MiRNA319 affects plant responses to drought and salinity stress and can be manipulated in transgenic plants to enhance performance under environmental stress.
Abstract
MicroRNA319 (miR319) is one of the first characterized and conserved microRNA families in plants and has been demonstrated to target TCP (for TEOSINTE BRANCHED/CYCLOIDEA/PROLIFERATING CELL FACTORS [PCF]) genes encoding plant-specific transcription factors. MiR319 expression is regulated by environmental stimuli, suggesting its involvement in plant stress response, although experimental evidence is lacking and the underlying mechanism remains elusive. This study investigates the role that miR319 plays in the plant response to abiotic stress using transgenic creeping bentgrass (Agrostis stolonifera) overexpressing a rice (Oryza sativa) miR319 gene, Osa-miR319a. We found that transgenic plants overexpressing Osa-miR319a displayed morphological changes and exhibited enhanced drought and salt tolerance associated with increased leaf wax content and water retention but reduced sodium uptake. Gene expression analysis indicated that at least four putative miR319 target genes, AsPCF5, AsPCF6, AsPCF8, and AsTCP14, and a homolog of the rice NAC domain gene AsNAC60 were down-regulated in transgenic plants. Our results demonstrate that miR319 controls plant responses to drought and salinity stress. The enhanced abiotic stress tolerance in transgenic plants is related to significant down-regulation of miR319 target genes, implying their potential for use in the development of novel molecular strategies to genetically engineer crop species for enhanced resistance to environmental stress.
Plant responses to drought and salt stresses have been studied extensively in understanding the physiological and molecular mechanisms underlying plant adaptation to abiotic stress (Munns, 2002; Wang et al., 2003; Chaves et al., 2009). At the physiological level, plant responses to salinity stress are generally divided into two phases: a rapid, osmotic phase inhibiting shoot growth and a slower, ionic phase accelerating the senescence of mature leaves (Munns and Tester, 2008). Plant responses to drought and salinity share many similarities, especially in the first phase. Plants subjected to drought and salinity experience a physiological water deficit that alters photosynthesis and cell growth and induces osmotic adjustment to maintain current water uptake and cell turgor (Chaves et al., 2009). However, under salinity stress, plants endure salt-specific effects, with very high Na+ or Cl− concentrations within cells leading to toxicity. To tolerate salt-specific effects, plants either minimize the uptake of salt or compartmentalize salt in the vacuoles (Munns, 2005). The current advances in plant biotechnologies for enhanced plant tolerance to drought (Garg et al., 2002; Capell et al., 2004; Chandra Babu et al., 2004; Lian et al., 2004; Oh et al., 2005; Wang et al., 2005; Hu et al., 2006; Fu et al., 2007) and salinity (Gaxiola et al., 2001; Rus et al., 2001, 2004; White and Broadley, 2001; Zhang and Blumwald, 2001; Zhang et al., 2001; Laurie et al., 2002; Shi et al., 2003; Flowers, 2004; Wu et al., 2004; Møller et al., 2009; Li et al., 2010) mainly concentrate on manipulating downstream genes, which function in the physiological responses discussed above (e.g. osmotic or ionic adjustment).
In recent years, the upstream regulatory networks of plant drought and salt stress responses, including hormones, transcription factors, protein kinases, protein phosphatases, and other signaling molecules such as calmodulin-binding proteins, have gradually been uncovered (Zhang et al., 2004; Shinozaki and Yamaguchi-Shinozaki, 2007; Nakashima et al., 2009). However, the fine details of these regulatory networks are still largely unknown, and questions about how different regulatory elements function together and are coordinated by master regulators posttranscriptionally or posttranslationally remain to be addressed. The discovery of plant microRNAs (miRNAs) largely involved in abiotic stress responses (Phillips et al., 2007; Sunkar et al., 2007; Lu and Huang, 2008; Mazzucotelli et al., 2008; Shukla et al., 2008; Lewis et al., 2009; Nakashima et al., 2009) shed light on these questions. It is known that miRNAs mediate plant abiotic stress responses through regulating their target genes, the majority of which are transcription factors constituting a complicated regulatory network, serving as key players in the gene regulation networks (Jones-Rhoades et al., 2006; Khraiwesh et al., 2012; Sunkar et al., 2012).
Plant miRNAs are approximately 20- to 24-nucleotide noncoding RNAs that specifically base pair to and induce the cleavage of target mRNAs or cause translational inhibition (Zhang et al., 2006; Shukla et al., 2008). They have diverse roles in plant development, such as phase transition, leaf morphogenesis, floral organ identity, developmental timing, and other aspects of plant development (Lu and Huang, 2008; Rubio-Somoza and Weigel, 2011). Genome-wide high-throughput sequencing and microarray profiling have identified stress-responsive miRNAs (Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004; Zhao et al., 2007; Ding et al., 2009; Yang et al., 2010; Wang et al., 2011), of which miR319 was found to respond to multiple stresses, such as up-regulation by dehydration, salt, and cold stress in Arabidopsis (Arabidopsis thaliana; Sunkar and Zhu, 2004; Liu et al., 2008), by cold stress in sugarcane (Saccharum officinarum) and rice (Oryza sativa; Lv et al., 2010; Thiebaut et al., 2012), and by water withholding at the tillering stage in rice (Zhou et al., 2010). As one of the first experimentally characterized and most conserved miRNA families (Axtell and Bowman, 2008), the miR319 targets, TCP (for TEOSINTE BRANCHED/CYCLOIDEA/PROLIFERATING CELL FACTORS [PCF]) genes encode plant-specific transcription factors sharing a conserved TCP domain with a basic helix-loop-helix structure. The TCP family is known to be largely involved in plant development such as the control of cell proliferation in leaf morphogenesis (Palatnik et al., 2003; Ori et al., 2007; Nag et al., 2009).
Although miR319-mediated changes in plant morphology have been well studied in dicots (Nath et al., 2003; Palatnik et al., 2003; Ori et al., 2007; Schommer et al., 2008; Nag et al., 2009), there have been no published reports studying monocots, especially perennial grass species. Moreover, although the involvement of miR319 in plant responses to drought and salinity stress has been suggested based on microarray data, experimental proof in support of the possible contribution of the miR319 family and the underlying molecular mechanisms are still lacking. In this study, transgenic plants of creeping bentgrass (Agrostis stolonifera) overexpressing a rice miR319 gene were generated to investigate the roles that miR319 plays in controlling plant development and plant responses to environmental stress. Through transgenic analysis, we seek to answer several questions. What impact may the miR319 gene family have in plant development in perennial grasses? Is miR319 involved in plant abiotic stress responses in perennial grasses? And what is the molecular mechanism of miR319-mediated plant tolerance to abiotic stress in perennial grasses?
RESULTS
Production and Molecular Characterization of Transgenic Creeping Bentgrass Plants Expressing the Rice miR319 Gene, Osa-miR319a
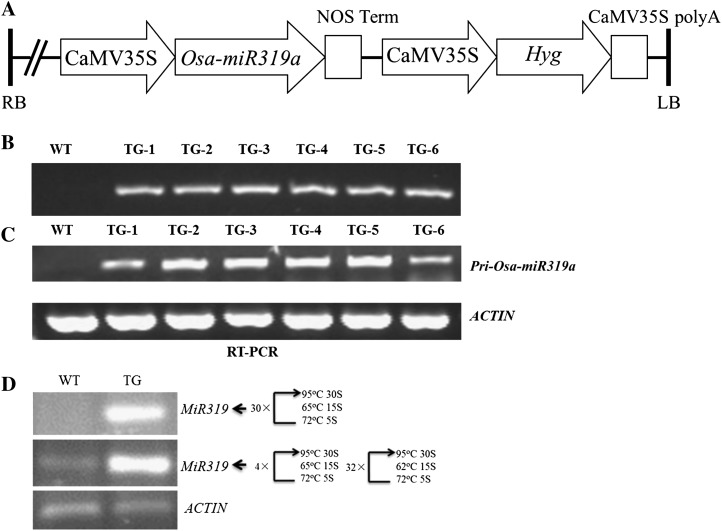
To study the potential of manipulating miR319 expression in perennial species for enhancing plant resistance to environmental stresses, we prepared a chimeric DNA construct, p35S-Osa-miR319a/p35S-hyg, containing one of the rice miR319 genes, Osa-miR319a (AK064418; Knowledge-based Oryza Molecular biological Encyclopedia; http://cdna01.dna.affrc.go.jp/cDNA/), under the control of the cauliflower mosaic virus (CaMV) 35S promoter (Fig. 1A). It was introduced into the creeping bentgrass ‘Penn A-4’ to produce a total of 15 independent transgenic lines ectopically expressing Osa-miR319a (Fig. 1, B and C). Under the higher stringency PCR conditions (30 cycles with an annealing temperature of 65°C), the high level of mature transcripts of miR319 could only be detected by stem-loop reverse transcription (RT)-PCR analysis in the transgenics, which accumulated in high quantities, suggesting that rice miR319 was successfully expressed in turfgrass plants and properly processed into mature miRNAs (Fig. 1D). However, the detection of mature miR319 transcripts in wild-type controls under lower stringency PCR conditions suggested that the transcripts of the turf endogenous miR319 were also amplified and shared sequence conservation with rice (Fig. 1D). The 15 transgenic lines were all morphologically indistinguishable and performed similarly in growth and response to various cultivation conditions when evaluated in the greenhouse. Five representative lines (TG1–TG4 and TG8) were selected for further analysis.
Figure 1.
Generation and molecular analysis of the transgenic lines expressing Osa-miR319a. A, Schematic diagram of the Osa-miR319a overexpression gene construct, p35S-Osa-miR319a /p35S-hyg, in which the Osa-miR319a gene is under the control of the CaMV 35S promoter. The CaMV 35S promoter-driven hyg gene is included for hygromycin resistance. LB, Left border; RB, right border. B, Example of PCR analysis of the hyg gene in wild-type (WT) and transgenic (TG) plants to detect transgene insertion into the host genome. C, Example of RT-PCR analysis of the primary Osa-miR319a transcripts in transgenics. Primary Osa-miR319a transcripts were detected in transgenic plants. D, Example of stem-loop RT-PCR analysis of the mature Osa-miR319a in wild-type and transgenic plants.
