Background: TGF-β induces apoptosis in Burkitt's lymphoma cells.
Results: PUMA is a direct target gene of TGF-β signaling and is required for rapid apoptosis.
Conclusion: TGF-β-mediated direct induction of PUMA contributes to apoptosis in human and murine c-Myc-driven lymphomas.
Significance: These studies link TGF-β signaling and transcriptional activation of PUMA, two factors with critical roles in regulating B-cell survival.
Keywords: Apoptosis, Bcl-2 Family Proteins, Lymphoma, Myc, Transforming Growth Factor Beta (TGFbeta), Burkitt's Lymphoma, PUMA
Abstract
c-Myc transformed human Burkitt's lymphoma (BL) cells are highly sensitive to TGF-β-induced apoptosis. Previously we demonstrated that TGF-β-mediated cell death in BL cells is regulated via the mitochondrial intrinsic apoptosis pathway, which is dependent on the activation of BAX and/or BAK. TGF-β directly induces transcription of the BH3-only protein BIK and represses expression of the pro-survival factor BCL-XL but has no effect on the direct BAX/BAK “activators” BIM or BID (tBID). Here we show that TGF-β induces the BH3-only activator PUMA to aid induction of the intrinsic cell death pathway. TGF-β also induced PUMA in normal germinal center CD77-positive centroblasts isolated from human tonsil tissue. PUMA was a direct TGF-β target gene in B-cells, and we identify a putative Smad-binding region within the human PUMA promoter that recruits Smad3 and Smad4 in cells in response to TGF-β signaling. Constitutive activity of the isolated Smad-binding region in luciferase reporter assays was dependent on Smad consensus sequences and was partially dependent on endogenous TGF-β signaling and Smad4. Knockdown of PUMA in BL cells using lentiviral shRNA resulted in slower kinetics of the TGF-β-mediated apoptotic response. Analysis of Eμ-Myc cell lines demonstrated that c-myc-driven murine lymphomas are also sensitive to TGF-β-mediated apoptosis. Moreover, Puma−/− Eμ-Myc lines demonstrated significantly delayed kinetics of the apoptotic response when compared with wild type lymphomas. TGF-β therefore induces a polygenic response in Myc-driven lymphomas involving transcription of PUMA, which is necessary for the rapid induction of cell death.
Introduction
TGF-β is a potent immune regulator, controlling the differentiation, function, and survival of hemopoietic cells including B- and T-cells (1). Control of B-cell homeostasis is mediated through the TGF-β-dependent regulation of cell cycle arrest and apoptosis. We have previously shown that TGF-β signaling contributes to “death by neglect” of spontaneously apoptosing human germinal center centroblasts, as well as inducing apoptosis of malignant lymphomas derived from the germinal center B-cells (c-Myc transformed Burkitt's lymphoma (BL)5 cells) (2, 3). This apoptosis program is induced by the intracellular signaling cascades activated by the binding of TGF-β to the high affinity TGF-β type II receptor. Binding of ligand to the TGF-β type II receptor enables the formation of a heterotetrameric complex between the constitutively active serine/threonine kinase type II receptor and the type I receptor, ALK5. The TGF-β type II receptor phosphorylates and activates ALK5, thereby inducing the canonical Smad pathway and/or several non-Smad pathways (4, 5). ALK5 activation results in C-terminal phosphorylation of the receptor-regulated Smads, Smad2 (Ser-465/476) and Smad3 (Ser-433/435), which then bind to the co-Smad, Smad4. The resulting heteroligomeric Smad complexes accumulate within the nucleus to control expression of genes involved in the regulation of cell survival and apoptosis (6).
In BL cells, TGF-β-induced apoptosis is associated with changes in expression levels of apoptosis regulators upstream of BAX and BAK. Activated BAX and BAK homo-oligomerize in the mitochondrial membrane to cause mitochondrial outer membrane permeabilization (7) and the release of pro-apoptotic factors. The function of BAX and BAK is tightly regulated by the BCL-2 family of proteins, including the pro-survival factors BCL-2 and its homologues, BCL-XL, MCL-1, BFL-1, BOO, and BCL-w. Other members of the family share one region of homology with BCL-2 (BH3-only proteins), are pro-apoptotic, and include BIK, PUMA, BID, NOXA, BIM, BAD, HRK, and BMF. The “activators” of BAX and BAK (8) include BIM, tBID, and PUMA (8–10), which are proposed to directly bind to BAX and BAK in response to apoptotic stimuli. The pro-survival factors such as BCL-2 prevent apoptosis by sequestering the activator proteins, but they themselves may be inhibited by selective interaction with BH3-only proteins (11). These proteins are therefore referred to as apoptosis “sensitizers,” which may either liberate activators from the pro-survival factors to enable BAX/BAK activation (12) or, alternatively, block the direct interaction of pro-survival factors with BAX and BAK (13).
Of these BCL-2 family members, TGF-β induces Smad-dependent transcription of the sensitizer BIK and down-regulation of the pro-survival factor BCL-XL in BL cells (3), however, no regulation of the activators BIM or tBID was evident during initiation of the apoptosis program. PUMA (p53-up-regulated modulator of apoptosis) is therefore the only other direct BAX/BAK activator that may have a role in promoting TGF-β-induced apoptosis in this system. PUMA physically associates with BAX via its BH3 domain (residues 127–150) to induce an apoptotic change in conformation of BAX (10), as well as promoting BAX-dependent apoptosis by dissociating BCL-XL from BAX (10, 14). PUMA is transcriptionally regulated by the p53 family (15–17), NF-κB (18), and E2F-1 (19), by activated FOXO3a following cytokine withdrawal (20) and in response to calcium pool depletion of the endoplasmic reticulum (21).
The tumor suppressor functions of p53 are rate-limiting for lymphomagenesis in Eμ-Myc murine models of BL. PUMA plays a major role in both p53-mediated and p53-independent cell death (22, 23) and is required for oncogene-induced cell death caused by c-Myc overexpression (22, 24). Consequently, genetic deletion of Puma accelerates the development of Eμ-Myc lymphomas (24), and the p53/PUMA pathway is often disrupted in Eμ-Myc and BL lymphomas, either by mutation or silencing of p53, by disruption of key p53 regulators (p19ARF and MDM2) (25), or by epigenetic silencing of PUMA itself (24). PUMA is also elevated in activated human and murine primary B-cells and is involved in the regulation of mature B-cell survival following T-cell-dependent B-cell response to antigen (26).
Because activation of PUMA effectively limits lymphomagenesis, in this study we investigate whether PUMA is a downstream effector of TGF-β-induced apoptosis of human and murine c-Myc driven lymphomas. We identify PUMA as a novel direct target of TGF-β signaling in B-cells that mediates rapid induction of apoptosis in response to TGF-β.
EXPERIMENTAL PROCEDURES
Cell Lines and Reagents
BL cell lines were maintained in RPMI 1640 (Invitrogen) supplemented with 5–10% (v/v) heat-inactivated FCS, 2 mm glutamine, and antibiotics. Murine Eμ-Myc lymphoma lines were generated as detailed previously (27) and maintained in DMEM (Invitrogen) supplemented with 10% heat-inactivated FCS, 2 mm glutamine (Invitrogen), 1 mm pyruvate (Invitrogen), 50 μm β-mercaptoethanol (Sigma), 200 μm asparagine (Sigma), and antibiotics. Analyses were performed on four cell lines derived from four independent Eμ-Myc lymphomas isolated from Puma+/+ mice and on four cell lines derived from four independent Eμ-Myc lymphomas isolated from Puma−/− mice. The cells were treated as required with 1–5 ng/ml TGF-β1 (Peprotech), 50 μm Z-VAD.FMK (Calbiochem), 5 μg/ml etoposide (Sigma), or 10 μm SB-431542 (Tocris). Protein synthesis inhibition was carried out by preincubation for 2 h with 50 μg/ml cycloheximide and 100 μm anisomycin (Sigma). Inhibition of transcription was carried out by pretreatment of cells for 1 h with 2.5 μg/ml actinomycin D (Sigma). Inhibition of c-Myc activity was carried out by treatment of cells with 25 μm of the c-Myc selective inhibitor 10058-F4 (5-[(4-ethylphenyl) methylene]-2-thioxo-4-thiazolidinone) (Sigma).
