Abstract
Aging liver is characterized by alterations of liver biology and by a reduction of many functions which are important for the maintenance of body homeostasis. The main dysfunctions include appearance of enlarged hepatocytes, impaired liver regeneration after partial hepatectomy (PH), development of hepatic steatosis, reduction of secretion of proteins and alterations in the hepatic sinusoid. RNA binding proteins are involved in the regulation of gene expression in all tissues including regulation of biological processes in the liver. This review is focused on the role of a conserved, multi-functional RNA-binding protein, CUGBP1, in the development of aging phenotype in the liver. CUGBP1 has been identified as a protein which binds to RNA CUG repeats expanded in Myotonic Dystrophy type 1 (DM1). CUGBP1 is highly expressed in the liver and regulates translation of proteins which are critical for maintenance of liver functions. In livers of young mice, CUGBP1 forms complexes with eukaryotic translation initiation factor eIF2 and supports translation of C/EBPβ and HDAC1 proteins, which are involved in liver growth, differentiation and liver cancer. Aging changes several signaling pathways which lead to the elevation of the CUGBP1-eIF2α complex and to an increase of translation of C/EBPβ and HDAC1. These proteins form multi-protein complexes with additional transcription factors and with chromatin remodeling proteins causing epigenetic alterations of gene expression in livers of old mice. It appears that CUGBP1-mediated translational elevation of HDAC1 is one of the key events in the epigenetic changes in livers of old mice, leading to the development of age- associated dysfunctions of the liver. This review will also discuss a possible role of CUGBP1 in liver dysfunction in patients affected with DM1.
Keywords: Aging, liver, CUGBP1, C/EBP, chromatin, epigenetic
1. Introduction
Liver is one of the largest tissues in the body and it provides essential functions for homeostasis of the organism. The liver is a highly complex tissue which consists of many distinct types of cells that interact with each other in a highly organized manner. The biological processes in quiescent liver and during development of liver are tightly regulated by many signaling pathways. The coordination of these signaling pathways is required at the stage when the liver starts development at yearly embryonic stages (Bort and Zaret, 2009) and at the stages when liver continues differentiation after birth reaching the differentiation status in 2 month-old mice (Grisham, 2009). In addition to providing homeostasis, the liver has enormous capabilities to regenerate itself in response to surgeries and liver injury (Michalopoulos, 2010). Prenatal and post-natal liver development and differentiation as well as liver regeneration are regulated by orchestrated expression of many genes and by co-operation of many signaling pathways (Bort and Zaret, 2009; Fausto and Campbell, 2009). The development of approaches for the analysis of global changes in expression of mRNAs sheds light on certain molecular mechanisms of liver development and regeneration (Michalopoulos, 2010). Although transcriptional regulation of genes is an important step in the maintenance of liver biology, it has become clear that post- transcriptional regulation of protein expression is also one of the critical steps in the regulation of liver biology. Increasing numbers of publications show that RNA binding proteins are important regulators of liver biology by control of RNA translation and stability. In this review, we have summarized studies on the role of an RNA binding protein, CUGBP1, in the regulation of liver biology and in age-associated liver dysfunctions.
2. Age-associated dysfunctions of the liver are the result of multiple alterations of signaling pathways
Effects of age on the liver morphology and liver functions have been well documented by numerous reports. Since these alterations are described in details in several reviews (Schmucker, 2005; Floreani, 2007; Timchenko, 2009; 2011), we will briefly summarize findings related to the subject of this review. It has been shown that liver size is reduced with age (Wynne et al., 1995) and that hepatic blood flow is also decreased with age (Zou et al., 1999; Wakabayashi et al., 2002). The liver is one of the biggest organs in the body and is responsible for secretions of many proteins into the blood. Examination of healthy human subjects showed a reduction of albumin in the blood of older individuals (Wakabayashi et al., 2002). It has also been shown that the concentrations of albumin are reduced in older patients after surgeries (Giovannini et al., 2006; Ishiko et al., 2008). Comparisons of serum parameters in the blood in young and old patients showed that serum levels of cholesterol, high-density lipoprotein cholesterol and triglycerides increase with age (Berrougui and Khalil, 2009). In addition, aging affects a number of different cells in the liver. It has been shown that the aging liver contains a much higher portion of macrohepatocytes with increased nuclei due to polyploidy (Chipchase et al., 2003). Aging also increases the accumulation of lipid droplets in the cytoplasm of hepatocytes (Kuk et al., 2009; Cree et al., 2004). It is also interesting that older patients with fatty livers have increased insulin resistance, similar to patients with Myotonic Dystrophy type 1 (Shieh et al., 2010). Non alcoholic fatty liver disease is a common liver disease observed in DM1 patients with increased insulin resistance (Shieh et al., 2010). Several studies have demonstrated age- associated changes in the sinusoids which include a reduction of the number of perfused sinusoids and the reduction of sinusoidal blood flow (Le Couteur et al., 2005; Ito et al., 2007). Caloric restriction corrects these abnormalities (Jamiesen et al., 2007). The age-associated liver dysfunctions are the result of multiple alterations of signaling pathways which are described in details in two recent reviews (Timchenko, 2009; 2011). In this review, we will briefly summarize known mechanisms of impaired liver regeneration since these pathways are closely associated with biological functions of CUGBP1.
3. Impaired liver regeneration in old organisms: the role of chromatin remodeling in the age-associated inhibition of liver regeneration
One of the unique properties of the liver in young organisms is its ability to completely regenerate after massive surgical resections. However, aging significantly reduces the ability of the liver to regenerate. Although this phenomenon was discovered by Bucher et al nearly 5 decades ago (Bucher et al., 1964), molecular mechanisms of this inhibition have only been partially elucidated within the last decade. The most commonly used animal model for these studies is liver regeneration after 2/3 partial hepatectomy (PH) in mice (Michalopoulos, 2010). Numerous studies of liver regeneration with young mice have shown that the mechanisms of liver regeneration are quite complex and include highly orchestrated co-operation of several signaling pathways (Michalopoulos, 2010). There are several critical steps of liver regeneration which occur in a highly orchestrated manner. One of these critical steps in liver regeneration is the activation of genes which are involved in the promotion of cell cycle division such as cyclin D1, DNA polymerase I, PCNA, cdc2, FoxM1B and cyclin E (Timchenko, 2009). Early examinations of liver regeneration in old rodents have shown that activation of DNA polymerase I, PCNA, cdc2 and FoxM1b are significantly reduced and delayed in older animals (Fry et al., 1984; Timchenko et al., 1997; Krupczak-Hollis et al., 2003). It has been found more recently that livers of older animals have an abundant multi-protein complex which contains transcription factors C/EBPα and E2F4, Rb and chromatin remodeling proteins Brm and HP1α (Iakova et al., 2003; Conboy et al., 2005; Wang et al., 2006; Wang et al., 2008a). These complexes will be further alluded to as the C/EBPα-Brm complex. The C/EBPα-Brm complex binds to the promoters of E2F-dependent genes (FoxM1B, cdc2, PCNA and DNA pol I) and reduces or blocks activation of these genes after PH in livers of old mice (Iakova et al., 2003, Wang et al., 2006). The C/EBPα-Brm complex seems to play a major role in the inhibition of liver regeneration in older mice because the reduction/elimination of this complex by a young systemic environment and by growth hormone increases liver proliferation in old mice (Conboy et al., 2005; Wang et al., 2006). The mechanisms of age-associated elevation of the C/EBPα- Brm complex involve alterations in several signaling pathways which lead to the hyper- phosphorylation of C/EBPα at S193. This phosphorylation is required for the interaction of C/EBPα with other components of the C/EBPα-Brm complex (Wang et al., 2006). The role of the phS193 isoform of C/EBPα in the inhibition of liver regeneration has recently been shown by the generation of knockin mice that express a S193D mutant of C/EBPα. Examination of liver regeneration in young S193D mice showed that this isoform of C/EBPα inhibits liver regeneration (Wang et al., 2010). Recent studies have shown that the RNA binding protein CUGBP1 is a key regulator of the expression of several components of the multi-protein complexes observed in livers of old mice (Jin et al., 2009a; 2009b; 2009c).
