Abstract
The nonstructural 3 (NS3) protein encoded by the hepatitis C virus possesses both an N-terminal serine protease activity and a C-terminal 3′–5′ helicase activity. This study examines the effects of the protease on the helicase by comparing the enzymatic properties of the full-length NS3 protein with truncated versions in which the protease is either deleted or replaced by a polyhistidine (His tag) or a glutathione S-transferase fusion protein (GST tag). When the NS3 protein lacks the protease domain it unwinds RNA more slowly and does not unwind RNA in the presence of excess nucleic acid that acts as an enzyme trap. Some but not all of the RNA helicase activity can be restored by adding a His tag or GST tag to the N terminus of the truncated helicase, suggesting that the effects of the protease are both specific and nonspecific. Similar but smaller effects are also seen in DNA helicase and translocation assays. While translocating on RNA (or DNA) the full-length protein hydrolyzes ATP more slowly than the truncated protein, suggesting that the protease allows for more efficient ATP usage. Binding assays reveal that the full-length protein assembles on single-stranded DNA as a higher order oligomer than the truncated fragment, and the binding appears to be more cooperative. The data suggest that hepatitis C virus RNA helicase, and therefore viral replication, could be influenced by the rotations of the protease domain which likely occur during polyprotein processing.
The epidemic caused by infection by the hepatitis C virus (HCV)1 is still a global crisis despite recent therapeutic advancements (1). Because HCV cannot be conventionally cultivated in cell culture and the only other host is the chimpanzee, the enzymes encoded by HCV have been studied intensely as targets for rational drug design. One key viral enzyme is the multifunctional nonstructural protein 3 (NS3), which possesses a serine protease activity and an ATPase function that fuels the ability of the protein to unwind RNA and DNA duplexes. Although it is clear that both the protease and helicase functions are necessary for viral replication (2), it is not clear whether the two functions, which reside in independent protein domains, cooperate in any manner (3).
To examine possible effects of the NS3 protease on its helicase function, the activities of the full-length NS3 protein were rigorously compared with the same protein lacking the protease and also with recombinant proteins in which the protease is replaced with other non-HCV peptides. The experiments were designed to uncover effects of the NS3 protease domain on its helicase function which are either specific or nonspecific. Nonspecific effects are defined as those that can be duplicated by peptides not derived from HCV NS3, whereas specific effects cannot.
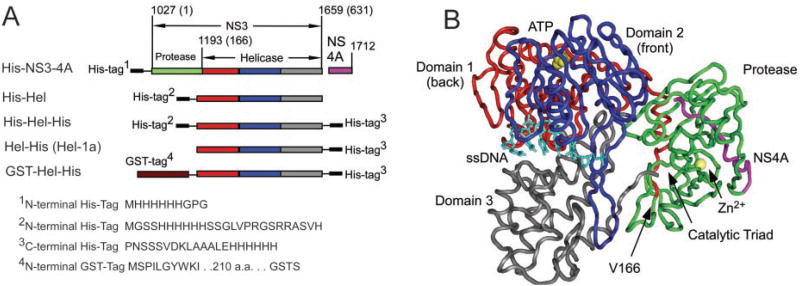
All recombinant proteins used in this study (Fig. 1) were derived from the same infectious clone of HCV genotype 1a (4). NS3 spans amino acids 1027-1659 of the ∼3,000-amino acid long polyprotein encoded by HCV. At the N terminus resides the protease, which is made of two subdomains. The active site with its chymotrypsin-like catalytic triad is formed in a shallow cleft between the two protease subdomains. The NS3 protease is activated by the NS4A protein, which is translated immediately after NS3. The NS3-4A complex cleaves itself at the NS3-4A junction, and NS4A remains associative with NS3 (Fig. 1) as the complex cleaves the rest of the viral polyprotein. To do so, the protease would likely need to move away from the back of the helicase so that its active site is accessible to other peptides (5).
Fig. 1. The NS3-4A helicase-protease complex of the HCV.
A, all recombinant proteins used in this study are derived from HCV genotype 1a isolate H77c (National Center for Biotechnology Information (NCBI) accession number AF011751). The NS3 protein extends from amino acid 1027 to 1659 of the HCV polyprotein, and NS4A encompasses amino acids 1660-1712. All of the helicase “fragments” begin with Val166, end at the NS3 C terminus, and possess fusion peptides as shown. The protease is coloredgreen, helicase domain 1 is red, domain 2 is blue, domain 3 is gray, and NS4A ispurple. B, the structure of the NS3-4A complex (5) (NCBI 1CU1) was aligned with the complex of the NS3 helicase domain with ssDNA (NCBI 1A1V (7)) using the Vector Alignment Search Tool (48) available at the NCBI. Shown is the ssDNA (light blue) from PDB 1A1V and a ribbon model of the backbone protein in 1CU1 colored as inA. Also shown in yellow are a zinc ion bound to the protease and a sulfate ion that marks the putative ATP binding site. In the complex, the C terminus of NS3 is bound in the active site of the protease, which is formed from a catalytic triad containing His57, Asp81, and Ser139. To cleave the rest of the polyprotein Yao et al. (5) propose that the protease domain swings away from the helicase via the flexible linker that connects the two domains.
