitosis. The studies with nucleic acid free purified coilin using a single substrate also strongly indicate that an integral RNA does not mediate the RNase activity of coilin. Rather, the coilin protein itself is responsible for the observed RNA degradation activity.
In terms of RNA degradation, the least active protein used in the studies presented here is the coilin deletion construct, d121–291. We have previously shown that this protein, relative to WT coilin, binds less RNA and has less RNA degradation activity, when using total RNA as a substrate (Broome and Hebert, 2013). The finding that the S489D and d121–291 proteins show a similar reduced RNA degradation activity for both the U2 snRNA and hTR substrates may indicate initially that they share a similar defect. However, mapping experiments demonstrate that the N-terminus of coilin, part of which is deleted in the d121–291 protein, contains the RNase activity (Broome and Hebert, 2013). Consequently, it is likely that the d121–291 protein is missing critical residues responsible for both RNA binding and degradation. In contrast, the S489D phosphomimic protein is not missing any of these amino acids, but may have a conformation that inhibits RNase activity. Prediction programs indicate that while the N-terminus and C-terminus of coilin may have structure, the intermediate 60% of coilin is predicted to be intrinsically disordered (Broome and Hebert, 2012). Although far from proven, the phosphorylation of S489 may impact the folding or accessibility of the N-terminal region of coilin responsible for RNA degradation. Additionally, S489 lies in the region of coilin known to contain a Tudor-like domain (Shanbhag et al., 2010), so it is also possible that S489 phosphorylation alters the structure of this domain.
Lastly, our somewhat surprising observations that coilin overexpression can reduce the RNA levels of several different transcripts (Fig. 2) implies that coilin may serve as a transcriptional regulator for these genes and/or takes part in the degradation or processing of these RNAs. The fact that coilin associates with U snRNA gene loci by ChIP assay also supports this hypothesis (Broome and Hebert, 2013). Among these RNAs are rRNAs generated by pol I. It is known that overexpression of coilin can result in nucleolar localization (Hebert and Matera, 2000). It is also known that endogenous coilin can be found within or around the nucleolus in certain conditions, such as coilin hypomethylation, DNA damage, or transcription inhibition (Carmo-Fonseca et al., 1992; Gilder et al., 2011; Tapia et al., 2010). We have previously reported an inverse relationship between coilin levels and the amount of pre-rRNA in unstressed cells: coilin overexpression reduces pre-rRNA levels while coilin knockdown increases pre-rRNA (Gilder et al., 2011). Since in untreated cells the 47/45S precursor rRNA can be found in a complex with coilin (Broome and Hebert, 2013), (Fig. 1), these findings collectively indicate that coilin is involved in rRNA regulation, either at the level of transcription or processing. Support for regulation of rRNA transcription comes from data showing that coilin knockdown increases RNA pol I occupancy of rDNA (Gilder et al., 2011). Noteworthy in its exception to the general trend of reduced transcript levels in response to coilin overexpression, U2 snRNA levels were not reduced upon overexpression of WT or S489D coilin in HeLa cells. In fact, the level of this snRNA was slightly, but significantly, increased in the presence of these ectopically expressed proteins. In stark contrast, expression of the phosphonull mutant S489A resulted in a 30% decrease of this RNA relative to that found in cells expressing GFP alone. Therefore, phosphorylation regulates this aspect of coilin function, but future studies will be needed before a clear understanding of the mechanism behind these results is obtained. Also of interest is the relationship between coilin and RNA pol III sites of transcription, as a recent report has shown in Xenopus that coilin accumulates in novel nuclear domains known as pearls (Nizami and Gall, 2012). In addition to coilin, pearls contain WRAP53 (WDR79/TCAB), scaRNAs and U3 snoRNA and are thought to participate in the processing of RNA pol III transcripts. Pearls associate at sites of RNA pol III transcription and disassemble upon RNA pol III inhibition. Hence it is possible that the nucleic acid binding and RNase activities of coilin impact transcripts derived from all three eukaryotic RNA polymerases.