4. A multi-functional CUG Triplet Repeat Binding protein, CUGBP1, is highly expressed in the liver
4. 1. CUGBP1 binds to a broad range of RNA sequences
CUGBP1 was discovered as a protein which interacts with RNA CUG repeats that are expanded in patients with DM1 (Timchenko et al., 1996a; 1996b). CUGBP1 is a multi-functional protein which regulates splicing, stability and translation of mRNAs (Schoser and Timchenko, 2010). Initial studies showed that CUGBP1 preferentially binds to pure RNA CUG repeats which are expanded in the 3′ UTR of the myotonic dystrophy protein kinase gene (Timchenko et al., 1996a; 1996b) suggesting a limited number of RNAs which might be controlled by this protein. Further studies of the translational functions of CUGBP1 have shown that CUGBP1 interacts with CUG, CCG and CGG (GCN) repeats located in the 5′ regions of the mRNAs whose translation is regulated by CUGBP1 (Timchenko et al., 1999; Welm et al., 2000). Mutations of GC islands within the 5′ region of C/EBPβ mRNA significantly reduce the interactions of CUGBP1 with the C/EBPβ mRNA (Welm et al., 2000). While the interactions of CUGBP1 with the 5′ regions of mRNAs are involved in the regulation of cap-dependent translation, CUGBP1 also interacts with Internal Ribosome Entry Sites or IRES. CUGBP1 represses expression of p27 mRNAs by binding to IRES (Zheng and Miskimins, 2011), but CUGBP1 also has an activating effect on the translation of other mRNAs by interactions with IRES (Woeller et al., 2007; Fox and Stover, 2009). Further studies of CUGBP1 targets have shown that CUGBP1 also interacts with an RNA CCUG repeat that is expanded in patients with Myotonic Dystrophy type 2 (DM2) (Salisbury et al., 2009). CCUG repeats change the activities of CUGBP1 leading to the development of DM2 pathology (Salisbury et al., 2009). Further experiments have revealed that, in addition to previously reported sequences, CUGBP1 binds to UGU-rich sequences (Marquis et al., 2006; Mori et al., 2008). New insights into the interactions of CUGBP1 with mRNA and into the biological functions of CUGBP1 have been revealed during the last three years by the studies of RNA sequences that stabilize mRNAs. Vlasova et al have shown that the consensus sequence UGUUUGUUUGU (GU-rich elements, GRE) is enriched in the 3′ UTR of many short- lived mRNAs and that CUGBP1 is the protein which binds to these sequences and causes degradation of these transcripts (Vlasova et al., 2008; Kim and Gorospe, 2008). These transcripts include several key regulators of cell biology such as mRNA coding for c-jun, jun B and the TNF receptor. The inhibition of CUGBP1 in HeLa cells stabilizes these mRNAs, while the insertion of the GREs from these transcripts into the stable β-globin mRNA reduces stability of β-globin mRNA (Vlasova et al., 2008). Analysis of CUGBP1 associated transcripts in HeLa cells has been performed by this group and has shown that CUGBP1 binds to hundreds of transcripts containing the GRE element (Rattenbacher et al., 2010; Vlasova-St Luis et al., 2011). Because CUGBP1 contains three RNA Binding Domains (RBD, see figure 1), further studies have examined the binding specificity of these domains and showed that the first two RBDs of CUGBP1 interact with GRE and CUG repeats (Edwards et al., 2011; Teplova et al., 2010). It is important to note that current knowledge of the CUGBP1 RNA-binding affinity to different RNA targets has been obtained by different groups using different cells or tissue source and different assays of the RNA-protein binding affinity. Some of these studies were performed in vitro and require verification of CUGBP1 interaction with its newly identified RNA targets in vivo. Since CUGBP1 binds to many mRNAs in other tissues, we predict that CUGBP1 might bind and regulate many mRNAs in the liver. Identification of the complete set of mRNAs bound by CUGBP1 in liver especially at the stages of liver regeneration and aging would help to determine all RNAs regulated by CUGBP1 during these processes. In summary, studies of the interactions of CUGBP1 with mRNAs suggest that CUGBP1 might regulate splicing, translation and stability of many mRNAs in liver, and might control liver-specific pathways through these interactions.
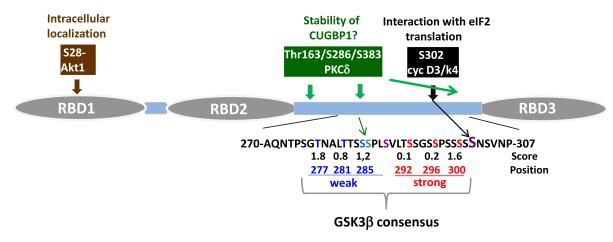
Figure 1. The structure and regulation of the biological activities of CUGBP1.
CUGBP1 consists of three RNA Binding Domains (RBD) separated by spacers. The key amino acids which regulate intracellular localization of CUGBP1 (S28), translational activity of CUGBP1 (S302), and stability of CUGBP1 are shown on the top. Note that the amino acids for the phosphorylation of CUGBP1 by PKCδ were found by the prediction program. Two consensuses (weak and strong) for phosphorylation by GSK3β are shown below.
4. 2. Post-translational regulation of the interaction of CUGBP1 with RNA and biological functions of CUGBP1
The majority of studies of CUGBP1 have been focused on the role of this protein in development of muscle pathology in DM1 and DM2 patients (Schoser and Timchenko, 2010; Lee and Cooper, 2009). These studies showed that the activity of CUGBP1 is mainly regulated on the level of phosphorylation and as a result, on the level of interactions of CUGBP1 with other proteins. CUGBP1 belongs to a family of RNA binding proteins which contain three RBDs. RBD1 and RBD2 are separated by a short spacer and are critical for the CUGBP1 interaction with all examined RNAs (Fig 1). RBD3 is separated from the first two RBDs by a long spacer, which seems to be important for the regulation of RNA binding activity of CUGBP1 and for regulation of interaction of CUGBP1 with other proteins. Recent observations have demonstrated that phosphorylation of CUGBP1 is involved in the control of intracellular localization of CUGBP1, stability of CUGBP1 and translational activity of CUGBP1. The phosphorylation of CUGBP1 at S28 is critical for the control of CUGBP1 levels in cytoplasm and in the nucleus, because the mutation of S28 to Ala resulted in a complete localization of the protein to the nucleus (Huichalaf et al., 2010). Because CUGBP regulates splicing in the nucleus and stability and translation of mRNAs in the cytoplasm, it appears that cells should keep a proper balance of S28-ph and S28-non-ph isoforms to support these different functions of CUGBP1. The phosphorylation of CUGBP1 at S302 is critical for the interactions of CUGBP1 with “active” translation initiation factor 2α, eIF2α, and subsequent activation of translation of target mRNAs (Timchenko et al., 2006; Wang et al., 2008a; 2008b). It has been shown that the interaction of CUGBP1 with “in-active” eIF2α (S51-ph) leads to the accumulation of CUGBP1-mRNA complexes in Stress Granules where the mRNAs are trapped and remain untranslated (Huichalaf et al., 2010). A recent report showed that phosphorylation of CUGBP1 is involved in the regulation of interactions of CUGBP1 with GU rich elements (GREs). Beisang et al have found that phosphorylation of CUGBP1 upon T cell activation leads to a dramatic reduction of the interaction of CUGBP1 with GRE containing transcripts (Beisang et al., 2011). The phosphorylation of CUGBP1 might also control stability of the protein. The phosphorylation of CUGBP1 by PKC stabilizes CUGBP1 in DM1 patients (Kuyumcu-Martinez et al., 2007). Since the residues responsible for this stabilization have not been shown, we have performed the search for PKC consensus sites and have identified three amino acids, Thr-163, Ser-285 and Ser-383, as possible sites for PKCδ binding (Figure 1). It is interesting that the site for cyclin D3-cdk4 (S302) and putative PKCδ sites are located in a spacer region which is very rich for serines and threonines. This region could perhaps be the subject of additional phosphorylations. For example, this region contains two consensuses for the GSK3β kinase (Figure 1) suggesting that alterations of GSK3β activity in cells might affect CUGBP1 activity and/or stability. Taken together, published observations indicate a critical role for phosphorylation of CUGBP1 in the regulation of intracellular localization of CUGBP1 as well as in the regulation of activity and stability of CUGBP1.