The helicase portion of NS3 is a three-domain Y-shaped molecule with two clefts separating the domains (6–8). The cleft between domains 1 and 2 is lined by several conserved motifs that are involved in ATP hydrolysis and are shared with other helicases. One strand of the nucleic acid substrate binds in the cleft that separates domain 3 from domains 1 and 2 (7). Fig. 1B shows the relative location of the protease when the C terminus of NS3 is bound in the protease active site (5). In this conformation, the protease active site and NS4A are far from the known RNA and ATP binding sites.
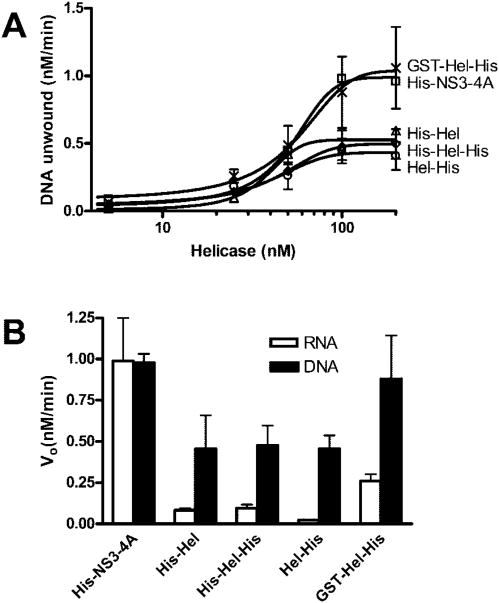
To analyze the helicase activities of the full-length protein, NS3 and NS4A were coexpressed as a single polypeptide that processes itself (9). The truncated helicase proteins lacking protease function (Fig. 1A) were based on those previously analyzed and include versions with N-terminal (10–12) and C-terminal (7, 13–16) polyhistidine tags (His tags) and a version in which the protease is replaced with a glutathione S-transferase (GST) protein (17). The presence of an intact protease domain dramatically enhances the ability of the protein to unwind RNA. However, the addition of an N-terminal His tag to the helicase fragment also enhanced RNA helicase activity, and the N-terminal GST-tagged protein unwound RNA almost as well as the full-length protein. Similar but smaller effects were seen in DNA unwinding assays and ssDNA translocation assays. A more specific effect of the protease was seen when RNA unwinding was analyzed in the presence of a DNA trap, which sequesters excess helicase not bound to the RNA substrate. More efficient unwinding correlates with a different mode of interaction seen in direct DNA binding assays, in which twice as many protein monomers bind the same amount of ssDNA. Finally, ATPase assays show that the full-length helicase hydrolyzes ATP more slowly than the truncated proteins (in the presence of DNA or RNA), suggesting that the protease domain allows the helicase to fuel RNA unwinding more efficiently with ATP hydrolysis.
Experimental Procedures
Materials
RNase-free reagents were purchased from Ambion (Austin, TX), and nucleotides and nucleic acids were treated with the RNAsecure reagent (Ambion). Poly(U) RNA with an average length of 2,500 nucleotides was purchased from Sigma. DNA oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA), and their concentrations were determined from provided extinction coefficients.
Protein Expression/Purification
The recombinant proteins utilized in this study were all derived from the same HCV isolate (4) and possess the viral regions indicated in Fig. 1. The proteins are designated according to the type and location of their fusion partners. After purification, the following extinction coefficients, derived using the method described by Pace et al. (18), were used to determine protein concentrations: His-NS3-4A, 77.3 mM−1 cm−1; His-Hel, 49.1 mM−1 cm−1; His-Hel-His, 49.1 mM−1 cm−1; Hel-His, 49.1 mM−1 cm−1; GST-Hel-His, 87.4 mM−1 cm−1. Cloning, expression, and purification of each are described below.
