Abstract
Background
Evidence suggests that insulin-like growth factor 2 messenger RNA binding protein 3 (IGF2BP3, also known as IMP3) represents a promising cancer biomarker. However, the clinical, pathological, molecular, and prognostic features of IGF2BP3-positive colorectal cancers remain uncertain.
Materials and methods
We evaluated IGF2BP3 expression by immunohistochemistry in 671 rectal and colon cancer cases that form part of a molecular pathological epidemiology database. Cox proportional hazards regression models were used to compute mortality hazard ratio (HR), adjusting for clinical, pathological, and molecular features, including microsatellite instability, the CpG island methylator phenotype, LINE-1 methylation, and KRAS, BRAF, and PIK3CA mutations.
Results
Among 671 colorectal cancers, 234 (35%) tumours were positive for IGF2BP3. In contrast, normal colorectal epithelium was negative for IGF2BP3 in all 403 specimens of normal mucosa adjacent to carcinoma. IGF2BP3 positivity was associated with poor differentiation (p=0.0003), stage III–IV disease (p=0.0081), BRAF mutation (p=0.031), and LINE-1 hypomethylation (p=0.020). IGF2BP3 positivity was significantly associated with shorter colorectal cancer-specific [log-rank p<0.0001; multivariate HR, 1.37; 95% confidence interval (CI), 1.02–1.84] and overall survival (log-rank p=0.0004; multivariate HR, 1.32; 95% CI, 1.05–1.66).
Conclusions
IGF2BP3 expression in colorectal cancer is associated with adverse clinical outcome. Our findings support a role for IGF2BP3 as a diagnostic and/or prognostic biomarker in colorectal cancer.
Keywords: adenocarcinoma, rectal cancer, cancer testis antigen, MAGE, carcinogenesis, diagnostic marker, pathology, therapeutic target, personalized medicine
Introduction
During embryogenesis, insulin-like growth factor 2 messenger RNA binding protein 3 (IGF2BP3, also known as IMP3) plays an important role in RNA trafficking and stabilization, cell growth, and cell migration.1,2 IGF2BP3 is expressed by a variety of malignant tumours including pancreatic, lung, oesophageal, thyroid, and gynaecological cancers, and melanomas.3 In addition, it has been shown that expression of IGF2BP3, determined by immunohistochemistry, has prognostic significance in a number of cancers,4–10 including colorectal cancer.11,12 Consequently, it has been proposed that IGF2BP3 represents a promising cancer biomarker.3 Nonetheless, no previous study has comprehensively examined the clinical, pathological, and molecular features that may influence IGF2BP3 expression in colorectal cancer. Furthermore, previous studies11,12 on IGF2BP3 expression and colorectal cancer survival have been limited by relatively small sample sizes (N=186 and 203), and by the inability to control for tumour molecular features, such as microsatellite instability (MSI), and KRAS and BRAF mutations, which may represent molecular confounders. Thus, the prognostic role of IGF2BP3 expression in colorectal cancer remains uncertain, and the molecular associations of IGF2BP3 expression, which are important in elucidating the role of IGF2BP3 in colorectal carcinogenesis, remain obscure.
We therefore examined the prognostic role, and clinical, pathological, and molecular associations of IGF2BP3 expression utilizing a molecular pathological epidemiology database13,14 containing 671 colorectal cancers from two prospective cohort studies. Because our database contains information on tumour variables, including KRAS, BRAF and PIK3CA mutations, MSI, the CpG island methylator phenotype (CIMP), and LINE-1 methylation, we could robustly evaluate associations between IGF2BP3 and these variables, as well as examine the prognostic association of IGF2BP3 independent of potential confounders.
Materials and Methods
Study population
We utilized a database of two U.S. nationwide prospective cohort studies, the Nurses’ Health Study (N=121,701 women followed since 1976), and the Health Professionals Follow-up Study (N=51,529 men followed since 1986).15 Colorectal cancer cases were ascertained by means of biennial questionnaire and the National Death Index, which was also used for ascertainment of death. The cause of death was determined through medical record review by study physicians. We collected formalin-fixed paraffin-embedded tissue blocks from hospitals where surgical resections had been performed.15 We collected diagnostic biopsy specimens from rectal cancer patients who had received preoperative treatment, to minimize treatment-induced bias or artefact. Based on tissue availability and IGF2BP3 expression data, we included 671 stage I–IV colorectal cancer cases diagnosed up to 2004 (Table 1). Patients were observed until death or January 1, 2011, whichever came first. Informed consent was obtained from all study subjects. This study was approved by the Human Subjects Committees of Harvard School of Public Health and Brigham and Women’s Hospital.
Table 1.