5. CUGBP1 promotes liver proliferation after PH in young mice through the translational activation of C/EBPβ-LAP, C/EBPβ-LIP and HDAC1
CUGBP1 is expressed at high levels in the liver of young mice. Although in cultured cells CUGBP1 is located in both the nucleus and cytoplasm, the major fraction of CUGBP1 in hepatocytes is observed in the cytoplasm (Timchenko et al., 1999; Welm et al., 2000). Investigations of CUGBP1 targets in liver have shown that CUGBP1 regulates translation of a member of the C/EBP family, the transcription factor C/EBPβ. C/EBPβ is encoded by an intron- less gene; however, the single C/EBPβ mRNA produces three isoforms (full length C/EBPβ–FL, C/EBPβ-LAP and C/EBPβ-LIP) through the alternative translation from in frame AUG codons. CUGBP1 is a key regulator of this alterative translation. CUGBP1 binds to the CUG and CCG repeats in the 5′ region of C/EBPβ mRNA and increases translation of the truncated isoform of C/EBPβ, C/EBPβ-LIP (Timchenko et al., 1999, Welm et al., 2000). CUGBP1 is involved in the up-regulation of C/EBPβ-LIP under several biological processes such as liver regeneration and during Acute Phase Response (Welm et al., 2000; Timchenko et al., 2005). Under these conditions, CUGBP1 is hyper-phosphorylated at S302 and forms CUGBP-eIF2 complexes which interact with C/EBPβ mRNA and deliver the C/EBPβ mRNA to polysomal fractions of hepatocytes leading to the increased translation of C/EBPβ-LIP (Timchenko et al., 2005). The causal role of CUGBP1 in up-regulation of C/EBPβ-LIP was shown by studies of livers of CUGBP1 transgenic mice. These investigations showed that CUGBP1 dramatically increases expression of C/EBPβ-LIP in the liver of young CUGBP1 TR mice (Timchenko et al., 2006).
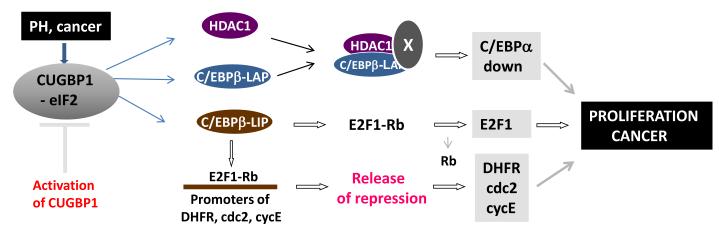
Because CUGBP1 binds to a large number of mRNAs, it has become clear that CUGBP1 could regulate translation of many mRNAs in the liver. One of these mRNAs codes for a chromatin remodeling protein, histone deacetylase I, HDAC1, which is a critical regulator of chromatin structure (Wang et al., 2008a; 2008b). In livers of young mice, translation of HDAC1 is increased after PH and this increase is required for proper liver regeneration (Wang et al., 2008b). Mechanisms for the elevation of HDAC1 in the liver after PH include activation of CUGBP1 and formation of CUGBP1-eIF2 complexes which bind to the 5′ region of HDAC1 mRNA and increase the translation of HDAC1 protein. Similar to proliferation of young liver after PH, proliferation of hepatocytes in human liver tumors is also associated with the activation of CUGBP1 followed by up-regulation of HDAC1 and C/EBPβ proteins: FL-C/EBPβ and C/EBPβ-LIP (Wang et al., 2008b). Figure 2 summarizes studies showing the critical roles of CUGBP1 in the regulation of liver proliferation in young mice and in liver cancer. Investigations of the mechanisms by which the CUGBP1-dependent elevation of HDAC1, C/EBPβ-LAP and C/EBPβ-LIP promotes liver proliferation have shown that there are at least two possible mechanisms. One mechanism includes formation of C/EBPβ-HDAC1 complex repressors and subsequent silencing of the promoters of select genes, products of which inhibit liver proliferation (Wang et al., 2008b). The second mechanism includes direct binding of C/EBPβ-LIP to the E2F consensuses within the promoters of DHFR, cdc2 and E2F1 genes and a replacement of Rb-E2F1 complexes from the promoters leading to the activation of transcription (Orellana et al., 2010). In addition to this mechanism, C/EBPβ-LIP directly interacts with Rb protein and disrupts E2F1-Rb complexes (Orellana et al., 2010). This causes C/EBPβ-LIP to reverse E2F1-Rb-mediated repression through replacement of E2F-Rb complexes on the promoters and by the disruption of the E2F1-Rb complexes (Figure 2). It is interesting to note that CUGBP1 also activates C/EBPβ-LIP in breast cancer, where C/EBPβ-LIP promotes the cancer perhaps by utilizing these mechanisms (Baldwin et al., 2004; Zhanow, 2009). Taken together, these findings show that, in young organisms, CUGBP1 plays a critical role in the promotion of liver proliferation after partial hepatectomy and during development of liver cancer (Figure 2).
Figure 2. CUGBP1 promotes liver proliferation in young mice after partial hepatectomy and in liver cancer.
CUGBP1 is activated in the mouse livers after PH and in liver cancer and forms complexes with eIF2. These complexes increase translation of HDAC1 and C/EBPβ isoforms LAP and LIP. These alterations lead to the promotion of liver proliferation via two distinct mechanisms: reduction of the proteins which inhibit liver proliferation (such as C/EBPα) and elimination of the Rb-E2F1-mediated repression of cell cycle proteins such as DHFR, cdc2 and cyclin E.
6. Age-associated alterations of CUGBP1 are involved in development of liver dysfunctions
6.1. The role of CUGBP1 in the age-associated elevation of the functional C/EBPa-Brm- HDAC1 complexes and in the inhibition of liver regeneration in old mice
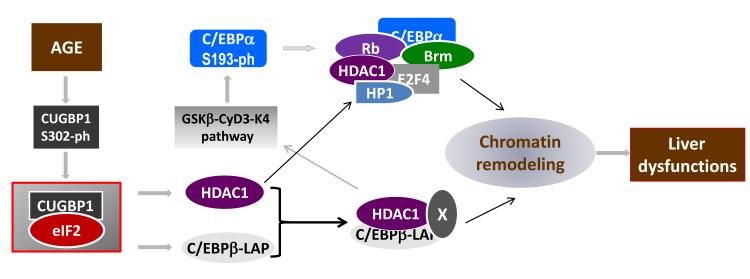
Although previous studies emphasized the critical role of multi-protein C/EBPα-Brm complexes in the inhibition of liver regeneration (Iakova et al., 2003; Conboy et al., 2005; Wang et al., 2006; Timchenko, 2009; 2011), the enzymatic activity of this complex has not been found. New insights into the mechanisms of formation of functional C/EBPα-Brm complexes come from studies of the biological role of CUGBP1 in the livers of old mice. Aging changes many pathways in the liver which are operating up-stream and downstream of CUGBP1. These pathways are shown in Figure 3. Aging increases the amounts of CUGBP1 in the liver and activates CUGBP1 by cyclin D3-cdk4 mediated phosphorylation at S302 (Wang et al., 2008a). These alterations further lead to the formation of a multi-protein complex of CUGBP1 with eIF2α, eIF2β, Grp78, eR99, CRT and eR60 (Timchenko et al., 2006). Interestingly, similar alterations of CUGBP1 and the formation of an identical complex were observed in skeletal muscle of patients with DM2 in which this complex up-regulates translation of proteins involved in muscle development (Salisbury et al., 2009). In livers of old mice, the multi-protein complex CUGBP1-eIF2α increases translations of C/EBPβ and HDAC1. The elevation of HDAC1 in the livers of old mice leads to the association of HDAC1 with the previously described C/EBPα- Brm complex. It has been shown that HDAC1 is a critical component of the C/EBPα-Brm- HDAC1 complex and that HDAC1 is required for the inhibition of E2F-dependent promoters by this complex (Wang et al., 2008a; 2008b). Since the C/EBPα-Brm-HDAC1 complex inhibits liver proliferation, these studies demonstrated that the age-associated elevation and activation of CUGBP1 are involved in the inhibition of liver regeneration after PH through the translational induction of HDAC1.