His-NS3-4A
PCR was used to amplify NS3-4A from the plasmid pCV-H77c, which was a generous gift from Dr. Jens Bukh (National Institutes of Health). The PCR primers added restriction sites to either end of the DNA and encoded the 5′-His tag depicted in Fig. 1. The sequence of the upstream primer was 5′-CAAGATCT ATG CAC CAT CAC CAT CAC CAT GGC CAC GGC GCG CCC ATC ACG GCG TAC GCC-3′ (BglII site underlined), and the sequence of the downstream primer was 5′-CAGAATTC TCA GCA CTC TTC CAT CTC ATC GAA C-3′ (EcoRI site underlined). The amplified DNA was digested with BglII and EcoRI and ligated into a similarly treated plasmid pVL1392 (Invitrogen). After verification of the appropriate DNA sequence, the resulting plasmid was recombined with the BacPAK system (Clontech) to generate a recombinant baculovirus. Baculovirus was plaque purified, amplified, and used for infection of Sf9 cells for protein purification. The protein was isolated from Sf9 cells using the protocol described by Sali et al. (9). Recombinant proteins were monitored during the purification procedure by Western blotting using antibodies raised against NS3 and NS4A.
Hel-His
The C-terminally His-tagged helicase fragment was based on the protein that was crystallized and characterized by Kim and co-workers (7, 13). Cloning, expression, and purification of Hel-His (which was previously designated Hel-1a) have been described (15). A detailed enzymatic characterization of Hel-1a was reported recently (16).
His-Hel-His
To generate a HCV helicase fragment with both Cterminal and N-terminal His tags, DNA encoding the helicase was excised from plasmid p24Hel-1a (15), which encodes Hel-His, and inserted into the vector pET33a (Novagen, Madison, WI). Plasmid p24Hel-1a was digested with NheI and BamHI. The resulting smaller DNA fragment was purified and inserted using T4 DNA ligase into a similarly treated pET33a to generate the vector p33Hel-1a. For expression, p33Hel-1a was used to transform strain BL21(DE3), which contains T7 polymerase under the control of a lac promoter. The recombinant protein is under control of a T7(lac) promoter, which contains a lac operator between the T7 promoter and the transcription start site (19). After cells had grown to mid log phase in Luria-Bertani broth, helicase expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside. Cells were harvested and lysed by two passes through a French pressure cell, and the clarified soluble extract was fractionated as described previously for Hel-His (15).
His-Hel
To generate a HCV helicase protein with a single His tag on its N terminus, plasmid p33Hel-1a was modified using site-directed mutagenesis to introduce a stop codon at the end of the HCV NS3 “gene.” The QuikChange site-directed mutagenesis kit (Stratagene) was used to modify p33Hel-1a using complementary mutagenic oligonucleotides: 5′-CGA CCT GGA GGT CGT CAC GTGAAC CCG GGA TCC G-3′ and 5′-CGG ATC CCG GGT TCA CGT GAC GAC CTC CAG GTC G-3′ (stop codon underlined). After the location of a stop codon was verified by DNA sequencing, the resulting plasmid (designated p33Helstop) was introduced into BL21(DE3) cells. Recombinant His-Hel was expressed and purified as described above for His-Hel-His.
GST-Hel-His
The GST-tagged HCV helicase was generated by excising the helicase encoding region from p24Hel-1a and inserting it into vector pET41a (Novagen). Plasmid p24Hel-1a was digested with the restriction enzymes NheI and BamHI. After the smaller of the two resulting DNA fragments was purified, it was ligated into pET41a that was digested with SpeI and BamHI (SpeI and NheI generate compatible cohesive ends). GST-Hel-His was then expressed in BL21(DE3) as described above. The GST-Hel-His protein was purified using the columns described for Hel-His (15) with the following modifications. After the nickel-nitrilotriacetic acid column, pooled fractions were loaded onto a 3-ml GST-bind™ column (Novagen). The column was washed with 50 ml of buffer A (50 mM Hepes, 1 mM EDTA, pH 7.5), and the protein was eluted with a gradient of the same buffer containing 0–20 mM reduced glutathione. Fractions containing HCV helicase were combined, precipitated with 60% ammonium sulfate, and purified further using gel filtration and DEAE chromatography as described previously for Hel-His (15).