Clinical, pathological or molecular characteristics according to IGF2BP3 status in colorectal cancer
| Clinical, pathological or molecular feature | Total N | IGF2BP3
|
p value | |
|---|---|---|---|---|
| negative | positive | |||
| All cases | 671 | 437 | 234 | |
| Sex | 0.61 | |||
| Male | 241 (36%) | 160 (37%) | 81 (35%) | |
| Female | 430 (64%) | 277 (63%) | 153 (65%) | |
| Mean age ± SD | 67.4 ± 8.4 | 67.5 ± 8.3 | 67.1 ± 8.5 | 0.58 |
| Body mass index (kg/m2) | 0.38 | |||
| <30 | 536 (80%) | 345 (79%) | 191 (82%) | |
| ≥30 | 133 (20%) | 91 (21%) | 42 (18%) | |
| Family history of colorectal cancer in any first degree relative | 0.70 | |||
| (−) | 536 (80%) | 351 (80%) | 185 (79%) | |
| (+) | 135 (20%) | 86 (20%) | 49 (21%) | |
| Year of diagnosis | 0.32 | |||
| Prior to 1997 | 336 (50%) | 225 (51%) | 111 (47%) | |
| 1997–2004 | 335 (50%) | 212 (49%) | 123 (53%) | |
| Tumour location | 0.58 | |||
| Proximal colon (cecum to transverse) | 323 (48%) | 204 (47%) | 119 (51%) | |
| Distal colon (splenic flexure to sigmoid) | 206 (31%) | 139 (32%) | 67 (29%) | |
| Rectum | 138 (21%) | 91 (21%) | 47 (20%) | |
| Disease stage | 0.0081 | |||
| I | 146 (22%) | 104 (24%) | 42 (18%) | |
| II | 212 (32%) | 142 (32%) | 70 (30%) | |
| III | 183 (27%) | 111 (25%) | 72 (31%) | |
| IV | 93 (14%) | 50 (11%) | 43 (18%) | |
| Unknown | 37 (5.5%) | 30 (6.9%) | 7 (3.0%) | |
| Tumour differentiation | 0.0003 | |||
| Well to moderate | 606 (91%) | 408 (94%) | 198 (85%) | |
| Poor | 63 (9.4%) | 28 (6.4%) | 35 (15%) | |
| Tumour growth pattern | 0.56 | |||
| Expansile | 201 (34%) | 134 (35%) | 67 (32%) | |
| Intermediate | 313 (53%) | 205 (53%) | 108 (52%) | |
| Infiltrative | 80 (13%) | 48 (12%) | 32 (15%) | |
| Mucinous component | 0.84 | |||
| 0% | 420 (63%) | 270 (62%) | 150 (64%) | |
| 1–50% | 174 (26%) | 116 (27%) | 58 (25%) | |
| >50% | 77 (11%) | 51 (12%) | 26 (11%) | |
| Signet ring cell component | 0.49 | |||
| 0% | 612 (91%) | 400 (91%) | 212 (91%) | |
| 1–50% | 53 (7.9%) | 32 (7.3%) | 21 (9.0%) | |
| >50% | 6 (0.9%) | 5 (1.1%) | 1 (0.4%) | |
| MSI status | 0.73 | |||
| MSI-low/MSS | 540 (82%) | 352 (83%) | 188 (82%) | |
| MSI-high | 115 (18%) | 73 (17%) | 42 (18%) | |
| CIMP status | 0.091 | |||
| CIMP-low/0 | 548 (83%) | 365 (85%) | 183 (80%) | |
| CIMP-high | 110 (17%) | 64 (15%) | 46 (20%) | |
| KRAS mutation | 0.13 | |||
| (−) | 424 (64%) | 268 (62%) | 156 (68%) | |
| (+) | 239 (36%) | 165 (38%) | 74 (32%) | |
| BRAF mutation | 0.031 | |||
| (−) | 557 (84%) | 372 (87%) | 185 (80%) | |
| (+) | 104 (16%) | 58 (13%) | 46 (20%) | |
| PIK3CA mutation | 0.84 | |||
| (−) | 506 (84%) | 332 (84%) | 174 (85%) | |
| (+) | 93 (16%) | 62 (16%) | 31 (15%) | |
| LINE-1 methylation level (Mean ± SD) | 61.3 ± 9.5 | 62.0 ± 8.9 | 60.1 ± 10.4 | 0.020 |
| TP53 expression | 0.33 | |||
| (−) | 388 (58%) | 259 (60%) | 129 (56%) | |
| (+) | 279 (42%) | 176 (40%) | 103 (44%) | |
| Nuclear CTNNB1 expression | 0.62 | |||
| (−) | 339 (53%) | 224 (54%) | 115 (52%) | |
| (+) | 299 (47%) | 192 (46%) | 107 (48%) | |
| PTGS2 expression | 0.33 | |||
| (−) | 249 (37%) | 168 (39%) | 81 (35%) | |
| (+) | 419 (63%) | 267 (61%) | 152 (65%) | |
| FASN expression | 0.17 | |||
| (−) | 249 (38%) | 170 (39%) | 79 (34%) | |
| (+) | 414 (62%) | 261 (61%) | 153 (66%) | |
(%) indicates the proportion of cases with a specific clinical, pathological or molecular feature among each expression group. A p value for significance was adjusted for multiple hypothesis testing to p=0.05/21=0.0024. Thus, a p value between 0.05 and 0.0024 should be regarded as of borderline significance.
CIMP, CpG island methylator phenotype; MSI, microsatellite instability; MSS, microsatellite stable; SD, standard deviation.
