Background: Talin mediates RIAM-dependent integrin activation and binds vinculin, which stabilizes adhesions.
Results: Structural and biochemical data show that vinculin inhibits RIAM binding to the compact N-terminal region of the talin rod, a region essential for focal adhesion assembly.
Conclusion: Talin·RIAM complexes activate integrins at the leading edge, whereas talin·vinculin promotes adhesion maturation.
Significance: Talin changes partners in response to force-induced conformational change.
Keywords: Adhesion, Cell Biology, Integrins, Nuclear Magnetic Resonance, Structural Biology, RIAM, Focal Adhesions, Talin, Vinculin
Abstract
Talin activates integrins, couples them to F-actin, and recruits vinculin to focal adhesions (FAs). Here, we report the structural characterization of the talin rod: 13 helical bundles (R1–R13) organized into a compact cluster of four-helix bundles (R2–R4) within a linear chain of five-helix bundles. Nine of the bundles contain vinculin-binding sites (VBS); R2R3 are atypical, with each containing two VBS. Talin R2R3 also binds synergistically to RIAM, a Rap1 effector involved in integrin activation. Biochemical and structural data show that vinculin and RIAM binding to R2R3 is mutually exclusive. Moreover, vinculin binding requires domain unfolding, whereas RIAM binds the folded R2R3 double domain. In cells, RIAM is enriched in nascent adhesions at the leading edge whereas vinculin is enriched in FAs. We propose a model in which RIAM binding to R2R3 initially recruits talin to membranes where it activates integrins. As talin engages F-actin, force exerted on R2R3 disrupts RIAM binding and exposes the VBS, which recruit vinculin to stabilize the complex.
Introduction
Cell-extracellular matrix interactions are fundamental to the development and homeostasis of multicellular organisms and involve the coordinated assembly and disassembly of the integrin family of heterodimeric receptors into adhesion complexes of varying complexity and stability (1). The cytoplasmic tails of integrin β-subunits bind a large number of cytoplasmic proteins with scaffolding, adaptor, regulatory, and mechanotransduction functions (2), and among these proteins, talin has been shown to play a pivotal role (3). Thus, talin promotes integrin clustering (4), the switching of integrins from a low to high affinity state (5), and provides a direct link between integrins and the actin cytoskeleton (6). Talin also acts as a scaffold for the recruitment of other proteins such as vinculin, which stabilizes cell-matrix junctions (focal adhesions; FA)5 (7).
Talin is a large 270-kDa protein (2541 amino acids) that consists of an N-terminal head (∼50 kDa) and a large flexible C-terminal rod (∼220 kDa) connected by an unstructured linker region. The talin head comprises a novel FERM domain with the F1–F3 domains in a linear rather than the usual clover leaf arrangement (8) and with an additional domain, F0, packed against F1 (9). The F3 PTB-like domain binds β-integrin cytoplasmic tails, but F0 and F1 are also required for maximal integrin activation (10). Moreover, a series of basic residues distributed along one face of the FERM domain that bind acidic membrane phospholipids are also essential for integrin activation (8–11). The talin rod is predicted to consist of 62 α-helices (12); we have previously shown that these are organized into a series of amphipathic helical bundles and have reported the structures of several such domains (13–18). The talin rod contains ∼11 vinculin binding sites (19), two actin-binding sites (20), as well as binding sites for β-integrin tails (15), the intermediate filament protein synemin (21) and the tumor suppressor DLC1, a RhoGAP localized in FAs (22).
We now report the domain boundaries and structures of the remaining talin rod domains, completing the structural characterization of all 18 domains that make up this large and complex molecule. This has enabled us to explore the interaction of correctly folded talin rod domains with vinculin and the Rap1-GTP-interacting adaptor molecule RIAM, a member of the MRL (Mig-10/RIAM/lamellipodin) family of adaptor proteins that is involved in integrin activation (23, 24). We show that vinculin and RIAM both bind to the N-terminal region of the talin rod but by different mechanisms and that binding is mutually exclusive. Ratio imaging in cells shows that RIAM/vinculin ratios are high at the leading edge, whereas vinculin predominates in FAs. We propose that talin·RIAM complexes are important in talin recruitment to the membrane, integrin activation, and the actin remodeling required for membrane protrusion, whereas in FAs, vinculin displaces RIAM, stabilizes talin in the activated state, and suppresses FA turnover and cell migration.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Talin constructs were synthesized by PCR using a mouse talin1 cDNA as template and cloned into pET-151/d-TOPO (Invitrogen). Constructs were expressed in Escherichia coli BL21 Star(DE3) cells cultured either in LB, or for preparation of 15N-labeled samples, in minimal media containing 1 g of 15N-ammonium chloride per liter. Recombinant His-tagged talin polypeptides were purified as described previously (16), and their concentrations were determined using extinction coefficients at 280 nm. Recombinant His-tagged chicken vinculin domain 1 (Vd1; residues 1–258) was expressed from a pET-15b plasmid and purified as described by Ref. 18. Constructs encoding RIAM residues 45–127, 147–174, and 1–127 were synthesized by PCR using a mouse RIAM cDNA as template and cloned into pET-151/d-TOPO. Purified RIAM wild type (6-30, EDIDQMFSTLLGEMDLLTQSLGVDT) and mutant (6-30-4E, EDIDQEESTEEGEMDLLTQSLGVDT) peptides were a kind gift from Mark Ginsberg (University of California, San Diego).
Vinculin Binding
Analytical gel filtration chromatography using either Superdex-75 (10/300) GL or Superdex-200 (10/300) (Amersham Biosciences) was used to assay binding of talin polypeptides to the vinculin Vd1 domain. Polypeptides were incubated at various temperatures for 30 min prior to gel filtration. Columns were pre-equilibrated and eluted with 20 mm Tris, pH 8.0, 150 mm NaCl, and 2 mm DTT at a flow rate of 0.8 ml/min at room temperature. Fractions were analyzed using SDS-PAGE.
Size Exclusion Chromatography Multiangle Laser Light Scattering (SEC-MALLS)
Proteins (0.5 ml) were separated on a Superdex 75 10/300 gel filtration column (GE Healthcare Life Sciences) equilibrated in 20 mm Tris (pH 7.4), 150 mm NaCl, 2 mm DTT at 0.7 ml/min. Elution was monitored by a Wyatt EOS 18-angle laser photometer (Wyatt Technology), an Optilab rEX refractive index detector, and a Jasco UV-2077 Plus UV/Vis spectrophotometer (Jasco). Molar mass determinations were performed using the Astra software (version 5.3.2.16) (Wyatt Technology).
Circular Dichroism (CD) Spectroscopy
CD spectra were recorded using a JASCO J-715 spectropolarimeter equipped with a JASCO PTC-348WI temperature control unit. Far-UV CD spectra were recorded at 20 °C over the wavelength range 200–250 nm in a quartz cell of 0.1-cm path length (scan rate, 50 nm·min−1). Proteins were dissolved in 20 mm sodium phosphate (pH 6.5), 50 mm NaCl at a concentration of 10 μm. For denaturation studies, the unfolding of α-helices was followed at 222 nm.
Isothermal Titration Calorimetry (ITC)
ITC data were collected using a VP-ITC microcalorimeter (MicroCal, Ltd., Northampton, MA) and analyzed by fitting to a single-site binding equation or a two-site binding equation using MicroCal Origin software.
NMR Spectroscopy
Proteins were prepared in 20 mm sodium phosphate (pH 6.5), 50 mm NaCl, 2 mm DTT with 10% (v/v) 2H2O. NMR spectra were obtained at 298 K using Bruker AVANCE DRX 600 or AVANCE AVII 800 spectrometers both equipped with CryoProbes. Resonance assignment and structure calculations were carried out as described previously (9, 25); the structural statistics for each domain are presented in supplemental Table 1.