Unwinding Assays
To generate substrates for helicase assays, two synthetic oligonucleotides were annealed by heating them to 95 °C and allowing them to cool slowly to room temperature. Before annealing, the shorter strand was 32P labeled using polynucleotide kinase. The duplex RNA substrate consisted of the RNA oligonucleotide 5′-UGGCGACGGCAGCGAGGC-UUUUUUUUUUUUUUUUUUUU-3′ annealed to the oligonucleotide 5′-[32P]GCCUCGCUGCCGUCGCCA-3′. The DNA substrate consisted of a shorter strand DNA oligonucleotide 5′–[32P]GCCTCGCTGCCGTC-GCCA-3′ annealed to a longer strand DNA oligonucleotide 5′-TGGCG-ACGGCAGCGAGGC TTTTTTTTTT TTTTTTTTTT-3.′ The resulting duplexes have a 3′-ssDNA (or ssRNA) tail. The DNA or RNA substrate (1 nM) and HCV helicase were incubated in reaction buffer (25 mM MOPS, pH 6.5, 3 mM MgCl2, 0.1% Tween 20) for 30 min. 10–μl reactions were initiated by the addition of 5 mM ATP and terminated with 2.5 μl of 5× stopping buffer after 20 s, 40 s, 1 min, 2 min, 3 min, 5 min, 7.5 min, and 10 min. 5× stopping buffer contained 250 mM Tris-Cl, pH 7.5, 20 mM EDTA, 0.5% SDS, 0.1% Nonidet P–40, 0.1% bromphenol blue, 0.1% xylene cyanol FF, 50% glycerol. 250 nM trap DNA was also added before electrophoresis to prevent reannealing. Products were analyzed by nondenaturing PAGE as described previously (15). In some assays, excess trap DNA, which consisted of the unlabeled shorter oligonucleotide, was added when the reactions were initiated. Assays were performed at 37 or 23 °C as indicated.
Translocation Assays
Translocation on ssDNA was assessed using an assay, developed by Raney and colleagues (20, 21), which measures the helicase catalyzed displacement of streptavidin from a biotinylated 32P-oligonucleotide. The streptavidin-bound biotinylated DNA substrate 5′-[32P]T(bio)GGCGACGGCAGCGAGGCTTTTTTTTTTTTTTTTTTTT-3′ (5 nM) and various concentrations of HCV helicase were incubated in reaction buffer (25 mM MOPS, pH 6.5, 3 mM MgCl2, 0.1% Tween 20, 37 °C) for 30 min. Reactions were initiated by the addition of 5 mM ATP and 6 μM biotin (to trap released streptavidin) and terminated at various times with the stopping buffer used in the unwinding assays. After electrophoresis on a 15% nondenaturing polyacrylamide gel, reactions were analyzed with a Molecular Dynamics PhosphorImager using ImageQuant software (Amersham Biosciences).
Nucleic Acid Binding Assay 1 (Intrinsic Protein Fluorescence)
Intrinsic protein fluorescence was measured by exciting the sample at 280 nm and reading the emission at 340 nm with a Varian Carey Eclipse fluorescence spectrophotometer. Aliquots of DNA were added to the helicase in 2 ml of 25 mM MOPS, pH 6.5, 3 mM MgCl2, and 0.2% Tween 20 at 25 °C. DNA oligonucleotide was either the same sequence as the short strand used in unwinding assay (18-mer) 5′-GCC TCG CTG CCG TCG CCA-3′ or the same oligonucleotide with a fluorescein moiety attached to its 3′-end (F18). Experiments were performed in the presence and absence of the nonhydrolyzable ATP analog ADP(BeF3), which was formed by including 0.1 mM ADP, 5 mM NaF, and 0.5 mM BeF2 in the reaction buffer (22). After correcting data for dilution and inner filter effects, fractional fluorescence (FF) was calculated by dividing the fluorescence values by the initial fluorescence of the protein in the absence of DNA. FF was fit to the total concentration of added ssDNA oligonucleotide ([DNA]T) calculated in terms of 3′-ends of DNA using Equation 1.
| (Eq. 1) |
In Equation 1, ΔFFMAX is the maximum change in fractional fluorescence resulting when all enzyme molecules are bound to nucleic acid. KD is the apparent dissociation constant, ET is total enzyme concentration, and n is the number of enzyme monomers bound to a single oligonucleotide at saturation. Data were fit to Equation 1 by nonlinear regression using Graphpad Prism version 4.0 (San Diego).