Histopathological evaluation
Haematoxylin and eosin-stained sections of all cases were examined by a pathologist (S.O.) unaware of other data. Tumour differentiation was categorized as well-moderate or poor (>50% vs. ≤50% gland formation). Tumour growth pattern at the tumour margin was categorized as expansile, intermediate, or infiltrative, as previously described.16 The presence and extent of mucinous and/or signet ring cell component was recorded.17 A subset of cases (N>100) was reviewed by a second pathologist (T.M.), and concordance was as follows (all p<0.0001): κ=0.72 for tumour differentiation; κ=0.73 for tumour growth pattern; Spearman r=0.87 for mucin (%); Spearman r=0.65 for signet ring cells (%).16
DNA extraction, Pyrosequencing of KRAS, BRAF and PIK3CA, and microsatellite instability (MSI) analyses
DNA was extracted from paraffin-embedded tissue. Mutation status for KRAS (codons 12 and 13),18 BRAF (codon 600),19 and PIK3CA (exons 9 and 20)20 was determined by Pyrosequencing. PCR-based MSI analysis was performed using 10 microsatellite markers (D2S123, D5S346, D17S250, BAT25, BAT26, BAT40, D18S55, D18S56, D18S67 and D18S487).21 MSI-high was defined as instability in ≥30% of the markers, and MSI-low/microsatellite stability (MSS) as instability in <30% of the markers.21
Methylation analyses for CpG islands and LINE-1
Using real-time PCR (MethyLight) on bisulfite-converted DNA, we quantified DNA methylation in eight CIMP-specific promoters (CACNA1G, CDKN2A, CRABP1, IGF2, MLH1, NEUROG1, RUNX3 and SOCS1).22 CIMP-high was defined as the presence of ≥6/8 methylated promoters, and CIMP-low/0 as 0–5/8 methylated promoters, in accordance with previously established criteria.22 In order to accurately quantify methylation changes within LINE-1, we used Pyrosequencing as previously described.23
Immunohistochemical analysis
Tissue microarrays (TMA) were constructed as previously described.24,25 Each tumour was represented by a minimum of two 0.6 mm tissue cores, which has previously been shown to be sufficient for assessment of IGF2BP3 expression.26 Immunohistochemistry was performed as previously described for TP53,27 CTNNB1 (β-catenin),15 PTGS2 (cyclooxygenase-2)24 and FASN.17 For IGF2BP3, antigen retrieval from deparaffinised tissue sections was achieved by microwaving in a pressure cooker for 10 minutes in Citra Antigen Retrieval Solution, pH 6 (BioGenex, San Ramon, CA). Tissue sections were incubated for 15 minutes with Peroxidase Blocking Reagent (Dako North America, Carpinteria, CA). A mouse monoclonal antibody against IGF2BP3 (IMP3; clone 69.1; Dako; 1:100 dilution) was applied, and the sections were incubated for 16 hours at 4°C. Visualization was achieved using EnVision™+/HRP (Dako), diaminobenzidine (Dako), and hematoxylin counterstain. Human placenta was used as a positive control tissue. Omission of primary antibody served as a negative control. Cytoplasmic staining for IGF2BP3 was scored as absent, weak, moderate, or intense. For subsequent analyses, absent or weak staining was categorized as negative, while moderate or intense staining was categorized as positive.
Each immunohistochemical maker was interpreted by one of the investigators (IGF2BP3 and CTNNB1 by T.M.; TP53, PTGS2 and FASN by S.O.) unaware of other data. For agreement studies, over 100 randomly-selected cases were examined for each marker by a second observer (IGF2BP3 by P.L.; CTNNB1 by S.O.; TP53, PTGS2 and FASN by T.M.) unaware of other data. The concordance between observers (all p<0.0001) was κ=0.70 for IGF2BP3, κ=0.78 for TP53, κ=0.80 for CTNNB1, κ=0.69 for PTGS2, κ=0.80 for FASN, indicating substantial agreement.
Statistical analysis
All statistical analyses were performed using SAS (Version 9.2, SAS Institute, Cary, NC). All p values were two-sided. Our primary hypothesis was that IGF2BP3 expression was associated with worse clinical outcome. When multiple hypothesis testing was performed for exploratory analyses of clinicopathological and molecular associations, or interactions, the p value for significance was adjusted by Bonferroni correction to p=0.0024 (=0.05/21). For categorical data, the chi-square test was performed. Mean age was compared using a t-test.
Kaplan-Meier method and log-rank test were used for survival analyses. For analyses of colorectal cancer-specific mortality, deaths as a result of other causes were censored. To control for confounding, multivariate Cox proportional hazards regression models were used. The multivariate model initially included sex, age at diagnosis (continuous), year of diagnosis (continuous), BMI (<30 vs. ≥30 kg/m2), family history of colorectal cancer in any first-degree relative (present vs. absent), tumour location (proximal vs. distal), tumour differentiation (well-moderate vs. poor), tumour growth pattern (expansile-intermediate vs. infiltrative), mucinous component (continuous), signet ring cell component (continuous), MSI (high vs. low/MSS), CIMP (high vs. low/0), KRAS, BRAF, and PIK3CA mutation, LINE-1 methylation (continuous), and expression of TP53, nuclear CTNNB1, PTGS2 and FASN. Because we observed non-proportionality in the hazards for disease stage over time, and to avoid overfitting, stage (I, II, III, IV or unknown) was used as a stratifying variable using the “strata” option in the SAS “proc phreg” command. A backward elimination was performed with a threshold of p=0.20, to avoid overfitting. Cases with missing information in any of the covariates were assigned to the majority category of the given covariate to avoid overfitting. We confirmed that excluding cases with missing covariate information did not substantially alter results (data not shown). The proportionality of hazards assumption was satisfied for all variables in the multivariate models by evaluating time-dependent variables, which were the cross-product of a given variable and survival time. Interaction was assessed by the Wald test on the cross product of IGF2BP3 and another variable of interest (without data-missing cases) in a multivariate Cox model.