Overexpression of miR319 Causes Pleiotropic Phenotypes in Transgenic Creeping Bentgrass Plants
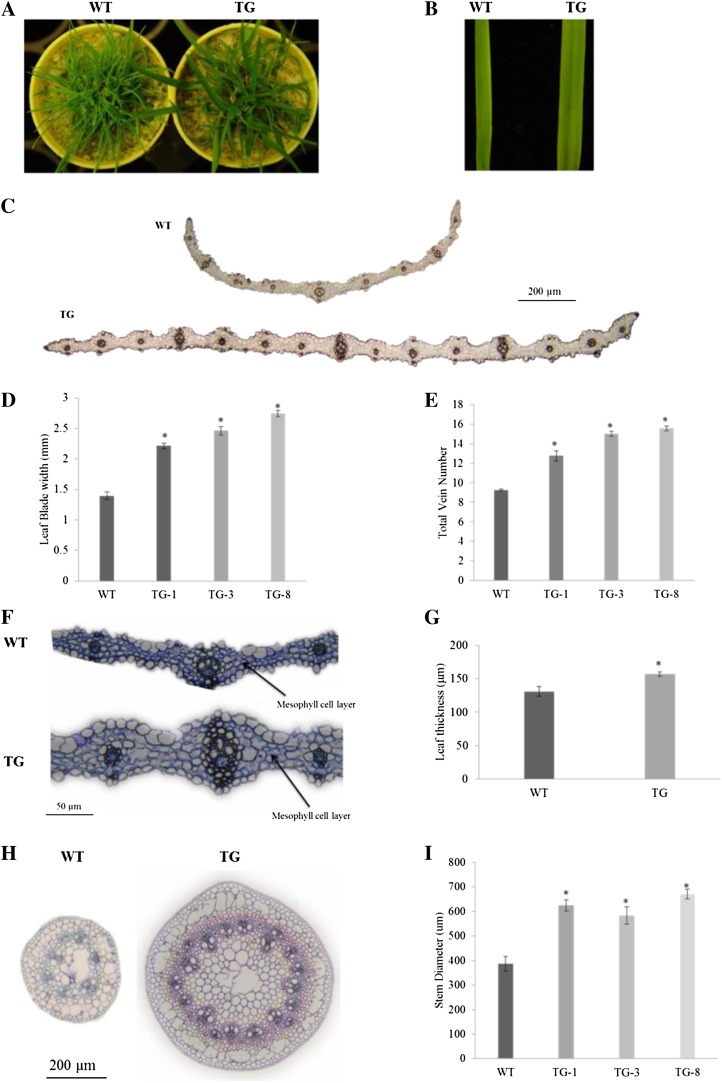
Analysis of transgenic creeping bentgrass plants constitutively expressing Osa-miR319a showed that Osa-miR319a plants displayed wider leaves and larger stems than the controls (Fig. 2, A–C and H). Microscopic analysis of plant leaf samples indicated that, compared with the controls, Osa-miR319a plants exhibited significantly greater leaf expansion (blade width and vein number) and thicker leaves (Fig. 2, C–G), which might be associated with increased mesophyll cell layers (Fig. 2F). Overexpression of miR319 also led to increased stem diameter in transgenic plants (Fig. 2, H and I).
Figure 2.
Morphology change in transgenic plants overexpressing Osa-miR319a. A, The transgenic plants (TG) exhibited wider leaves than wild-type controls (WT). B, A closer look at the wild-type control and Osa-miR319a transgenic plants. The representative transgenic plant leaf is wider. C, Leaf sectioning images of wild-type and transgenic plants. D, Statistical analysis of leaf blade width in wild-type and transgenic plants (n = at least 20). E, Statistical analysis of total vein number in wild-type and transgenic plants (n = at least 8). F, Examples of representative sectioning images of wild-type and transgenic plant leaves. G, Statistical analysis of leaf thickness in wild-type and transgenic plants (n = 10). H, Representative sectioning images of wild-type and transgenic plant stems. I, Statistical analysis of stem diameter in wild-type and transgenic plants (n = at least 6). Statistical analysis of leaf blade width, total vein number, leaf thickness, and stem diameter was conducted on wild-type control plants and various transgenic lines. Data are presented as means ± se, and error bars represent se. Asterisks indicate significant differences between transgenic and control plants at P < 0.05 by Student’s t test.
Leaf and stem sizes are dependent on both the number and the size of cells in the organ. To determine whether the enlargement of Osa-miR319a plant leaves and stems was associated with increased cell number, cell size, or a combination of these two parameters, we compared control and Osa-miR319a plants when they were fully developed after mowing. The numbers of cells in both leaves and stems were different between control and Osa-miR319a plants (Fig. 2, C, F, and H), with the latter having many more than the former, suggesting that an increase in cell proliferation may have contributed to the wide-leaf and large-stem phenotypes observed in transgenic plants. Moreover, compared with wild-type controls, an expansion of at least certain cell types, if not all, also appeared to have taken place in transgenic plants (Fig. 2, C, F, and H). Further study with additional quantitative analysis of various individual cells should provide more information about the role that cell expansion might play in miR319-mediated plant morphology change in turfgrass.
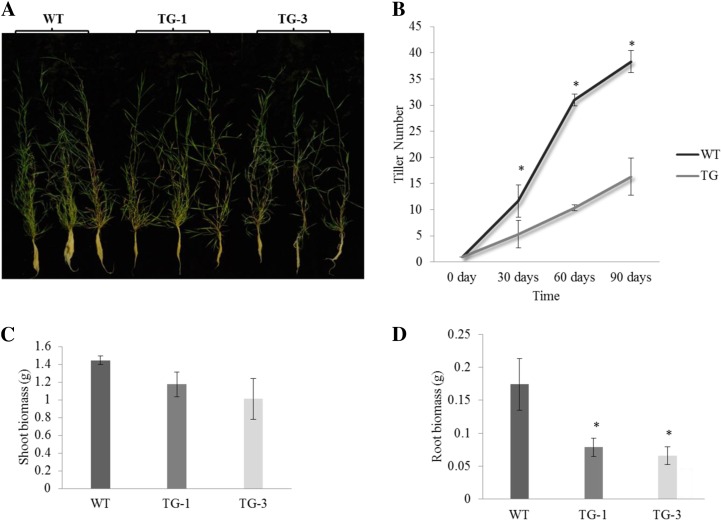
To elucidate any potential impact of constitutively expressed miR319 on other aspects of plant development, we grew plants starting from a single tiller and studied plant growth and development by monitoring changes in plant tiller numbers at different stages and measuring shoot biomass as represented by dry weight in control and Osa-miR319a plants. As shown in Figure 3, A and B, Osa-miR319a transgenics had fewer tillers than control plants. However, no significant difference in shoot biomass between control and Osa-miR319a plants was observed (Fig. 3C), suggesting that increased leaf expansion in Osa-miR319a plants might have compensated for the loss in biomass caused by decreased tillering. In contrast, the root biomass of Osa-miR319a plants was significantly lower than that of the controls (Fig. 3, A and B).
Figure 3.
Tillering and plant development under normal growth conditions. Both wild-type (WT) and transgenic (TG) plants initiated from a single tiller. A, Tillering and shoot and root development in wild-type and transgenic plants 90 d after initiation from a single tiller of the same size. The transgenic plants have fewer tillers and less root growth than wild-type controls. B, Comparison of tiller numbers in wild-type and transgenic plants 30, 60, and 90 d after initiation from a single tiller of the same size (n = at least 3). C, Biomass of wild-type and transgenic shoots 90 d after initiation from a single tiller of the same size (n = at least 4). D, Biomass of wild-type and transgenic roots 90 d after initiation from a single tiller of the same size (n = 5). Statistical analysis of tiller number and shoot and root biomass was conducted on wild-type control plants and various transgenic lines. Data are presented as means ± se, and error bars represent se. Asterisks indicate significant differences between transgenic and control plants at P < 0.05 by Student’s t test.
Overexpression of miR319 Improves Salt Tolerance in Transgenic Plants
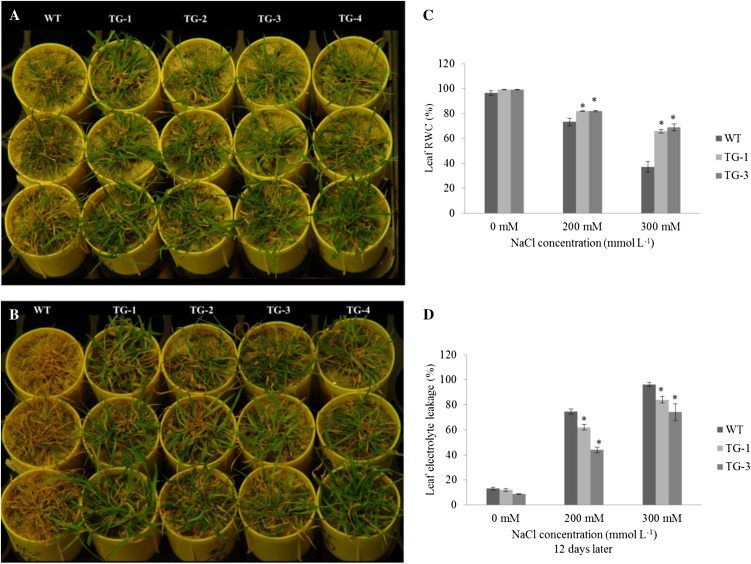
The involvement of many miRNAs in plant resistance to abiotic stress, such as drought and salinity, has been suggested by their up- or down-regulation in the presence of various environmental stimuli (Sunkar and Zhu, 2004; Liu et al., 2008; Zhou et al., 2010; Thiebaut et al., 2012). Using stem-loop RT-PCR analysis, we found that creeping bentgrass plants subjected to salinity or drought stress exhibited an elevated accumulation of mature miR319 transcripts compared with those grown under normal conditions (Supplemental Fig. S1). However, direct evidence about the role that miR319 plays in plant stress responses is still lacking. To investigate this, we first examined control and Osa-miR319a transgenic plants under salinity stress (200 mm NaCl). As shown in Figure 4A, the growth of control plants without Osa-miR319a was severely hampered, and serious tissue damage was observed 16 d after salt treatment, whereas salt-elicited damage was much less pronounced in Osa-miR319a plants. Upon release from the stress, the Osa-miR319a transgenics sustained the treatment, whereas only a few tillers in control plants survived (Fig. 4B). Similar results were also obtained with plants subjected to more severe salinity stress (Supplemental Fig. S2A).
Figure 4.
Responses of wild-type (WT) and transgenic (TG) plants to salt stress. A, The fully developed wild-type and transgenic plants clonally propagated from individual tillers were grown in sand and subjected to 200 mm NaCl treatment. The images show differences in damage elicited by salinity in wild-type and transgenic plants 16 d after treatment. B, Performance of wild-type and transgenic plants 12 d after recovery from a 16-d salt treatment. C, RWC of wild-type and transgenic plants under salt stress (n = 3). Fully developed wild-type and transgenic plants were watered daily with 200 μg mL−1 20:10:20 fertilizer supplemented with 0, 200, and 300 mm NaCl, as indicated. Leaf tissues were carefully excised 12 d after NaCl treatment and used for measuring RWC. D, EL of leaf cells of wild-type and transgenic plants under normal and various salinity conditions (n = 3). Leaf tissues from wild-type and transgenic plants were carefully excised 12 d after NaCl treatment and used for measuring EL. Data are presented as means ± se, and error bars represent se. Asterisks indicate significant differences between transgenic and control plants at P < 0.05 by Student’s t test.