Nucleic Acid Binding Assay 2 (Fluorescently Labeled Oligonucleotide)
Fluorescence of the fluorescein-labeled DNA oligonucleotide F18 increased about 2-fold upon binding to HCV helicase when monitored at an excitation wavelength of 492 and an emission of 518. This fluorescence change was used to measure helicase DNA interactions as follows. Various amounts of F18 ranging from 0.5 to 2 nM were titrated with recombinant helicases. Titrations were performed in 25 mM MOPS, pH 6.5, 3 mM MgCl2, and 0.2% Tween 20 at 25 °C. After correcting for dilution and negligible inner filter effects, the corrected fluorescence (Fc) at each enzyme concentration was fit to total enzyme concentration ([E]T) using Equation 2
| (Eq. 2) |
In Equation 2, Fi is the initial fluorescence of F18 in the absence of protein, and ΔFMAX is the maximum fluorescence change of each titration. This equation assumes that the observed fluorescence is proportional to the concentration of bound DNA divided by the total concentration of DNA [DNA]T. Bound DNA is calculated from [DNA]T and total enzyme ([E]T) at each point in the titration from estimates of the apparent dissociation constant. The factor n again represents the number of protein monomers bound to a single oligonucleotide. KD and n were calculated by globally fitting five data sets obtained at different [DNA]T values to Equation 2 using Graphpad Prism.
ATPase Assay
Initial rates of ATP hydrolysis were measured as described previously (15). Reactions were performed at 37 °C in 25 mM MOPS, pH 6.5, 5 mM MgCl2 0.1% Tween 20, ATP, and 10–100 nM helicase. Data were fit to the Michaelis-Menten equation by nonlinear regression to calculate the Km and Vmax in the presence and absence of nucleic acids. To determine the constant KDNA, defined as the concentration of DNA that supports a half-maximum rate of catalysis, reactions were performed in the presence of 4 mM ATP with various concentrations of DNA, and the data were fit to Equation 3.
| (Eq. 3) |
Results
Although the HCV helicase has been analyzed both as an isolated fragment and as part of the full-length NS3 protein, the few studies that directly compared the HCV helicase with and without its attached protease have not noted clear differences (11, 23, 24). There are nevertheless several noteworthy differences between studies that utilize full-length NS3 and those that use recombinant HCV helicase lacking the protease domain. For example, as discussed below, Pang et al. (25) showed that the full-length protein is a processive RNA helicase, whereas a recent study by Lam et al. (16) concluded that the isolated domain unwinds only DNA processively. Unfortunately, no firm conclusions can be reached by comparing such studies because different research groups use different HCV isolates as their source for recombinant NS3 proteins; any apparent differences among the studies could simply result from genetic variation. Helicases isolated from different HCV genotypes display different properties (15), and point mutations in the NS3 helicase influence RNA replication as observed using both HCV replicons (26–29) and chimpanzees (30). In this study, the properties of the full-length NS3-4A helicase are compared with truncated helicases, derived from the same viral isolate, which either lack the protease domain or have it replaced with fusion peptides.
Expression and Purification of the Recombinant Proteins
The proteins used in this study are shown in Fig. 1A. The source of each protein was the cDNA from the infectious clone H77c, an isolate of HCV genotype 1a (4). Although the full length construct (His-NS3-4A) encoded all of NS3 and NS4A, the truncated proteins lack the first 166 amino acids of the N-terminal protease domain of the mature NS3 protein and the protease cofactor NS4A. All helicases with the exception of the full-length NS3 were expressed in the same cell line (BL21(DE3)) and purified using the same basic protocol.
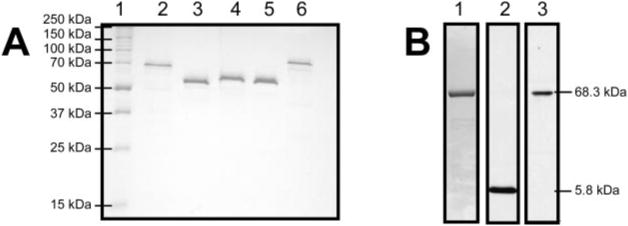
Repeated attempts to express the full-length protein in numerous Escherichia coli strains failed, resulting in the formation of primarily insoluble inclusion bodies. Therefore, His-NS3-4A was purified from insect cells infected with recombinant baculovirus containing an open reading frame encoding His-NS3-4A. The protein was expressed and purified using the procedure described for the same protein isolated from the similar HCV genotype 1a H strain (9). The purity of the recombinant HCV helicase proteins is shown in Fig. 2. Although the smaller NS4A protein is only faintly visible when His-NS3-4A is stained with Coomassie Blue, it is detectable with antibodies raised against NS4A peptide (Fig. 2B).
Fig. 2. Purified recombinant proteins.
A, a 10% polyacrylamide gel containing 1% SDS with 1 μg each of the proteins used in this study: lane 2, His-NS3-4A, 5.786 kDa (NS4A), 68.350 kDa (His-NS3); lane 3, His-Hel, 52.697 kDa; lane 4, His-Hel-His, 55.464 kDa; lane 5, Hel-His, 52.731 kDa; lane 6, GST-Hel-His, 79.682 kDa. B, to visualize the NS4A protein not seen in the 10% gel, 5 μg of the His-NS3-4A complex was separated on a 20% SDS gel and transferred to nitrocellulose membranes for Western analysis. Lane 1 shows a gel stained with Coomassie Blue, lane 2 shows a membrane probed with antibodies raised against NS4A, and lane 3 shows an identical membrane stained with antibodies raised against NS3.