Figure 3. The activation of CUGBP1 in livers of old mice leads to the translational elevation of HDAC1 which causes chromatin remodeling and alteration of liver functions.
Ageing liver increases amounts and activity of CUGBP1 leading to the elevation of CUGBP1- eIF2 complex. The CUGBP1-eIF2 complex increases translation of HDAC1 and C/EBPβ-LAP leading to the appearance of two complexes containing chromatin remodeling proteins: C/EBPα- Brm-HDAC1 and C/EBPβ-HDAC1 complexes. These complexes cause epigenetic alterations which lead to age-associated dysfunctions of the liver.
6.2. The age-associated activation of CUGBP1 in the liver is involved in epigenetic down- regulation of SIRT1 and GSK3β
Similar to the activation of CUGBP1in livers of young mice after PH, the activation of CUGBP1 in livers of old mice leads to the elevation HDAC1 and to the formation of HDAC1-C/EBPβ complexes (Jin et al., 2009a; Jin et al., 2011). In an opposing function to the HDAC1-C/EBPα- Brm complex, C/EBPβ-HDAC1 complex binds to the C/EBPβ and C/EBPα-dependent promoters. Because C/EBP proteins regulate critical biological processes in the liver, CUGBP1- dependent elevation of the C/EBPβ-HDAC1 complex in livers of old mice is likely to repress expression of many C/EBP-dependent genes. One target of this complex in livers of old mice is Glycogen Synthase Kinase beta, GSK3β. GSK3β is a serine-threonine kinase which regulates a variety of biological processes (Rayasam et al., 2009). The promoter region of the GSK3β gene contains three C/EBP binding sites and the elevation of the HDAC1-C/EBPβ complex in livers of old mice leads to the binding of this complex to and repression of the GSK3β promoter. As a result, the amounts of GSK3β are significantly reduced in livers of old mice (Jin et al., 2009a). This reduction of GSK3β leads to the stabilization and elevation of cyclin D3 which is involved in the inhibition of liver regeneration through hyper-phosphorylation of C/EBPα at S193 (Jin et al., 2009a; Wang et al., 2006). Since the S193-ph isoform of C/EBPα inhibits liver proliferation, CUGBP1-mediated elevation of C/EBPβ-HDAC1 complexes contributes to the inhibition of liver proliferation in livers of old mice. GSK3β regulates a variety of biological processes in the liver; therefore, the reduction of this enzyme is likely to affect additional pathways in the liver of old mice.
The second target of C/EBPβ-HDAC1 complexes in livers of old mice is SIRT1. SIRT1 belongs to a family of histone deacetylases (sirtuins) and it controls expression and activities of proteins mainly through two mechanisms: epigenetic regulation of the promoters and acetylation of proteins (Herranz and Serrano, 2010). SIRT1 is one of the more well characterized proteins which plays a critical role in the development of the aging phenotype in several organisms (Herranz and Serrano, 2010; Erion et al., 2009). Although the reduction of SIRT1 in tissues of old mice has been described (Ramsey et al., 2008), the mechanisms of this down-regulation in the livers of old mice were not understood until recently. Promoters of both the human and mouse SIRT1 gene contain several binding sites for C/EBP proteins and are positively regulated by C/EBPβ in livers of young organisms. However, the SIRT1 promoter is repressed in livers of old mice by the C/EBPβ-HDAC1 complexes and this repression is involved in the inhibition of glucose and triglycerides metabolism in serum of old mice (Jin et al., 2011). Thus, the age- associated activation of CUGBP1 is also involved in the development of liver dysfunctions through the down-regulation of SIRT1. Further work will likely identify additional targets of the HDAC1-C/EBPβ complex as well as new targets for the complexes of HDAC1 with other transcription factors.
Age-dependent activation of CUGBP1 results in the appearance/elevation of two multi- protein complexes: C/EBPα-Brm-HDAC1 and C/EBPβ-HDAC1. These complexes contain chromatin remodeling proteins HDAC1, Brm and HP1α (Wang et al., 2008b; and see fig 3). Recent studies have examined biological consequences of the accumulation of C/EBP-HDAC1 complexes in liver biology (Wang et al., 2010, Jin et al., 2010). In these investigations, the C/EBPα gene was replaced with a S193D mutant, which is a product of the CUGBP1-GSK3β- cyclinD3/cdk4 pathway. Therefore, these C/EBPα-S193D knockin mice (S193D mice) reflect the age-associated alterations on several pathways including CUGBP1-dependent pathways. Examination of young S193D knockin mice showed a dramatic alteration of chromatin structure with the appearance of heterochromatin structures which contain histone H3 trimethylated at K9 (Jin et al., 2010). This alteration in chromatin structure is similar to that observed in livers of old mice (Jin et al., 2010; Kreiling et al., 2011). Studies with these S193D mice and old WT mice showed that alterations of the chromatin structure are caused by C/EBPα-HDAC1 and C/EBPβ- HDAC1 complexes and that this epigenetic change is the major cause of age-associated liver dysfunctions. Chromatin remodeling-dependent dysfunctions include inhibition of liver proliferation, elevation of the number of large hepatocytes, elevation of glycogen in the liver, increased sensitivity to the development of liver cancer, and increased levels of ALT/AST and triglycerides in the blood (Wang et al., 2010; Jin et al., 2010). The C/EBPα-HDAC1 complexes in S193D mice and the C/EBPβ-HDAC1 complexes in livers of old mice seem to be the major cause of this epigenetic change since inhibition of the complexes by siRNA to HDAC1 corrects chromatin structure and liver functions in young S193D and in old WT mice. Taken together, these studies revealed that age-associated activation of the RNA binding protein CUGBP1 is involved in the development of liver dysfunctions through a CUGBP1-dependent increase in translation of key regulators of liver biology.
6.3. The elevation of CUGBP1 in pre-adipocytes with age leads to impaired adipogenesis and might be involved in liver dysfunctions
Although activation of CUGBP1 in liver seems to be a major cause of liver dysfunctions, there are several reports which suggest that liver functions might be affected by the elevation of CUGBP1 in other tissues. One of the age-associated liver dysfunctions is hepatic steatosis, which is characterized by accumulation of fat droplets in the cytoplasm of hepatocytes (Timchenko, 2009; 2010). The mechanisms of hepatic steatosis are poorly understood, but it is likely that age- dependent hepatic steatosis is partially the result of alterations in functions of adipose tissue. It has been shown that fat deposits decrease with age leading to redistribution of fat to other tissues such as liver (Tchkonia et al., 2010; Sepe et al., 2011). The age-associated decline of adipose functions is the result of alterations of several pathways. Particularly, expression of two key regulators of adipogenesis (targets of CUGBP1), C/EBPα and C/EBPβ, are altered in adipose tissue by age. Searching for mechanisms by which age alters expression of C/EBPα and C/EBPβ, Kirkland’s group has shown that tumor necrosis factor alpha (TNFα) is increased in adipose tissue of old mice and that TNFα increases levels of CUGBP1 and RNA binding activity of CUGBP1 (Karagiannides et al., 2006). The elevation of CUGBP1, in turn, increases translation of a truncated C/EBPβ isoform, C/EBPβ-LIP, and reduces levels of C/EBPα. Since C/EBPβ-LIP works as a dominant negative molecule and blocks activities of the full length C/EBPα and C/EBPβ proteins, the CUGBP1-mediated elevation of C/EBPβ-LIP leads to impaired adipogenesis (Karagiannides et al., 2006). It is interesting that the inhibition of CUGBP1 in preadipocytes from old animals leads to enhanced lipid accumulation (Karagiannides et al., 2006). In agreement with these observations, Bae and Kim have shown that oltipraz, a cancer- chemopreventive agent, inhibits adipogenesis through activation of CUGBP1 and subsequent elevation of C/EBPβ-LIP (Bae and Kim 2005). Taken together, these studies show that CUGBP1 is also a key regulator of adipogenesis and that age changes adipose functions by elevation of CUGBP1. It is likely that the elevation of CUGBP1 in adipose tissue is involved in age- dependent reduction of fat deposits and subsequent redistribution of fat droplets to the liver.