Results
IGF2BP3 status in colorectal cancer and normal colorectal mucosa
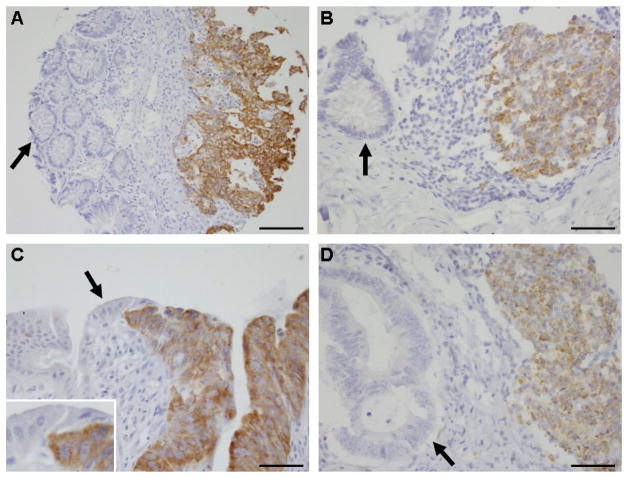
Among the database of 671 stage I–IV colorectal cancers in the two prospective cohort studies, 234 (35%) showed IGF2BP3 positivity (Figure 1). In contract, IGF2BP3 was negative in normal colorectal epithelial cells in all the 403 adjacent normal mucosal specimens that were evaluable. In normal colorectal tissue, only lymphocytes in the germinal centres of mucosa-associated lymphoid tissue stained positively for IGF2BP3 (Figure 1). As demonstrated in Table 1, IGF2BP3 positivity was significantly associated with poor differentiation (p=0.0003). Because multiple hypothesis testing required a Bonferroni corrected significance level of p=0.0024, the relationships between IGF2BP3 positivity and stage III–IV disease (p=0.0081), BRAF mutation (p=0.031), and LINE-1 hypomethylation (p=0.020) were considered of borderline significance; these findings require validation in an independent dataset.
Figure 1.
(A) IGF2BP3-positive colon cancer (right). Adjacent normal glands are negative for IGF2BP3 (left, arrrow). Bar, 100 μm. (B) Germinal center lymphocytes in normal colonic mucosa are positively stained for IGF2BP3 (right). Normal glands are negative for IGF2BP3 (left, arrow). Bar, 50 μm. (C) IGF2BP3-positive colon cancer (right). Adjacent normal colonic epithelial cells are negative for IGF2BP3 (left, arrow). Inset, high-power view of the boundary between normal epithelial cells and cancer cells. Bar, 50 μm. (D) IGF2BP3-negative colon cancer (left, arrow). Germinal center lymphocytes are positive for IGF2BP3 (right). Bar, 50 μm.
IGF2BP3 positivity and survival of colorectal cancer patients
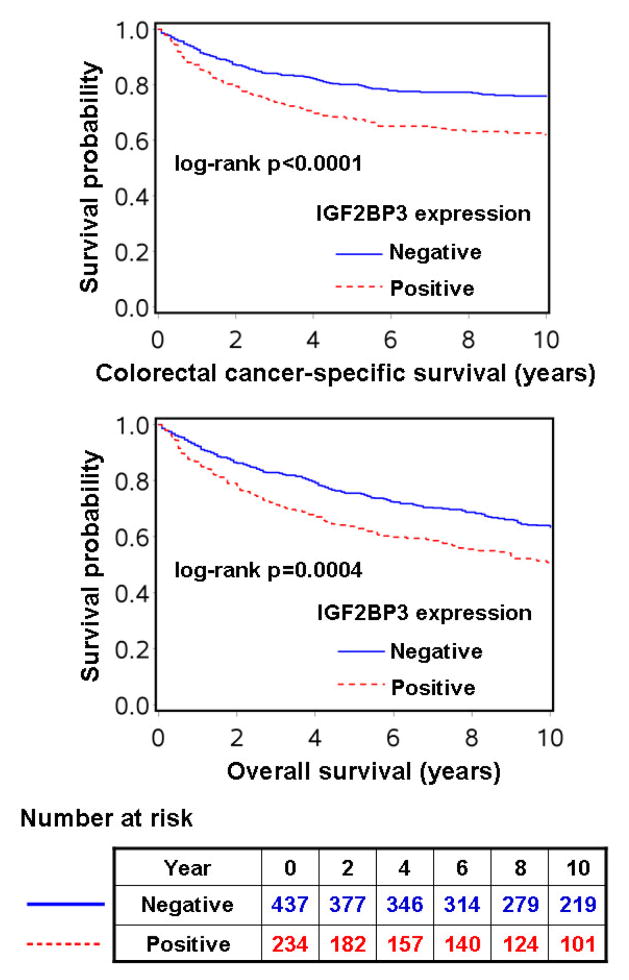
Among the 671 patients, there were 328 deaths, including 194 colorectal cancer-specific deaths, during a median follow-up of 160 months (interquartile range, 123–207 months) for censored cases. In Kaplan-Meier analysis, IGF2BP3 positivity was significantly associated with shorter colorectal cancer-specific (log-rank p<0.0001) and overall survival (log-rank p=0.0004) (Figure 2).