Osa-miR319a Transgenics Exhibit Better Water Retention and Cell Membrane Integrity Than Controls under Salt Stress
To further elucidate the physiological mechanism of enhanced salt tolerance in Osa-miR319a plants, we investigated the water status of transgenic lines in comparison with control plants without Osa-miR319a. As shown in Figure 4C, results from two representative transgenic lines indicate no significant difference in leaf relative water content (RWC) between control and Osa-miR319a plants under normal growth conditions. When exposed to various concentrations of NaCl, the leaf RWC in all tested plants declined. However, this decline was more significant in control plants than in Osa-miR319a transgenics, especially when high-salinity stress (300 mm NaCl) was applied, indicating greater water retention capacity in Osa-miR319a transgenics than in control plants without the Osa-miR319a gene.
Next, we examined cell membrane integrity in both control and Osa-miR319a transgenic plants. To do this, we measured leaf cell electrolyte leakage (EL) in plants grown under normal and salinity conditions. Both control and Osa-miR319a plants exhibited low levels of cell EL, which were not significantly different from each other under normal growth conditions (Fig. 4D). In contrast, the leaf EL significantly increased in both control and Osa-miR319a plants upon exposure to various concentrations of NaCl. Increasing concentrations of the salt used for plant treatment resulted in higher levels of leaf cell EL (Fig. 4D). However, the increase of leaf cell EL was significantly more pronounced in control plants than in Osa-miR319a transgenics (Fig. 4D), indicating that the control plants without Osa-miR319a are more prone to salt-elicited cell membrane damage than plants overexpressing Osa-miR319a.
Osa-miR319a Transgenics Accumulate Less Na+ Than Control Plants under Salinity Conditions
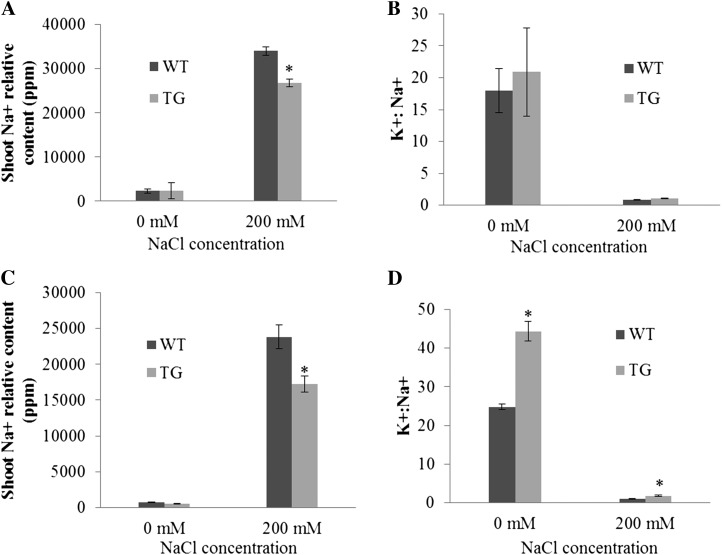
To investigate how Osa-miR319a transgenics perform in Na+ uptake compared with control plants without Osa-miR319a, we measured shoot Na+ content in plants grown under normal and salt stress conditions. As shown in Figure 5A, no significant difference in shoot Na+ content was observed between Osa-miR319a transgenics and control plants under normal growth conditions. Both Osa-miR319a transgenic and control plants accumulated low levels of Na+ (Fig. 5A). Sodium accumulations increased drastically in both control and Osa-miR319a plants when treated with 200 mm NaCl. However, the increase in Na+ accumulation was significantly more pronounced in controls than in Osa-miR319a transgenics (P = 0.01; Fig. 5, A and C), suggesting that the enhanced salt tolerance in Osa-miR319a plants might be the result of less Na+ accumulation in the cytoplasm of the cell and, consequently, reduced toxicity. Interestingly, Osa-miR319a plants appeared to accumulate more K+ than wild-type controls in both normal and salinity conditions, especially when plants were grown in soil (presented as K+:Na+ ratio in Fig. 5, B and D). The discrepancy in significance level for K+ uptake in plants grown in sand and soil might be attributed to the characteristics of the potting mixture soil used for plant maintenance. The soil itself might be rich in nutrients, facilitating the plant uptake of ions. Overexpression of the Osa-miR319a might have led to modified ion transport in transgenic plants, resulting in reduced uptake of sodium but enhanced uptake of potassium. This is not surprising considering that miR319 controls the expression of multiple targets (transcriptional factors) that could be involved in regulating various downstream biological pathways, including ion channels. These channels implicated in the uptake of various ions could be differentially regulated, thereby positively or negatively impacting the transport efficiency of different ions. Further studies analyzing the activities of various ion channels in Osa-miR319a plants would provide information for a better understanding of the molecular mechanisms underlying miR319-mediated plant ion uptake. It should be noted that the accumulation of phosphate, calcium, and magnesium was also higher in Osa-miR319a plants than in the controls when plants were propagated in soil and treated with 200 mm NaCl (data not shown). The enhanced K+:Na+ discrimination has been reported to be strongly associated with greater salt tolerance (Asch et al., 2000; Munns et al., 2000; Colmer et al., 2006; Møller et al., 2009). The higher K+:Na+ ratio observed in Osa-miR319a plants (Fig. 5D) provides additional evidence supporting their improved salt tolerance.
Figure 5.
Mineral contents in wild-type (WT) and transgenic (TG) plants under normal and salinity conditions. Shoot tissues from wild-type and transgenic plants were carefully excised 12 d after NaCl treatment and used for measuring mineral contents. A, Shoot sodium contents in wild-type and transgenic plants propagated and grown in sand under normal conditions and 200 mm NaCl treatment (n = at least 3). B, Shoot K+/Na+ ratio in wild-type and Osa-miR319a transgenic plants propagated and grown in sand under normal conditions and 200 mm NaCl treatment (n = at least 3). C, Shoot sodium contents in wild-type and transgenic plants propagated and grown in soil under normal conditions and 200 mm NaCl treatment (n = at least 3). D, Shoot K+/Na+ ratio in wild-type and Osa-miR319 transgenic plants propagated and grown in soil under normal conditions and 200 mm NaCl treatment (n = at least 3). Statistical analysis of shoot sodium content and shoot K+/Na+ ratio was conducted on wild-type control plants and various transgenic lines. Data are presented as means ± se, and error bars represent se. Asterisks indicate significant differences between transgenic and control plants at P < 0.05 by Student’s t test.
Overexpression of miR319 Improves Drought Tolerance in Transgenic Plants That Is Associated with Enhanced Water Retention and Cell Membrane Integrity and Well-Maintained Photosynthesis
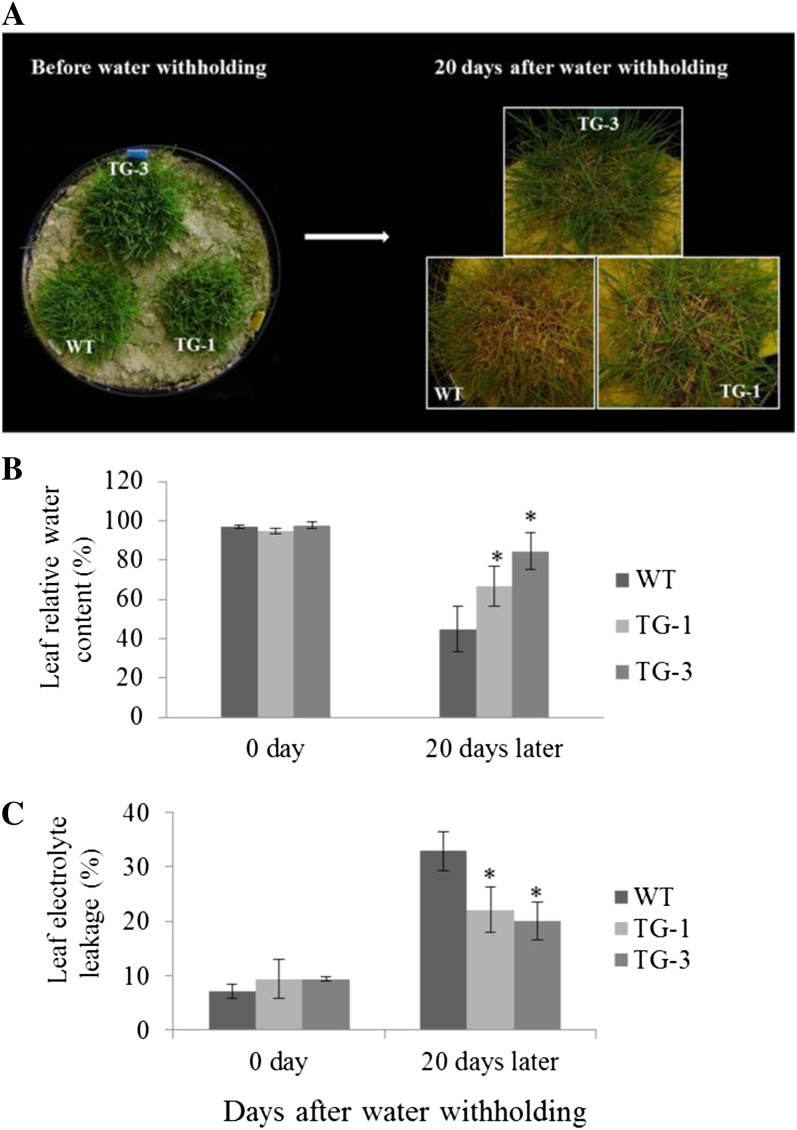
To study whether the overexpression of miR319 impacts plant responses to water stress, we examined both control and Osa-miR319a transgenic plants subjected to drought stress. Fifteen days after water withholding, control plants without Osa-miR319a started to display dehydration symptoms, such as loss of turgor and wilting, whereas Osa-miR319a transgenics remained healthy without obvious damage (data not shown). Twenty days after water withholding, control plants showed serious tissue damage while Osa-miR319a transgenics remained largely turgid and green (Fig. 6A). Osa-miR319a transgenics also outperformed control plants in recovery from desiccation damage (Supplemental Fig. S2B). Further investigation of plant water status and cell membrane integrity revealed that both control and Osa-miR319a plants had similar RWC under normal growth conditions, whereas under dehydration stress, water loss and drought-elicited cell membrane damage in Osa-miR319a plants were significantly less than in controls (Fig. 6, B and C), suggesting an enhanced water retention capacity and cell membrane integrity in Osa-miR319a plants.
Figure 6.
Responses of wild-type (WT) and transgenic (TG) plants to drought stress. A, Fully developed wild-type and transgenic plants clonally propagated from individual tillers were grown in big pots (33 × 44.7 cm) and subjected to water withholding. The performance of wild-type and two independent transgenic lines before water withholding and 20 d after water stress is shown. B, Leaf RWC of wild-type and transgenic plants 20 d after water withholding (n = 3). C, Leaf EL of wild-type and transgenic plants 20 d after water withholding (n = at least 3). Statistical analysis of RWC and EL was conducted on wild-type control plants and the two transgenic lines. Data are presented as means ± se, and error bars represent se. Asterisks indicate significant differences between transgenic and control plants at P < 0.05 by Student’s t test.