7. Liver dysfunctions in Myotonic Dystrophy: possible role of CUGBP1
Although the main pathological alterations in DM1 are observed in skeletal muscle and heart, mutant DMPK mRNA is expressed in a range of tissues, including liver, and might affect liver biology (Sarkar et al., 2004). Early investigations of a cohort of 53 patients with DM1 showed that at least one of the six liver enzymes was affected in each DM1 patient (Achiron et al., 1998). A recent examination of a group of thirty-six patients with DM1 showed that 44% of DM1 patients have developed nonalcoholic fatty liver disease (NAFLD) as it was shown by abnormal liver chemistry and by evidence of steatosis (Shieh et al., 2010). Particularly, these patients have increased levels of ALT and AST, elevated triglycerides and elevated total cholesterol. The authors found that DM1 patients with higher insulin resistance develop stronger NAFLD. Since the pathogenesis of NAFLD is associated with insulin resistance, these findings suggest that NAFLD may progress in DM1 causing serious liver disease. It has been shown that serum gamma-glutamyltransferase (GGT), a sensitive biomarker of liver disease, is elevated in DM1 patients suggesting that the secretion functions of the liver might be affected by the expansion of CTG repeats (Franzine et al., 2010).
Mechanistic studies into the role of expanded CTG repeats in liver function are limited; however, several reports show that CTG repeat-mediated elevation of CUGBP1 or overexpression of CUGBP1 (mimicking the elevation of CUGBP1 in DM1) change molecular pathways in the liver. It is known that somatic CTG repeat expansions are unstable in proliferating and in post-mitotic tissues and have the ability to increase in length with age. It appears that the rate of CTG repeat instability is significantly increased in mouse liver hepatocytes. Van der Broek et al have studied the instability of CTG repeat expansions in tissues from a CTG knock-in mouse model (Van der Broek et al., 2007). The authors found that CTG repeat expansions were increasing in length almost uniquely in polyploid hepatocytes, showing high nuclearity, and that in liver, CTG repeat instability was increased with age (Van der Broek et al., 2007). This increased DNA ploidy is one of the main characteristics of aged livers (Chipchase et al., 2003). Thus, it is predicted that the length of CTG repeat expansions are particularly increased in DM1 liver and that CTG instability in liver of DM1 patients is increased with age. It is also possible that expanded CTG repeats increase DNA ploidy in liver hepatocytes, which might result in premature liver aging. We suggest that CTG repeats change hepatocyte morphology and function through CUGBP1, since CUGBP1 is elevated in the liver of DM1 patients. One of functions of CUGBP1 is regulation of splicing of mRNAs including insulin receptor (IR) mRNA (Lee and Cooper, 2009). Examination of insulin receptor isoforms in a mouse model of DM1 showed that the de-regulation of IR splicing occurs in all tissues including liver (Guiraud-Dogan et al., 2007). Interestingly, in this study, the authors have found that the expression of the DMPK transcript with expanded CTG repeats is significantly increased in aged liver suggesting that livers might be more affected in older DM1 patients (Guiraud-Dogan et al., 2007). Since CUGBP1 is elevated in DM1 and since IR is one of the splicing targets of CUGBP1, mis-splicing of IR in DM1 tissues, including liver, might be associated with insulin resistance in DM1.
Additional evidence for the role of CUGBP1 in liver dysfunctions has been obtained through the study of CUGBP1 transgenic mice. Initial studies of CUGBP1 TR mice, in which CUGBP1 was mainly overexpressed in skeletal muscle and in heart, showed that the elevation of CUGBP1 inhibits myogenesis in skeletal muscle through the translational activation of C/EBPβ- LIP, MEF2 and p21 proteins (Timchenko et al., 2004). Further studies of the liver biology in these mice showed that CUGBP1 is also elevated in liver and that at least some molecular dysfunctions in the livers of CUGBP1 TR mice are similar to those observed in livers of old WT mice. First, it has been shown that the elevation of CUGBP1 in livers of young CUGBP1 TR mice leads to the formation of multi-protein CUGBP1-eIF2 complexes, the composition of which is identical to that observed in old WT mice (Timchenko et al., 2006). Similar to livers of old mice, the CUGBP1-eIF2 complex increases translation of the dominant negative C/EBPβ- LIP isoform in the liver of young CUGBP1 TR mice. Second, the CUGBP1-eIF2 complex also increases translation of HDAC1 (Wang et al., 2008b) and this increase leads to the elevation of HDAC1-C/EBPβ complexes in livers of young CUGBP1 TR mice (Wang et al., 2008b). In livers of young CUGBP1 TR mice, the HDAC1-C/EBPβ complex binds to the C/EBPα promoter and inhibits expression of C/EBPα. As a result, the livers of young CUGBP1 TR mice have an increased rate of proliferation (Wang et al., 2008b). These studies also suggested that activation of CUGBP1 in liver tumors is one of the critical steps for development of liver cancer.
A growing number of reports shows that the elevation of CUGBP1 is the main feature of Myotonic Dystrophy type 2 (DM2), a disease, similar to but distinct from DM1 (Salisbury et al., 2009; Jones et al., 2011; Schoser and Timchenko, 2010). It has been shown that the expansion of CCTG repeats in a mouse model of DM2 (CCTG transgenic mice) also increases levels of CUGBP1 (Salisbury et al., 2009). This increase of CUGBP1 in CCTG transgenic mice by non- coding CCUG repeats leads to the formation of translational complexes of CUGBP1 with eIF2. Interestingly, the composition of the translational CUGBP1-eIF2 complexes in DM2 muscle cells and in the DM2 mouse model (CCTG TR mice) is identical to those in livers of old WT mice (Salisbury et al., 2009; Timchenko et al., 2006). These studies suggested pathways by which CUGBP1 might affect liver functions in patients with DM and in normal liver during aging.
8. Remarking Conclusions
Age-associated liver dysfunctions are mediated by a complex alteration in many signaling pathways which change biological functions of transcription factors, chromatin remodeling proteins, proteins of translational apparatus and RNA binding proteins. In this review, we have summarized several of the main studies showing a critical role of RNA binding protein, CUGBP1, in liver biology. The functions of CUGBP1 in livers of young mice are quite different from those in livers of older mice. While CUGBP1 is a positive regulator of liver proliferation in young mice, the age-associated alterations of CUGBP1 are involved in liver dysfunctions which occur with age and in liver dysfunctions observed in DM1 patients. Figure 3 presents a simplified diagram with key steps by which aging activates CUGBP1 and how this activation changes expression of downstream targets of CUGBP1. The final outcome of the activation of CUGBP1 is the appearance of two multi-protein chromatin remodeling complexes and subsequent alteration of the chromatin structure. As a result, epigenetic control of key regulators of liver functions is changed leading to dysfunctions in aged liver. It is interesting that epigenetic alterations seem to be the main cause of replicative senescence in human fibroblasts (Sedivy et al., 2008). At this stage in research, the CUGBP1 protein is implicated in the development of aging phenotype in the liver. However, it is likely that aging might affect several other RNA binding proteins, such as HuR, in the liver and that these proteins might also contribute to liver dysfunctions. The role of CUGBP1 in the regulation of liver biology has been confirmed by several animal models including CUGBP1 transgenic mice and long-lived Little mice where CUGBP1 is activated. Because conventional CUGBP1 KO mice do not live long (Kress et al., 2007), studies of age-associated liver dysfunctions are not possible with these mice. It would be an important next step to generate liver-specific CUGBP1 KO mice and examine liver biology in these mice. There are also several interesting questions about the role of CUGBP1 in aging liver that remain unanswered and require further studies. Current knowledge of the role of CUGBP1 in liver functions was obtained mainly in investigations of known targets of CUGBP1. These studies showed that CUGBP1 regulates liver growth and differentiation through translational activation of HDAC1 and C/EBPβ. However, systemic examination of CUGBP1-dependent pathways in livers of young and old mice has not been performed. Given the multiple RNA targets of CUGBP1 in cultured cells, it is likely that this protein might also regulate stability of GRE containing mRNAs in the liver and that CUGBP1 might be involved in regulation of additional biological processes through stabilization of mRNAs. Therefore, important future studies should be performed to compare sets of mRNA targets of CUGBP1 in liver of young and old mice as well as identify all targets of the HDAC1-C/EBPβ complex. These studies will provide a more complete picture of the global changes in aged liver caused by the activation of CUGBP1.