Materials and Methods
Cell culture and DNA constructs
Human cervical carcinoma (HeLa) and normal human fetal lung fibroblast (WI38) cells were purchased from the American Type Culture Collection (Manassas, VA, USA). Cell lines were cultured as described previously (Sun et al., 2005), and GFP expression vectors have been described (Hebert and Matera, 2000). GST-coilin wild types (GST-WT and WT) were cloned into pGEX vectors as described previously (Broome and Hebert, 2012). GST-Δ121–291 was generated via the Quick Change Mutagenesis kit from Stratagene (Santa Clara, CA, USA) using pGEX-2T-coilin as template with the following primers: 5′-ATCGTCGCAGGATCCGACAAACTGGCTATAAAACTTGGC-3′ and 5′-GGATCCTGCGACGATTTCACCCTCCTCTAACTGAAATGC-3′. GST-coilin S489D was generated using the Quick Change Mutagenesis kit from GST-coilin wild type. GFP-coilin S489D and GFP-coilin S489A phosphomutants were generated by Quick Change Mutagenesis (Carrero et al., 2011) from a GFP-coilin WT precursor (Hebert and Matera, 2000). pBluescript KS U2 used for generation of the 820 nt U2 RNA transcript has been described previously (Broome and Hebert, 2012). The pBluescript KS hTR expression vector was generated by first amplifying HeLa genomic DNA with the following primers that contain engineered sites for SacI and EcoRI (underlined) designed by using the human TERC NG_016363.1 sequence: 5′-GCCGAGCTCCGGGTTGCGGAGGGTGGGCCTGGGAG-3′ and 5′-GCCGAATTCCCATTGCCGGCGAGGGGTGACGGATG-3′. Following amplification, the PCR product was cloned into a PCR4-TOPO vector using the TOPO TA Cloning kit from Invitrogen (Carlsbad, CA, USA). The SacI/EcoRI digested hTR insert was then cloned into SacI/EcoRI digested pBluescript KS vector. The sequence was verified against NG_016363.1 and found to contain one C to T mutation at nucleotide 5087 of NG_016363.1.
Transfections, RNAi and drug treatment
Cells were transfected with GFP expression vectors using Fugene HD Transfection Reagent (Promega, Madison, WI, USA) following the manufacturer's suggested protocol. Coilin siRNA (Toyota et al., 2010) and the non-targeting control siRNA (Carrero et al., 2011) have been described previously. The siRNA transfections were carried out using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. Nocodazole treatment was for 16 hours at 0.4 µg/ml.
RNA IP
Approximately 20 million cells were lysed in 1 ml RIPA buffer (50 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 0.1% SDS, 1 mM EDTA) containing protease inhibitor cocktail (Roche, Indianapolis, IN, USA) for RNA IP experiments. Cell lysates were sonicated four times for 5 seconds each at 4 W using the Fisher Scientific Sonic Dismembrator Model 100, then centrifuged at 16000 g for 5 minutes at 4°C. Sonicate from ∼20 million cells was used for each RNA IP reaction. Antibody and 45 µl 50% Protein G Sepharose beads (GE Healthcare, Pittsburgh, PA, USA) were added to the sonicate and incubated at 4°C for ∼18 hours on a multi-tube rotator. Normal rabbit IgG (sc-2027) and α-coilin H300 rabbit polyclonal (sc-32860) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA); 5 µg of each antibody was used per IP. Following the IP incubation, antibody/bead complexes were pelleted at 16000 g for 1 minute, the supernatant removed, and complexes washed in 1 ml RIPA buffer 3 times. RNA was then isolated from the antibody/bead complex using a modified Ambion RNAqueous kit protocol (Life Technologies, Carlsbad, CA, USA). Briefly, 350 µl lysis buffer and 350 µl 64% ethanol was added to the aspirated antibody/bead complex and the subsequent steps performed per the manufacturer's suggested protocol. RNA was eluted in 100 µl. Following elution, samples were incubated at 37°C for 30 minutes with TURBO DNase (Life Technologies, Carlsbad, CA, USA), then equal volumes of isolated RNA were used for the qRT-PCR analysis.
Bacterially expressed proteins
GST-tagged coilin proteins containing co-isolated nucleic acid were purified to SDS-PAGE single band homogeneity as described previously (Broome and Hebert, 2012). For nucleic acid-free proteins, the protocol was modified as follows: protein was incubated at 37°C for 30 minutes with DNase I and RNase A/T1 cocktail before separation by SDS-PAGE. Following electro-elution from the gel slices, the protein was put over a Pierce detergent removal column (Thermo Scientific (Waltham, MA, USA) for SDS-removal then concentrated using a centrifugal filter unit with 50K molecular weight cutoff (Millipore, Billerica, MA, USA). The protein was then incubated on ice for 30 minutes at a final concentration of 21 mM α-cyclodextrin (Otzen and Oliveberg, 2001) and 250 mM NaCl in order to remove residual protein-bound SDS, followed by centrifugation for 5 minutes at 16000 g. The resulting supernatant was then dialyzed against 1× PBS high salt buffer containing 10 mM Na2HPO4, 1.8 mM NaH2PO4 and 250 mM NaCl using a 10 K MWCO Pierce Slide-A-Lyzer Dialysis Cassette from Thermo Scientific following the manufacturer's suggested protocol. Concentration of dialyzed proteins was estimated using Pierce BCA Protein Assay Kit (Thermo Scientific, Waltham, MA, USA) following the manufacturer's protocol.