Figure 2.
Kaplan-Meier curves for colorectal cancer-specific survival (upper) and overall survival (lower) according to IGF2BP3 status in colorectal cancer. The table (bottom) shows the number of patients who remained alive, and at risk, at each time point after the diagnosis of colorectal cancer. The number at risk is applicable to both colorectal cancer-specific and overall survival.
We used Cox multivariate regression models to assess the prognostic influence of IGF2BP3 positivity independent of clinical, pathological, and other tumour molecular features (Table 2). Univariate hazard ratio (HR) for IGF2BP3 positivity was 1.76 [95% confidence interval (CI), 1.32–2.33] for colorectal cancer-specific survival, and 1.48 (95% CI, 1.19–1.85) for overall survival. The multivariate HR for IGF2BP3 positivity was 1.37 (95% CI, 1.02–1.84) for colorectal cancer-specific survival, and 1.32 (95% CI, 1.05–1.66) for overall survival. The attenuation of the effect of IGF2BP3 positivity in the multivariate analysis occurred largely as a result of adjusting for disease stage. When adjusted for disease stage alone, the HR for IGF2BP3 positivity was 1.39 (95% CI, 1.04–1.85) for colorectal cancer-specific survival, and 1.30 (95% CI, 1.04–1.63) for overall survival (Table 2). No other major confounder was present.
Table 2.
IGF2BP3 positivity in colorectal cancer and patient mortality
| Total N (%) | Colorectal cancer-specific mortality
|
Overall mortality
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Number of events | Univariate HR (95% CI) | Stage-stratified HR (95% CI) | Multivariate stage-stratified HR (95% CI) | Number of events | Univariate HR (95% CI) | Stage-stratified HR (95% CI) | Multivariate stage-stratified HR (95% CI) | ||
| IGF2BP3 (−) | 437 (65%) | 105 | 1 (referent) | 1 (referent) | 1 (referent) | 194 | 1 (referent) | 1 (referent) | 1 (referent) |
| IGF2BP3 (+) | 234 (35%) | 89 | 1.76 (1.32–2.33) | 1.39 (1.04–1.85) | 1.37 (1.02–1.84) | 134 | 1.48 (1.19–1.85) | 1.30 (1.04–1.63) | 1.32 (1.05–1.66) |
| p value | <0.0001 | 0.028 | 0.039 | 0.0005 | 0.022 | 0.016 | |||
We tested the specific study hypothesis on the prognostic role of IGP2BP3 positivity. Thus, a p value for significance was set at p=0.05. The multivariate, stage-stratified Cox regression model initially included the IGF2BP3 variable (negative or positive), age, sex, year of diagnosis, body mass index, family history of colorectal cancer in any first degree relative, tumour location, tumour differentiation, tumour growth pattern, mucinous component, signet ring cell component, microsatellite instability, CpG island methylator phenotype, KRAS, BRAF, PIK3CA, LINE-1 methylation, TP53, nuclear CTNNB1, PTGS2 and FASN. A backward elimination with threshold of p=0.20 was used to select variables in the final models.
CI, confidence interval; HR, hazard ratio.
Given that stage appeared to be a mediator of the association between IGF2BP3 expression and survival, we performed Kaplan-Meier analyses stratified by disease stage (Supplementary figures 1 and 2). A significant difference in survival time distribution according to IGF2BP3 expression was observed only for colorectal cancer-specific survival in stage II disease (Supplementary figure 1, B; log-rank p=0.0005), and with borderline significance for stage II overall survival (Supplementary figure 2, B; log-rank p=0.071). Similarly, in stage-specific Cox regression, IGF2BP3 was significantly associated with colorectal cancer-specific survival only in stage II disease (multivariate HR=2.50; 95% CI, 1.14–5.47). We did not, however, observe a significant interaction between disease stage and IGF2BP3 positivity, indicating that the prognostic association of IGF2BP3 did not differ significantly by stage.
Exploratory analyses of interactions between IGF2BP3 and other covariates
We further examined whether any of the clinical, pathological, or molecular variables modified the influence of IGF2BP3 positivity on cancer-specific mortality. We found no significant interaction between IGF2BP3 and any of the covariates examined (all Pinteraction>0.10). Notably, the effect of IGF2BP3 positivity did not significantly differ between the two cohort studies (Pinteraction=0.33).
Discussion
Here, we report our findings on IGF2BP3 expression in colorectal cancer in relation to patient survival as well as clinical, pathological, and molecular features, in two large prospective cohort studies. We found significant associations between IGF2BP3 positivity, poor differentiation, and increased mortality, independent of patient characteristics or tumour molecular variables. Our results suggest that IGF2BP3 positivity may define a colorectal cancer phenotype with more aggressive biological behaviour.