Highlights.
CUGBP1 regulates splicing, translation and stability of mRNAs
CUGBP1 increases translation of key mediators of liver biology
Aging increases CUGBP1 in livers and activates CUGBP1 by phosphorylation
The activation of CUGBP1 in liver of old mice increases C/EBPβ and HDAC1
CUGBP1-mediated increase of HDAC1 changes chromatin structure of hepatocytes
Acknowledgements
NAT is supported by NIH grants GM551888, CA100070, AG039885, AG028865, AG039885, and CA159942. LT is supported by NIH grants AR052791 and AR044387. KJ is supported by NIH grant HL007676.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Achiron A, Barak Y, Magal N, Shonat M, Cohen M, Barar R, Gadoth N. Abnormal liver test in Myotonic Dystrophy. Journal of Clinical Gastroenterology. 1998;26:292–295. doi: 10.1097/00004836-199806000-00016. [DOI] [PubMed] [Google Scholar]
- Bae EJ, Kim SG. Enhanced CCAAT/enhancer-binding protein beta-liver-enriched inhibitory protein production by Oltipraz, which accompanies CUG repeat-binding protein-1 (CUGBP1) RNA-binding protein activation, leads to inhibition of preadipocyte differentiation. Molecular Pharmacology. 2005;3:660–669. doi: 10.1124/mol.105.012997. [DOI] [PubMed] [Google Scholar]
- Baldwin BR, Timchenko NA, Zhanow CA. EGF receptor stimulation activates the RNA binding protein, CUGBP1, and increases expression of C/EBPβ-LIP in mammary epithelial cells. Molecular and Cellular Biology. 2004;24:3682–3691. doi: 10.1128/MCB.24.9.3682-3691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisang D, Rattenbacher B, Vlasova-St Louis IA, Bohjanen P. Regulation of CUG-Binding Protein 1 (CUGBP1) Binding to Target Transcripts upon T Cell Activation. Journal of Biological Chemistry. 2011 doi: 10.1074/jbc.M111.291658. doi/10.1074/jbc.M111.291658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrougui H, Khalil A. Age-Associated Decrease of High-Density Lipoprotein-Mediated Reverse Cholesterol Transport activity. Rejuvenation Research. 2009;12:117–126. doi: 10.1089/rej.2009.0840. [DOI] [PubMed] [Google Scholar]
- Bort R, Zaret KS. Chapter 2, Embryonic development of the liver. In: Arias M, editor. The liver: Pathology and Pathobiology. fifth edition Wiley-Blackwell, Wiley & Sons, LTD production; 2009. pp. 17–27. [Google Scholar]
- Bucher NLR, Glinos MN, Di Troi JF. The influence of age upon the incorporation of thymidine-2C14 into the DNA of regenerating rat liver. Cancer Research. 1964;24:509–512. [PubMed] [Google Scholar]
- Chipchase MD, O’Neill M, Melton DW. Characterization of premature liver polyploidy in DNA repair (Ercc1)-deficient mice. Hepatology. 2003;38:968–966. doi: 10.1053/jhep.2003.50421. [DOI] [PubMed] [Google Scholar]
- Conboy IM, Conboy MJ, Wagers AJ, Girma ER, Weisman IL, Rando TA. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature. 2005;43:760–764. doi: 10.1038/nature03260. [DOI] [PubMed] [Google Scholar]
- Cree MG, Newcomer BR, Katsanos CS, Sheffield-Moore M, Chinkes D, Aarsland A, Urban R, Wolee RR. Intramuscular and liver triglycerides are increased in elderly. Journal of Clinical Endocrinology and Metabolism. 2004;89:3864–3871. doi: 10.1210/jc.2003-031986. [DOI] [PubMed] [Google Scholar]
- Edwards J, Malaurie E, Kondratov A, Long J, de Moor C, Searle MS, Emsey J. Sequence determinants for the tandem recognition of UGU and CUG rich RNA elements by two N-terminal RRMs of SELF1. Nucleic Acids Research. 2011;39:8638–8650. doi: 10.1093/nar/gkr510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erion DM, Yonemitsu S, Nie Y, Nagai Y, Gillum MP, Hsiao JJ, Iwasaki T, Stark R, Weismann D, Yu XX, Murray SF, Bhanot S, Monia BP, Hovath TL, Gao Q, Samuel VT, Shilman GI. SirT1 knockdown in liver decreases basal hepatic glucose production and increases hepatic insulin responsiveness in diabetic rats. Proceedings of the National Academy Science of the United States. 2009;106:11288–11293. doi: 10.1073/pnas.0812931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fausto N, Campbell JS. Chapter 36. Liver regeneration. In: Arias M, editor. The liver: Pathology and Pathobiology. fifth edition Wiley-Blackwell, Wiley & Sons, LTD production; 2009. pp. 549–565. [Google Scholar]
- Floreani A. Liver diseases in the elderly: an update. Digestive Diseases. 2007;25:138–143. doi: 10.1159/000099478. [DOI] [PubMed] [Google Scholar]
- Fox JT, Stover PJ. Mechanism of the internal ribosome entry site-mediated translation of serine hydroxymethyltransferase 1. Journal of Biological Chemistry. 2009;284:31085–31089. doi: 10.1074/jbc.M109.035576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzini M, Fornaciari I, Siciliano G, Volpi L, Ricci G, Marchi S, Gagliardi G, Baggiani A, Torraca F, Fierabracci V, Miccoli M, Pompella A, Emdin M, Paolicchi A. Serum gamma glutamyltransferase fractions in Myotonic Dystrophy type 1: differences with healthy subjects and patients with liver disease. Clinical Biochemistry. 2010;43:1246–1248. doi: 10.1016/j.clinbiochem.2010.07.018. [DOI] [PubMed] [Google Scholar]
- Fry M, Silbe J, Loeb LA, Martin GM. Delayed and reduced cell replication and diminishing levels of DNA-polymerase alpha in regenerating liver of aging mice. Journal of Cellular Physiology. 1984;118:225–232. doi: 10.1002/jcp.1041180302. [DOI] [PubMed] [Google Scholar]
- Giovannini I, Chiarla C, Giuliante F, Vellone M, Ardito F, Nuzzo G. The relationship between albumin, other plasma proteins and variables, and age in the acute phase response after liver resections in man. Amino Acids. 2006;31:463–469. doi: 10.1007/s00726-005-0287-5. [DOI] [PubMed] [Google Scholar]
- Grisham JW. Chapter 1, Organisational principles of the liver. In: Arias M, editor. The liver: Pathology and Pathobiology. fifth edition Wiley-Blackwell, Wiley & Sons, LTD production; 2009. pp. 6–16. [Google Scholar]
- Guiraud-Dogan C, Huguet A, Gomes-Pereira M, Brisson E, Bassez G, Junien C, Gourdon G. DM1 CTG expansions affect insulin receptor isoforms expression in various tissues of transgenic mice. Biochimica et Biophisica Acta. 2007;1772:1183–1191. doi: 10.1016/j.bbadis.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Herranz D, Serrano M. Impact of Sirt1 on mammalian aging. Aging. 2010;2:315–316. doi: 10.18632/aging.100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huichalaf C, Sakai K, Jin B, Jones K, Wang G-L, Schoser B, Schnider, Gold C, Sarkar P, Pereira-Smith O, Timchenko NA, Timchenko LT. Expansion of CUG RNA repeats causes stress and inhibition of translation in Myotonic Dystrophy 1 (DM1) cells. FASEB Journal. 2010;10:3706–3719. doi: 10.1096/fj.09-151159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson HA, Hilmer SN, Cogger VC, Warren A, Cheluvappa R, Abermethy DR, Everit AV, Fraser R, De Cabo R, Le Couteur DG. Caloric restriction reduces age-related pseodocapillarization of the hepatic sinusoid. Experimental Gerontology. 2007;42:374–378. doi: 10.1016/j.exger.2006.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Iakova P, Wang G-L, Shi X, Haefliger S, Finegold M, Timchenko NA. Epigenetic changes play critical role in age-associated dysfunction of the liver. Aging Cell. 2010;9:895–910. doi: 10.1111/j.1474-9726.2010.00617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Iakova P, Jiang J, Medrano EE, Timchenko NA. The reduction of SIRT1 in livers of old mice leads to impaired body homeostasis and to inhibition of liver proliferation. Hepatology. 2011;54:898–998. doi: 10.1002/hep.24471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Wang G-L, Shi X, Darlington GJ, Timchenko NA. The age-associated decline of GSK3β plays a critical role in the inhibition of liver regeneration. Molecular and Cellular Biology. 2009a;29:3867–3880. doi: 10.1128/MCB.00456-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Wang G-L, Salisbury E, Timchenko LT, Timchenko NA. GSK3β-cyclin D3-CUGBP1-eIF2 pathway in aging and in Myotonic Dystrophy. Cell Cycle. 2009b;15:2356–2359. doi: 10.4161/cc.8.15.9248. [DOI] [PubMed] [Google Scholar]