Protein Crystallization
Vinculin Vd1(1–258) was co-crystallized with RIAM residues 1–32 (2:1 peptide excess) in 0.05 m Tri-sodium citrate, 1.2 m ammonium sulfate, 3% isobutanol. Crystals were vitrified in the mother liquor containing 20% glycerol and 10% isobutanol. Data were collected to 2.3 Å at base line I24 at the Diamond Light Source. Data were processed and integrated using MOSFLM (26) and SCALA (27). Molecular replacement was performed using PHASER and the template structure (PDB ID 1XWJ) and refined using REFMAC (28). Statistics of the refinement are presented in supplemental Table 2.
Cell Culture, Transfection, Immunofluorescence, and Imaging
Human umbilical vein endothelial cells (HUVEC) were grown according to the manufacturer's instructions (PromoCell). For siRNA knockdown of human talin1, Stealth Select RNAiTM oligo 804 (Invitrogen catalog no. 1299003) was used and a Universal Stealth RNATM siRNA was used as control (6). Cells (6 × 106 per ml) were electroporated (Microporator; Invitrogen) with 80 pmol of talin1 siRNA or control RNA and grown on tissue culture plastic for 72 h. Cells (4 × 104 cells) were then replated on 22-mm glass coverslips and analyzed 4 h later. Where appropriate, cells were co-transfected with 10 μg of plasmid DNA encoding GFP or GFP-tagged mouse talin1. Cells were fixed, stained for paxillin (AB-88 Sigma) or vinculin (monoclonal antibody F9, a gift from V. Koteliansky), imaged, and analyzed as described previously (6).
Ratio Imaging
Vinculin null mouse embryonic fibroblasts were cultured in Dulbecco's Modified Eagle's Medium (DMEM), supplemented with penicillin/streptomycin, 10% FCS, 2 mm l-glutamine, non-essential amino acids, and β-mercaptoethanol (all from Sigma-Aldrich). Cells were transfected with GFP-vinculin Vd1 and RIAM-mCherry (kind gift from Dr. Maddy Parsons; Kings College, London, UK) using Lipofectamine Plus (Invitrogen) according to the manufacturer's instructions and plated 3 h post transfection on glass-bottomed dishes (MatTek Corp.) coated with 10 μg/ml bovine plasma fibronectin (Sigma-Aldrich). After fixation with 3% paraformaldehyde at 24–36 h after transfection, cells were imaged using an inverted microscope (IX70; Olympus) controlled by a Deltavision system (Applied Precision) equipped with a Coolsnap HQ CCD (Photometrics) camera, suitable filter sets (Chroma), and a 100× numerical aperture 1.35 Uplan Apo objective. Ratio images of two components were calculated as described previously (29). Images showing localization of GFP-vinculin Vd1 and RIAM-mCherry were aligned, and ratio values were calculated for each pixel above threshold in both GFP and mCherry channels. Ratio images were displayed using a spectral color look-up table ranging from 0.2 (blue) to 5 (red). To compensate for different expression levels and the different photon yields of GFP and mCherry fusion proteins, the ratios were normalized by a constant that shifted their average values toward 1. For the presentation of ratio images were “smoothed” by averaging 2 × 2 pixels.
RESULTS
The Overall Structure of the Talin Rod
The 2,059 residues of the talin rod are predicted to form 62 α-helices separated by short unstructured regions (12). Using a combination of limited proteolysis, NMR, and x-ray crystallography, we previously identified and solved the structures of two adjacent domains at the N terminus (residues 482–786) (18) and seven sequential domains (residues 1357–2482) at the C terminus of the talin rod (13, 15–17, 19). All of these domains form compact four- or five-helix bundles stabilized by hydrophobic cores. However, the domain organization of residues 787–1356 (18 predicted helices) proved far more difficult to establish, and stably folded polypeptides could be obtained with coordinates incompatible with those of established neighboring domains (17, 30, 31). We therefore expressed multiple fragments containing either four or five helices (supplemental Fig. S1) and assessed their fold by NMR. Only the four non-overlapping fragments 787–911 (four helices), 913–1044 (four helices), 1046–1206 (five helices), and 1206–1357 (five helices) gave 1H,15N HSQC spectra with high chemical shift dispersion and the uniform line widths characteristic of stable globular domains (supplemental Fig. S2). The four domains identified in this way complete the domain mapping of the talin rod, which consists of 13 globular domains (R1 to R13), terminating in a single helix responsible for talin dimerization (Fig. 1A). The structures of the four new domains R3–R6 are shown in Fig. 1, B–E, and supplemental Fig. S3 and are described in the supplemental data.
FIGURE 1.
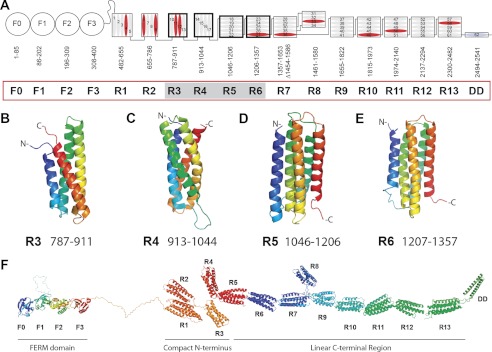
Domain structure of talin. A, schematic diagram of talin showing the N-terminal FERM domain (F0, F1, F2, and F3 domains) linked via an unstructured region to the 13 amphipathic helical bundles of the talin rod, which terminates in a single helix, the dimerization domain (DD). Residue numbers for each domain (R1–R13) are shown. Helices are numbered and vinculin-binding sites are colored red. Domains corresponding to the new structures reported here are shaded. B–E, NMR structures of the talin R3, R4, R5, and R6 rod domains. Ribbon diagrams of representative low-energy structures show the overall topology of each bundle. F, model of talin showing the structures of all 18 domains.
The talin rod begins with a five-helix bundle (R1) followed by three four-helix bundles (R2–R4). R2 makes extensive hydrophobic interactions with R1 to form a double-domain (18). Although no interactions have been detected between R2, R3, and R4, the R1–R4 region will have a compact arrangement (Fig. 1F) because the N and C termini of four-helix bundles are at the same end of the domain (e.g. see Fig. 1, B and C). The three four-helix bundles are followed by a series of eight five-helix bundles in which the N and C termini are positioned at opposite ends of the bundle (e.g. see Fig. 1, D and E), creating a linear chain (Fig. 1, A and F). The four-helix bundle R8 does not disrupt the chain because it is inserted into a loop in the R7 five-helix bundle (14). Importantly, four-helix and five-helix bundles are expected to differ in their response to mechanical stretch. Unfolding five-helix bundles by pulling on the termini positioned at the opposite ends of the bundle is restricted by extensive contacts throughout the length of the helices and requires a gradual breaking of hydrogen bonds. In four-helix bundles, the termini are at the same end of the bundle, and the applied force acts on the weak hydrophobic contacts, peeling helices away from the bundle. Molecular dynamics simulations on the N-terminal fragments of the talin rod demonstrate that the R1 five-helix bundle is more resistant to stretch than the four-helix bundle R2 and the bundle made by helices H9–H12 (32). Thus, the uneven distribution of four- and five-helix bundles in the talin rod creates two distinctly different zones: a compact N-terminal region sensitive to stretch and a linear C- terminal part that is optimal for force transmission.