It has been reported that IGF2BP3 represents a novel biomarker that can differentiate normal from cancerous tissue in a variety of organ systems.3 In the present study, IGF2BP3 positivity was observed in approximately 35% of colorectal cancer cases, while normal colorectal epithelial cells were always negative for IGF2BP3. Thus, IGF2BP3 positivity in small biopsy specimens may provide a pathologist with greater confidence in the diagnosis of colorectal cancer, especially when substantial crush artefact is present. The frequency of IGF2BP3 positivity in colorectal adenomata and specimens with inflammation or reactive atypia must, however, be determined in future studies.
We found that germinal centre lymphocytes in gut-associated lymphoid tissue also stained positively for IGF2BP3. Consistent with this observation, IGF2BP3 positivity in germinal centre B cells has been reported in normal lymphoid tissues including lymph node, tonsil, spleen, and thymus.28 To the best of our knowledge, IGF2BP3 positivity in gut-associated lymphoid tissue has not previously been reported. This pattern of IGF2BP3 positivity may generate a potential diagnostic pitfall, and pathologists should be aware of it, especially where a nest of IGF2BP3-positive cells lacking glandular structure occurs in a biopsy specimen. Careful histological evaluation together with immunohistochemistry for epithelial and lymphocytic markers would help confirm the correct diagnosis in such cases.
It has recently been reported that IGF2BP3 protein expression is associated with a poorer prognosis in colorectal cancer patients.11,12 However, both of the two previous studies11,12 on IGF2BP3 expression and colorectal cancer survival were limited by relatively small sample sizes (N=186 and 203). Furthermore, neither of these studies11,12 took into account the potential confounding effects of established colorectal cancer molecular biomarkers. In the present study, IGF2BP3 positivity appeared to be associated with BRAF mutation and LINE-1 hypomethylation, both of which are associated with poor survival.29–33 We have shown that IGF2BP3 positivity is associated with poorer survival in patients with colorectal cancer, independent of these molecular features.
Experimental data generally support an oncogenic role for IGF2BP3.10,34–37 Overexpression of IGF2BP3 in lung and breast cancer cell lines enhances tumour cell invasiveness.35 Similarly, in a mouse melanoma model, Igf2bp3 overexpression in melanoma cells enhances tumour growth, angiogenesis, and metastasis, resulting in poorer survival.37 In leukaemia,34 hepatocellular carcinoma,35 melanoma,36 glioma,37 and cervical cancer10 cell lines in vitro, small interfering RNA-mediated knockdown of IGF2BP3 attenuates malignant characteristics, including cell proliferation, migration, anchorage-independent growth, and invasion 10,34–37. Our observational data certainly support a role of IGF2BP3 in colorectal cancer progression.
Since the prognostic association of IGF2BP3 overexpression might be related to biological effects on invasion and metastasis,35,36 it might have been illuminating to evaluate the influence of IGF2BP3 expression on disease-free survival or time to metastasis; unfortunately, these data were not available for our cohorts. Further studies are therefore needed to determine exactly how IGF2BP3 influences the biological behaviour of colorectal cancer cells.
IGF2BP3 is not only a diagnostic/prognostic marker, but also a potential therapeutic target.38 Cytotoxic T-cell clones, generated by in vitro exposure to an HLA-A24-restricted peptide epitope derived from IGF2BP3, exert a considerable cytotoxic response against lung and oesophageal cancer cells that express endogenous IGF2BP3.39 Moreover, in a phase I study, vaccination therapy using an IGF2BP3-derived peptide, together with two other peptides, induced clinical response in five out of 10 oesophageal cancer patients who were resistant to chemotherapy and/or radiotherapy.40 Tissue expression of IGF2BP3 in colorectal cancer might serve as a predictive biomarker, and could be used to identify patients who may benefit from vaccination-based therapies that target IGF2BP3.
In summary, IGF2BP3 is a promising diagnostic biomarker candidate for colorectal cancer and its expression is independently associated with poorer prognosis. Our findings may have broader clinical implications by facilitating the evolution of IGF2BP3 as a therapeutic target. Further studies are required to validate our findings and elucidate the molecular mechanisms and pathways through which IGF2BP3 affects the biological phenotype of colorectal cancer cells.
Supplementary Material
Kaplan-Meier curves for colorectal cancer-specific survival stratified by disease stage: A, stage I; B, stage II; C, stage III; D, stage IV.
Kaplan-Meier curves for overall survival stratified by disease stage: A, stage I; B, stage II; C, stage III; D, stage IV.
Acknowledgments
Funding: This work was supported by U.S. National Institute of Health (NIH) grants P01 CA87969 (to S. Hankinson), P01 CA55075 (to W.C. Willett), P50 CA127003 (to C.S.F.), and R01 CA151993 (to S.O.) and by grants from the Bennett Family Fund and the Entertainment Industry Foundation through National Colorectal Cancer Research Alliance. P.L. is a Scottish Government Clinical Academic Fellow and was supported by a Harvard University Frank Knox Memorial Fellowship. T.M. was supported by a fellowship grant from the Japan Society for Promotion of Science. The content is solely the responsibility of the authors and does not necessarily represent the official views of NCI or NIH.