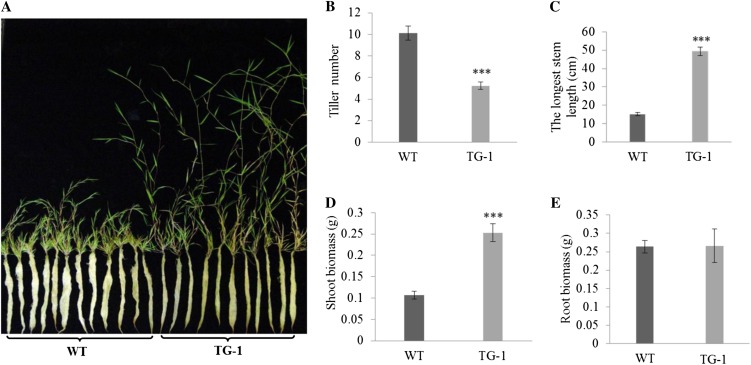
Osa-miR319a and control plants were also examined when subjected to limited water supply treatment. As exemplified in Figure 7A, while the growth of control plants was completely arrested and plants displayed severely disturbed morphology with shortened and deformed leaves and stems, Osa-miR319a plants were impacted less and continued their growth (Fig. 7, A and C; Supplemental Fig. S3). Although the tiller number of control plants was still higher than that of Osa-miR319a transgenics (Fig. 7B), the shoot biomass of control plants was significantly reduced (Fig. 7D). In addition, no significant difference in root biomass was observed between them under water deficit conditions, indicating that root development in Osa-miR319a plants was impacted less by the stress than in control plants (Fig. 7, A and E).
Figure 7.
Tillering and plant development of wild-type (WT) and transgenic (TG) plants under drought stress for 60 d after 30 d of normal development starting from a single tiller. A, Tillering and plant growth in wild-type and transgenic plants developed from a single tiller of the same size for 30 d and then subjected to 60 d of water stress (limited water supply). B, Statistical analysis of tiller numbers in wild-type and transgenic plants counted 60 d after water stress (n = at least 13). C, Length of the longest stem of wild-type and transgenic plants (n = at least 13). D, Biomass of wild-type and transgenic shoots weighed 60 d after initiation of drought stress (n = at least 12). E, Biomass of wild-type and transgenic roots weighed 60 d after initiation of drought stress (n = at least 7). Statistical analysis of tiller number and shoot and root biomass was conducted on wild-type control plants and various transgenic lines. Data are presented as means ± se, and error bars represent se. Asterisks indicate significant differences between transgenic and control plants at P < 0.001 by Student’s t test.
Photosynthesis is one of the primary processes affected when plants are subjected to environmental stress (Munns et al., 2006; Chaves et al., 2009). An increase in stomatal conductance and maintenance of photosynthesis has been demonstrated to be positively correlated to plant performance under stress (Nelson et al., 2007). Our study on Osa-miR319a transgenic and control plants revealed that under normal growth conditions, they were not significantly different in photosynthesis rate and stomatal conductance. However, when subjected to drought stress, Osa-miR319a transgenics exhibited significantly higher stomatal conductance (104.66 ± 24.41 mmol m−2 s−1) than controls without Osa-miR319a (50.08 ± 12.65 mmol m−2 s−1) and higher photosynthesis rate (2.93 ± 0.58 μmol m−2 s−1) than controls (0.75 ± 0.80 μmol m−2 s−1).
Overexpression of miR319 Results in Increased Leaf Weight-Area Ratio and Total Wax Coverage in Transgenic Plants
Plants adapted to dry and saline soil possess a common feature (i.e. their leaves have a higher weight-area ratio), which means that their transpiration efficiency is higher (more carbon fixed per water lost; Munns, 2005). Although Osa-miR319a plants were not significantly different from controls in shoot biomass (Fig. 3, A and C), they exhibited drastically reduced tillering (Fig. 3, A and B), increased leaf expansion (Fig. 2, A–D), and increased leaf thickness (Fig. 2, F and G). This may lead to a change in plant weight-to-surface area ratio. Indeed, Osa-miR319a transgenic plants displayed a significantly higher weight-area ratio (13.7 ± 0.24 mg cm−2) than the controls (10.17 ± 1.30 mg cm−2).
Wax coverage on the leaf cuticle is positively correlated with plant performance under dehydration conditions. When subjected to drought stress, plants may be triggered for increased wax production as one of the avoidance mechanisms to reduce water loss (Kosma et al., 2009). To investigate whether miR319 overexpression impacts leaf epicuticle wax content in transgenic plants, we conducted gas chromatography analysis of leaf cuticles from both control and transgenic plants grown under normal conditions. Although wax accumulation per unit weight in Osa-miR319a transgenics was not significantly different from that in controls, the total leaf wax coverage per unit surface area in Osa-miR319a plants (28.61 ± 1.99 µg cm−2) was significantly higher than that in control plants (19.52 ± 2.52 µg cm−2). It should be noted that wax composition is highly species specific. There has been no report on the composition of wax in creeping bentgrass so far. The identity of a couple of components detected remains to be determined.
The changes in physical parameters, such as weight-area ratio and leaf wax content, resulting from the overexpression of miR319 in transgenic plants might contribute to better plant adaptation to physiological water deficit elicited by drought or salinity (Kosma et al., 2009; Zhou et al., 2009).
Osa-miR319a Regulates the Expression of Putative Target Genes in Transgenic Creeping Bentgrass Plants
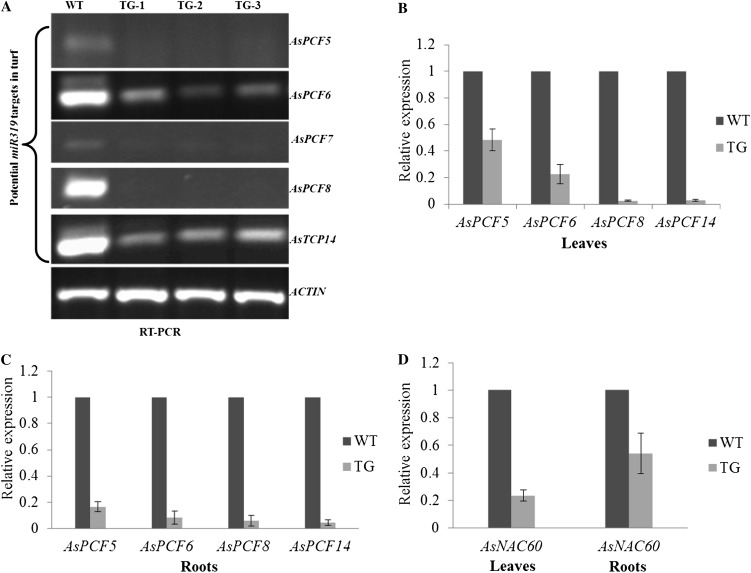
To investigate the molecular mechanism of miR319-mediated alterations in plant development and plant responses to environmental stress, we searched for putative miR319 target genes in plants for further characterization. Bioinformatics analysis of rice genome sequences allowed the identification of five putative miR319 target genes, OsPCF5, OsPCF6, OsPCF7, OsPCF8, and OsTCP14 (C. Yang, D. Li , D. Mao, X. Liu, C. Ji, X. Li, X. Zhao, C. Chen, and L. Zhu, unpublished data), all belonging to the TCP gene family of plant-specific transcription factors (Palatnik et al., 2003; Schommer et al., 2008). Based on sequence information for these putative miR319 targets in rice, we designed PCR primers to partially amplify and clone the corresponding creeping bentgrass homologs, AsPCF5 (JX570754), AsPCF6 (JX570757), AsPCF7 (JX570759), AsPCF8 (JX570755), and AsTCP14 (JX570758), and identified the highly complementary putative miR319 target sites (Fig. 8). Semiquantitative and real-time RT-PCR analyses demonstrated that the putative miR319 target genes were all down-regulated in both leaves and roots of the Osa-miR319a transgenic creeping bentgrass plants (Fig. 9, A–C). It should be noted that although results from semiquantitative PCR suggested down-regulated expression of the AsPCF7 gene (Fig. 9A), our attempt to amplify AsPCF7 by real-time RT-PCR was unsuccessful, probably due to specifically required experimental conditions that need to be further optimized.
Figure 8.
Comparison of target sites in the five putative miR319 target genes in rice and creeping bentgrass with the mature sequence of Osa-miR319. The complementary sequence of the mature Osa-miR319 was used to facilitate the comparison. Asterisks indicate identical sequences.
Figure 9.
Expression levels of the five putative miR319 target genes (AsPCF5, AsPCF6, AsPCF7, AsPCF8, and AsTCP14) and AsNAC60 in wild-type (WT) and Osa-miR319a transgenic (TG) plants by semiquantitative and real-time RT-PCR. A, Semiquantitative RT-PCR analysis of the expression levels of the five putative miR319 target genes in wild-type and transgenic plants. B, Real-time RT-PCR analysis of the expression levels of the four putative miR319 target genes in wild-type leaves and transgenic leaves. C, Real-time RT-PCR analysis of the expression levels of the four putative miR319 target genes in wild-type and transgenic roots. D, Real-time RT-PCR analysis of AsNAC60 expression in wild-type and transgenic leaves and roots. The ΔΔCt method was used for real-time RT-PCR analysis. Three biological replicates and three technical replicates for each biological replicate were used for data analysis followed by Student’s t test. ACTIN was used as the endogenous control. Error bars indicate se (n = 9).
Taken together, our data indicate a negative regulation of target genes by miR319 and suggest the potential direct involvement of individual miR319 targets in modulating plant development and plant responses to environmental stresses.
The Impact of miR319 on the Expression of Other Stress-Related Genes
To examine how other stress-related genes are affected in Osa-miR319a transgenic plants, we studied a transcription factor gene, AsNAC60, a homolog of the rice NAC-like gene ONAC60 (Os12g41680) whose expression has been shown to be dramatically impacted by the overexpression of Osa-miR319 (C. Yang, D. Li, and L. Zhu, unpublished data). ONAC60 has been shown to be the target gene of miR164 in rice (Wu et al., 2009), and in Arabidopsis, TCP genes regulate miR164 expression (Koyama et al., 2010). It would be interesting to test whether the overexpression of miR319 can impact ONAC60 in turfgrass through a TCP-mediated miR164 regulation pathway. Partial sequence of the AsNAC60 complementary DNA (cDNA; JX570756) was cloned from creeping bentgrass and found to be highly homologous to ONAC60. Real-time RT-PCR analyses demonstrated that AsNAC60 expression was down-regulated in Osa-miR319a transgenic creeping bentgrass plants in both leaves and roots (Fig. 9D), indicating that miR319 indeed indirectly regulates AsNAC60 expression.