Transient Transfection of CA46 BL Cells
5 × 106 CA46 BL cells were transfected in triplicate with 3 μg of firefly luciferase reporter plasmid and 1 μg of Renilla luciferase or β-galactosidase expression plasmids as internal controls for transfection efficiency using an Amaxa nucleofector (solution T, program A-23) as recommended by the manufacturer. Treatment with SB-431542 (10 μm) or solvent vehicle was carried out after transfection if required. Transfected cells were incubated overnight before being treated with TGF-β (5 ng/ml) for 6 h. The ratio of firefly/Renilla luciferase activity was determined using the dual luciferase assay system (Promega) and expressed as the means ± S.D. ratio relative to untreated control samples. The Smad4 shRNA plasmid (TRC 40028) was obtained from Open Biosystems.
Isolation of Centroblasts
CD77+ve cells were purified as described previously (3). Following isolation, the cells were incubated with or without TGF-β in the presence of z-VAD-fmk.
Immunoblotting and Antibodies
Radioimmune precipitation assay lysates were analyzed by SDS-PAGE. Antibodies used in Western blotting were mouse monoclonals against PARP (BD Biosciences); Smad2/3 (BD Biosciences); actin (Sigma); rabbit polyclonal antibodies recognizing phospho-Smad 2 (Ser-465/476), phospho-Smad3 (Ser-433/435), and PUMA (Cell Signaling/Enzo Life Sciences); and a rabbit monoclonal against Tubulin (Cell Signaling). Secondary HRP-conjugated antibodies were obtained from Dako/GE Biosciences. Bound immunocomplexes were detected by enhanced chemiluminescence (ECL; Amersham Biosciences) or Immobilon western chemiluminescent substrate (Millipore).
Analysis of Apoptosis by Flow Cytometry
The cells were fixed in 80% ethanol, labeled with propidium iodide and analyzed by flow cytometry for less than 2 n DNA content. Murine lymphoma lines were stained with FITC-conjugated annexin V (1 μg/ml, kindly provided by Dr. P. Duriez) and propidium iodide (5 μg/ml) in binding buffer (10 mm HEPES, pH 7.4, 140 mm NaCl, 2.5 mm CaCl2) and analyzed by flow cytometry.
Lentiviral Infection of BL Cell Lines
HEK293FT cells were co-transfected with lentiviral packaging plasmids and pLKO.1-based shRNA constructs (TRC 33610 or TRC 33613; Open Biosystems, termed sh1 and sh2). Lentivirus-containing supernatants were collected, filtered, pooled, and used to infect BL cells. After 48 h, the cells were selected in 0.4 μg/ml puromycin. The resulting stably transduced BL30 cells were routinely passaged in growth medium (RPMI 1640 plus 20% FCS) containing 0.6 μg/ml puromycin. Experiments to analyze TGF-β-induced apoptosis were carried out in the absence of selection medium.
Retroviral Infection of Murine Eμ-Myc Lymphoma Lines
Ecotropic Phoenix cells were treated with 5 μm chloroquine 30 min prior to calcium: phosphate precipitation-mediated transfection with the pMIH vector (modified pMIG, GFP cassette exchanged for hygromycin resistance) encoding a human BCL-2 transgene, or vector alone, alongside the helper vector pCL.Eco. After 48 h, virus-containing supernatants were removed and filtered, and Polybrene was added to 4 μg/ml. Eμ-Myc lymphomas were plated at 6 × 105 cells/well in a 12-well plate, and 1.5 ml of virus-containing medium was applied. The plates were centrifuged at 480 × g at 37 °C for 45 min. Selection was then carried out using 250 μg/ml hygromycin for 7 days. Subsequently, BCL-2 expression levels were analyzed by flow cytometry and Western blot.
qRT-PCR
Total cell RNA was isolated using TRIzol as recommended by the manufacturer. cDNA and qRT-PCRs were prepared using SYBR green two-step qRT-PCR kits (Finnzymes) and specific primers for each gene (Qiagen). Amplified products were analyzed by Chromo4 continuous fluorescence detector and Opticon Monitor3 software. The relative amount of RNA for each gene is expressed after normalization to the amount of 18 S rRNA in each sample. RNA isolated from murine lymphomas was converted to cDNA using a first strand synthesis RT-PCR kit (Invitrogen). Quantitative PCRs were performed using qPCR Supermix-UND (Invitrogen) and TaqMan gene-specific primers (Applied Biosystems). Amplification and analysis was achieved via use of a C1000 thermal cycler fitted with a CFX96 fluorescence detector (Bio-Rad) and CFX96 manager software (Bio-Rad).
ChIP Assay
2 × 107 CA46 cells were treated with 1 ng/ml TGF-β for 1 h and fixed in 1% formaldehyde for 15 min. The formaldehyde was quenched with glycine, the cells were washed in PBS, and nuclear extracts were prepared. The extracts were sonicated to fragment the DNA and incubated overnight with 20 ng of goat anti-Smad3 or 4 (Santa Cruz) or isotype-matched control antibodies. Immune complexes were captured using anti-goat IgG-coated agarose beads, the samples were washed, and the DNA was eluted off the beads. DNA cross-links were reversed by incubation at 65 °C in 0.3 m NaCl, and the samples were then digested with proteinase K. DNA was extracted using phenol/chloroform and ethanol-precipitated. PCRs using specific primers for human PUMA (5′-AGACTTGCTTGAACCCAGGA-3′ and 5′-AATCGCCTCACCAGAGTGAC-3′) were performed for 35–50 cycles, and the PCR products were analyzed on 2% agarose gels stained with SYBR Green.
RESULTS
TGF-β Induces the BH3-only Activator PUMA
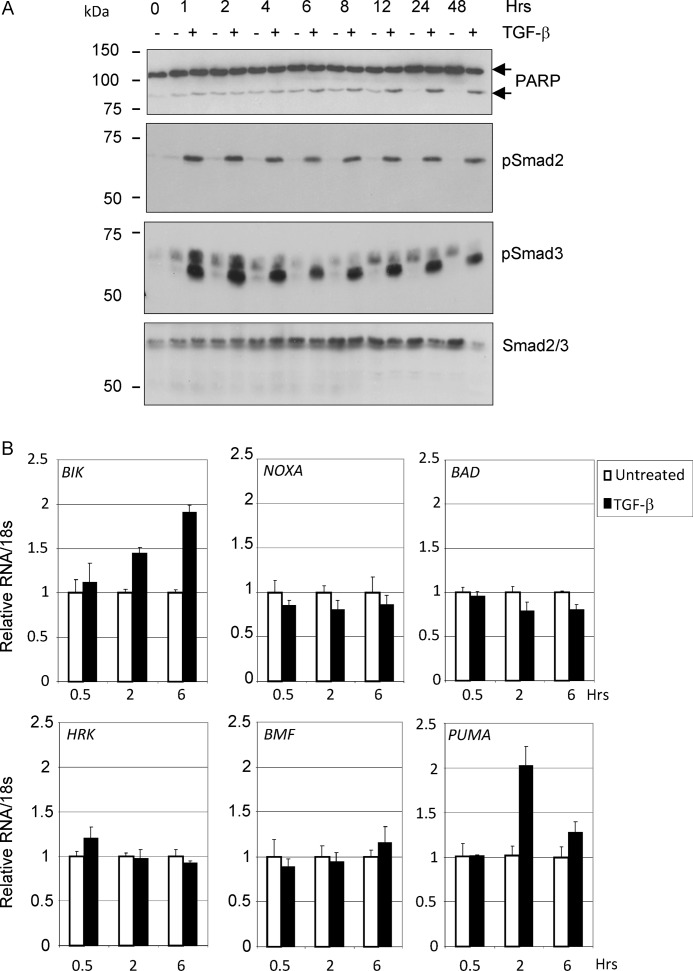
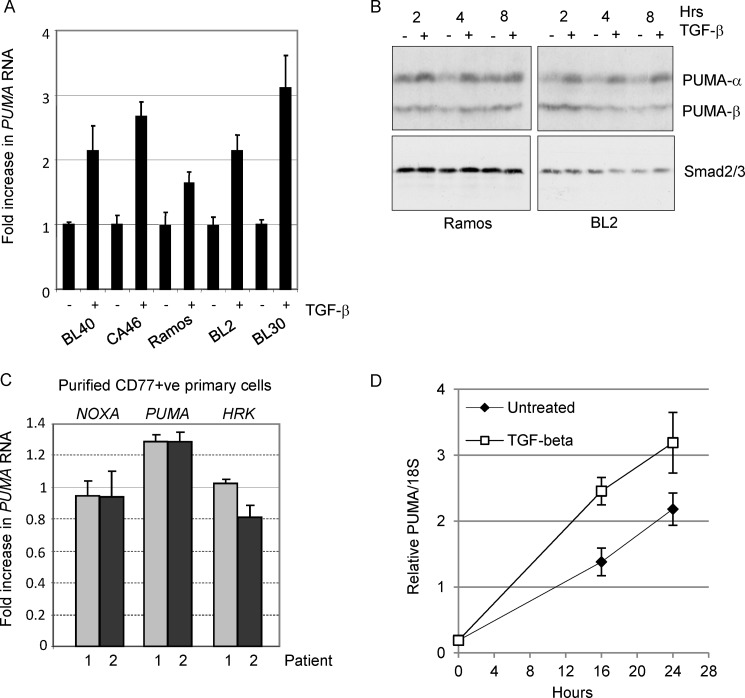
We previously showed that TGF-β treatment of BL cells results in the transcriptional regulation of members of the BCL-2 family of apoptosis regulators including BIK and BCL-XL (3). Analysis of other BH3-only proteins that might contribute to the apoptotic program induced by TGF-β was carried out on subsequent TGF-β treatment time courses (Fig. 1). Apoptosis was evident, as detected by Western blotting for cleavage of the caspase-3 substrate PARP, as early as 6–8 h after TGF-β addition (Fig. 1A). During this period, the induction of BIK mRNA was confirmed; however, we also detected a transient increase in transcripts of the apoptosis activator PUMA (Fig. 1B). There was no concurrent increase in expression levels of the apoptosis sensitizers, NOXA, BAD, HRK, or BMF. The induction of PUMA mRNA in response to exogenous TGF-β addition was confirmed across a panel of BL cell lines (Fig. 2A) and at the level of protein expression (Fig. 2B). The Western blot analysis shown in Fig. 2B indicates that the α isoform but not the β isoform of PUMA was induced in response to TGF-β signaling.