Vinculin Binding to the Compact N-terminal Region of the Talin Rod
The talin rod contains multiple vinculin binding sites (VBS), with each defined by hydrophobic residues on a single helix that are normally buried within the helical bundles that make up the rod (Fig. 1A) (19). Thus, vinculin binding requires domain unfolding (33), and domain destabilization by force has been shown to promote binding (34). VBS are found in both four- and five-helix bundles, and when present, there is generally just one VBS per domain. However, the two four-helix bundles R2 and R3 are atypical, with each containing two VBS. This creates a cluster of VBS in the compact N-terminal region of the talin rod optimal for activation by force (Fig. 1, A and F).
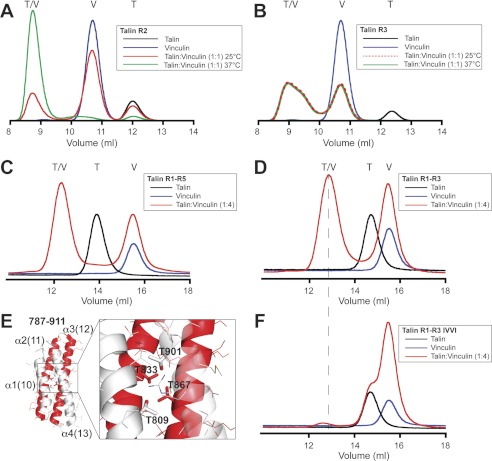
Gel filtration experiments show that R3 binds the vinculin Vd1 domain (residues 1–258) readily at 25 °C, whereas R2 requires incubation at 37 °C for maximal binding (Fig. 2, A and B). The multidomain fragments R1–R3 and R1–R5 bind vinculin at 25 °C to the same extent as R3 alone (Fig. 2, C and D), demonstrating that the vinculin binding properties of R3 are not affected by adjacent domains. Size-exclusion chromatography multiangle laser light scattering data show that at 25 °C, R2R3 forms a 1:2 complex with vinculin, indicating that both VBS in R3 are engaged (supplemental Fig. S4). Incubation at 40 °C led to formation of a 1:4 complex, indicating that all the VBS in both R2 and R3 are now occupied.
FIGURE 2.
Vinculin binding to the talin rod. A and B, vinculin Vd1 (V) was incubated at a 1:1 molar ratio with talin (T) R2 (A) and R3 polypeptides (B) at 25 or 37 °C as indicated, and complex (T/V) formation was analyzed on a Superdex-75 (10/300) GL gel filtration column. C and D, vinculin Vd1 was incubated (25 °C) with talin R1–R5 (C) and talin R1–R3 (D) at a molar ratio of 4:1, and complex formation was analyzed by gel filtration on a Superdex-200 (10/300). The results show that the VBS in the N-terminal region of talin are active even in larger fragments. E, structure of R3; the two vinculin-binding helices are shown in red. The enlarged region shows the four threonine residues embedded within the hydrophobic core. F, mutation of the four threonines in R3 to hydrophobic residues (T809I/T833V/T867V/T901I; the IVVI mutation) abolished Vd1 binding to the R1–R3 IVVI talin polypeptide at 25 °C.
Our new structural data reveal that R3 contains four threonines within its hydrophobic core creating a potentially weak point in the core of this domain (Fig. 2E). Replacing these threonines with larger hydrophobic residues (T809I/T833V/T867V/T901I, the IVVI mutation) markedly stabilized R3 with minimal effects on its overall structure (supplemental Fig. S5, A and B). Remarkably, the R3 IVVI mutant was no longer able to bind Vd1 at 25 °C (supplemental Fig. S5, C and D) although it bound efficiently at 45 °C (supplemental Fig. S5D), confirming that the mutations do not inactivate the VBS. Furthermore, introducing the IVVI mutation into R1–R3 (Fig. 2F) and R1–R5 rendered these larger N-terminal fragments unable to bind vinculin. We conclude that the presence of the four threonines in R3 enhances its ability to bind vinculin and that the VBS in R3 will be among the first to bind vinculin in response to force exerted on talin.
Multisite Interactions between Talin and RIAM
The Rap1 effector RIAM has been implicated in talin and thereby integrin activation (23, 24). The minimal talin-binding site (TBS) in RIAM has been mapped to residues 6–30 (TBS1), and a TBS1 peptide was shown to be sufficient to bind talin (24). The TBS1 sequence is highly conserved and is predicted to contain an amphipathic helix (24). To map the RIAM binding sites in the talin rod, we used the TBS1 peptide to conduct an NMR-based screen for possible interactions with each of the 13 authentic talin rod domains and identified four domains (R2, R3, R8, and R11) that bound RIAM-TBS1 (Fig. 3A and supplemental Fig. S6). Control NMR titrations showed that a mutant RIAM-TBS1 peptide (M11E/F12E/L15E/L16E), which does not bind full-length talin (24), did not bind any of these four domains, indicating that the interactions observed are specific.
FIGURE 3.
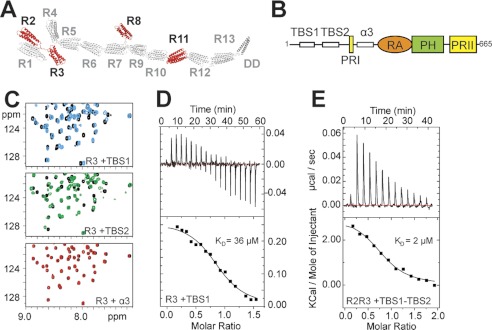
Characterization of the RIAM binding sites in the talin rod. A, schematic of the talin1 rod. Domains that interact with a RIAM(6–30) peptide as determined by NMR are in red. B, schematic of RIAM showing the two N-terminal talin-binding helices (TBS1 and TBS2), the proline-rich region (PRI), the additional helix (α3), the Ras association (RA) domain, the pleckstrin homology (PH) domain, and the second proline-rich region (PRII). C, 1H,15N HSQC spectra of 100 μm 15N-labeled talin R3 rod domain in the absence (black) or presence of RIAM TBS1 (6-30, blue), RIAM TBS2 (45–127; green) or RIAM α3 (147–174; red) at a ratio of 1:3. D and E, ITC characterization of RIAM binding to talin rod domains at 25°C. D, talin R3 titration with RIAM(1–32) (TBS1): 500 μm R3 in the cell and 5 mm TBS1 in the syringe. The lower panel shows fitting to a single-site binding model; KD, 36 μm. E, talin R2R3 titration with RIAM(1–127) (TBS1-TBS2); 18 μm R2R3 in the cell and 180 μm TBS1-TBS2 in the syringe. The lower panel shows fitting to a single-site binding model; KD, 2 μm.
In addition to RIAM-TBS1 (residues 6–30), two more regions in the N-terminal part of RIAM (residues 50–85, TBS2, and 149–173, α3) are predicted to form helical structures (Fig. 3B and supplemental Fig. S7A). We therefore tested constructs (45–127 and 147–174) containing these regions for an interaction with the talin rod domains that bind RIAM-TBS1 (R2, R3, R8, or R11). RIAM(147–174) containing α-helix 3 had no effect on the NMR spectra of any of these rod domains (Fig. 3C). However, RIAM(45–127) induced spectral changes that were similar to those caused by RIAM(6–30) (Fig. 3C), indicating comparable interactions. We conclude that the N-terminal region of RIAM contains two sequential talin binding sites TBS1 (residues 6–24) and TBS2 (residues 50–85) (numbers based on the secondary structure prediction).