Expression of miR319 Target Genes Alters in Response to Salt and Drought Stresses in Creeping Bentgrass Plants
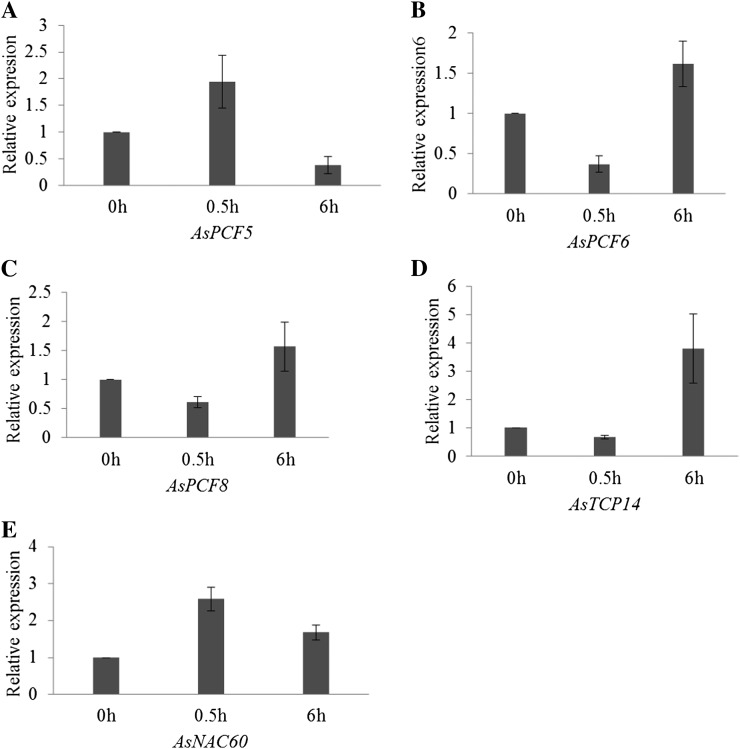
To investigate whether the five putative miR319 target genes are directly involved in miR319-mediated plant responses to abiotic stress, we first examined the expression levels of these genes in wild-type creeping bentgrass plants treated with 200 mm NaCl. Real-time RT-PCR analyses showed an increase in AsPCF5 transcript 0.5 h after salinity stress and then a decline at 6 h (Fig. 10A). Real-time RT-PCR analysis also suggested a trend of up-regulation of the other three putative target genes AsPCF6 (1.16-fold), AsPCF8 (1.56-fold), and AsTCP14 (3.79-fold) at 6 h after exposure to salt stress, but this was not statistically significant (Fig. 10, B–D).
Figure 10.
Expression pattern of putative miR319 target genes in wild-type plants under 200 mm NaCl treatment demonstrated by real-time RT-PCR analysis. A, Real-time RT-PCR analysis of AsPCF5 gene expression 0, 0.5, and 6 h after exposure to salt stress. B, Real-time RT-PCR analysis of AsPCF6 gene expression 0, 0.5, and 6 h after exposure to salt stress. C, Real-time RT-PCR analysis of AsPCF8 gene expression 0, 0.5, and 6 h after exposure to salt stress. D, Real-time RT-PCR analysis of AsTCP14 gene expression 0, 0.5, and 6 h after exposure to salt stress. E, Real-time RT-PCR analysis of AsNAC60 gene expression 0, 0.5, and 6 h after exposure to salt stress. The ΔΔCt method was used for real-time RT-PCR analysis. ACTIN was used as the endogenous control. Error bars indicate se (n = 3).
We also analyzed AsNAC60 expression in plants exposed to 200 mm NaCl. Real-time RT-PCR analyses revealed a significant increase in AsNAC60 expression level at time point 0.5 h (2.5-fold) and also a significant decrease at time point 6 h (1.67-fold; Fig. 10E).
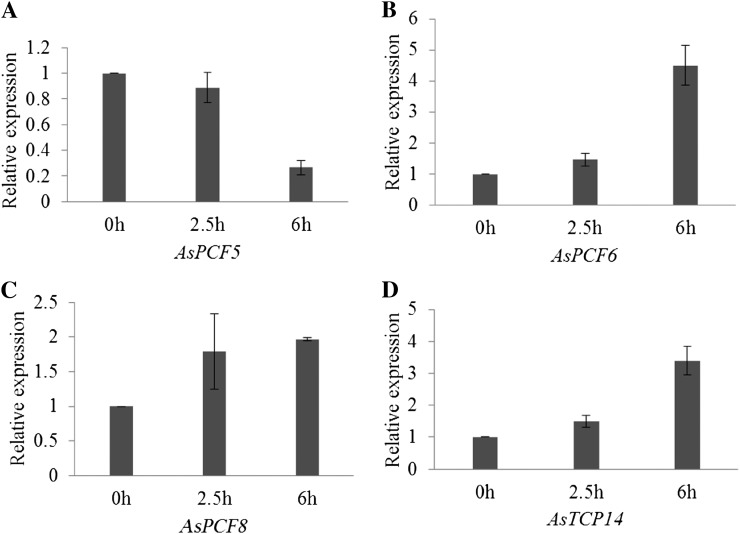
We then studied miR319 target TCP gene activities in response to dehydration stress. Wild-type plants collected from pots were placed on filter paper for desiccation treatment. RNA was extracted 2.5 and 6 h after treatment. Real-time RT-PCR results suggested that AsPCF6, AsPCF8, and AsTCP14 but not AsPCF5 were up-regulated 2.5 h after exposure to desiccation stress (Fig. 11), but the expression level change is statistically insignificant. However, at 6 h upon desiccation, the expression of all three of these genes was significantly up-regulated (Fig. 11, B–D).
Figure 11.
Expression pattern of putative miR319 targets in creeping bentgrass plants under dehydration stress. A, Real-time RT-PCR analysis of AsPCF5 gene expression in plants 0, 2.5, and 6 h after exposure to dehydration stress. B, Real-time RT-PCR analysis of AsPCF6 gene expression level in plants 0, 2.5, and 6 h after exposure to dehydration stress. C, Real-time RT-PCR analysis of AsPCF8 gene expression level in plants 0, 2.5, and 6 h after exposure to dehydration stress. D, Real-time RT-PCR analysis of AsTCP14 gene expression level in plants 0, 2.5, and 6 h after exposure to dehydration stress. The ΔΔCt method was used for real-time RT-PCR analysis. ACTIN was used as the endogenous control. Error bars indicate se (n = 3).
DISCUSSION
Leaf Morphology and Plant Abiotic Stress Tolerance
Leaves are very important plant organs responsible for trapping solar energy through photosynthesis for plant growth and development (Tsukaya, 2005). Transgenic turfgrass plants overexpressing Osa-miR319a exhibit wider and thicker leaves, bigger stems, less tillering, increased weight-area ratio, and increased total leaf cuticle wax coverage, of which thicker leaves, increased weight-area ratio, and increased total wax coverage are thought to be positively correlated with plant abiotic stress tolerance (Bondada et al., 1996; Zhang et al., 2005; Zhou et al., 2009), whereas leaf width is thought to be negatively correlated with plant abiotic stress tolerance (Deák et al., 2011) due to the increased transpiration area. Despite the observation that leaf width frequently correlates with plant abiotic stress responses, the underlying physiological mechanisms remain largely unknown.
A recent study in rice showed that a mutant with a defective zinc finger transcription factor, dst (for drought and salt tolerance), had remarkably wider leaves than controls (Huang et al., 2009). However, the dst mutant plants exhibited significantly enhanced tolerance to drought and salinity stress compared with wild-type controls, which was associated with lower stomata density and less stomata aperture, resulting in reduced water loss and increased water content in the dst mutant. Moreover, sodium uptake of the dst mutants was significantly less than that of control plants when subjected to 100 mm NaCl treatment. The introduction of a wild-type genomic DNA fragment of DST into the dst mutant restores the wild-type phenotype (i.e. narrow leaves and sensitivity to drought and salt stress; Huang et al., 2009).
In this study, although transgenic turfgrass plants overexpressing Osa-miR319a exhibited wider leaves than wild-type controls, no significant differences in stomata opening, stomata conductance, and stomata density were observed between transgenics and wild-type controls (data not shown). However, the Osa-miR319a transgenic plants had thicker leaves, increased weight-area ratio, and increased wax contents, which most likely helped reduce water loss, thus contributing to the enhanced plant resistance to drought stress. Although wider leaves are more likely to be beneficial for plant photosynthesis and thinner leaves would be more efficient in gas exchange (oxygen, carbon dioxide, and water), and thus also desirable for photosynthesis (Tsukaya, 2005), these two features are less desirable for plant responses to water stress. However, other characteristics of leaves, such as stomatal density, stomatal aperture, increased weight-area ratio, and increased wax content, may override the cost of increased leaf width. Thus, like the dst mutant plants, a better adaptation of Osa-miR319a transgenic plants to abiotic stress conditions might be the result of a good balance of the costs, benefits, and associated tradeoffs for each individual morphological/physiological trait.
As reported in the dst mutant (Huang et al., 2009), sodium uptake of the Osa-miR319a transgenic plants was also significantly less than that in the wild-type controls when subjected to salinity stress, indicating the important role that the salt-exclusion mechanism may play in plant salinity resistance. Although our data suggest the possible involvement of some of the putative miR319 target genes in plant responses to both drought and salt stress (see below), further study investigating the molecular mechanisms underlying miR319-mediated plant resistance to abiotic stress will shed light on how plants cope with adverse environmental conditions through the coordinated functions of various regulatory networks.
Root Architecture and Plant Abiotic Stress Tolerance
As one of the very first plant organs that sense many environmental changes, the plant root system plays an important role in plant response to abiotic stress. In many circumstances, a more robust root system with increased root length or root biomass has been considered a positive feature under drought conditions (de Dorlodot et al., 2007; Tardieu, 2012). For example, in fields with deep soil, increased root biomass and length would enable plants to reach deeper soil layers and explore a wider soil area, thus leading to more water and nutrient uptake (Javaux et al., 2008; Schroder et al., 2008; Tardieu, 2012). However, new plant cultivars with enhanced drought tolerance generated in several breeding programs exhibited decreased root biomass (Bolaños and Edmeades, 1993; Bruce et al., 2002; Campos et al., 2004; Tardieu, 2012). When grown in relatively poor conditions with limited soil depth, it makes more sense for plants to invest photosynthetic products in sinks other than roots. Under such circumstances, enhanced root biomass or length may not be the key factor impacting plant water uptake. Data from our research show that Osa-miR319a transgenic plants display a less robust root system but normal shoot growth compared with controls under normal conditions (Fig. 3). However, when subjected to water stress, transgenic plants maintain their growth in both shoot and root, displaying less stress-elicited impact (Fig. 7). This suggests that under normal conditions, Osa-miR319a transgenic plants mainly invest photosynthetic products to the aboveground parts for shoot development, whereas under stressed conditions, more photosynthates are distributed to the underground parts for root development, while there is still an ample reservation of carbohydrates in the leaves, ensuring aboveground plant growth (Fig. 7).