FIGURE 1.
Transcripts of PUMA and BIK are elevated in TGF-β-treated Ramos BL cells. A, time course of TGF-β-induced apoptosis in human BL cells (Ramos). The cells were treated with 1 ng/ml TGF-β for the times indicated. Radioimmune precipitation assay lysates were analyzed by Western blotting for TGF-β signaling (phosphorylation of Smad2 and Smad3) and apoptosis (cleavage of PARP). A Western blot for total Smad2/3 is included as a loading control. B, quantitative real time qRT-PCR analysis of BH3-only members of the BCL-2 family in TGF-β-treated Ramos cells. The mean (± S.D.) fold increases relative to untreated cells (set at 1) are shown.
FIGURE 2.
PUMA is induced by TGF-β in human BL cell lines and in normal tonsil CD77-positive B-cells. A, real time qRT-PCR analysis of PUMA in a panel of TGF-β responsive BL cell lines following 2 h of treatment. The mean (± S.D.) fold increase above background (set at 1 in untreated cells) is shown. B, Western blot analysis of radioimmune precipitation assay extracts from BL cells either untreated or treated with TGF-β for 2–8 h. Immunoreactive bands showing both the TGF-β-inducible PUMA-α (23 kDa) and PUMA-β isoforms are indicated. A Western blot for total Smad2/3 is included as a loading control. C and D, real time qRT-PCR analysis of RNA isolated from human primary CD77-positive tonsil B-cells treated with z-VAD.FMK and TGF-β for 2 h (C) and over an extended 24-h time course of TGF-β treatment (D). The mean (± S.D.) fold TGF-β-induced increase in normalized RNA expression compared with untreated cells (set at 1) is shown (C). The mean (± S.D.) relative levels of PUMA RNA normalized to 18 S are shown in D.
BLs are derived from centroblasts within the germinal centers of secondary lymphoid tissue. By isolating CD77-positive centroblasts from human tonsil tissue (as described in Ref. 3), we were able to demonstrate a small but consistent increase in PUMA mRNA expression in TGF-β treated cells relative to untreated controls. No comparable increase in either NOXA or HRK mRNA expression was evident (Fig. 2C). Increased abundance of PUMA RNA transcripts in the presence of TGF-β was also evident throughout a 24-h time course of treatment of CD77-positive primary cells (Fig. 2D). PUMA can therefore be induced in response to TGF-β in both BL cell lines and in the cell of origin isolated from primary tissue. Induction of PUMA mRNA in response to exogenous TGF-β addition could also be detected in the pro-B-cell line RS4;11, but not in non-B-cell lineages including mouse embryo fibroblasts and HaCaT epithelial cells (data not shown).
PUMA Is a Direct Target Gene of TGF-β Signaling in Human B-cells
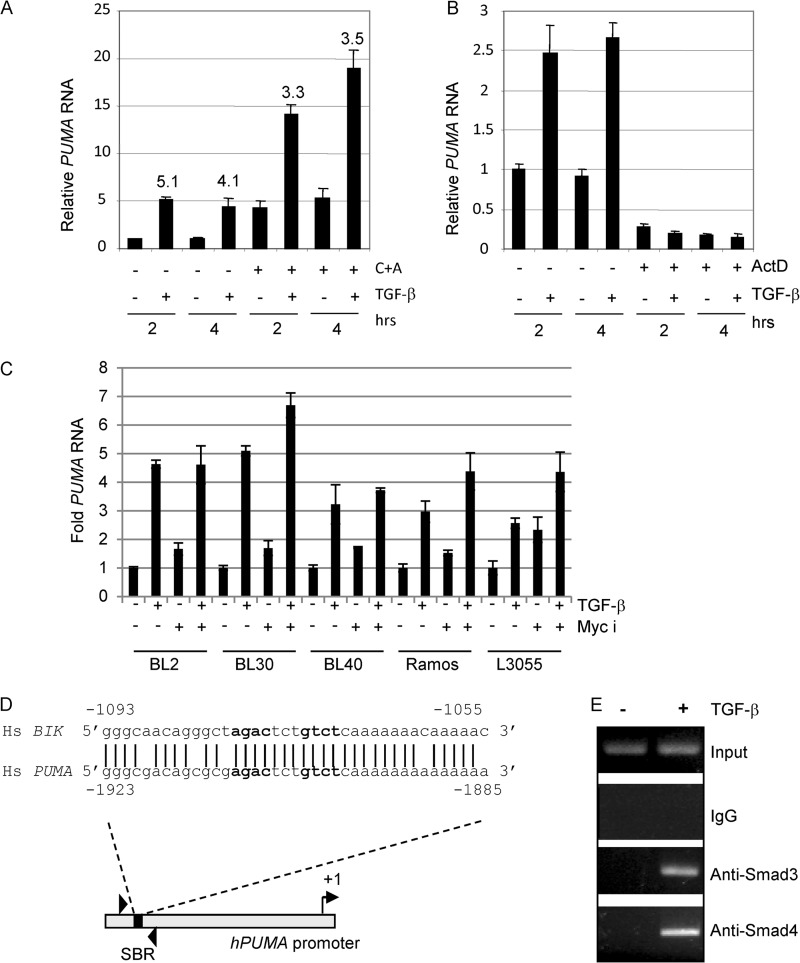
We next investigated the mechanism of TGF-β-mediated regulation of PUMA expression first by testing whether PUMA is an immediate early target of TGF-β signaling. BL cells were pretreated with cycloheximide and anisomycin and were subsequently treated with exogenous TGF-β for the times indicated. Induction of PUMA occurred even in the absence of de novo protein synthesis (Fig. 3A), but blocking transcription with actinomycin D resulted in the complete ablation of PUMA mRNA induction (Fig. 3B). c-Myc overexpression is characteristic of BL cells, and c-Myc itself is known to repress PUMA transcription following growth factor stimulation (28). We tested the effect of c-Myc overexpression on TGF-β-induced activation of PUMA by pretreating BL cells with the c-Myc inhibitor 10058-F4 at concentrations previously shown to induce BL cell death (29). Inhibition of c-Myc modestly increased basal PUMA transcription (especially in L3055 cells) but did not significantly affect PUMA induction in response to TGF-β (Fig. 3C). Our data indicate that PUMA is a direct target of TGF-β signaling and, as such, implies that PUMA transcription is likely to be regulated by Smad complexes binding to a cognate binding element within the gene promoter. Smad-binding regions (SBRs) frequently contain two copies of the Smad-binding element (SBE) sequence 5′-GTCT-3′ or its reverse complement 5′-AGAC-3′ (30). We identified a potential SBR (5′-agactctgtct-3′) (Fig. 3D) ∼1.9 kb upstream of the PUMA transcription start site, contained within a region of the promoter bearing striking homology to the SBR we previously identified within the human BIK promoter (Fig. 3D and Ref. 3). ChIP analysis using primers spanning this region demonstrated TGF-β-dependent binding of Smads 3 and 4 to the endogenous PUMA promoter in vivo (Fig. 3E).