We would like to thank the participants and staff of the Nurses’ Health Study and the Health Professionals Follow-Up Study, as well as the U.S. state cancer registries, for their valuable contributions and assistance.
Role of the funding source
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Conflict of interest: The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mueller-Pillasch F, Pohl B, Wilda M, et al. Expression of the highly conserved RNA binding protein KOC in embryogenesis. Mech Dev. 1999;88:95–9. doi: 10.1016/s0925-4773(99)00160-4. [DOI] [PubMed] [Google Scholar]
- 2.Nielsen J, Christiansen J, Lykke-Andersen J, et al. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol. 1999;19:1262–70. doi: 10.1128/mcb.19.2.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Findeis-Hosey JJ, Xu H. The use of insulin like-growth factor II messenger RNA binding protein-3 in diagnostic pathology. Hum Pathol. 2011;42:303–14. doi: 10.1016/j.humpath.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Hoffmann NE, Sheinin Y, Lohse CM, et al. External validation of IMP3 expression as an independent prognostic marker for metastatic progression and death for patients with clear cell renal cell carcinoma. Cancer. 2008;112:1471–9. doi: 10.1002/cncr.23296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeng YM, Wang TH, Lu SH, et al. Prognostic significance of insulin-like growth factor II mRNA-binding protein 3 expression in gastric adenocarcinoma. Br J Surg. 2009;96:66–73. doi: 10.1002/bjs.6438. [DOI] [PubMed] [Google Scholar]
- 6.Kobel M, Xu H, Bourne PA, et al. IGF2BP3 (IMP3) expression is a marker of unfavorable prognosis in ovarian carcinoma of clear cell subtype. Mod Pathol. 2009;22:469–75. doi: 10.1038/modpathol.2008.206. [DOI] [PubMed] [Google Scholar]
- 7.Walter O, Prasad M, Lu S, et al. IMP3 is a novel biomarker for triple negative invasive mammary carcinoma associated with a more aggressive phenotype. Hum Pathol. 2009;40:1528–33. doi: 10.1016/j.humpath.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 8.Schaeffer DF, Owen DR, Lim HJ, et al. Insulin-like growth factor 2 mRNA binding protein 3 (IGF2BP3) overexpression in pancreatic ductal adenocarcinoma correlates with poor survival. BMC Cancer. 2010;10:59. doi: 10.1186/1471-2407-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen ST, Jeng YM, Chang CC, et al. Insulin-like growth factor II mRNA-binding protein 3 expression predicts unfavorable prognosis in patients with neuroblastoma. Cancer Sci. 2011;102:2191–8. doi: 10.1111/j.1349-7006.2011.02100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu D, Yang X, Jiang NY, et al. IMP3, a New Biomarker to Predict Progression of Cervical Intraepithelial Neoplasia Into Invasive Cancer. Am J Surg Pathol. 2011;35:1638–45. doi: 10.1097/PAS.0b013e31823272d4. [DOI] [PubMed] [Google Scholar]
- 11.Yuan RH, Wang CC, Chou CC, et al. Diffuse expression of RNA-binding protein IMP3 predicts high-stage lymph node metastasis and poor prognosis in colorectal adenocarcinoma. Ann Surg Oncol. 2009;16:1711–9. doi: 10.1245/s10434-009-0446-0. [DOI] [PubMed] [Google Scholar]
- 12.Li D, Yan D, Tang H, et al. IMP3 is a novel prognostic marker that correlates with colon cancer progression and pathogenesis. Ann Surg Oncol. 2009;16:3499–506. doi: 10.1245/s10434-009-0648-5. [DOI] [PubMed] [Google Scholar]
- 13.Ogino S, Stampfer M. Lifestyle factors and microsatellite instability in colorectal cancer: the evolving field of molecular pathological epidemiology. J Natl Cancer Inst. 2010;102:365–7. doi: 10.1093/jnci/djq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ogino S, Chan AT, Fuchs CS, et al. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60:397–411. doi: 10.1136/gut.2010.217182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morikawa T, Kuchiba A, Yamauchi M, et al. Association of CTNNB1 (beta-catenin) alterations, body mass index, and physical activity with survival in patients with colorectal cancer. JAMA. 2011;305:1685–94. doi: 10.1001/jama.2011.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morikawa T, Kuchiba A, Qian ZR, et al. Prognostic significance and molecular associations of tumor growth pattern in colorectal cancer. Ann Surg Oncol. 2012;19:1944–53. doi: 10.1245/s10434-011-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogino S, Brahmandam M, Cantor M, et al. Distinct molecular features of colorectal carcinoma with signet ring cell component and colorectal carcinoma with mucinous component. Mod Pathol. 2006;19:59–68. doi: 10.1038/modpathol.3800482. [DOI] [PubMed] [Google Scholar]