It also has been argued that plant root capacity in water uptake, one of the key factors contributing to the plant response to water stress, is determined by root spatial distribution rather than root biomass or length, even when plants are grown in better field conditions with deep soil layers (Tardieu et al., 1992; Manschadi et al., 2006, 2008; Tardieu, 2012). Thus, it is not surprising to observe that with a less robust root system than wild-type controls, Osa-miR319a transgenic creeping bentgrass plants still perform better under drought stress. Further field study analyzing root spatial distribution in both control and Osa-miR319a plants grown under normal and stress conditions would provide information for a better understanding of the physiological mechanisms of miR319-mediated morphological changes in roots and their impact on plant responses to environmental stress.
Leaf Senescence and Abiotic Stress Tolerance
In some cases, early leaf senescence is thought to serve as one of the stress-avoidance mechanisms induced to avoid cellular stresses by decreasing water demand (Tardieu, 1996, 2012). Younger leaves are conserved under stress by sacrificing older leaves (Munné-Bosch and Alegre, 2004; Munns, 2005). On the other hand, senescence is also assumed to serve as a type of cell death program that could be induced during drought and salinity that imposes negative effects on plants. Suppression of this kind of induced leaf senescence would make plants maintain higher water contents and better photosynthetic activity during the stress (Lutts et al., 1996; Rivero et al., 2007).
In this study, when wild-type plants started displaying damage caused by sodium toxicity 5 d after 200 mm NaCl treatment, transgenic plants did not show any typical salinity stress-elicited symptoms (wilting and loss of cell turgor), as observed in wild-type controls, but appeared to show slight senescence symptoms earlier than wild-type controls (Supplemental Fig. S4). However, the progression of leaf senescence in transgenic plants was much slower than that in wild-type plants (Fig. 4). Considering that miR319 has been reported to positively regulate plant leaf senescence through the jasmonic acid (JA) biosynthesis pathway (Schommer et al., 2008), it is hypothesized that overexpression of miR319 in transgenic creeping bentgrass plants impaired the function of the miR319 target, TCP, resulting in less accumulation of JA and, consequently, delay in leaf senescence in general. However, this hypothesis cannot explain the earlier leaf senescence that appeared to take place in the transgenic plants. As JA is not the only pathway that regulates senescence (Schommer et al., 2008), it is likely that other factors triggered by miR319 overexpression may be involved, contributing to this particular response in transgenic plants. Another possibility would be that better sodium exclusion in Osa-miR319a plants (Fig. 5) may contribute to their overall delayed leaf senescence and slower ionic phase, minimizing sodium toxicity.
Molecular Basis of Salt-Exclusion Mechanisms in Osa-miR319a Transgenic Plants
A strong correlation between salt exclusion and salt tolerance has been reported in many plant species, including rice, durum wheat (Triticum durum), bread wheat (Triticum aestivum), barley (Hordeum vulgare), pearl millet (Pennisetum americanum), Hordeum spp., tall wheatgrass, and Triticum tauschii (Yeo and Flowers, 1983; Läuchli, 1984; Tardieu et al., 1992; Bolaños and Edmeades, 1993; Bruce et al., 2002; Munns and James, 2003; Munns et al., 2003; Tester and Davenport, 2003; Campos et al., 2004; Manschadi et al., 2006, 2008; Tardieu, 2012). Salt-sensitive cultivars tend to have higher accumulations of Na+. Therefore, lower accumulation of Na+ in plants as a positive trait has been used to select superior genotypes in different breeding programs (Munns and James, 2003; Munns et al., 2003). Our results demonstrated that Osa-miR319a transgenics accumulated less Na+ than control plants (Fig. 5). The low Na+ accumulation in transgenic plants should lead to less cell damage and might contribute to the enhanced salt tolerance observed in Osa-miR319a plants. What is the mechanism of miR319-mediated salt exclusion? In wheat, it has been demonstrated that low and high Na+ accumulation is controlled by two major genes, Nax1 and Nax2. Nax1 was located on chromosome 2A (Lindsay et al., 2004) and was identified as an Na+ transporter of the HKT family (HKT7 and ortholog HKT1;4 in Triticum monococcum; Huang et al., 2006). Nax2 was located on chromosome 5A and was identified as HKT8 (with ortholog HKT1;5 in rice; Byrt et al., 2007). It is deduced that Nax1 correlates with the loading of Na+ in root xylem, while Nax2 correlates with the relocation of Na+ in the upper parts of plants (Munns and James, 2003; Munns, 2005). Genetic engineering of a salt-exclusion mechanism by tissue-specific overexpression of the Na+ transporter HKT1;1 in Arabidopsis reduced 37% to 64% Na+ accumulation in the shoots (Møller et al., 2009). However, the detailed mechanism of how HKT transporters mediate sodium management is still unknown. In transgenic rice plants overexpressing miR319, the Na+ transporter gene OsHKT2 was also found to be highly up-regulated (C. Yang, D. Li, and L. Zhu, unpublished data). In this study, we observed that some of the putative miR319 target genes were up-regulated upon salt stress (Fig. 10). It is likely that these target genes are involved in the plant response to salt stress via participation in controlling various downstream biological pathways, including Na+ transporter production and activity. This is further supported by salt-induced up-regulation of the creeping bentgrass AsNAC60, a stress-responsive rice homolog gene that is not a miR319 target (Fig. 9D). Modification of miR319 expression interferes with this regulatory network, leading to an enhanced salt-resistance phenotype in transgenic plants. Further characterization of the miR319 target genes will provide information for a better understanding of the molecular mechanisms underlying miR319-mediated plant resistance to salt stress. Additionally, our data also demonstrate the possible involvement of the putative miR319 target genes in plant responses to drought stress (Fig. 11). Therefore, studies of stress-responsive miR319 targets would help elucidate key molecular pathways governing plant responses to abiotic stresses.
It should be noted that the behavior of miR319 in response to stress can be complicated. For example, miR319 was first up-regulated after a 24-h exposure to cold stress and then down-regulated 48 h after the treatment (Thiebaut et al., 2012). The expression level change of miR319 target genes in response to stress could also be impacted by other signals, such as hormones. The ultimate expression level of target genes would be a balance of various influences. We speculate that, in our study, the strong and constitutive expression of miR319 in transgenic plants may have overridden the effects of other regulators, causing the down-regulation of potential target genes contributing to the enhanced drought and salt tolerance.
In summary, overexpression of miR319 impacts plant development and enhances plant drought and salt tolerance. The miR319-mediated down-regulation of target genes in transgenic plants may have caused changes in various biological processes, including those associated with water retention capacity, leaf wax synthesis, and salt uptake beneficial to plants responding to salinity and water deficiency. The manipulation of miR319 target genes provides novel molecular strategies to genetically engineer crop species for enhanced resistance to environmental stress.
MATERIALS AND METHODS
Cloning of the Osa-miR319a Gene and Construction of the Plant Expression Vectors
The full-length rice (Oryza sativa) Osa-miR319a cDNA (AK064418; Knowledge-based Oryza Molecular biological Encyclopedia; http://cdna01.dna.affrc.go.jp/cDNA/) containing a stem-loop structure was cloned to the binary vector pZH01 (Xiao et al., 2003), producing p35S-Osa-miR319a/p35S-hyg. The construct contains the CaMV 35S promoter driving Osa-miR319a and the CaMV 35S promoter driving the hyg gene for hygromycin resistance as a selectable marker. The construct was transferred into Agrobacterium tumefaciens strain LBA4404 for plant transformation.
Plant Materials and Transformation
A commercial creeping bentgrass (Agrostis stolonifera ‘Penn A-4’; supplied by HybriGene) was used for transformation. Transgenic plants overexpressing Osa-miR319a were produced and maintained as described previously (Luo et al., 2004a, 2004b).
Plant Propagation, Maintenance, and Abiotic Stress Treatments
The Osa-miR319a transgenic plants and controls (untransformed plants either geminated from seeds or regenerated from tissue culture and transgenic lines harboring backbone vector) initially maintained in the greenhouse were transferred to a growth room with a 14-h photoperiod for propagation. Both transgenic and control plants were clonally propagated from tillers and grown in small cone-tainers (4.0 × 20.3 cm; Dillen Products), middle pots (15 × 10.5 cm; Dillen Products), or big pots (33 × 44.7 cm; Dillen Products) using only silica sand. Shoots were clipped weekly to achieve uniform plant growth. Illumination in the growth room was 350 to 450 μmol m−2 s−1 provided by AgroSun Gold 1,000-W sodium/halide lamps (Maryland Hydroponics). Temperatures and humidity were maintained at 25°C and 30% during the day and 17°C and 60% at night. Plants were watered every other day with 200 μg mL−1 of water-soluble fertilizer (20:10:20 nitrogen:phosphorus:potassium; Peat-Lite Special; Scotts).
For plants grown in cone-tainers, salinity treatments were conducted by injecting 10 mL of 200 mm NaCl supplemented with 200 μg mL−1 fertilizer twice every day. For plants grown in the middle pots, salinity treatments were conducted by immersing the whole pots in 1 L of 200 mm NaCl supplemented with 200 μg mL−1 fertilizer and changing the solution every day. Shoots were harvested 12 d later and used for measuring mineral contents and other analyses. Plant recovery from salt stress by watering with 200 μg mL−1 fertilizer was documented by photography.
Water withholding or limited water supply (10 mL for each plant every 2 weeks) was applied for drought treatment.
For measuring leaf net photosynthesis rate and stomatal conductance, plants were propagated and maintained in nutrient-rich soil (3-B Mix; Fafard) without trimming to achieve maximum leaf growth to facilitate measurements.
Isolation of Plant DNA, RNA, and cDNA Synthesis
Plant genomic DNA was extracted as described previously (Luo et al., 2005). Plant total RNA was isolated with Trizol reagent and reverse transcribed using SuperScript III Reverse Transcriptase (Invitrogen) following the manufacturer’s instructions.
Stem-Loop RT and Real-Time RT-PCR Analysis
Stem-loop RT was preformed after a previous protocol (Varkonyi-Gasic et al., 2007). The Osa-miR319a stem-loop RT primer and forward primer were 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGGGAGC-3′ and 5′-CGGCGGTTGGACTGAAGGGT-3′. Based on DNA sequences of the PCR products, real-time RT-PCR primers were designed. Each PCR contained 12.5 μL of iQ SYBR Green Supermix. The reaction was performed using the Bio-Rad iQ5 real-time detection system, and the fluorescence data were collected using the iQ5 Optical System Software version 2.0 (Bio-Rad Laboratories). One reference gene, AsACT1 (JX644005), was used as an endogenous control. The ΔΔCt method was used for real-time PCR analysis. ΔCt values were calculated by first normalizing threshold cycle (Ct) values of targets to the endogenous control (ΔCt = Cttarget − Ctreference) and subsequently calculating ΔΔCt values using the ΔCt value of 0 h as a reference (ΔΔCt = ΔCttest − ΔCt0h). Relative expression level was calculated using the formula 2−ΔΔCt = normalized expression ratio.