FIGURE 3.
TGF-β directly regulates hPUMA transcription and induces recruitment of activated Smad complexes to the hPUMA promoter. A, real time qRT-PCR for PUMA expression in CA46 cells treated with or without TGF-β and the protein synthesis inhibitors cycloheximide and anisomycin (C+A) as indicated. The cells were pretreated with cycloheximide and anisomycin for 2 h, and TGF-β was added at 0 h. The mean (± S.D.) PUMA RNA expression levels relative to the 2-h untreated sample is shown (2-h untreated sample value was set at 1). The fold induction relative to the untreated sample at each time point is shown. B, real time qRT-PCR for PUMA expression in CA46 cells untreated or pretreated for 1 h with the transcription inhibitor actinomycin D (ActD), followed by a time course of TGF-β treatment. RNA expression is expressed relative to the levels detected in the 2-h untreated sample (assigned a value of 1). C, a panel of BL cell lines was pretreated for 30 min with 25 μm of the c-Myc inhibitor (Myc i) 10058-F4 followed by 2 h of TGF-β treatment (1 ng/ml). RNA was extracted and analyzed by qPCR for PUMA expression. Expression levels were normalized to the internal standard 18 S RNA and are shown as the mean (± S.D.) fold expression level relative to untreated controls (set at 1). D, sequence of a known SBR within the human BIK promoter (3) compared with a putative Smad-binding element within the human PUMA promoter. The putative PUMA SBR at position −1923 to −1885 was analyzed by ChIP using specific primers encompassing this region. E, ChIP assay for Smad recruitment to the endogenous PUMA promoter in CA46 cells cultured with and without TGF-β for 1 h. Input lanes are from 10% of samples used in the IPs performed with control IgG, Smad3, and Smad4 antibodies.
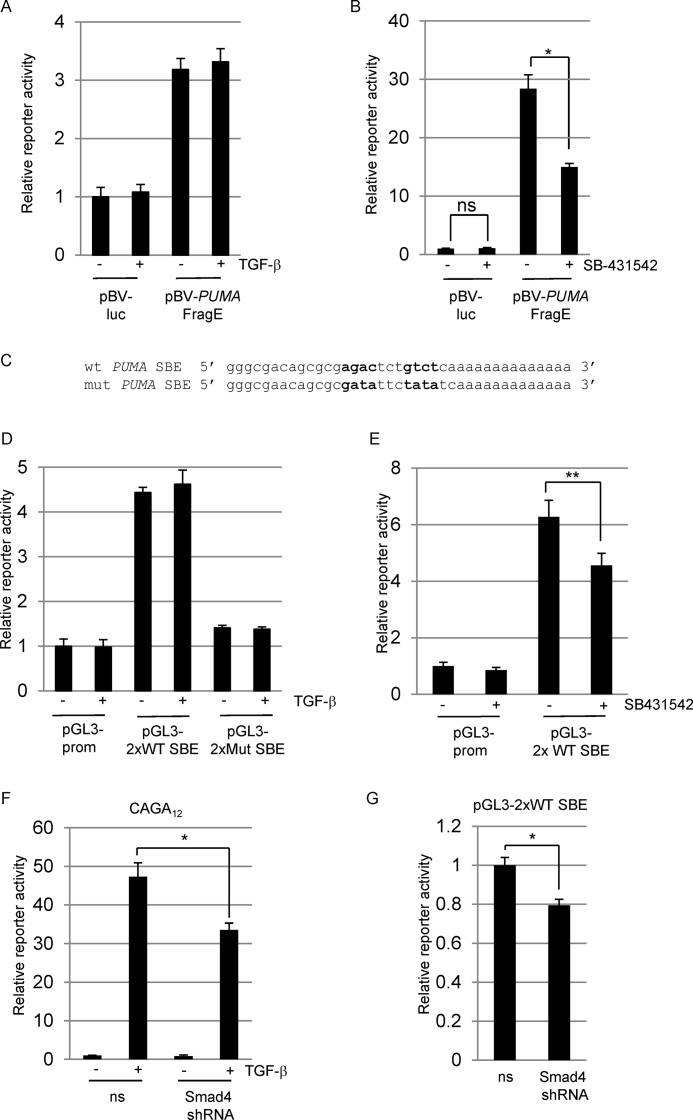
We next analyzed the activity of a luciferase promoter reporter construct containing ∼2 kb of the 5′ region upstream of the PUMA transcription start site (31). The PUMA promoter construct was constitutively active in CA46 BL cells and could not be further induced by the addition of exogenous TGF-β (Fig. 4A), suggesting that the endogenous level of TGF-β signaling in these cells may be sufficient for maximal activity of the isolated promoter region. We therefore tested whether inhibition of basal TGF-β signaling using 10 μm of the ALK5 inhibitor SB-431542 would affect PUMA promoter activity (Fig. 4B). We observed an approximate 50% decrease in luciferase activity following SB-431542 treatment, indicating that TGF-β signaling is required for maximal promoter activity.
FIGURE 4.
hPUMA promoter SBR luciferase reporter constructs. A and B, transient transfection of CA46 BL cells with control reporter vector pBV-luciferase (pBV-luc) or pBV containing the human PUMA promoter sequence spanning ∼2 kb upstream from the transcription start site (pBV-PUMA FragE (31)). Transfected cells were treated with 5 ng/ml TGF-β (A) or 10 μm SB-431542 (B) as indicated. C, wild type and mutant SBE reporter plasmids were generated by cloning 2× concatemerized oligonucleotides of the sequences shown into the pGL3-promoter vector (Promega). D and E, the wild type SBE (pGL3–2×WT SBE) and the mutant SBE (pGL3–2xMut SBE) reporter constructs were assayed following exogenous TGF-β addition (D) or addition of SB-431542 (E) as indicated. F and G, CAGA12 (F) or pGL3–2×WT SBE (G) constructs were transfected into CA46 cells with either nonsilencing vector (ns) or shRNA vector targeting Smad4. After 48 h, the transfections were left untreated or treated with 5 ng/ml TGF-β. Transfection results throughout are expressed as the means (± S.D.) (n = 3) luciferase reporter activity relative to untreated control samples (set at 1). All data shown are normalized to either co-transfected Renilla luciferase or β-galactosidase activity.
To examine the activity of the potential Smad-binding region more directly, we generated firefly luciferase reporter constructs containing concatemerized (2×) versions of wild type and mutant sequences shown in Fig. 4C. Consistent with the 2-kb promoter fragment, the 2× wild type SBE sequence was active in CA46 cells. This activity was dependent on the consensus SBEs because mutation of the sequence agactctgtct to atattctatat reduced reporter activity to near basal levels (Fig. 4D). Inhibition of endogenous TGF-β signaling also reduced activity of the 2× wild type SBE reporter (Fig. 4E).
We next transiently transfected a shRNA construct targeting Smad4 to determine whether Smad4 is required for PUMA promoter activity. Because BL cells are renowned for their low transfection rate, we monitored efficacy of Smad4 knockdown by inhibition of the Smad4 dependent CAGA12-luciferase reporter response to TGF-β addition (Fig. 4F). We were able to achieve a partial inhibition of CAGA12 activity and likewise observed a small but significant decrease in PUMA activity (Fig. 4G). Taken together, these findings indicate that regulation of the PUMA promoter by TGF-β signaling is most likely dependent on Smad binding at the SBR at positions −1923 to −1885 and that proper regulation of the endogenous gene depends on the correct repressive chromatin context.