- Jin J, Wang G-L, Timchenko LT, Timchenko NA. GSK3β and aging liver. Aging. 2009c;6:582–585. doi: 10.18632/aging.100060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K, Jin B, Huichalaf C, Sarkar P, Schneider-Gold C, Schoser B, Meola G, Shyi A-B, Timchenko NA, Timchenko LT. RNA foci, CUGBP1 and ZNF9 are the primary targets of the mutant CUG and CCUG repeats expanded in Myotonic Dystrophies type 1 and type 2. American Journal of Pathology. 2011;5:2475–2489. doi: 10.1016/j.ajpath.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakova P, Awad SS, Timchenko NA. Aging reduces proliferative capacities of liver by switching pathways of C/EBPα -mediated growth arrest. Cell. 2003;113:495–506. doi: 10.1016/s0092-8674(03)00318-0. [DOI] [PubMed] [Google Scholar]
- Ishiko T, Inomata Y, Beppu T, Asonuma K, Okajima H, Takeitchi T, Baba H. Age and donor safety in living-donor liver transplantation in 110 consecutive cases at 1 institute. Experimental and Clinical Transplantation. 2008;6:190–193. [PubMed] [Google Scholar]
- Ito Y, Sorensen K, Bethea NW, Svistounov D, McCuskey M, Smedsrod BH, McCuskey RS. Age-related changes in the hepatic microcirculation in mice. Experimental Gerontology. 2007;42:789–797. doi: 10.1016/j.exger.2007.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagiannides I, Thomou T, Tchkonia T, Pirtshalava T, Kypreos K, Cartwright A, Dalagiorgou D, Lash TL, Farmer SR, Timchenko NA, Kirkland JL. Increased CUG triplet repeat-binding protein-1 predisposes to impaired adipogenesis with aging. Journal of Biological Chemistry. 2006;281:23025–23033. doi: 10.1074/jbc.M513187200. [DOI] [PubMed] [Google Scholar]
- Kim HH, Gorospe M. GU-rich RNA: expanding CUGBP1 function, broadening mRNA turnover. Molecular Cell. 2008;29:151–152. doi: 10.1016/j.molcel.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiling JA, Tamamori-Adachi M, Sexton AN, Jeyapalan JC, Munoz-Najar U, Peterson AL, Manivannan J, Rogers ES, Pchelintsev NA, Adams PD, Sedivy JM. Age-associated increase in heterochromatic marks in murine and primate tissues. Aging Cell. 2011;10:292–304. doi: 10.1111/j.1474-9726.2010.00666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress C, Gautier-Courteille C, Osborne HB, Babinet C, Paillard L. Inactivation of CUG-BP1/CELF1 causes growth, viability and spermatogenesis defects in mice. Molecular and Cellular Biology. 2007;27:1146–1157. doi: 10.1128/MCB.01009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupczak-Hollis K, Wang X, Dennewitz MB, Costa RH. Growth hormone stimulates proliferation of old-aged regenerating liver through forkhead box m1b. Hepatology. 2003;38:1552–1562. doi: 10.1016/j.hep.2003.08.052. [DOI] [PubMed] [Google Scholar]
- Kuk JL, Saunders TJ, Davidson LE, Toss R. Age-related changes in total and regional fat distribution. Ageing Research Reviews. 2009;8:339–348. doi: 10.1016/j.arr.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Kuyumcu-Martinez NM, Wang G-S, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy are due to PKC-mediated hyperphosphorylation. Molecular Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Couteur DG, Warren A, Cogger V, Smedsrod B, Sorensen KK, De Cabo R, Fraser R, McCuskey R. Old age and the hepatic sinusoid. The Anatomical Record. 2008;291:672–683. doi: 10.1002/ar.20661. [DOI] [PubMed] [Google Scholar]
- Lee JE, Cooper TA. Pathogenic mechanisms of myotonic dystrophy. Biochemical Society Transactions. 2009;37:1281–1286. doi: 10.1042/BST0371281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquis J, Paillard L, Audic Y, Cosson B, Danos O, Le Bec C, Osborne HB. CUGBP1/SELF1 requires UGU-rich sequences for high affinity binding. Biochemical Journal. 2006;4000:291–301. doi: 10.1042/BJ20060490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalopoulos G. Liver Regeneration after Partial Hepatectomy. American Journal of Pathology. 2010;176:2–12. doi: 10.2353/ajpath.2010.090675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori D, Sasagawa N, Kino Y, Ushiura S. Quantitative analysis of CUGBP1 binding to RNA repeats. Journal of Biochemistry. 2008;153:377–383. doi: 10.1093/jb/mvm230. [DOI] [PubMed] [Google Scholar]
- Orellana D, Liu X, Wang G-L, Jin J, Iakova P, Timchenko NA. Calmodulin controls liver proliferation via interactions with C/EBPβ-LAP and C/EBPβ-LIP. Journal of Biological Chemistry. 2010;285:23444–23456. doi: 10.1074/jbc.M110.129825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey KM, Mills KF, Satoh A, Imai S. Age-associated loss of Sirt1-mediated enhancement of glucose-stimulated insulin secretion in beta cell-specific Sirt1-overexpressing (BESTO) mice. Aging Cell. 2008;7:78–88. doi: 10.1111/j.1474-9726.2007.00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattenbacher B, Beisang D, Wiesner DL, Jeschke JC, von Hohenberg M, St Louis-Vlasova IA, Bohjanen PR. Analysis of CUGBP1 targets identifies GU-repeat sequences that mediate rapid mRNA decay. Molecular and Cellular Biology. 2010;30:3970–3980. doi: 10.1128/MCB.00624-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayasam GV, Tulasi VK, Sodhi R, Davis JA, Ray A. Glycogen synthase kinase 3: more than a namesake. British Journal of Pharmacology. 2009;156:885–898. doi: 10.1111/j.1476-5381.2008.00085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury E, Schoser B, Schneider-Gold C, Wang G-L, Huichalaf C, Jin B, Sirito M, Sarkar P, Krahe R, Timchenko NA, Timchenko LT. Expression of RNA CCUG repeats dysregulates translation and degradation of proteins in DM2 patients. American Journal of Pathology. 2009;175:748–762. doi: 10.2353/ajpath.2009.090047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar PS, Han J, Reddy S. In situ hybridization analysis of Dmpk mRNA in adult mouse tissues. Neuromuscular disorders. 2006;14:497–506. doi: 10.1016/j.nmd.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Schmucker DL. Age-related changes in liver structure and functions: Implications for disease? Experimental Gerontology. 2005;40:650–659. doi: 10.1016/j.exger.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Schoser B, Timchenko LT. Myotonic Dystrophies 1 and 2: Complex Diseases with Complex Mechanisms. Current Genomics. 2010;11:79–90. doi: 10.2174/138920210790886844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedivy JM, Banumathy G, Adams PD. Aging by epigenetics - a consequence of chromatin damage? Experimental Cell Research. 2008;314:1909–1919. doi: 10.1016/j.yexcr.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepe A, Tchkonia T, Thomou T, Zamboni M, Kirkland JL. Aging and regional differences in fat cell progenitors. Gerontology. 2011;57:66–75. doi: 10.1159/000279755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh K, Gilchrist JM, Promrat K. Frequency and predictors of nonalcoholic fatty liver diseases in myotonic dystrophy. Muscle Nerve. 2010;41:197–201. doi: 10.1002/mus.21484. [DOI] [PubMed] [Google Scholar]