Measurement of Mineral Contents, Leaf RWC, and EL
Upon salt treatment, plant leaves were collected and dried for 48 h at 80°C for dry weights. Sodium contents were determined following previous protocols (Haynes, 1980; Plank, 1992; Li et al., 2010). Leaf RWC and EL were measured as described previously (Li et al., 2010).
Leaf Sections and Microscopic Analysis
Fully expanded leaves from the top one-third of the plants were collected and fixed in formalin-acetic-alcohol, followed by dehydration through graded ethanol and infiltration in catalyzed resin (1.25 g of benzoyl peroxide per 100 mL of immunobed monomer A). Samples were then embedded, polymerized at room temperature, and put in a desiccator under vacuum until ready to block. Photographs were taken using a MEIJI EM-5 microscope connected with a 35-mm SLR camera body (Canon).
Measurement of Photosynthesis Parameters
Single-leaf net photosynthesis rate was measured on at least two leaves from plants selected from three pots using a PLC-6 narrow leaf chamber connected to a CIRAS-2 (PP Systems). Leaf chamber carbon dioxide concentration was maintained constantly at 375 μmol mol−1. Light intensity was controlled at 400 μmol m−2 s−1 with the light-emitting diode light sources installed in the leaf chamber of the CIRAS-2 system. Leaf chamber temperature was maintained at equilibrium to the ambient temperature.
Cuticular Wax Analysis
Leaf wax composition was measured after leaves were submerged in hexane for 45 s (Kosma et al., 2009; F. Bethea and H. Liu, unpublished data). Wax extracts were evaporated under nitrogen gas and derivatized by heating at 100°C for 15 min in N,O-bis(trimethylsily)trifluoroacetamide (Supelco). Silylated samples were analyzed by gas chromatography with a Hewlett-Packard 5890 series II gas chromatograph equipped with a flame ionization detector and a 12-m, 0.2-mm HP-1 capillary column with helium as the carrier gas. Quantification was based on flame ionization detector peak areas relative to the internal standard tetrococine.
The total amount of cuticular wax was expressed as unit weight per unit of leaf surface area (μg cm−2). Leaf areas were determined by ImageJ software (http://rsb.info.nih.gov/ij/) using digital images of flattened leaves.
Statistical Analysis
Student’s t test was used for data analysis. P < 0.05 was considered to be statistically significant.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers JX570754, JX570755, JX570756, JX570757, JX570758, JX570759, and JX644005.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Creeping bentgrass miR319 expression analysis in wild-type plants under drought and salinity stresses.
Supplemental Figure S2. Performance of wild-type and transgenic plants under extreme salinity and drought stress.
Supplemental Figure S3. A close look at the leaves and stems of wild-type and transgenic plants subjected to 60-d limited water supply.
Supplemental Figure S4. Performance of wild-type and transgenic plants 5 d after 200 mm NaCl treatment.
Acknowledgments
We thank Dr. Halina Knap and Nancy Korn for help on sectioning, Dr. Haibo Liu and Frank Bethea for help in wax analysis, and Dr. Guido Schnabel, Dr. Chinfu Chen, Dr. William R. Marcotte, Dr. Julia Frugoli, and Elise Schnabel for helpful discussions. We also thank Dr. Emerson Shipe for critically reading the manuscript.
Glossary
- miRNA
micro RNA
- CaMV
cauliflower mosaic virus
- RT
reverse transcription
- RWC
relative water content
- EL
electrolyte leakage
- JA
jasmonic acid
- cDNA
complementary DNA
References
- Asch F, Dingkuhn M, Dörffling K, Miezan K. (2000) Leaf K/Na ratio predicts salinity induced yield loss in irrigated rice. Euphytica 113: 109–118 [Google Scholar]
- Axtell MJ, Bowman JL. (2008) Evolution of plant microRNAs and their targets. Trends Plant Sci 13: 343–349 [DOI] [PubMed] [Google Scholar]
- Bolaños J, Edmeades G. (1993) Eight cycles of selection for drought tolerance in lowland tropical maize. II. Responses in reproductive behavior. Field Crops Res 31: 253–268 [Google Scholar]
- Bondada BR, Oosterhuis DM, Murphy JB, Kim KS. (1996) Effect of water stress on the epicuticular wax composition and ultrastructure of cotton (Gossypium hirsutum L.) leaf, bract, and boll. Environ Exp Bot 36: 61–69 [Google Scholar]
- Bruce WB, Edmeades GO, Barker TC. (2002) Molecular and physiological approaches to maize improvement for drought tolerance. J Exp Bot 53: 13–25 [PubMed] [Google Scholar]
- Byrt CS, Platten JD, Spielmeyer W, James RA, Lagudah ES, Dennis ES, Tester M, Munns R. (2007) HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1. Plant Physiol 143: 1918–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos H, Cooper M, Habben J, Edmeades G, Schussler J. (2004) Improving drought tolerance in maize: a view from industry. Field Crops Res 90: 19–34 [Google Scholar]
- Capell T, Bassie L, Christou P. (2004) Modulation of the polyamine biosynthetic pathway in transgenic rice confers tolerance to drought stress. Proc Natl Acad Sci USA 101: 9909–9914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra Babu R, Zhang J, Blum A, David Ho TH, Wu R, Nguyen HT. (2004) HVA1, a LEA gene from barley confers dehydration tolerance in transgenic rice (Oryza sativa L.) via cell membrane protection. Plant Sci 166: 855–862 [Google Scholar]
- Chaves MM, Flexas J, Pinheiro C. (2009) Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann Bot (Lond) 103: 551–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmer TD, Flowers TJ, Munns R. (2006) Use of wild relatives to improve salt tolerance in wheat. J Exp Bot 57: 1059–1078 [DOI] [PubMed] [Google Scholar]
- Deák C, Jäger K, Fábián A, Nagy V, Albert Z, Miskó A, Barnabás B, Papp I. (2011) Investigation of physiological responses and leaf morphological traits of wheat genotypes with contrasting drought stress tolerance. Acta Biologica Szegediensis 55: 69–71 [Google Scholar]
- de Dorlodot S, Forster B, Pagès L, Price A, Tuberosa R, Draye X. (2007) Root system architecture: opportunities and constraints for genetic improvement of crops. Trends Plant Sci 12: 474–481 [DOI] [PubMed] [Google Scholar]
- Ding D, Zhang L, Wang H, Liu Z, Zhang Z, Zheng Y. (2009) Differential expression of miRNAs in response to salt stress in maize roots. Ann Bot (Lond) 103: 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers TJ. (2004) Improving crop salt tolerance. J Exp Bot 55: 307–319 [DOI] [PubMed] [Google Scholar]
- Fu D, Huang B, Xiao Y, Muthukrishnan S, Liang GH. (2007) Overexpression of barley hva1 gene in creeping bentgrass for improving drought tolerance. Plant Cell Rep 26: 467–477 [DOI] [PubMed] [Google Scholar]
- Garg AK, Kim JK, Owens TG, Ranwala AP, Choi YD, Kochian LV, Wu RJ. (2002) Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc Natl Acad Sci USA 99: 15898–15903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaxiola RA, Li J, Undurraga S, Dang LM, Allen GJ, Alper SL, Fink GR. (2001) Drought- and salt-tolerant plants result from overexpression of the AVP1 H+-pump. Proc Natl Acad Sci USA 98: 11444–11449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes RJ. (1980) A comparison of two modified Kjeldahl digestion techniques for multi-element plant analysis with conventional wet and dry ashing methods. Commun Soil Sci Plant Anal 11: 459–467 [Google Scholar]
- Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L. (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103: 12987–12992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Spielmeyer W, Lagudah ES, James RA, Platten JD, Dennis ES, Munns R. (2006) A sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiol 142: 1718–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XY, Chao DY, Gao JP, Zhu MZ, Shi M, Lin HX. (2009) A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev 23: 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javaux M, Schroder T, Vanderborght J, Vereecken H. (2008) Use of a three-dimensional detailed modeling approach for predicting root water uptake. Vadose Zone J 7: 1079–1088 [Google Scholar]
- Jones-Rhoades MW, Bartel DP. (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14: 787–799 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57: 19–53 [DOI] [PubMed] [Google Scholar]
- Khraiwesh B, Zhu J-K, Zhu J. (2012) Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta 1819: 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosma DK, Bourdenx B, Bernard A, Parsons EP, Lü S, Joubès J, Jenks MA. (2009) The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiol 151: 1918–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. (2010) TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22: 3574–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Läuchli A. (1984) Salt exclusion: an adaptation of legumes for crops and pastures under saline conditions. In RC Staples, GH Toenniessen, eds, Salinity Tolerance in Plants—Strategies for Crop Improvement. Wiley, New York, pp 171–187 [Google Scholar]
- Laurie S, Feeney KA, Maathuis FJM, Heard PJ, Brown SJ, Leigh RA. (2002) A role for HKT1 in sodium uptake by wheat roots. Plant J 32: 139–149 [DOI] [PubMed] [Google Scholar]
- Lewis R, Mendu V, Mcnear D, Tang G. (2009) Roles of microRNAs in plant abiotic stress. In SM Jain, DS Brar, eds, Molecular Techniques in Crop Improvement, Ed 2. Springer, Cambridge, pp 357–372 [Google Scholar]
- Li Z, Baldwin CM, Hu Q, Liu H, Luo H. (2010) Heterologous expression of Arabidopsis H+-pyrophosphatase enhances salt tolerance in transgenic creeping bentgrass (Agrostis stolonifera L.). Plant Cell Environ 33: 272–289 [DOI] [PubMed] [Google Scholar]
- Lian HL, Yu X, Ye Q, Ding XS, Kitagawa Y, Kwak SS, Su WA, Tang ZC. (2004) The role of aquaporin RWC3 in drought avoidance in rice. Plant Cell Physiol 45: 481–489 [DOI] [PubMed] [Google Scholar]
- Lindsay MP, Lagudah ES, Hare RA, Munns R. (2004) A locus for sodium exclusion (Nax1), a trait for salt tolerance, mapped in durum wheat. Funct Plant Biol 31: 1105–1114 [DOI] [PubMed] [Google Scholar]
- Liu HH, Tian X, Li YJ, Wu CA, Zheng CC. (2008) Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14: 836–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X-Y, Huang X-L. (2008) Plant miRNAs and abiotic stress responses. Biochem Biophys Res Commun 368: 458–462 [DOI] [PubMed] [Google Scholar]
- Luo H, Hu Q, Nelson K, Longo C, Kausch A (2004a) Controlling transgene escape in genetically modified grasses. In A Hopkins, Z-Y Wang, R Mian, M Sledge, R Barker, eds, Molecular Breeding of Forage and Turf, Vol 11. Springer, Dordrecht, The Netherlands, pp 245–254 [Google Scholar]