PUMA Knockdown Retards TGF-β-induced Apoptosis
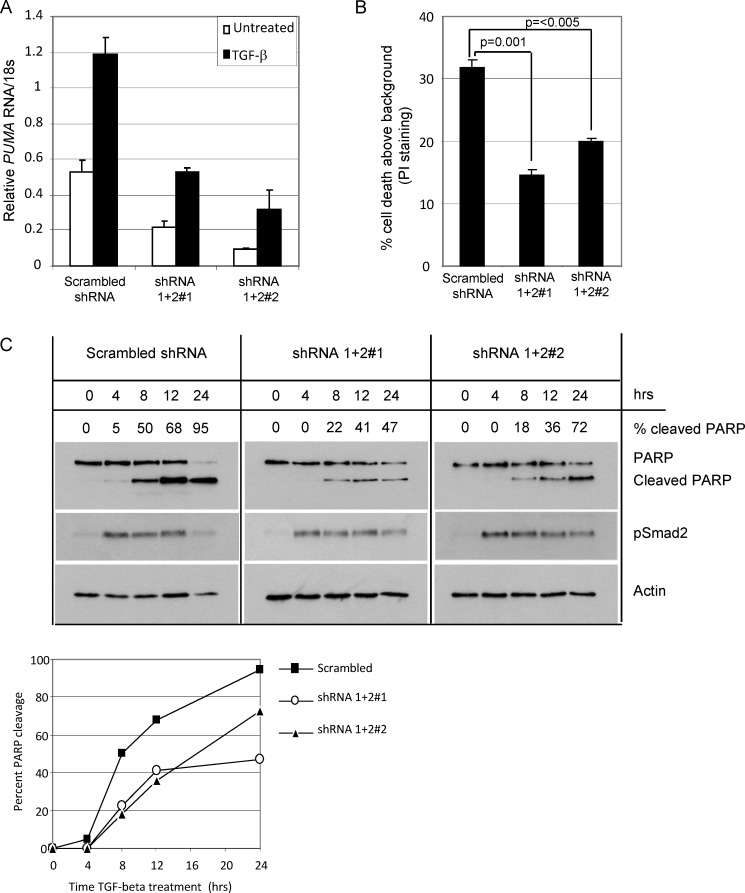
To determine the functional consequence of PUMA induction by TGF-β, we used lentiviral transduction of shRNA constructs to stably knockdown PUMA in BL30 cells. Transducing BL30 cells with a combination of two commercially available shRNA constructs (shRNA1 + 2) reduced the basal and TGF-β-inducible PUMA levels to below the background levels observed in the control line (Fig. 5A). Two PUMA knockdown stable cell pools (shRNA1 + 2#1 and shRNA1 + 2#2) were analyzed for apoptosis induction by TGF-β using propidium iodide staining and flow cytometry. Knockdown of PUMA resulted in a significant reduction in the amount of apoptosis induction by TGF-β after 24 h of treatment (Fig. 5B). We noted, however, that at later time points (48 h) there was little discernable difference between apoptosis induction in control versus PUMA knockdown cell lines (data not shown). We therefore carried out a more detailed kinetic analysis of early apoptosis induction using PARP cleavage as a measure (Fig. 5C, top panel). The Western blots revealed that although all lines retained equivalent TGF-β signaling, shown by equal levels of phosphorylation of the receptor regulated Smad2, the apoptotic response in PUMA knockdown lines was retarded when compared with the control line, 22 and 18% PARP cleavage in the knockdown lines after 8 h of treatment compared with 50% (summarized in the graph shown in Fig. 5C, bottom panel).
FIGURE 5.
Knockdown of PUMA retards TGF-β-induced apoptosis in BL lines. A, independent stable cell lines expressing nonsilencing (Scrambled) or shRNAs targeted against PUMA (shRNA1 + 2#1 and shRNA1 + 2#2) were generated by lentiviral infection of BL30 cells. Real time qPCR was used to demonstrate the efficacy of knockdown of PUMA in stable BL30 cells in untreated cells and cells treated for 2 h with TGF-β. Mean relative amounts of PUMA RNA are expressed after normalization to 18 S rRNA levels. B, determination of apoptosis by propidium iodide (PI) staining and flow cytometry following 24 h TGF-β treatment of the cell lines described in A. Statistical analysis was performed using Student's t test. C, Western blot analysis of apoptosis (PARP) and TGF-β signaling (p-Smad2) in BL30 stable cell lines described in A and B during a time course of TGF-β treatment. A Western blot for actin is included as a loading control. The amount of cleaved PARP as a percentage of total PARP determined by densitometry is indicated above each lane and is summarized in the graph shown below.
TGF-β Induces Apoptosis and PUMA Expression in Eμ-Myc Murine Lymphoma Lines
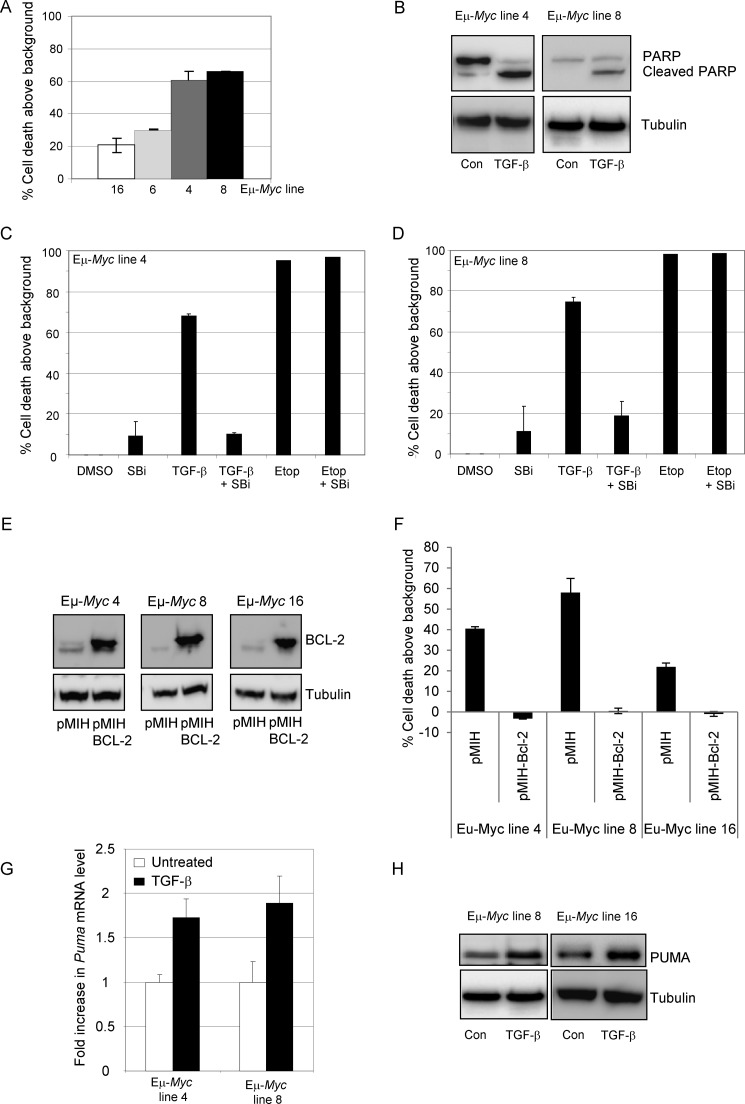
Given that PUMA expression levels in the stable knockdown cell lines were reduced to below the basal levels detected in untreated controls (Fig. 5A), the lack of any significant effect on the percentage of apoptotic cells at later time points is unlikely to be a consequence of the incomplete inhibition of PUMA induction in response to TGF-β. However, to exclude this possibility, we extended our study to compare wild type and Puma null murine B-lymphoma cells. Eμ-Myc murine lymphoma lines are a model of BL. We first determined whether isolated wild type Eμ-Myc lymphoma cells undergo apoptosis in response to TGF-β addition in vitro. Annexin V staining of Eμ-Myc cells revealed TGF-β-induced cell death after 24 h of treatment (Fig. 6A). Apoptosis was confirmed by cleavage of PARP in TGF-β-treated wild type lymphoma cells (Fig. 6B). Cell death in response to TGF-β was dependent on the TGF-β type I receptor ALK5 because pretreatment of lymphoma cells with SB-431542 inhibited death induced by TGF-β but had no effect on the cellular response to the DNA damage-inducing agent etoposide (Fig. 6, C and D). Apoptosis was inhibited by overexpression of BCL-2 in murine lymphoma lines, indicating the involvement of the intrinsic apoptosis pathway (Fig. 6, E and F). In addition, TGF-β treatment resulted in induction of Puma shown by quantitative RT-PCR (Fig. 6G) and by Western blot (Fig. 6H). These data indicate that wild type murine Eμ-Myc lymphoma cells mimic the response of human BL cell lines to TGF-β and undergo ALK5-dependent intrinsic apoptosis involving PUMA induction.
FIGURE 6.