The identification of two adjacent TBS in RIAM raises the possibility that simultaneous engagement of both sites might enhance binding to the talin rod. Moreover, two of the four RIAM binding sites in the talin rod are in adjacent domains (R2 and R3). Binding affinities determined by ITC show that R3 binds RIAM-TBS1 with a KD of 36 μm (Fig. 3D), whereas the interaction with R2 was at least an order of magnitude weaker as judged by the relative chemical shift changes in the HSQC spectra of 15N-labeled R2 and R3 induced by RIAM-TBS1. In contrast, a RIAM polypeptide (residues 1–127) containing both TBS1 and TBS2 bound >10× more tightly to a talin R2R3 double domain fragment (KD of 2 μm determined by ITC) (Fig. 3E). The number of binding sites derived from the ITC data (n = 0.8) indicates formation of a 1:1 talin R2R3·RIAM(1–127) complex. This was confirmed by size-exclusion chromatography multiangle laser light scattering; the molecular weight of the complex was 40 kDa, consistent with the calculated molecular weight (41.3 kDa) of a 1:1 complex (supplemental Fig. S7B). The HSQC spectrum of 15N-labeled R2R3 mixed 1:1 with RIAM(1–127) shows severe broadening of the resonances of both R2 and R3 (supplemental Fig. S7C), indicating that both domains are simultaneously engaged in the interaction with RIAM, restricting their relative mobility.
Competition between RIAM and Vinculin for Talin
Interestingly, all four talin rod domains that bind RIAM also contain VBS (Fig. 4A). However, vinculin and RIAM have markedly different modes of binding to talin. The talin binding sites in RIAM are single helices that interact with a folded talin domain, as shown by the localized chemical shift changes in the NMR spectra of R2, R3, R8, and R11 following addition of a RIAM(6–24) peptide (Fig. 3C and supplemental Fig. S6). The positions of many talin resonances do not show large changes even at high RIAM/talin ratios, indicating that no major structural rearrangements occur in regions not in direct contact with RIAM. In contrast, vinculin binding requires domain unfolding (33) and is therefore likely to be incompatible with RIAM binding (Fig. 4B). The different modes of binding are demonstrated by experiments with the IVVI mutation that stabilizes the R3 bundle. Although this mutation dramatically reduced vinculin binding (Fig. 2F), it had minimal effects on RIAM binding as determined by NMR (supplemental Fig. S8, A and B). Overall, the data suggest that RIAM and vinculin must compete for binding to the talin rod. Indeed, gel filtration experiments show that addition of RIAM(1–127) to a 1:1 mixture of Vd1/talin R2R3 results in the disappearance of the talin·vinculin complex and the appearance of a lower molecular weight peak close to the position of the talin·RIAM complex (Fig. 4C). This clearly demonstrates that vinculin and RIAM cannot bind to talin simultaneously and suggests that RIAM has a higher affinity for R2R3 than vinculin.
FIGURE 4.
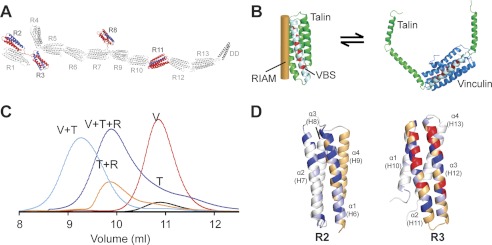
RIAM and vinculin binding to talin is mutually exclusive. A, schematic of the talin1 rod. Domains that interact with a RIAM(6–30) peptide as determined by NMR are shown in red. VBS within these domains are shown in blue. B, left, model of single RIAM helix (orange) bound to a talin rod helical bundle (green). A single vinculin binding helix is shown in white, and residues that interact with vinculin are in red. Right, model of the Vd1 domain of vinculin (blue) bound to the VBS in a talin helical bundle illustrating that binding involves domain unfolding. C, analytical gel filtration (Superdex 75/10 300 GL) of talin R2R3 1:1 with vinculin Vd1 (V+T; cyan); talin R2R3 1:1 with RIAM 1–127 (T+R; orange); talin R2R3 1:1:1 with RIAM(1–127) and vinculin Vd1 (V+T+R; purple); Vd1 alone (V; Red); R2R3 alone (T; black). D, effects of RIAM(6–30) binding to R2 and R3 as detected by NMR. Weighted chemical shift differences, determined as described previously (16), are shown on ribbon representations of the R2 and R3 structures; peaks that broaden are shown in red, shifts > 0.13 ppm are in dark blue, and shifts > 0.07 ppm are in light blue. The VBS helices are shown in orange.
The NMR spectral changes in R3 following RIAM binding map predominantly to the sequential VBS helices that form a hairpin structure (Fig. 4D). We conclude that the RIAM helix likely binds to the groove on the surface of the R3 bundle made by the two contacting VBS helices. This type of interaction is likely to shield the VBS in R3 from vinculin, enhancing vinculin-RIAM competition for talin. In R2, the RIAM interactions map predominantly to the first helix that is also a VBS. Additional perturbations on the neighboring helix 2 and lack of perturbations on helix 4 suggest that RIAM occupies the groove between helices 1 and 2. Significantly, this surface is not involved in the close contact between the R1 and R2 domains (18), leaving it exposed in the R1R2 fragment, and NMR experiments show that R1R2 is able to interact with RIAM (supplemental Fig. S8C).
Interaction between RIAM and Vinculin
Although addition of a RIAM(1–127) peptide disrupts the talin·vinculin complex, a peak corresponding to free vinculin was not observed in the gel filtration experiments (Fig. 4C), raising the possibility that the displaced vinculin might bind RIAM. Indeed, the gel filtration profile of a 1:1 mixture of Vd1 and RIAM(1–127) clearly demonstrates complex formation (Fig. 5A). Furthermore, a Vd1·RIAM(1–32) complex readily crystallized under conditions similar to those used for the Vd1·talin peptide complex (18, 19); its structure was solved by molecular replacement using the Vd1/talin structure and refined to 2.3 Å (Fig. 5B and supplemental Table 2). The conformation of vinculin Vd1 is very similar in both talin and RIAM complexes (supplemental Fig. S9A). As in the Vd1/talin VBS peptide structures, the RIAM TBS1 helix is embedded in the hydrophobic groove formed by α-helices 1 and 2 of the Vd1 four-helix bundle (Fig. 5, C and D). The position and register of the RIAM helix is well defined by the electron density (supplemental Fig. S9, B and C), and its interaction with Vd1 is mediated by the hydrophobic side chains on one face of the amphipathic RIAM helix. These residues align well with the talin VBS residues contacting Vd1 (Fig. 5E), but most of the RIAM residues have bulkier hydrophobic side chains than those found in the talin VBS consensus sequence. As a result, the RIAM helix is positioned further away from the Vd1 core than talin VBS helices, reducing the packing density. In addition, Leu1965 of the VBS in talin R10, which embeds in the Vd1 hydrophobic core, corresponds to Thr23 in RIAM that makes no significant contacts with Vd1.
FIGURE 5.
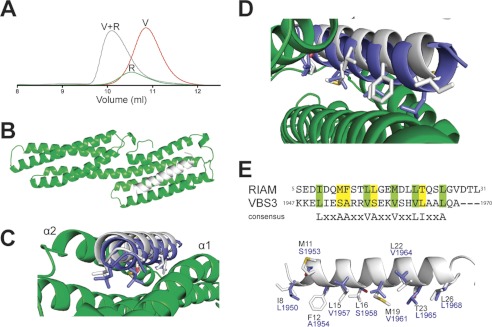
RIAM binding to vinculin. A, analytical gel filtration (Superdex 75/10 300 GL) of RIAM(1–127) mixed with vinculin Vd1. Protein samples are vinculin Vd1 1:1 with RIAM(1–127) (V+R; gray); RIAM1–127 alone (R; green); and Vd1 alone (V; red). All proteins were loaded at 50 μm. B, crystal structure of RIAM TBS1(1–32) (white) bound to vinculin Vd1 (green). C, top down view of B. The position of the talin R10 VBS (VBS3; blue) complexed to Vd1 (Protein Data Bank code 1XWJ) is shown for comparison. D, close up showing the similarity in binding of RIAM TBS1 and talin VBS3 to vinculin Vd1 and the overall displacement of RIAM TBS1 helix relative to talin VBS. E, sequence alignment of RIAM(1–32) and talin VBS3 plus the consensus VBS sequence. Lower panel, overlay of RIAM(1–32) and the VBS3 in talin R10. Side chains of residues contacting vinculin in the complex are shown in stick representation and labeled.