RNase assays
RNase assays for agarose gel end point analysis were conducted with 500 ng HeLa RNA isolated using the Macherey–Nagel NucleoSpin RNA II kit (Clontech, Mountain View, CA, USA) and up to 500 ng purified protein. Reactions were incubated at 37°C for 30 minutes, loaded into a 1% agarose gel and visualized by ethidium bromide. Densitometry of gel bands was measured using Quantity One software. U2 and hTR transcripts were generated from their respective pBluescript vectors using the MaxiScriptT7 kit (Ambion Life Technologies, Carlsbad, CA, USA) per the manufacturer's suggested protocol. RNase assays with U2 and hTR transcription reactions were conducted with 100 nM RNA and either 200 nM (300 ng) coilin WT or S489D or 300 ng GST-WT or GST-d121–291 protein at 37°C for 30 minutes. Following incubation, reactions were treated with TURBO DNase (Ambion Life Technologies, Carlsbad, CA, USA) for 20 minutes at 37°C. Equal volumes of the reactions were used for qRT-PCR.
qRT-PCR
Brilliant II SYBR Green qRT-PCR Master Mix Kit from Agilent (Santa Clara, CA, USA) was used for analysis of RNA from RNA IP experiments, transfected cells and U2 and hTR RNase assays and the manufacturer's suggested thermal cycling parameters, primer concentrations and RNA amounts were used. Primers for GAPDH, 47/45S pre-rRNA, U1, pre-U1, U2 and pre-U2 have been described (Broome and Hebert, 2012; Gilder et al., 2011; Hearst et al., 2009). Primer sequences are: 5.8S rRNA: 5′-CGGCTCGTGCGTCGAT-3′ forward and 5′-CCGCAAGTGCGTTCGAA-3′ reverse; telomerase RNA (hTR): 5′-AAATGTCAGCTGCTGGCCCGTTCG-3′ forward and 5′-ACCCGCGGCTGACAGAGCCCAAC-3′ reverse; pre-processed telomerase RNA (pre-hTR): 5′-AGGTTCAGGCCTTTCAGGCCGCAG-3′ forward and 5′-GACGGATGCGCACGATCGGCGTTC-3′ reverse. The Stratagene Mx3000P Real-Time PCR System was used with Agilent MxPro software for real-time analysis using Windows Excel for post hoc statistical analysis. The Student's t-test for independent variables with unequal variance was used for calculation of P-values.
Acknowledgments
This work was supported by The National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM081448. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Competing interests: The authors have no competing interests to declare.
References
- Andrade L. E. C., Chan E. K. L., Raska I., Peebles C. L., Roos G., Tan E. M. (1991). Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J. Exp. Med. 173, 1407–1419 10.1084/jem.173.6.1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillat D., Hakimi M. A., Näär A. M., Shilatifard A., Cooch N., Shiekhattar R. (2005). Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 123, 265–276 10.1016/j.cell.2005.08.019 [DOI] [PubMed] [Google Scholar]
- Beausoleil S. A., Jedrychowski M., Schwartz D., Elias J. E., Villén J., Li J., Cohn M. A., Cantley L. C., Gygi S. P. (2004). Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. USA 101, 12130–12135 10.1073/pnas.0404720101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broome H. J., Hebert M. D. (2012). In vitro RNase and nucleic acid binding activities implicate coilin in U snRNA processing. PLoS ONE 7, e36300 10.1371/journal.pone.0036300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broome H. J., Hebert M. D. (2013). Coilin displays differential affinity for specific RNAs in vivo and is linked to telomerase RNA biogenesis. J. Mol. Biol. 425, 713–724 10.1016/j.jmb.2012.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo–Fonseca M., Pepperkok R., Carvalho M. T., Lamond A. I. (1992). Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J. Cell Biol. 117, 1–14 10.1083/jcb.117.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo–Fonseca M., Ferreira J., Lamond A. I. (1993). Assembly of snRNP-containing coiled bodies is regulated in interphase and mitosis—evidence that the coiled body is a kinetic nuclear structure. J. Cell Biol. 120, 841–852 10.1083/jcb.120.4.841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrero Z. I., Velma V., Douglas H. E., Hebert M. D. (2011). Coilin phosphomutants disrupt Cajal body formation, reduce cell proliferation and produce a distinct coilin degradation product. PLoS ONE 6, e25743 10.1371/journal.pone.0025743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier S., Pendle A., Boudonck K., van Rij T., Dolan L., Shaw P. (2006). A distant coilin homologue is required for the formation of cajal bodies in Arabidopsis. Mol. Biol. Cell 17, 2942–2951 10.1091/mbc.E05-12-1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. (2008). A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA 105, 10762–10767 10.1073/pnas.0805139105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S., O'Reilly D., Chapman R. D., Taylor A., Tanzhaus K., Pitts L., Eick D., Murphy S. (2007). Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 318, 1777–1779 10.1126/science.1145989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustino N. A., Cooper T. A. (2003). Pre-mRNA splicing and human disease. Genes Dev. 17, 419–437 10.1101/gad.1048803 [DOI] [PubMed] [Google Scholar]
- Frey M. R., Matera A. G. (1995). Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc. Natl. Acad. Sci. USA 92, 5915–5919 10.1073/pnas.92.13.5915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey M. R., Matera A. G. (2001). RNA-mediated interaction of Cajal bodies and U2 snRNA genes. J. Cell Biol. 154, 499–509 10.1083/jcb.200105084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey M. R., Bailey A. D., Weiner A. M., Matera A. G. (1999). Association of snRNA genes with coiled bodies is mediated by nascent snRNA transcripts. Curr. Biol. 9, 126–136 10.1016/S0960-9822(99)80066-9 [DOI] [PubMed] [Google Scholar]
- Gilder A. S., Do P. M., Carrero Z. I., Cosman A. M., Broome H. J., Velma V., Martinez L. A., Hebert M. D. (2011). Coilin participates in the suppression of RNA polymerase I in response to cisplatin-induced DNA damage. Mol. Biol. Cell 22, 1070–1079 10.1091/mbc.E10-08-0731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearst S. M., Gilder A. S., Negi S. S., Davis M. D., George E. M., Whittom A. A., Toyota C. G., Husedzinovic A., Gruss O. J., Hebert M. D. (2009). Cajal-body formation correlates with differential coilin phosphorylation in primary and transformed cell lines. J. Cell Sci. 122, 1872–1881 10.1242/jcs.044040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert M. D., Matera A. G. (2000). Self-association of coilin reveals a common theme in nuclear body localization. Mol. Biol. Cell 11, 4159–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády B. E., Darzacq X., Tucker K. E., Matera A. G., Bertrand E., Kiss T. (2003). Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. EMBO J. 22, 1878–1888 10.1093/emboj/cdg187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam Y. W., Lyon C. E., Lamond A. I. (2002). Large-scale isolation of Cajal bodies from HeLa cells. Mol. Biol. Cell 13, 2461–2473 10.1091/mbc.02-03-0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeflang E. P., Liu W.–M., Hashimoto C., Choudary P. V., Schmid C. W. (1992). Phylogenetic evidence for multiple Alu source genes. J. Mol. Evol. 35, 7–16 10.1007/BF00160256 [DOI] [PubMed] [Google Scholar]
- Lemm I., Girard C., Kuhn A. N., Watkins N. J., Schneider M., Bordonné R., Lührmann R. (2006). Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol. Biol. Cell 17, 3221–3231 10.1091/mbc.E06-03-0247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. L., Wu Z., Nizami Z., Deryusheva S., Rajendra T. K., Beumer K. J., Gao H., Matera A. G., Carroll D., Gall J. G. (2009). Coilin is essential for Cajal body organization in Drosophila melanogaster. Mol. Biol. Cell 20, 1661–1670 10.1091/mbc.E08-05-0525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machyna M., Heyn P., Neugebauer K. M. (2013). Cajal bodies: where form meets function. Wiley Interdiscip Rev RNA 4, 17–34 10.1002/wrna.1139 [DOI] [PubMed] [Google Scholar]
- Mahmoudi S., Henriksson S., Weibrecht I., Smith S., Söderberg O., Strömblad S., Wiman K. G., Farnebo M. (2010). WRAP53 is essential for Cajal body formation and for targeting the survival of motor neuron complex to Cajal bodies. PLoS Biol. 8, e1000521 10.1371/journal.pbio.1000521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesic D., Tanackovic G., Krämer A. (2004). A role for Cajal bodies in the final steps of U2 snRNP biogenesis. J. Cell Sci. 117, 4423–4433 10.1242/jcs.01308 [DOI] [PubMed] [Google Scholar]