A protein without fusion tags was not analyzed because when His-Hel was digested with thrombin, which removes the N-terminal His tag, the helicase became very insoluble, and only a small amount could be isolated. When analyzed, the protein had also lost much of its specific activity. Therefore, effects of the His tags were instead determined by comparing the properties of the various proteins. For example, the effect of the C-terminal His tag can be seen by simply comparing His-Hel with His-Hel-His, and likewise, the effects of the N-termi-nal His tag can be uncovered by comparing His-Hel-His with Hel-His. Others have shown that an N-terminal His tag on full-length NS3 has no detectable effects on the activity of that protein (25, 31).
The Protease Domain Aids RNA Unwinding
Previous studies using Hel-His have shown that it unwinds RNA poorly relative to DNA (15, 16), and when excess DNA is added upon initiation to reactions containing Hel-His, virtually no RNA unwinding is detected (15). Although a preference for DNA has been long established for HCV helicase (32), the absence of RNA helicase activity in the presence of an enzyme trap (such as excess DNA) starkly contrasted results published previously by other authors (25, 31). The ability of a helicase to unwind DNA (or RNA) in the presence of a trap is indicative of a processive reaction because if enzyme molecules dissociate after only a few base pairs are unwound then no reaction will be detected. The trap prevents enzymes not initially bound to substrate from participating in the reaction (33). Thus, our previous results suggest that Hel-His is a processive DNA helicase but not a processive RNA helicase (15, 16), which is a very different conclusion than that of Pang et al. (25), who showed the HCV helicase is processive on both DNA and RNA. The study by Pang et al. (25) differed from ours (16) in two ways. First, the helicases were isolated from different HCV strains, and second, Pang et al. (25) used full-length NS3 proteins, whereas we used the truncated protein Hel-His (16). This study was therefore initiated using proteins with no genotypic variation to test whether the protease in fact aids RNA unwinding.
The ability of four of the proteins studied here to unwind duplex RNA is shown in Fig. 3. RNA unwinding by Hel-His was analyzed previously (15). The RNA helicase assays in Fig. 3 are done in the absence of a trap but under conditions in which no RNA reannealing occurs. The duplex RNA substrate consists of a 5′-32P-labeled 18-nucleotide-long RNA oligonucleotide annealed to a 38-nucleotide-long oligonucleotide in such a manner to create a substrate with a 3′-ssRNA tail. This tail is required for unwinding. None of the helicases studied here unwinds substrates with either blunt ends or with only a 5′-ssRNA tail. In these assays, the RNA and helicase are combined and incubated for 30 min to allow efficient substrate loading. Reactions are initiated with ATP, quenched at various times, and analyzed on nondenaturing polyacrylamide gels.
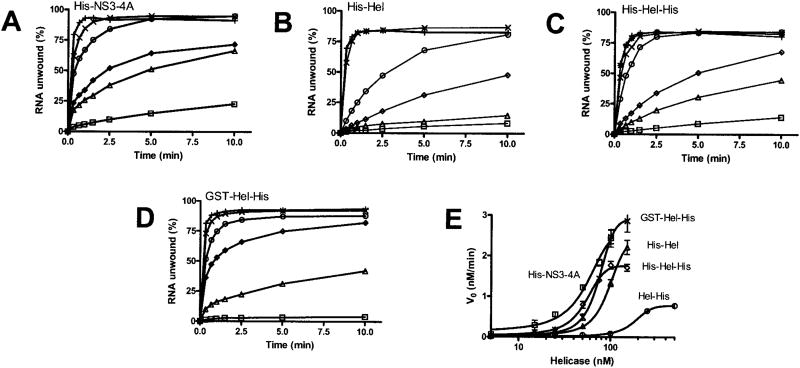
Fig. 3. RNA unwinding.