Isothermal titration calorimetry (ITC) data (supplemental Fig. S10) demonstrate that vinculin Vd1 has a significantly lower affinity for RIAM (1–32) (KD, 5 μm) compared with the VBS (residues 820–844) in talin R3 (KD, 0.8 μm). The lower affinity for RIAM is associated with a higher enthalpy of interaction than for talin (−1.5 and −5.9 kcal, respectively). This correlates with the displacement of the RIAM helix further away from the Vd1 bundle relative to the talin VBS, which reduces Vd1 contacts with RIAM.
The Balance between RIAM and Vinculin Binding to Talin in Cells
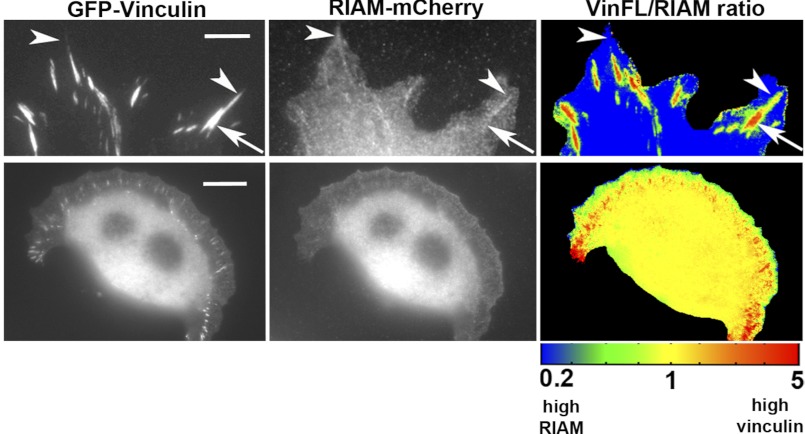
To explore whether our in vitro data on competition between RIAM and vinculin for talin applies in cells, we co-expressed RIAM-mCherry and GFP-vinculin fusion constructs in vinculin null mouse embryonic fibroblasts. In stationary cells, both vinculin and RIAM localization overlapped in mature FAs (Fig. 6, upper panels). Ratio imaging revealed that RIAM preferentially localized to the distal tip of FAs, whereas vinculin intensity was high in the proximal area. In cells with a migratory arc-shaped phenotype, the rim of the protruding cell edge was particularly rich in RIAM, whereas vinculin was prominent in FAs behind the leading edge, suggesting that RIAM binding to talin precedes the binding of vinculin to talin during FA formation (Fig. 6, lower panels).
FIGURE 6.
Differential localization of vinculin and RIAM in FA. Vinculin (−/−) cells were transfected with full-length GFP-vinculin and RIAM-mCherry. Upper panel, RIAM and vinculin localize to FAs. Ratio images displayed in a spectral color scale ranging from 0.2 (blue, high in RIAM) to 5 (red, high in vinculin), reveal that FA areas with high vinculin intensity (arrows) have low amounts of RIAM, and areas with high RIAM intensity (arrowheads) have low amounts of vinculin. Scale bar, 3 μm. Lower panel, in a migrating cell, RIAM localizes strongly to the leading edge in front of developing FAs, which are high in vinculin. Scale bar,10 μm.
The N-terminal Region of the Talin Rod Is Required for FA Formation
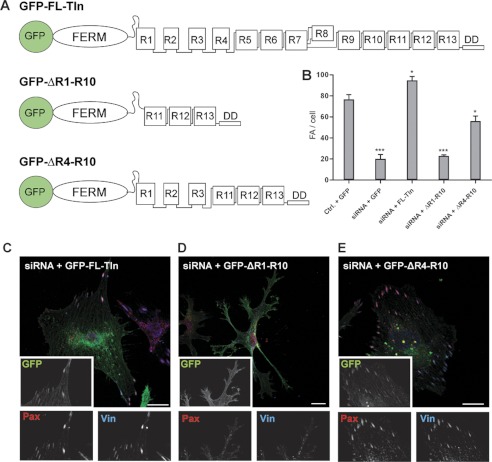
Although both the RIAM- and vinculin-binding sites are distributed throughout the talin rod (supplemental Fig. S11), the unusual features of the RIAM and vinculin binding sites located in the compact N-terminal region of the rod suggest that these may play a unique role in FA assembly. To explore this possibility, we used our detailed knowledge of the structures and domain boundaries of talin1 to design two mini-talin1 constructs (Fig. 7A). The first contained just the N-terminal FERM domain (residues 1–405), which includes the integrin binding and activation domains, coupled via the authentic unstructured linker region (residues 405–482) to the C-terminal part of the talin rod (residues 1974–2541; R11–R14). This region contains the C-terminal actin-binding site and dimerization domain, plus several RIAM and vinculin binding sites, and has recently been shown to be sufficient to support the signaling pathways that regulate cell cycle progression (35). The GFP-tagged mouse TlnΔR1-R10 construct was then tested for its ability to rescue defects in cell spreading and FA assembly in human endothelial cells depleted of talin1. However, although GFP-TlnΔR1-R10 was expressed to about the same extent as GFP-talin1 (supplemental Fig. S12A), it was unable to support efficient cell spreading on glass coverslips within a 4-h time frame, although the cells displayed numerous protrusions (Fig. 7, B–D, and supplemental Fig. S12B). Moreover, the cells contained very few paxillin- or vinculin-staining FAs, and GFP-TlnΔR1-R10 was diffusely distributed throughout the cytosol (Fig. 7D). In marked contrast, a second GFP-tagged mini-talin1 construct (GFP-TlnΔR4-R10) that includes the vinculin and RIAM binding sites in R2R3 (Fig. 7A) supported assembly of abundant paxillin- and vinculin-containing FAs (Fig. 7, B and E) and partially rescued cell spreading (supplemental Fig. S12, B and C). These results clearly establish that the N-terminal part of the talin rod plays a key role in cell spreading and the assembly of vinculin-containing FAs.
FIGURE 7.
Talin R1–R3 are essential for FA assembly. Human endothelial cells were electroporated with a talin1 siRNA or control RNA (Ctrl.) plus constructs encoding either GFP alone, full-length GFP-talin1, or GFP-mini-talin1 constructs (mouse) containing the rod domain deletions shown in A. Cells were grown on collagen-coated plastic for 72 h and then replated on uncoated glass coverslips and fixed/stained 4 h later. B, quantitative analysis shows that knockdown of endogenous talin1 markedly reduced FA numbers/cell and that the phenotype is rescued by full-length talin (FL-Tln) and in a large part by the mini-talin1 (GFP-ΔR4–R10) containing the R2R3 rod domains. However, the GFP-ΔR1-R10 construct lacking R2R3 failed to do so. This is unlikely to be due to incorrect folding because the expressed protein was stable (supplemental Fig. 12C) and both the isolated talin head (10) and rod fragment (35) contained in this construct are biologically active when expressed in cells. A two-tailed paired Student's t test was performed to test for significance. Results significantly different from control + GFP are indicated; t test p < 0.05 (*) and p < 0.001 (***). C–E, epifluorescence images showing the localization of GFP-talin1 constructs indicated (GFP), paxillin (Pax), and vinculin (Vin). Bar, 15 microns.