Murine Eμ-Myc lines are sensitive to TGF-β-induced apoptosis. A, murine Eμ-Myc cell lines were analyzed by annexin V staining and flow cytometry for their response to TGF-β treatment (24 h). B, Western blot analysis of lysates from Eμ-Myc cell lines showing cleavage of PARP following 12 h of treatment with TGF-β. C and D, murine Eμ-Myc line 4 (C) and line 8 (D) were treated for 24 h with TGF-β or etoposide (5 μg/ml) in the presence or absence of a TGF-β RI (ALK5) inhibitor, SB-431542 (SBi, 10 μm). Apoptosis induction was determined by annexin V staining and flow cytometry and is expressed as the mean (± S.D.) percentage induction above background levels. E and F, Eμ-Myc cell lines stably transfected with empty vector (pMIH) or Bcl-2 expressing vector (pMIH-Bcl-2) were analyzed by Western blot for Bcl-2 expression (E) and by annexin V staining (F) for susceptibility to TGF-β induced apoptosis following 24 h of treatment. G, wild type Eμ-Myc cell lines were left untreated or treated with TGF-β for 2 h and analyzed by qRT-PCR for Puma mRNA expression levels. The results are expressed as the mean fold mRNA level relative to the untreated control. H, Western blot analysis of PUMA expression in untreated (Con) wild type Eμ-Myc lines or lines treated for 6 h with TGF-β. A blot for tubulin is included as a loading control. DMSO, dimethyl sulfoxide.
PUMA Is Required for Efficient Apoptosis Induction in Eμ-Myc Lymohoma Lines
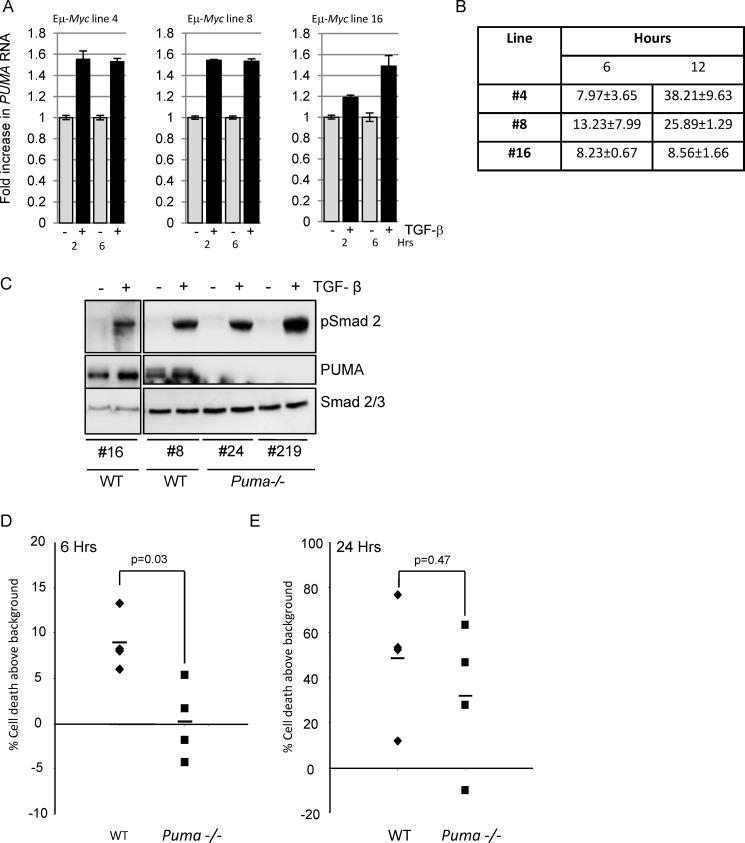
shRNA knockdown of PUMA in human BL cell lines delayed the onset of apoptosis in response to TGF-β (Fig. 5C). We observed a similar correlation in the murine Eμ-Myc line 16, which had slower Puma RNA induction after TGF-β addition (Fig. 7A) and lower levels of apoptosis by 12 h of TGF-β treatment (Fig. 7B). However, to definitively investigate the role of PUMA in this response, we compared the response of wild type Eμ-Myc cell lines with Puma null lines (Puma−/−) to TGF-β. All lines responded to TGF-β because they showed similar levels of Smad2 phosphorylation following exogenous TGF-β addition (Fig. 7C). When the effects of TGF-β treatment were directly compared in wild type versus Puma null cells, there were significantly fewer apoptotic Puma null cells than apoptotic wild type cells at early time points following exposure to TGF-β (Fig. 7D) but not at later time points (Fig. 7E), thus confirming the data obtained in human BL lines. PUMA is therefore selectively required for efficient, rapid TGF-β-induced apoptosis.
FIGURE 7.
Eμ-Myc cell lines derived from Puma−/− mice exhibit a delayed apoptotic response to TGF-β. A, kinetics of TGF-β-mediated Puma RNA induction in wild type Eμ-Myc lymphoma lines as determined by qPCR. B, early kinetics of apoptosis induction in wild type Eμ-Myc lymphoma lines as measured by annexin V staining. C, Western blot analysis of TGF-β signaling (pSmad2) and PUMA expression in WT and Puma null (Puma−/−) Eμ-Myc lymphoma lines in uninduced or induced conditions (6 h of treatment with TGF-β). A blot for total Smad2/3 is included as a loading control. D and E, apoptosis induction above background in lymphoma lines derived from WT (n = 4) and homozygous Puma null (Puma−/−) (n = 4) mice following 6 h (D) and 24 h (E) of treatment with TGF-β. Apoptosis induction above background was determined by annexin V staining and flow cytometry. The mean apoptosis values are shown as bars, and the p values were determined by Mann-Whitney statistical analysis.
DISCUSSION
In this study we demonstrate a direct link between pro-apoptotic TGF-β signaling and transcriptional up-regulation of the apoptosis activator PUMA in c-Myc-driven B-cell lymphomas. PUMA was required for the effective, early induction of apoptosis in both human and murine lymphomas, but its induction does not require c-Myc activity. The involvement of PUMA in this process was demonstrated by stable knockdown of PUMA in human BL, but to be confident that we could assess the effect of complete lack of PUMA on TGF-β-induced apoptosis, we extended our analysis to compare Eμ-Myc lymphomas derived from Puma null and wild type mice. We believe this is the first demonstration that murine Eμ-Myc lymphomas are susceptible to TGF-β-induced apoptosis. Stroma-derived TGF-β from infiltrating immune cells has previously been reported to mediate a tumor suppressor function in vivo by inducing senescence of Eμ-Myc lymphoma cells (32). Our data suggest that in addition to senescence, TGF-β expressed in the tumor microenvironment would also be likely to induce ALK5-dependent apoptosis of malignant cells.
We report that a complete loss of PUMA in knock-out Eμ-Myc cells and knockdown of PUMA in human BL cells delay cell death but do not ultimately prevent cells from undergoing apoptosis. This observation is consistent with the idea that PUMA has a role in priming cells for apoptosis as part of a polygenic apoptotic response that may have a role in “sensing” the apoptotic signal (33). As well as PUMA activation, the apoptotic program involves the early activation of BIK in human cells followed by down-regulation of BCL-XL (3). Such changes in expression levels of the other BCL-2 family members presumably are sufficient to induce apoptosis, albeit with slower kinetics. As predicted previously, we have seen no TGF-β-dependent activation of BIK in mouse cells because the Smad-binding element identified within the human BIK promoter is not conserved (Ref. 3 and Fig. 3). BCL-XL, however, is down-regulated in response to TGF-β in murine Eμ-Myc lymphoma cells.6 This suggests that loss of BCL-XL function may ultimately be sufficient in murine lymphoma to induce apoptosis. but we cannot exclude the possibility that other BH3-only sensitizers might also be induced in response to TGF-β in the mouse.
The mechanism of PUMA induction by TGF-β in this system requires some further analysis. We have determined that PUMA is induced even in the presence of protein synthesis inhibitors, demonstrating that de novo protein synthesis is not required for PUMA transcriptional up-regulation in BL cells and also that in vivo, activated Smad3/Smad4 is recruited directly to a SBR within the endogenous promoter. Constitutive activity of the isolated putative Smad-binding region was dependent on consensus SBE sequences in transient transfection assays, and it was partially inhibited by blocking endogenous TGF-β signaling and lowering Smad4 levels; however, exogenous addition of ligand did not induce its activity. These data suggest that chromatin structure and/or transcriptional repressors may be important for correct regulation of the endogenous gene.