- Nizami Z. F., Gall J. G. (2012). Pearls are novel Cajal body-like structures in the Xenopus germinal vesicle that are dependent on RNA pol III transcription. Chromosome Res. 20, 953–969 10.1007/s10577-012-9320-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otzen D. E., Oliveberg M. (2001). A simple way to measure protein refolding rates in water. J. Mol. Biol. 313, 479–483 10.1006/jmbi.2001.5039 [DOI] [PubMed] [Google Scholar]
- Shanbhag R., Kurabi A., Kwan J. J., Donaldson L. W. (2010). Solution structure of the carboxy-terminal Tudor domain from human Coilin. FEBS Lett. 584, 4351–4356 10.1016/j.febslet.2010.09.034 [DOI] [PubMed] [Google Scholar]
- Smith K. P., Carter K. C., Johnson C. V., Lawrence J. B. (1995). U2 and U1 snRNA gene loci associate with coiled bodies. J. Cell. Biochem. 59, 473–485 10.1002/jcb.240590408 [DOI] [PubMed] [Google Scholar]
- Spector D. L., Lark G., Huang S. (1992). Differences in snRNP localization between transformed and nontransformed cells. Mol. Biol. Cell 3, 555–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern J. L., Zyner K. G., Pickett H. A., Cohen S. B., Bryan T. M. (2012). Telomerase recruitment requires both TCAB1 and Cajal bodies independently. Mol. Cell. Biol. 32, 2384–2395 10.1128/MCB.00379-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzelecka M., Oates A. C., Neugebauer K. M. (2010a). Dynamic control of Cajal body number during zebrafish embryogenesis. Nucleus 1, 96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzelecka M., Trowitzsch S., Weber G., Lührmann R., Oates A. C., Neugebauer K. M. (2010b). Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat. Struct. Mol. Biol. 17, 403–409 10.1038/nsmb.1783 [DOI] [PubMed] [Google Scholar]
- Sun J., Xu H., Subramony S. H., Hebert M. D. (2005). Interactions between coilin and PIASy partially link Cajal bodies to PML bodies. J. Cell Sci. 118, 4995–5003 10.1242/jcs.02613 [DOI] [PubMed] [Google Scholar]
- Tapia O., Bengoechea R., Berciano M. T., Lafarga M. (2010). Nucleolar targeting of coilin is regulated by its hypomethylation state. Chromosoma 119, 527–540 10.1007/s00412-010-0276-7 [DOI] [PubMed] [Google Scholar]
- Theimer C. A., Jády B. E., Chim N., Richard P., Breece K. E., Kiss T., Feigon J. (2007). Structural and functional characterization of human telomerase RNA processing and cajal body localization signals. Mol. Cell 27, 869–881 10.1016/j.molcel.2007.07.017 [DOI] [PubMed] [Google Scholar]
- Toyota C. G., Davis M. D., Cosman A. M., Hebert M. D. (2010). Coilin phosphorylation mediates interaction with SMN and SmB′. Chromosoma 119, 205–215 10.1007/s00412-009-0249-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker K. E., Berciano M. T., Jacobs E. Y., LePage D. F., Shpargel K. B., Rossire J. J., Chan E. K., Lafarga M., Conlon R. A., Matera A. G. (2001). Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J. Cell Biol. 154, 293–307 10.1083/jcb.200104083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski K. T., Shu M. D., Kukoyi A., Steitz J. A. (2009). A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol. Cell 34, 47–57 10.1016/j.molcel.2009.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher A. S., Abreu E. B., Meng Z., McCann K. E., Terns R. M., Veenstra T. D., Terns M. P., Artandi S. E. (2009). A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 323, 644–648 10.1126/science.1165357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker M. P., Tian L., Matera A. G. (2009). Reduced viability, fertility and fecundity in mice lacking the cajal body marker protein, coilin. PLoS ONE 4, e6171 10.1371/journal.pone.0006171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P. J., Le T. T., thi Man N., Burghes A. H. M., Morris G. E. (2000). The relationship between SMN, the spinal muscular atrophy protein, and nuclear coiled bodies in differentiated tissues and cultured cells. Exp. Cell Res. 256, 365–374 10.1006/excr.2000.4858 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Tomlinson R. L., Lukowiak A. A., Terns R. M., Terns M. P. (2004). Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol. Biol. Cell 15, 81–90 10.1091/mbc.E03-07-0525 [DOI] [PMC free article] [PubMed] [Google Scholar]