DISCUSSION
The results reported here, together with our earlier work, define the structures of all 18 domains in talin, and this has enabled us to develop a structural model of the entire molecule spanning >2500 amino acids (Fig. 1F and supplemental Fig. S11). The structural features of talin are entirely consistent with a role in linking integrins to F-actin. Thus, the N-terminal FERM domain (head) binds synergistically to acidic membrane phospholipids and β-integrin tails shifting the integrin conformational equilibrium toward the activated state (11, 36). The talin rod, which is joined to the head by a large unstructured region (37), comprises 13 α-helical bundles (R1–R13) terminating in a single helix (DD) responsible for talin dimerization (Fig. 1A). The dimeric R13-DD double domain contains the C-terminal actin-binding site ABS3 (13), which is essential for FA assembly (6). VBS are distributed throughout the length of the rod (19) (Fig. 1A), and vinculin binding to talin stabilizes FAs (7, 38).
Superimposed onto these basic features is a complex mode of regulation. First, much of talin exists in a cytosolic autoinhibited form (39), and flexible linkers facilitate intramolecular interactions between the head and rod masking the integrin-binding site in the head (16, 40) and the plasma membrane association site in the rod (39). The Rap1 effector RIAM recruits talin to the plasma membrane (23, 24, 39, 41), and we show here that four of the talin rod domains (R2, R3, R8, and R11) bind RIAM. Whether binding of RIAM to these sites is directly involved in talin activation remains to be established, and binding of anionic phospholipids such as PIP2 to the talin head (4, 8) has also been implicated in talin activation (40).
Second, the domain organization of the talin rod indicates that it has the potential to act as a mechanotransducer. The three sequential four-helix bundles (R2–R4) in the N-terminal region of the rod define a compact structure likely to change conformation in response to force, whereas the C-terminal region with its succession of five-helix bundles forms a linear chain perfectly suited to force transmission (Fig. 1F). The R2 and R3 talin rod domains are unusual in that each contains two VBS. Vinculin binding requires domain unfolding, and we show here that the cluster of four threonines buried within the hydrophobic core of R3 destabilizes the bundle and makes this region of the talin rod ideally suited to recruit vinculin in response to the initial weak forces exerted on nascent adhesions at the leading edge. Elegant single molecule experiments on talin 482–889 (domains R1, R2, and most of R3) demonstrate that the VBS therein can be activated by force (34), and in vivo studies show that recruitment of vinculin to FAs is myosin II-dependent (42). Moreover, talin undergoes repeated actomyosin-dependent stretching in the direction of actin flow from a length of 50–60 nm to 100–350 nm (43). The importance of the compact N-terminal region of the talin rod in adhesion assembly is clearly demonstrated by our experiments with mini-talin constructs showing that the N-terminal region of the talin rod is essential for robust cell spreading and the assembly of vinculin- and paxillin-containing FAs.
Talin R2 and R3 also contain binding sites for RIAM, and we have identified two adjacent amphipathic helices in the N-terminal region of RIAM that bind talin. Importantly, we demonstrate that a talin R2R3 double domain fragment binds synergistically to the two sequential talin binding sites (TBS1 and TBS2) in RIAM. However, the binding mechanism is fundamentally different from that for vinculin, whereas vinculin binding requires domain unfolding, RIAM only binds to folded R2R3 domains and will be disrupted by domain unfolding either induced by force or by vinculin binding. Interestingly, we have found that vinculin also binds directly to RIAM, and the crystal structure of the complex shows that RIAM-TBS1 docks into the same hydrophobic groove in vinculin Vd1 as that occupied by the talin VBS (18, 19, 44). Indeed, we observe competitive interactions between talin, vinculin and RIAM in gel filtration experiments.
The larger size of the RIAM hydrophobic side chains that contact vinculin compared with talin (Fig. 5E) results in looser packing of the RIAM helix against the Vd1 surface. Consequently, the affinity of vinculin for RIAM TBS1 is lower than for the isolated VBS helices of talin. However, the interaction of vinculin with talin helical bundles is weaker than with isolated VBS, making the vinculin affinities for talin bundles and RIAM comparable. Moreover, the interaction between RIAM and talin R2R3 is synergistic and the simultaneous engagement of the two RIAM binding sites in R2R3 and the two TBS in RIAM is sufficient to inhibit the talin-vinculin interaction as detected in gel-filtration experiments (Fig. 4C). This suggests that the talin-RIAM interaction must be disrupted before a talin·vinculin complex can be formed. The synergy between talin R2R3 and RIAM TBS1 and TBS2 also means that disruption of just one of the sites will strongly reduce the overall affinity. Thus, RIAM binding is likely to be exquisitely sensitive to force-induced conformational changes leading to the disruption of the compact N-terminal region of the talin rod. In agreement with this model, we observe a progressive decrease in RIAM and an increase in vinculin in adhesion complexes as they undergo force-dependent maturation into FAs (Fig. 6). The ability of vinculin to bind RIAM will also affect the RIAM-talin interaction. However, vinculin exists in both activated and autoinhibited forms (45), and pathways that regulate vinculin activation will also impact on the stability of RIAM·talin complexes.
On the basis of the above data, we propose a model in which competition between RIAM and vinculin for talin regulates the assembly and maturation of integrin-containing adhesion complexes (Fig. 8). Signaling pathways that activate Rap1 lead to the recruitment of RIAM to the plasma membrane, and the recent crystal structure of a RIAM RA-PH double domain shows that they form a single structural unit that acts as a proximity detector for activated Rap1 and PIP2 (46). In the absence of force, we propose that talin R2R3 binds synergistically to TBS1 and TBS2 in RIAM, recruiting talin to the plasma membrane where it is activated by an as yet undefined mechanism. Here, the talin FERM domain engages both β-integrin tails and anionic phospholipids such as PIP2, and both interactions are required for integrin activation (36). The talin·RIAM complex may also recruit proteins that regulate actin polymerization to the leading edge promoting lamellipodial protrusion (47, 48). The C-terminal actin-binding site in talin then captures the retrograde flow of actin filaments, generating tension across the talin rod. This destabilizes the four-helix bundles in the compact N-terminal region of the talin rod, reducing its affinity for RIAM while enhancing the affinity for activated vinculin, which then cross-links talin to F-actin. Vinculin also binds RIAM, further inhibiting the interaction of RIAM with the talin rod. Overall, the effect is to promote the transition from transient RIAM-positive nascent adhesions to more stable FAs enriched in vinculin. The stronger forces exerted on talin by actomyosin contraction may subsequently activate additional VBS in the talin rod, increasing the number of vinculin molecules bound to talin, strengthening its connection to F-actin. Indeed, vinculin binding locks talin and thereby integrins into an active conformation (7). Force exerted on talin has the potential to stretch the whole molecule, including the long unstructured linker between the head and rod, enabling talin to bridge the gap between the membrane and actin filaments some 40 nm away (49). The model highlights the role of the R2R3 region of the talin rod in regulation of adhesion progression. This prediction is validated by our structure-based talin engineering, which demonstrates that much of the functionality of talin is retained in a construct that contains the integrin-binding head region, the regulatory RIAM/vinculin binding region R1–R3, and the C-terminal actin binding site R11-DD.
FIGURE 8.