In all reactions, 1 nM duplex [32P]RNA substrate was preincubated with the indicated amounts of HCV helicase. Reactions were initiated with ATP, incubated at 37 °C for various times, terminated, and analyzed using native polyacrylamide gels. A, reactions contained His-NS3-4A at 5 nM (□), 15 nM (Δ), 25 nM (◊), 50 nM (○), 75 nM (×), or 100 nM (+). B, reactions contained His-Hel at 5 nM (□), 15 nM (Δ), 25 nM (◊), 50 nM (○), 100 nM (×), or 150 nM (+). C, reactions contained His-Hel-His at 5 nM (□), 15 nM (Δ), 25 nM (◊), 50 nM (○), 75 nM (×), 100 nM (+), and 150 nM (*). D, reactions contained GST-Hel-His at 5 nM (□), 25 nM (Δ), 50 nM (◊), 75 nM (○), 100 nM (×), or 150 nM (+). E, initial rates of RNA unwinding in reactions catalyzed by His-NS3-4A (□), His-Hel (Δ), His-Hel-His (◊), and GST-Hel-His (×), are compared with the data obtained with Hel-His (data are from Ref. 16) (○). Data are fit to a sigmoidal dose-response curve with variable slope using nonlinear regression (using Graphpad Prism, version 4.0). The resulting maximum velocities and EC50 values are listed in Table I.
All proteins with peptides attached to the N terminus of domain 1 of HCV helicase unwind RNA better than Hel-His, which lacks an N-terminal attachment (Fig. 3). When the initial rates of RNA unwinding by the N-terminally tagged enzymes (Fig. 3, A–D) are compared with rates derived from the same experiments conducted with Hel-His, dramatic differences are apparent (Fig. 3E). Less of these proteins are required to initiate unwinding, and RNA is unwound faster and almost to completion. The initial rates of RNA unwinding can be analyzed to yield a maximum unwinding rate (Vmax) and a concentration of helicase which leads to 50% maximum activity. Estimates of these parameters can be obtained by fitting the data in Fig. 3E to the Michaelis-Menten equation. How ever, because the data better describe sigmoid curves, such an analysis would be misleading. The data were therefore fit to a sigmoidal dose-response curve with a variable slope. The top of these curves provides an estimate of Vmax and the midpoint provides an EC50 value. Such an analysis (Table I) reveals that His-NS3-4A unwinds RNA most efficiently with an EC50 of 54 nM and a Vmax of 2.6 nM/min. GST-Hel-His unwinds RNA at a slightly faster maximum rate but with a higher EC50. This suggests that the protease is structurally important for RNA unwinding, but it can be essentially replaced by a protein of entirely different origin. Surprisingly, even the proteins in which the protease is replaced with a short His tag unwind RNA almost as well as the full-length NS3 protein (Table I).
Table I. Kinetic analysis of helicase activity.
Initial rates of RNA unwinding (Fig. 3) and DNA unwinding (Fig. 5) were fit to protein concentration to determine a maximum unwinding rate (Vmax) and a concentration of helicase which leads to a half-maximal rate of unwinding (EC50). Also shown is the ratio of the initial rate of DNA unwinding to the rate of RNA unwinding for reactions containing 100 nM each helicase at 23 °C.
| Protein | RNA(37 °C) | DNA(23 °C) | DNA preference (23 °C) | ||
|---|---|---|---|---|---|
|
|
|
||||
| Vmax | EC50 | Vmax | EC50 | ||
| nM/min | μM | nM/min | μM | DNA ν0: RNA ν0 | |
| His-NS3–4A | 2.6 ± 0.2 | 54 ± 5 | 1.0 ± 0.2 | 52 ± 3 | 1.0 |
| His-Hel | 2.4 ± 0.1 | 95 ± 3 | 0.5 ± 0.1 | 38 ± 5 | 5.5 |
| His-Hel-His | 1.8 ± 0.1 | 53 ± 2 | 0.5 ± 0.1 | 42 ± 1 | 4.9 |
| Hel-His | 0.8 ± 0.1 | 182 ± 5 | 0.4 ± 0.1 | 38 ± 6 | 19.6 |
| GST-Hel-His | 2.9 ± 0.1 | 74 ± 5 | 1.0 ± 0.2 | 54 ± 5 | 3.4 |
Unlike Hel-His, the other proteins analyzed here unwind RNA in the presence of a DNA trap (Fig. 4). In these assays, reactions were initiated with ATP and various amounts of a DNA oligonucleotide that consists of the same sequence as the shorter strand in the RNA duplex (except with Ts replacing Us). The trap binds both free enzyme not bound to substrate and enzyme that dissociates from the substrate before the reaction is terminated. In other words, only a single reaction cycle of enzyme and substrate should be theoretically observed in the presence of trap DNA. In the presence of 20 times more trap than enzyme, all of the helicases except Hel-His could unwind a significant amount of RNA. The initial rates of RNA unwinding for all proteins were affected by trap (Fig. 4F), although even in the presence of 20-fold excess trap, both His-NS3-4A and GST-Hel-His unwound with rates 5–10-fold faster than the helicases possessing an N-terminal His tag (His-Hel and His-Hel-His).