The induction of PUMA-mediated apoptosis by TGF-β superfamily members has been described previously in oligodendrocytes (induced by activin A but not by TGF-β) and by TGF-β in a gastric cancer cell line. In both cases, however, elevated PUMA levels were dependent on p53 family members. A p53 family inhibitor pifithrin-α blocked activin A-induced apoptosis of oligodendrocytes (34), whereas TGF-β-induced apoptosis of SNU-16 gastric cancer cells was due to the induction of p73, which presumably bound p53 consensus sequences on the PUMA promoter (35). In these studies, there was no evidence that Smads were recruited directly to the PUMA promoter as detected in BL cells. However, it is possible that a p53 family member might still be involved in TGF-β-induced regulation of PUMA in our system by acting as co-activators with the Smad proteins. Smads are regulated by numerous post-translational modifications and cooperate with many other transcription factors (including the p53 family) to regulate gene transcription, thus ensuring that their function is highly context dependent (6). p53 and p73, for example, directly bind Smads 2 and 3 and can stabilize Smad-DNA complexes on specific gene promoters (36). It would seem unlikely that p53 is involved in TGF-β/Smad activation of the PUMA promoter because all but one of the BL cell lines used in this study express mutant p53 (BL2 cells express wild type p53 (37)). Transcripts of p73 are expressed in BL cells but are not increased by TGF-β treatment.6
So far, direct activation of PUMA by TGF-β has been demonstrated in B-cell subsets (lymphomas and human primary centroblasts) but not in murine embryonic fibroblasts or HaCaT epithelial cells. The reason for the selective response in certain cell types is currently unclear. It is possible that the proteome of B-cells uniquely allows PUMA activation in response to TGF-β. The panel of BL cells examined here lack expression of SLUG, a transcription factor involved in the repression of PUMA transcription.6 Expression of SLUG in hematopoietic progenitor cells is responsible for protecting progenitor cells from DNA damage-induced apoptosis by repressing p53-mediated PUMA transcription (38). TGF-β may be able to induce PUMA in B-cells by virtue of its lack of repression. Another possibility is that PUMA is regulated by a B-cell-specific transcription factor in conjunction with TGF-β-activated Smads.
A study published during the preparation of this manuscript implicates the activation status of B-cells as a factor in PUMA production, with PUMA being involved in the regulation of antigen specific memory B-cells both in vitro and in vivo. PUMA protein expression was detected in histological sections of human lymph nodes, potentially as a result of antigenic stimulation, because stimulation of cells in vitro with the mitogens Staphylococcus aureus Cowan or LPS, or alternatively following ligation of the B-cell receptor or CD40, all resulted in up-regulation of PUMA. Provided sufficient pro-survival signals are received to maintain BCL-XL levels, the effect of PUMA induction in this system is negated (26). The mechanism of PUMA induction was not identified by Clybouw et al. (26) but was determined to be independent of p53 because p53−/− B-cells produced similar levels of PUMA to wild type cells.
Given our finding that PUMA is induced in vitro by TGF-β treatment of centroblasts (Fig. 2) and that active, phosphorylated Smad2 is detectable throughout the germinal center (3), we can speculate that PUMA detected in the tonsil sections could result from TGF-β signaling. This may arise as a result of the establishment of an activation-induced TGF-β-autocrine feedback loop (39). There are several lines of evidence to support this hypothesis. TGF-β mRNA is detectable in both resting and activated B-cells (40). Stimulation of human tonsil cells with S. aureus Cowan does not significantly affect TGF-β mRNA levels but does result in a 7-fold increase in TGF-β protein secretion (41). Similarly, LPS treatment of murine splenocytes induces the production of bioactive TGF-β (42). The induction of TGF-β in activated B-cells is thought to limit the expansion of the activated population. Without this autoregulatory feedback loop severe autoimmunity develops (43, 44). Our data presented here indicate that regulation of PUMA plays a role in the apoptotic response of B-cells exposed to TGF-β and suggest that this may play a role in the efficient control of activated B-cell proliferation. This notion may provide an explanation for the elevated levels of IgA observed in Puma null mice (45) because IgA class switching is promoted by TGF-β (46). The intriguing possibility that activation-induced PUMA production occurs because of an autocrine TGF-β/ALK5/Smad pathway and the relative role, mechanisms, and kinetics of regulation of BCL-XL and other BCL-2 family members in B-cell survival warrants further investigation.
Acknowledgments
We acknowledge Dr. P. Duriez (Cancer Research UK Protein Core Facility, Southampton) for the preparation and provision of annexin V-FITC (following the kind gift of the annexin V plasmid from Prof. S. Martin), as well as Dr. C. Scott and Prof. A. Strasser (Walter and Eliza Hall Institute) for the provision of murine lymphoma material, K. Cox and Dr. E. Williams for help with the generation and characterization of the Eμ-Myc cell lines, and Dr. G. V. Helgason for advice on the preparation of lentivirus.
This work was supported, in whole or in part, by National Institutes of Health Grants CA129829 and U01-DK085570 (to J. Y.).
L. C. Spender, unpublished observations.
- BL
- Burkitt's lymphoma
- SBR
- Smad-binding region
- SBE
- Smad-binding element
- Z-VAD.FMK
- benzyloxycarbonyl-Val-Ala-Asp (OMe) fluoromethylketone
- qRT-PCR
- quantitative RT-PCR
- PARP
- poly(ADP-ribose) polymerase
- PUMA
- p53-up-regulated modulator of apoptosis.
REFERENCES
- 1. Li M. O., Wan Y. Y., Sanjabi S., Robertson A. K., Flavell R. A. (2006) Transforming growth factor-β regulation of immune responses. Annu. Rev. Immunol. 24, 99–146 [DOI] [PubMed] [Google Scholar]
- 2. Inman G. J., Allday M. J. (2000) Apoptosis induced by TGF-β1 in Burkitt's lymphoma cells is caspase 8 dependent but is death receptor independent. J. Immunol. 165, 2500–2510 [DOI] [PubMed] [Google Scholar]
- 3. Spender L. C., O'Brien D. I., Simpson D., Dutt D., Gregory C. D., Allday M. J., Clark L. J., Inman G. J. (2009) TGF-β induces apoptosis in human B cells by transcriptional regulation of BIK and BCL-XL. Cell Death Differ. 16, 593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shi Y., Massagué J. (2003) Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 113, 685–700 [DOI] [PubMed] [Google Scholar]
- 5. Derynck R., Zhang Y. E. (2003) Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature 425, 577–584 [DOI] [PubMed] [Google Scholar]
- 6. Massagué J., Seoane J., Wotton D. (2005) Smad transcription factors. Genes Dev. 19, 2783–2810 [DOI] [PubMed] [Google Scholar]
- 7. George N. M., Evans J. J., Luo X. (2007) A three-helix homo-oligomerization domain containing BH3 and BH1 is responsible for the apoptotic activity of Bax. Genes Dev. 21, 1937–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Letai A., Bassik M. C., Walensky L. D., Sorcinelli M. D., Weiler S., Korsmeyer S. J. (2002) Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell 2, 183–192 [DOI] [PubMed] [Google Scholar]
- 9. Kim H., Rafiuddin-Shah M., Tu H. C., Jeffers J. R., Zambetti G. P., Hsieh J. J., Cheng E. H. (2006) Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat. Cell Biol. 8, 1348–1358 [DOI] [PubMed] [Google Scholar]
- 10. Gallenne T., Gautier F., Oliver L., Hervouet E., Noël B., Hickman J. A., Geneste O., Cartron P. F., Vallette F. M., Manon S., Juin P. (2009) Bax activation by the BH3-only protein Puma promotes cell dependence on antiapoptotic Bcl-2 family members. J. Cell Biol. 185, 279–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen L., Willis S. N., Wei A., Smith B. J., Fletcher J. I., Hinds M. G., Colman P. M., Day C. L., Adams J. M., Huang D. C. (2005) Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol. Cell 17, 393–403 [DOI] [PubMed] [Google Scholar]