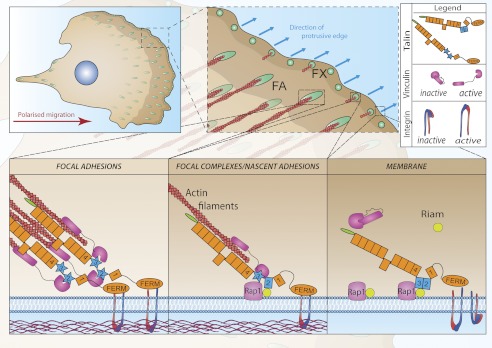
Role of RIAM and vinculin binding to talin in the assembly of focal adhesions at the front of a migrating cell. Small dynamic cell-matrix contacts (focal complexes; FX) form at the protrusive edge of cells; some disassemble, whereas others engage cytoskeletal actin and mature into larger streak-shaped FAs. Our data support a model in which Rap1-GTP recruits RIAM to the plasma membrane. RIAM binds synergistically to the R2R3 domains (blue) of the talin rod recruiting talin to the membrane. Here, talin binds to and activates integrins and also engages the actin cytoskeleton, which exerts force on talin. This changes the conformation of the talin R2R3 rod domains reducing their affinity for RIAM and increasing their affinity for vinculin. Vinculin also binds F-actin re-enforcing the link between talin and the acto-myosin contractile apparatus, promoting integrin clustering and the maturation of focal complexes into FAs.
Acknowledgments
We are grateful to Mark Ginsberg (University of California, San Diego) for supplying the RIAM(6–30) peptide, Dr. Maddy Parsons (Kings College, London, UK) for the RIAM-mCherry plasmid, Kees Straatman (Leicester) and the Bioimaging Core Facility of the University of Manchester for help with microscopy, Mohamed Bouaouina (Yale) for assistance with the statistical analysis on FAs, Alex Carisey for support in producing the artwork in Fig. 8, and to Marjarie Howard and Tom Jowette (University of Manchester) for help with size-exclusion chromatography multiangle laser light scattering experiments. The x-ray data were collected at Diamond Light Source under Liverpool BAG allocation.
This work was supported by National Institutes of Health Cell Migration Consortium Grant U54 GM64346 from NIGMS, the Wellcome Trust, BBSRC, and Cancer Research UK.

This article contains supplemental Tables 1 and 2, Figs. S1–S12, and additional references.
The atomic coordinates and structure factors (codes 2L7A, 2LQG, 2L7N, 2L10, and 3ZDL) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Resonance assignments have been deposited in the BioMagResBank (http://www.bmrb.wisc.edu) with the accession numbers 17332 (787–911; R3), 18313 (913–1044; R4), 17363 (1046–1207; R5), and 17070 (1206–1357; R6).
- FA
- focal adhesion
- Vd1
- vinculin domain 1 (residues 1–258)
- ABS3
- third actin-binding site
- RIAM
- Rap1-GTP-interacting adaptor molecule
- TBS
- talin-binding site(s)
- ITC
- isothermal titration calorimetry
- FERM
- 4.1 ezrin radixin moesin
- VBS
- vinculin-binding site(s).
REFERENCES
- 1. Zaidel-Bar R., Geiger B. (2010) The switchable integrin adhesome. J. Cell Sci. 123, 1385–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Legate K. R., Fässler R. (2009) Mechanisms that regulate adaptor binding to β-integrin cytoplasmic tails. J. Cell Sci. 122, 187–198 [DOI] [PubMed] [Google Scholar]
- 3. Critchley D. R. (2009) Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Annu. Rev. Biophys. 38, 235–254 [DOI] [PubMed] [Google Scholar]
- 4. Saltel F., Mortier E., Hytönen V. P., Jacquier M. C., Zimmermann P., Vogel V., Liu W., Wehrle-Haller B. (2009) New PI(4,5)P2- and membrane proximal integrin-binding motifs in the talin head control β3-integrin clustering. J. Cell Biol. 187, 715–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shattil S. J., Kim C., Ginsberg M. H. (2010) The final steps of integrin activation: the end game. Nat. Rev. Mol. Cell Biol. 11, 288–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kopp P. M., Bate N., Hansen T. M., Brindle N. P., Praekelt U., Debrand E., Coleman S., Mazzeo D., Goult B. T., Gingras A. R., Pritchard C. A., Critchley D. R., Monkley S. J. (2010) Studies on the morphology and spreading of human endothelial cells define key inter- and intramolecular interactions for talin1. Eur J. Cell Biol. 89, 661–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Humphries J. D., Wang P., Streuli C., Geiger B., Humphries M. J., Ballestrem C. (2007) Vinculin controls focal adhesion formation by direct interactions with talin and actin. J. Cell Biol. 179, 1043–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Elliott P. R., Goult B. T., Kopp P. M., Bate N., Grossmann J. G., Roberts G. C., Critchley D. R., Barsukov I. L. (2010) The Structure of the talin head reveals a novel extended conformation of the FERM domain. Structure 18, 1289–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goult B. T., Bouaouina M., Elliott P. R., Bate N., Patel B., Gingras A. R., Grossmann J. G., Roberts G. C., Calderwood D. A., Critchley D. R., Barsukov I. L. (2010a) Structure of a double ubiquitin-like domain in the talin head: a role in integrin activation. EMBO J. 29, 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bouaouina M., Lad Y., Calderwood D. A. (2008) The N-terminal domains of talin cooperate with the phosphotyrosine binding-like domain to activate β1 and β3 integrins. J. Biol. Chem. 283, 6118–6125 [DOI] [PubMed] [Google Scholar]
- 11. Anthis N. J., Wegener K. L., Ye F., Kim C., Goult B. T., Lowe E. D., Vakonakis I., Bate N., Critchley D. R., Ginsberg M. H., Campbell I. D. (2009) The structure of an integrin/talin complex reveals the basis of inside-out signal transduction. EMBO J. 28, 3623–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McLachlan A. D., Stewart M., Hynes R. O., Rees D. J. (1994) Analysis of repeated motifs in the talin rod. J. Mol. Biol. 235, 1278–1290 [DOI] [PubMed] [Google Scholar]
- 13. Gingras A. R., Bate N., Goult B. T., Hazelwood L., Canestrelli I., Grossmann J. G., Liu H., Putz N. S., Roberts G. C., Volkmann N., Hanein D., Barsukov I. L., Critchley D. R. (2008) The structure of the C-terminal actin-binding domain of talin. EMBO J. 27, 458–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gingras A. R., Bate N., Goult B. T., Patel B., Kopp P. M., Emsley J., Barsukov I. L., Roberts G. C., Critchley D. R. (2010) Central region of talin has a unique fold that binds vinculin and actin. J. Biol. Chem. 285, 29577–29587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gingras A. R., Ziegler W. H., Bobkov A. A., Joyce M. G., Fasci D., Himmel M., Rothemund S., Ritter A., Grossmann J. G., Patel B., Bate N., Goult B. T., Emsley J., Barsukov I. L., Roberts G. C., Liddington R. C., Ginsberg M. H., Critchley D. R. (2009) Structural determinants of integrin binding to the talin rod. J. Biol. Chem. 284, 8866–8876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goult B. T., Bate N., Anthis N. J., Wegener K. L., Gingras A. R., Patel B., Barsukov I. L., Campbell I. D., Roberts G. C., Critchley D. R. (2009) The structure of an interdomain complex that regulates talin activity. J. Biol. Chem. 284, 15097–15106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goult B. T., Gingras A. R., Bate N., Barsukov I. L., Critchley D. R., Roberts G. C. (2010b) The domain structure of talin: residues 1815–1973 form a five-helix bundle containing a cryptic vinculin-binding site. FEBS Lett. 584, 2237–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Papagrigoriou E., Gingras A. R., Barsukov I. L., Bate N., Fillingham I. J., Patel B., Frank R., Ziegler W. H., Roberts G. C., Critchley D. R., Emsley J. (2004) Activation of a vinculin-binding site in the talin rod involves rearrangement of a five-helix bundle. EMBO J. 23, 2942–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gingras A. R., Ziegler W. H., Frank R., Barsukov I. L., Roberts G. C., Critchley D. R., Emsley J. (2005) Mapping and Consensus Sequence Identification for Multiple Vinculin Binding Sites within the Talin Rod. J. Biol. Chem. 280, 37217–37224 [DOI] [PubMed] [Google Scholar]