- Luo H, Hu Q, Nelson K, Longo C, Kausch AP, Chandlee JM, Wipff JK, Fricker CR. (2004b) Agrobacterium tumefaciens-mediated creeping bentgrass (Agrostis stolonifera L.) transformation using phosphinothricin selection results in a high frequency of single-copy transgene integration. Plant Cell Rep 22: 645–652 [DOI] [PubMed] [Google Scholar]
- Luo H, Kausch A, Hu Q, Nelson K, Wipff J, Fricker C, Owen T, Moreno M, Lee J, Hodges T. (2005) Controlling transgene escape in GM creeping bentgrass. Mol Breed 16: 185–188 [Google Scholar]
- Lutts S, Kinet J, Bouharmont J. (1996) NaCl-induced senescence in leaves of rice (Oryza sativa L.) cultivars differing in salinity resistance. Ann Bot (Lond) 78: 389–398 [Google Scholar]
- Lv D-K, Bai X, Li Y, Ding X-D, Ge Y, Cai H, Ji W, Wu N, Zhu Y-M. (2010) Profiling of cold-stress-responsive miRNAs in rice by microarrays. Gene 459: 39–47 [DOI] [PubMed] [Google Scholar]
- Manschadi AM, Christopher J, Hammer GL. (2006) The role of root architectural traits in adaptation of wheat to water-limited environments. Funct Plant Biol 33: 823–837 [DOI] [PubMed] [Google Scholar]
- Manschadi AM, Hammer GL, Christopher JT, DeVoil P. (2008) Genotypic variation in seedling root architectural traits and implications for drought adaptation in wheat (Triticum aestivum L.). Plant Soil 303: 115–129 [Google Scholar]
- Mazzucotelli E, Mastrangelo AM, Crosatti C, Guerra D, Stanca AM, Cattivelli L. (2008) Abiotic stress response in plants: when post-transcriptional and post-translational regulations control transcription. Plant Sci 174: 420–431 [Google Scholar]
- Møller IS, Gilliham M, Jha D, Mayo GM, Roy SJ, Coates JC, Haseloff J, Tester M. (2009) Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 21: 2163–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munné-Bosch S, Alegre L. (2004) Die and let live: leaf senescence contributes to plant survival under drought stress. Funct Plant Biol 31: 203–216 [DOI] [PubMed] [Google Scholar]
- Munns R. (2002) Comparative physiology of salt and water stress. Plant Cell Environ 25: 239–250 [DOI] [PubMed] [Google Scholar]
- Munns R. (2005) Genes and salt tolerance: bringing them together. New Phytol 167: 645–663 [DOI] [PubMed] [Google Scholar]
- Munns R, Hare RA, James RA, Rebetzke GJ. (2000) Genetic variation for improving the salt tolerance of durum wheat. Aust J Agric Res 51: 69–74 [Google Scholar]
- Munns R, James RA. (2003) Screening methods for salinity tolerance: a case study with tetraploid wheat. Plant Soil 253: 201–218 [Google Scholar]
- Munns R, James RA, Läuchli A. (2006) Approaches to increasing the salt tolerance of wheat and other cereals. J Exp Bot 57: 1025–1043 [DOI] [PubMed] [Google Scholar]
- Munns R, Rebetzke GJ, Husain S, James RA, Hare RA. (2003) Genetic control of sodium exclusion in durum wheat. Aust J Agric Res 54: 627–635 [Google Scholar]
- Munns R, Tester M. (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Nag A, King S, Jack T. (2009) miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc Natl Acad Sci USA 106: 22534–22539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Ito Y, Yamaguchi-Shinozaki K. (2009) Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol 149: 88–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath U, Crawford BCW, Carpenter R, Coen E. (2003) Genetic control of surface curvature. Science 299: 1404–1407 [DOI] [PubMed] [Google Scholar]
- Nelson DE, Repetti PP, Adams TR, Creelman RA, Wu J, Warner DC, Anstrom DC, Bensen RJ, Castiglioni PP, Donnarummo MG, et al. (2007) Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc Natl Acad Sci USA 104: 16450–16455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SJ, Song SI, Kim YS, Jang HJ, Kim SY, Kim M, Kim YK, Nahm BH, Kim JK. (2005) Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol 138: 341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N, Cohen AR, Etzioni A, Brand A, Yanai O, Shleizer S, Menda N, Amsellem Z, Efroni I, Pekker I, et al. (2007) Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet 39: 787–791 [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Phillips JR, Dalmay T, Bartels D. (2007) The role of small RNAs in abiotic stress. FEBS Lett 581: 3592–3597 [DOI] [PubMed] [Google Scholar]
- Plank CO. (1992) Plant analysis reference procedures for the southern region of the United States. Southern Cooperative Series Bulletin No. 368, http://www.ncagr.gov/agronomi/pdffiles/sera368.pdf (December 18, 2008) [Google Scholar]
- Rivero RM, Kojima M, Gepstein A, Sakakibara H, Mittler R, Gepstein S, Blumwald E. (2007) Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc Natl Acad Sci USA 104: 19631–19636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Somoza I, Weigel D. (2011) MicroRNA networks and developmental plasticity in plants. Trends Plant Sci 16: 258–264 [DOI] [PubMed] [Google Scholar]
- Rus A, Estan M, Gisbert C, Garcia-Sogo B, Serrano R, Caro M, Moreno V, Bolarin M. (2001) Expressing the yeast HAL1 gene in tomato increases fruit yield and enhances K+/Na+ selectivity under salt stress. Plant Cell Environ 24: 875–880 [Google Scholar]
- Rus A, Lee BH, Muñoz-Mayor A, Sharkhuu A, Miura K, Zhu JK, Bressan RA, Hasegawa PM. (2004) AtHKT1 facilitates Na+ homeostasis and K+ nutrition in planta. Plant Physiol 136: 2500–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Nath U, Weigel D. (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder T, Javaux M, Vanderborght J, Korfgen B, Vereecken H. (2008) Effect of local soil hydraulic conductivity drop using a three-dimensional root water uptake model. Vadose Zone J 7: 1089–1098 [Google Scholar]
- Shi H, Lee BH, Wu SJ, Zhu JK. (2003) Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat Biotechnol 21: 81–85 [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58: 221–227 [DOI] [PubMed] [Google Scholar]
- Shukla LI, Chinnusamy V, Sunkar R. (2008) The role of microRNAs and other endogenous small RNAs in plant stress responses. Biochim Biophys Acta 1779: 743–748 [DOI] [PubMed] [Google Scholar]
- Sunkar R, Chinnusamy V, Zhu J, Zhu JK. (2007) Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci 12: 301–309 [DOI] [PubMed] [Google Scholar]
- Sunkar R, Li Y-F, Jagadeeswaran G. (2012) Functions of microRNAs in plant stress responses. Trends Plant Sci 17: 196–203 [DOI] [PubMed] [Google Scholar]
- Sunkar R, Zhu JK. (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16: 2001–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardieu F. (1996) Drought perception by plants: do cells of droughted plants experience water stress? Plant Growth Regul 20: 93–104 [Google Scholar]
- Tardieu F. (2012) Any trait or trait-related allele can confer drought tolerance: just design the right drought scenario. J Exp Bot 63: 25–31 [DOI] [PubMed] [Google Scholar]
- Tardieu F, Bruckler L, Lafolie F. (1992) Root clumping may affect the root water potential and the resistance to soil-root water transport. Plant Soil 140: 291–301 [Google Scholar]
- Tester M, Davenport R. (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot (Lond) 91: 503–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebaut F, Rojas CA, Almeida KL, Grativol C, Domiciano GC, Lamb CRC, Engler JdeA, Hemerly AS, Ferreira PCG. (2012) Regulation of miR319 during cold stress in sugarcane. Plant Cell Environ 35: 502–512 [DOI] [PubMed] [Google Scholar]
- Tsukaya H. (2005) Leaf shape: genetic controls and environmental factors. Int J Dev Biol 49: 547–555 [DOI] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. (2007) Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang FZ, Wang QB, Kwon SY, Kwak SS, Su WA. (2005) Enhanced drought tolerance of transgenic rice plants expressing a pea manganese superoxide dismutase. J Plant Physiol 162: 465–472 [DOI] [PubMed] [Google Scholar]
- Wang T, Chen L, Zhao M, Tian Q, Zhang WH. (2011) Identification of drought-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. BMC Genomics 12: 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Vinocur B, Altman A. (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218: 1–14 [DOI] [PubMed] [Google Scholar]
- White PJ, Broadley MR. (2001) Chloride in soils and its uptake and movement within the plant: a review. Ann Bot (Lond) 88: 967–988 [Google Scholar]
- Wu CA, Yang GD, Meng QW, Zheng CC. (2004) The cotton GhNHX1 gene encoding a novel putative tonoplast Na+/H+ antiporter plays an important role in salt stress. Plant Cell Physiol 45: 600–607 [DOI] [PubMed] [Google Scholar]
- Wu L, Zhang Q, Zhou H, Ni F, Wu X, Qi Y. (2009) Rice MicroRNA effector complexes and targets. Plant Cell 21: 3421–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Wang Y, Liu D, Wang W, Li X, Zhao X, Xu J, Zhai W, Zhu L. (2003) Functional analysis of the rice AP3 homologue OsMADS16 by RNA interference. Plant Mol Biol 52: 957–966 [DOI] [PubMed] [Google Scholar]
- Yang S, Vanderbeld B, Wan J, Huang Y. (2010) Narrowing down the targets: towards successful genetic engineering of drought-tolerant crops. Mol Plant 3: 469–490 [DOI] [PubMed] [Google Scholar]
- Yeo A, Flowers T. (1983) Varietal differences in the toxicity of sodium ions in rice leaves. Physiol Plant 59: 189–195 [Google Scholar]
- Zhang B, Pan X, Cobb GP, Anderson TA. (2006) Plant microRNA: a small regulatory molecule with big impact. Dev Biol 289: 3–16 [DOI] [PubMed] [Google Scholar]
- Zhang HX, Blumwald E. (2001) Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol 19: 765–768 [DOI] [PubMed] [Google Scholar]
- Zhang HX, Hodson JN, Williams JP, Blumwald E. (2001) Engineering salt-tolerant Brassica plants: characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc Natl Acad Sci USA 98: 12832–12836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JY, Broeckling CD, Blancaflor EB, Sledge MK, Sumner LW, Wang ZY. (2005) Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). Plant J 42: 689–707 [DOI] [PubMed] [Google Scholar]
- Zhang JZ, Creelman RA, Zhu JK. (2004) From laboratory to field: using information from Arabidopsis to engineer salt, cold, and drought tolerance in crops. Plant Physiol 135: 615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Liang R, Ge L, Li W, Xiao H, Lin H, Ruan K, Jin Y. (2007) Identification of drought-induced microRNAs in rice. Biochem Biophys Res Commun 354: 585–590 [DOI] [PubMed] [Google Scholar]
- Zhou L, Liu Y, Liu Z, Kong D, Duan M, Luo L. (2010) Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J Exp Bot 61: 4157–4168 [DOI] [PubMed] [Google Scholar]
- Zhou X-J, Zhao H-B, Ma C-C, Li Q-F. (2009) Effects of exogenous silicon on leaf structure and water-holding capacity of cucumber plants. Chin J Ecol 28: 556–559 [Google Scholar]