- 18.Ogino S, Kawasaki T, Brahmandam M, et al. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn. 2005;7:413–21. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogino S, Kawasaki T, Kirkner GJ, et al. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8:582–8. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao X, Morikawa T, Lochhead P, et al. Prognostic role of PIK3CA mutation in colorectal cancer: cohort study and literature review. Clin Cancer Res. 2012;18:2257–68. doi: 10.1158/1078-0432.CCR-11-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogino S, Kawasaki T, Kirkner GJ, et al. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9:305–14. doi: 10.2353/jmoldx.2007.060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irahara N, Nosho K, Baba Y, et al. Precision of pyrosequencing assay to measure LINE-1 methylation in colon cancer, normal colonic mucosa, and peripheral blood cells. J Mol Diagn. 2010;12:177–83. doi: 10.2353/jmoldx.2010.090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan AT, Ogino S, Fuchs CS. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. N Engl J Med. 2007;356:2131–42. doi: 10.1056/NEJMoa067208. [DOI] [PubMed] [Google Scholar]
- 25.Ogino S, Brahmandam M, Kawasaki T, et al. Combined analysis of COX-2 and p53 expressions reveals synergistic inverse correlations with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Neoplasia. 2006;8:458–64. doi: 10.1593/neo.06247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eckel-Passow JE, Lohse CM, Sheinin Y, et al. Tissue microarrays: one size does not fit all. Diagn Pathol. 2010;5:48. doi: 10.1186/1746-1596-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morikawa T, Kuchiba A, Liao X, et al. Tumor TP53 expression status, body mass index, and prognosis in colorectal cancer. Int J Cancer. 2012 doi: 10.1002/ijc.26495. in press (published online: Oct 31, 2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King RL, Pasha T, Roullet MR, et al. IMP-3 is differentially expressed in normal and neoplastic lymphoid tissue. Hum Pathol. 2009;40:1699–1705. doi: 10.1016/j.humpath.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 29.Ogino S, Nosho K, Kirkner GJ, et al. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst. 2008;100:1734–8. doi: 10.1093/jnci/djn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhee YY, Kim MJ, Bae JM, et al. Clinical Outcomes of Patients with Microsatellite-Unstable Colorectal Carcinomas Depend on L1 Methylation Level. Ann Surg Oncol. 2012 doi: 10.1245/s10434-012-2410-7. [DOI] [PubMed] [Google Scholar]
- 31.Ahn JB, Chung WB, Maeda O, et al. DNA methylation predicts recurrence from resected stage III proximal colon cancer. Cancer. 2011;117:1847–54. doi: 10.1002/cncr.25737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farina-Sarasqueta A, van Lijnschoten G, Moerland E, et al. The BRAF V600E mutation is an independent prognostic factor for survival in stage II and stage III colon cancer patients. Ann Oncol. 2010;21:2396–402. doi: 10.1093/annonc/mdq258. [DOI] [PubMed] [Google Scholar]
- 33.Yokota T, Ura T, Shibata N, et al. BRAF mutation is a powerful prognostic factor in advanced and recurrent colorectal cancer. Br J Cancer. 2011;104:856–62. doi: 10.1038/bjc.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao B, Hu Y, Herrick DJ, et al. The RNA-binding protein IMP-3 is a translational activator of insulin-like growth factor II leader-3 mRNA during proliferation of human K562 leukemia cells. J Biol Chem. 2005;280:18517–24. doi: 10.1074/jbc.M500270200. [DOI] [PubMed] [Google Scholar]
- 35.Jeng YM, Chang CC, Hu FC, et al. RNA-binding protein insulin-like growth factor II mRNA-binding protein 3 expression promotes tumor invasion and predicts early recurrence and poor prognosis in hepatocellular carcinoma. Hepatology. 2008;48:1118–27. doi: 10.1002/hep.22459. [DOI] [PubMed] [Google Scholar]
- 36.Kabbarah O, Nogueira C, Feng B, et al. Integrative genome comparison of primary and metastatic melanomas. PLoS One. 2010;5:e10770. doi: 10.1371/journal.pone.0010770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suvasini R, Shruti B, Thota B, et al. Insulin growth factor-2 binding protein 3 (IGF2BP3) is a glioblastoma-specific marker that activates phosphatidylinositol 3-kinase/mitogen-activated protein kinase (PI3K/MAPK) pathways by modulating IGF-2. J Biol Chem. 2011;286:25882–90. doi: 10.1074/jbc.M110.178012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang T, Fan L, Watanabe Y, et al. L523S, an RNA-binding protein as a potential therapeutic target for lung cancer. Br J Cancer. 2003;88:887–94. doi: 10.1038/sj.bjc.6600806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suda T, Tsunoda T, Daigo Y, et al. Identification of human leukocyte antigen-A24-restricted epitope peptides derived from gene products upregulated in lung and esophageal cancers as novel targets for immunotherapy. Cancer Sci. 2007;98:1803–1808. doi: 10.1111/j.1349-7006.2007.00603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kono K, Mizukami Y, Daigo Y, et al. Vaccination with multiple peptides derived from novel cancer-testis antigens can induce specific T-cell responses and clinical re sponses in advanced esophageal cancer. Cancer Sci. 2009;100:1502–9. doi: 10.1111/j.1349-7006.2009.01200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Kaplan-Meier curves for colorectal cancer-specific survival stratified by disease stage: A, stage I; B, stage II; C, stage III; D, stage IV.
Kaplan-Meier curves for overall survival stratified by disease stage: A, stage I; B, stage II; C, stage III; D, stage IV.