- 12. Certo M., Del Gaizo Moore V., Nishino M., Wei G., Korsmeyer S., Armstrong S. A., Letai A. (2006) Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell 9, 351–365 [DOI] [PubMed] [Google Scholar]
- 13. Willis S. N., Fletcher J. I., Kaufmann T., van Delft M. F., Chen L., Czabotar P. E., Ierino H., Lee E. F., Fairlie W. D., Bouillet P., Strasser A., Kluck R. M., Adams J. M., Huang D. C. (2007) Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science 315, 856–859 [DOI] [PubMed] [Google Scholar]
- 14. Ming L., Wang P., Bank A., Yu J., Zhang L. (2006) PUMA dissociates Bax and Bcl-XL to induce apoptosis in colon cancer cells. J. Biol. Chem. 281, 16034–16042 [DOI] [PubMed] [Google Scholar]
- 15. Yu J., Zhang L., Hwang P. M., Kinzler K. W., Vogelstein B. (2001) PUMA induces the rapid apoptosis of colorectal cancer cells. Mol. Cell 7, 673–682 [DOI] [PubMed] [Google Scholar]
- 16. Nakano K., Vousden K. H. (2001) PUMA, a novel proapoptotic gene, is induced by p53. Mol. Cell 7, 683–694 [DOI] [PubMed] [Google Scholar]
- 17. Pyati U. J., Gjini E., Carbonneau S., Lee J. S., Guo F., Jette C. A., Kelsell D. P., Look A. T. (2011) p63 mediates an apoptotic response to pharmacological and disease-related ER stress in the developing epidermis. Dev. Cell 21, 492–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang P., Qiu W., Dudgeon C., Liu H., Huang C., Zambetti G. P., Yu J., Zhang L. (2009) PUMA is directly activated by NF-κB and contributes to TNF-α-induced apoptosis. Cell Death Differ. 16, 1192–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hershko T., Ginsberg D. (2004) Up-regulation of Bcl-2 homology 3 (BH3)-only proteins by E2F1 mediates apoptosis. J. Biol. Chem. 279, 8627–8634 [DOI] [PubMed] [Google Scholar]
- 20. You H., Pellegrini M., Tsuchihara K., Yamamoto K., Hacker G., Erlacher M., Villunger A., Mak T. W. (2006) FOXO3a-dependent regulation of Puma in response to cytokine/growth factor withdrawal. J. Exp. Med. 203, 1657–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luo X., He Q., Huang Y., Sheikh M. S. (2005) Transcriptional upregulation of PUMA modulates endoplasmic reticulum calcium pool depletion-induced apoptosis via Bax activation. Cell Death Differ. 12, 1310–1318 [DOI] [PubMed] [Google Scholar]
- 22. Jeffers J. R., Parganas E., Lee Y., Yang C., Wang J., Brennan J., MacLean K. H., Han J., Chittenden T., Ihle J. N., McKinnon P. J., Cleveland J. L., Zambetti G. P. (2003) Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell 4, 321–328 [DOI] [PubMed] [Google Scholar]
- 23. Villunger A., Michalak E. M., Coultas L., Müllauer F., Böck G., Ausserlechner M. J., Adams J. M., Strasser A. (2003) p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 302, 1036–1038 [DOI] [PubMed] [Google Scholar]
- 24. Garrison S. P., Jeffers J. R., Yang C., Nilsson J. A., Hall M. A., Rehg J. E., Yue W., Yu J., Zhang L., Onciu M., Sample J. T., Cleveland J. L., Zambetti G. P. (2008) Selection against PUMA gene expression in Myc-driven B-cell lymphomagenesis. Mol. Cell. Biol. 28, 5391–5402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Eischen C. M., Weber J. D., Roussel M. F., Sherr C. J., Cleveland J. L. (1999) Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 13, 2658–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Clybouw C., Fischer S., Auffredou M. T., Hugues P., Alexia C., Bouillet P., Raphael M., Leca G., Strasser A., Tarlinton D. M., Vazquez A. (2011) Regulation of memory B-cell survival by the BH3-only protein Puma. Blood 118, 4120–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Happo L., Cragg M. S., Phipson B., Haga J. M., Jansen E. S., Herold M. J., Dewson G., Michalak E. M., Vandenberg C. J., Smyth G. K., Strasser A., Cory S., Scott C. L. (2010) Maximal killing of lymphoma cells by DNA damage-inducing therapy requires not only the p53 targets Puma and Noxa, but also Bim. Blood 116, 5256–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Amente S., Zhang J., Lavadera M. L., Lania L., Avvedimento E. V., Majello B. (2011) Myc and PI3K/AKT signaling cooperatively repress FOXO3a-dependent PUMA and GADD45a gene expression. Nucleic Acids Res. 39, 9498–9507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Spender L. C., Inman G. J. (2012) Phosphoinositide 3-kinase/AKT/mTORC1/2 signaling determines sensitivity of Burkitt's lymphoma cells to BH3 mimetics. Mol. Cancer Res. 10, 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang Y. C., Piek E., Zavadil J., Liang D., Xie D., Heyer J., Pavlidis P., Kucherlapati R., Roberts A. B., Böttinger E. P. (2003) Hierarchical model of gene regulation by transforming growth factor beta. Proc. Natl. Acad. Sci. U.S.A. 100, 10269–10274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ming L., Sakaida T., Yue W., Jha A., Zhang L., Yu J. (2008) Sp1 and p73 activate PUMA following serum starvation. Carcinogenesis 29, 1878–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reimann M., Lee S., Loddenkemper C., Dörr J. R., Tabor V., Aichele P., Stein H., Dörken B., Jenuwein T., Schmitt C. A. (2010) Tumor stroma-derived TGF-β limits myc-driven lymphomagenesis via Suv39h1-dependent senescence. Cancer Cell 17, 262–272 [DOI] [PubMed] [Google Scholar]
- 33. Ramjaun A. R., Tomlinson S., Eddaoudi A., Downward J. (2007) Upregulation of two BH3-only proteins, Bmf and Bim, during TGF β-induced apoptosis. Oncogene 26, 970–981 [DOI] [PubMed] [Google Scholar]
- 34. Schulz R., Vogel T., Dressel R., Krieglstein K. (2008) TGF-β superfamily members, ActivinA and TGF-β1, induce apoptosis in oligodendrocytes by different pathways. Cell Tissue Res. 334, 327–338 [DOI] [PubMed] [Google Scholar]
- 35. Yamamura Y., Lee W. L., Goh M. X., Ito Y. (2008) Role of TAp73α in induction of apoptosis by transforming growth factor-β in gastric cancer cells. FEBS Lett. 582, 2663–2667 [DOI] [PubMed] [Google Scholar]
- 36. Cordenonsi M., Dupont S., Maretto S., Insinga A., Imbriano C., Piccolo S. (2003) Links between tumor suppressors. p53 is required for TGF-β gene responses by cooperating with Smads. Cell 113, 301–314 [DOI] [PubMed] [Google Scholar]
- 37. Farrell P. J., Allan G. J., Shanahan F., Vousden K. H., Crook T. (1991) p53 is frequently mutated in Burkitt's lymphoma cell lines. EMBO J. 10, 2879–2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu W. S., Heinrichs S., Xu D., Garrison S. P., Zambetti G. P., Adams J. M., Look A. T. (2005) Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell 123, 641–653 [DOI] [PubMed] [Google Scholar]
- 39. Gros M. J., Naquet P., Guinamard R. R. (2008) Cell intrinsic TGF-β1 regulation of B cells. J. Immunol. 180, 8153–8158 [DOI] [PubMed] [Google Scholar]
- 40. Spender L. C., Cornish G. H., Rowland B., Kempkes B., Farrell P. J. (2001) Direct and indirect regulation of cytokine and cell cycle proteins by EBNA-2 during Epstein-Barr virus infection. J. Virol. 75, 3537–3546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kehrl J. H., Roberts A. B., Wakefield L. M., Jakowlew S., Sporn M. B., Fauci A. S. (1986) Transforming growth factor β is an important immunomodulatory protein for human B lymphocytes. J. Immunol. 137, 3855–3860 [PubMed] [Google Scholar]
- 42. Snapper C. M., Waegell W., Beernink H., Dasch J. R. (1993) Transforming growth factor-β1 is required for secretion of IgG of all subclasses by LPS-activated murine B cells in vitro. J. Immunol. 151, 4625–4636 [PubMed] [Google Scholar]
- 43. Yaswen L., Kulkarni A. B., Fredrickson T., Mittleman B., Schiffman R., Payne S., Longenecker G., Mozes E., Karlsson S. (1996) Autoimmune manifestations in the transforming growth factor-β1 knockout mouse. Blood 87, 1439–1445 [PubMed] [Google Scholar]
- 44. Cazac B. B., Roes J. (2000) TGF-beta receptor controls B cell responsiveness and induction of IgA in vivo. Immunity 13, 443–451 [DOI] [PubMed] [Google Scholar]
- 45. Erlacher M., Labi V., Manzl C., Böck G., Tzankov A., Häcker G., Michalak E., Strasser A., Villunger A. (2006) Puma cooperates with Bim, the rate-limiting BH3-only protein in cell death during lymphocyte development, in apoptosis induction. J. Exp. Med. 203, 2939–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Borsutzky S., Cazac B. B., Roes J., Guzmán C. A. (2004) TGF-β receptor signaling is critical for mucosal IgA responses. J. Immunol. 173, 3305–3309 [DOI] [PubMed] [Google Scholar]