- 20. Hemmings L., Rees D. J., Ohanian V., Bolton S. J., Gilmore A. P., Patel B., Priddle H., Trevithick J. E., Hynes R. O., Critchley D. R. (1996) Talin contains three actin-binding sites each of which is adjacent to a vinculin-binding site. J. Cell Sci. 109, 2715–2726 [DOI] [PubMed] [Google Scholar]
- 21. Sun N., Critchley D. R., Paulin D., Li Z., Robson R. M. (2008) Identification of a repeated domain within mammalian α-synemin that interacts directly with talin. Exp. Cell Res. 314, 1839–1849 [DOI] [PubMed] [Google Scholar]
- 22. Li G., Du X., Vass W. C., Papageorge A. G., Lowy D. R., Qian X. (2011) Full activity of the deleted in liver cancer 1 (DLC1) tumor suppressor depends on an LD-like motif that binds talin and focal adhesion kinase (FAK). Proc. Natl. Acad. Sci. U.S.A. 108, 17129–17134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Han J., Lim C. J., Watanabe N., Soriani A., Ratnikov B., Calderwood D. A., Puzon-McLaughlin W., Lafuente E. M., Boussiotis V. A., Shattil S. J., Ginsberg M. H. (2006) Reconstructing and Deconstructing Agonist-Induced Activation of Integrin αIIbβ3. Curr. Biol. 16, 1796–1806 [DOI] [PubMed] [Google Scholar]
- 24. Lee H. S., Lim C. J., Puzon-McLaughlin W., Shattil S. J., Ginsberg M. H. (2009) RIAM activates integrins by linking talin to ras GTPase membrane-targeting sequences. J. Biol. Chem. 284, 5119–5127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goult B. T., Gingras A. R., Bate N., Roberts G. C., Critchley D. R., Barsukov I. L. (2008) NMR assignment of the C-terminal actin-binding domain of talin. Biomol. NMR Assign. 2, 17–19 [DOI] [PubMed] [Google Scholar]
- 26. Leslie A. G. (1992) Recent changes to the MOSFLM package for processing film and image plate data. Jnt CCP4 + ESF-EAMCB Newsl on Protein Crystallography 26, [Google Scholar]
- 27. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 28. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 29. Zamir E., Katz B. Z., Aota S., Yamada K. M., Geiger B., Kam Z. (1999) Molecular diversity of cell-matrix adhesions. J. Cell Sci. 112, 1655–1669 [DOI] [PubMed] [Google Scholar]
- 30. Fillingham I., Gingras A. R., Papagrigoriou E., Patel B., Emsley J., Critchley D. R., Roberts G. C., Barsukov I. L. (2005) A vinculin binding domain from the talin rod unfolds to form a complex with the vinculin head. Structure 13, 65–74 [DOI] [PubMed] [Google Scholar]
- 31. Gingras A. R., Vogel K. P., Steinhoff H. J., Ziegler W. H., Patel B., Emsley J., Critchley D. R., Roberts G. C., Barsukov I. L. (2006) Structural and dynamic characterization of a vinculin binding site in the talin rod. Biochemistry 45, 1805–1817 [DOI] [PubMed] [Google Scholar]
- 32. Hytönen V. P., Vogel V. (2008) How force might activate talin's vinculin binding sites: SMD reveals a structural mechanism. PLoS Comput. Biol. 4, e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roberts G. C., Critchley D. R. (2009) Structural and biophysical properties of the integrin-associated cytoskeletal protein talin. Biophys. Rev. 1, 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. del Rio A., Perez-Jimenez R., Liu R., Roca-Cusachs P., Fernandez J. M., Sheetz M. P. (2009) Stretching single talin rod molecules activates vinculin binding. Science 323, 638–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang P., Ballestrem C., Streuli C. H. (2011) The C-terminus of talin links integrins to cell cycle progression. J. Cell Biol. 195, 499–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Anthis N. J., Campbell I. D. (2011) The tail of integrin activation. Trends Biochem. Sci. 36, 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bate N., Gingras A. R., Bachir A., Horwitz R., Ye F., Patel B., Goult B. T., Critchley D. R. (2012) Talin contains a C-terminal calpain2 cleavage site important in focal adhesion dynamics. PLoS One 7, e34461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Saunders R. M., Holt M. R., Jennings L., Sutton D. H., Barsukov I. L., Bobkov A., Liddington R. C., Adamson E. A., Dunn G. A., Critchley D. R. (2006) Role of vinculin in regulating focal adhesion turnover. Eur. J. Cell Biol. 85, 487–500 [DOI] [PubMed] [Google Scholar]
- 39. Banno A., Goult B. T., Lee H., Bate N., Critchley D. R., Ginsberg M. H. (2012) Subcellular localization of talin is regulated by inter-domain interactions. J. Biol. Chem. 287, 13799–13812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Goksoy E., Ma Y. Q., Wang X., Kong X., Perera D., Plow E. F., Qin J. (2008) Structural basis for the autoinhibition of talin in regulating integrin activation. Mol. Cell 31, 124–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Watanabe N., Bodin L., Pandey M., Krause M., Coughlin S., Boussiotis V. A., Ginsberg M. H., Shattil S. J. (2008) Mechanisms and consequences of agonist-induced talin recruitment to platelet integrin αIIbβ3. J. Cell Biol. 181, 1211–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pasapera A. M., Schneider I. C., Rericha E., Schlaepfer D. D., Waterman C. M. (2010) Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J. Cell Biol. 188, 877–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Margadant F., Chew L. L., Hu X., Yu H., Bate N., Zhang X., Sheetz M. (2011) Mechanotransduction in vivo by repeated talin stretch-relaxation events depends upon vinculin. PLoS biology 9, e1001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Izard T., Evans G., Borgon R. A., Rush C. L., Bricogne G., Bois P. R. (2004) Vinculin activation by talin through helical bundle conversion. Nature 427, 171–175 [DOI] [PubMed] [Google Scholar]
- 45. Ziegler W. H., Liddington R. C., Critchley D. R. (2006) The structure and regulation of vinculin. Trends Cell Biol. 16, 453–460 [DOI] [PubMed] [Google Scholar]
- 46. Wynne J. P., Wu J., Su W., Mor A., Patsoukis N., Boussiotis V. A., Hubbard S. R., Philips M. R. (2012) Rap1-interacting adapter molecule (RIAM) associates with the plasma membrane via a proximity detector. J. Cell Biol. 199, 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bear J. E., Gertler F. B. (2009) Ena/VASP: towards resolving a pointed controversy at the barbed end. J. Cell Sci. 122, 1947–1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lafuente E. M., van Puijenbroek A. A., Krause M., Carman C. V., Freeman G. J., Berezovskaya A., Constantine E., Springer T. A., Gertler F. B., Boussiotis V. A. (2004) RIAM, an Ena/VASP and Profilin ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion. Dev. Cell 7, 585–595 [DOI] [PubMed] [Google Scholar]
- 49. Kanchanawong P., Shtengel G., Pasapera A. M., Ramko E. B., Davidson M. W., Hess H. F., Waterman C. M. (2010) Nanoscale architecture of integrin-based cell adhesions. Nature 468, 580–584 [DOI] [PMC free article] [PubMed] [Google Scholar]