- Tchkonia T, Morbeck DE, Von Zglinicki T, Van Deursen J, Lustgarten J, Scrable H, Khosia S, Jensen MD, Kirkland JL. Fat tissue, aging, and cellular senescence. Aging Cell. 2010;9:667–684. doi: 10.1111/j.1474-9726.2010.00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplova M, Song J, Gaw HY, Teplov A, Patel DJ. Structural Insights into RNA recognition by the alternate-splicing regulator CUG-Binding Protein 1. Structure. 2010;18:1364–1377. doi: 10.1016/j.str.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko NA. Aging and liver regeneration. Trends in Endocrinology and Metabolism. 2009;20:171–176. doi: 10.1016/j.tem.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Timchenko NA. Chapter 19. In: Monga SP, editor. Molecular Pathology of the Liver Diseases. Springer Science + Business Media, LLC; New York: Senescent Liver: 2011. pp. 279–290. [Google Scholar]
- Timchenko NA, Harris TE, Wilde M, Bilyeu TA, Burgess-Beusse BL, Finegold MJ, Darlington GJ. CCAAT/enhancer binding protein α regulates p21 protein and hepatocyte proliferation in newborn mice. Molecular and Cellular Biology. 1997;17:353–7361. doi: 10.1128/mcb.17.12.7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko NA, Wang G-L, Timchenko LT. CUG triplet repeat binding protein, CUGBP1, increases translation of C/EBPβ isoform, LIP, by interacting with the α and β subunits of eIF2. Journal of Biological Chemistry. 2005;280:20549–20557. doi: 10.1074/jbc.M409563200. [DOI] [PubMed] [Google Scholar]
- Timchenko NA, Patel R, Iakova P, Cai Z-J, Quan L, Timchenko LT. Overexpression of CUG triplet repeat binding protein, CUGBP1, inhibits myogenesis. Journal of Biological Chemistry. 2004;279:13129–13139. doi: 10.1074/jbc.M312923200. [DOI] [PubMed] [Google Scholar]
- Timchenko LT, Timchenko NA, Caskey CT, Roberts R. Novel proteins with binding specificity for DNA CTG repeats and RNA CUG repeats: implications for myotonic dystrophy. Human Molecular Genetics. 1996a;5:115–123. doi: 10.1093/hmg/5.1.115. [DOI] [PubMed] [Google Scholar]
- Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, Roberts R, Caskey CT, Swanson MS. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Research. 1996b;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko LT, Salisbury E, Wang G-L, Nguyen H-D, Albrecht JH, Hershey JWB, Timchenko NA. Age-specific CUGBP1-eIF2 complex increases translation of C/EBPβ in old liver. Journal of Biological Chemistry. 2006;281:32806–32819. doi: 10.1074/jbc.M605701200. [DOI] [PubMed] [Google Scholar]
- Timchenko NA, Welm A, Lu X, Timchenko LT. CUG repeat binding protein (CUGBP1) interacts with the 5′region of C/EBPβ mRNA and regulates translation of C/EBPβ isoforms. Nucleic Acids Research. 1999;27:4517–4525. doi: 10.1093/nar/27.22.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Broek WJ, Wansink DG, Wieringa B. Somatic CTG-CAG repeat instability in a mouse model for myotonic dystrophy type 1 is associated with changes in cell nuclearity and DNA ploidy. BMC Molecular Biology. 2007;8:1–8. doi: 10.1186/1471-2199-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova IA, Tahoe NM, Fan D, Larsson O, Rattenbacher B, Sternjohn JR, Vasdewani J, Karypis G, Reilly CS, Bitterman PB, Bohjanen PR. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Molecular Cell. 2008;29:263–270. doi: 10.1016/j.molcel.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova-St Louis I, Bohjanen PR. Coordinate regulation of mRNA decay networking by GU-rich elements and SELF1. Current Opinion in Genetics & Development. 2011;21:444–451. doi: 10.1016/j.gde.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi H, Nishiyama Y, Ushiyama T, Maeba T, Maeta H. Evaluation of the effect of age on functioning hepatocyte mass and liver blood flow using liver scintigraphy in preoperative estimations for surgical patients: comparison with CT volumetry. Journal of Surgical Research. 2002;1006:246–253. doi: 10.1006/jsre.2002.6462. [DOI] [PubMed] [Google Scholar]
- Wang G-L, Shi X, Salisbury E, Sun Y, Albrecht JH, Smith RG, Timchenko NA. Cyclin D3 maintains growth inhibitory activity of C/EBPα by stabilizing C/EBPα-cdk2 and C/EBPα-Brm complexes. Molecular and Cellular Biology. 2006;26:2570–2582. doi: 10.1128/MCB.26.7.2570-2582.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G-L, Salisbury E, Shi X, Timchenko LT, Medrano EE, Timchenko NA. HDAC1 cooperates with C/EBPα in the inhibition of liver proliferation in old mice. Journal of Biological Chemistry. 2008a;283:26196–26178. doi: 10.1074/jbc.M803544200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G-L, Salisbury E, Shi X, Timchenko LT, Medrano EE, Timchenko NA. HDAC1 promotes liver proliferation in young mice via interaction with C/EBPβ. Journal of Biological Chemistry. 2008b;283:26179–26187. doi: 10.1074/jbc.M803545200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G-L, Shi X, Haefliger S, Jin J, Major A, Iakova P, Finegold M, Timchenko NA. Elimination of C/EBPα through the ubiquitin-proteasome system promotes the development of liver cancer in mice. Journal of Clinical Investigation. 2010;120:2549–2562. doi: 10.1172/JCI41933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welm AL, Mackey SL, Timchenko LT, Darlington GJ, Timchenko NA. Translational induction of LIP during acute phase response leads to repression of C/EBP alpha mRNA. Journal of Biological Chemistry. 2000;275:27406–27413. doi: 10.1074/jbc.M002343200. [DOI] [PubMed] [Google Scholar]
- Woeller CF, Fox JT, Perry C, Stover PJ. A ferritin-responsive internal ribosome entry site regulates folate metabolism. Journal of Biological Chemistry. 2007;282:29927–29935. doi: 10.1074/jbc.M706264200. [DOI] [PubMed] [Google Scholar]
- Wynne HA, Cope LH, Kelly P, Whittimgham T, Edwards C, Kamali R. The influence of age, liver size and anantiomer concentrations of warfarin requirements. British Journal of Clinical Pharmacology. 1995;40:203–207. [PMC free article] [PubMed] [Google Scholar]
- Zahnow CA. CCAAT/enhancer-binding protein β: its role in breast cancer and associations with receptor tyrosine kinases. Expert Reviews in Molecular Medicine. 2009;11:1–34. doi: 10.1017/S1462399409001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Miskimins WK. CUG-binding protein represses translation of p27Kip1 mRNA through its internal ribosomal entry site. RNA Biology. 2011;8:365–371. doi: 10.4161/rna.8.3.14804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou M, Magalotti D, Bianchi G, Gueli C, Orlandini C, Grimaldi M, Marchesini G. Total functional hepatic blood flow decrease in parallel with ageing. Age and Ageing. 1999;28:29–33. doi: 10.1093/ageing/28.1.29. [DOI] [PubMed] [Google Scholar]