Fig. 4. Effect of a DNA trap on RNA unwinding.
Unwinding assays were performed at 37 °C using a [32P]RNA substrate in which the shorter strand is labeled. Reactions were initiated by adding ATP and various amounts of the shorter DNA strand, lacking a radiolabel, which acts as an enzyme trap. Reactions contained no trap (□), 100 nM (Δ), 250 nM (◊), 500 nM (○), 1000 nM (×), or 2000 nM (+) trap oligonucleotide and 100 nM either His-NS3-4A (A), His-Hel (B), His-Hel-His (C), Hel-His (D), or GST-Hel-His (E). F, the initial rates of RNA unwinding are plotted against trap concentration for reactions containing His-NS3-4A (□), His-Hel (Δ), His-Hel-His (◊), Hel-His (○), and GST-Hel-His (×).
Another way to analyze these data is to examine the final amount of RNA that is unwound in the presence of trap. Theoretically, this final amplitude should be proportional to reaction processivity if the reaction is a pseudo-first order process. Based on the demonstration that the amplitudes of reaction catalyzed by the E. coli UvrD helicase correlate with processiv-ity (33), an analysis of reaction amplitudes was used previously to demonstrate processivity of HCV helicase (25). If such an analysis is applied to the data in Fig. 4, one would conclude that the protease domain is essential for helicase processivity. Such a bold conclusion might not be warranted, however. The reaction catalyzed by HCV helicase takes place over a relatively long time frame, and final reaction amplitude might depend on other parameters, such as the depletion of ATP needed to fuel helicase action. Nevertheless, His-NS3-4A is clearly less sensitive to trap concentration than all the other proteins, highlighting a specific role for the protease in RNA unwinding.
DNA Unwinding
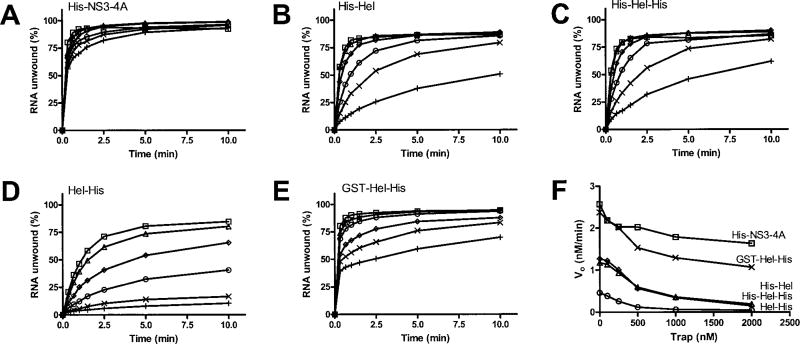
Hel-His unwinds DNA much more efficiently than RNA. In fact at 37°C, the temperature at which RNA unwinding was analyzed (Figs. 3 and 4), Hel-His unwinds all DNA in a helicase assay in less than 1 min (16). Therefore, to analyze DNA unwinding by the proteins studied here, DNA helicase assays were instead performed at 23°C, where the reaction is slower (Fig. 5A). As with the RNA unwinding experiments, initial DNA unwinding rates fit best to sigmoid curves. About the same amount of each protein results in a 50% maximum rate of unwinding, but His-NS3-4A and GST-Hel-His unwind DNA at a maximum rate that is about twice as fast as the other proteins (Table I).
Fig. 5. DNA unwinding compared with RNA unwinding at 23 °C.
Because DNA unwinding occurs too fast to measure accurately at 37 °C, DNA unwinding assays were performed at 23 °C. In all reactions, 1 nM duplex [32P]DNA substrate was preincubated with the indicated amounts of HCV helicase. Reactions were initiated with ATP, incubated at 23 °C for various times, terminated, and analyzed using native polyacrylamide gels. A, initial rates of DNA unwinding at various concentrations of His-NS3-4A (□), His-Hel (Δ), His-Hel-His (◊), Hel-His (○), and GST-Hel-His (×). Data are fit to a sigmoidal dose-response curve with variable slope using nonlinear regression. The resulting maximum velocities and EC50 values are listed in Table I. B, reactions with 100 nM helicase were repeated at 23 °C using a [32P]RNA substrate, and the resulting initial rates of RNA unwinding are compared with the initial rates of DNA unwinding.
To compare properly the efficiency with which each protein unwinds DNA or RNA, RNA unwinding assays were repeated at 23°C (Fig. 5B). Each of the proteins clearly prefers DNA except His-NS3-4A, again highlighting a specific role for the protease in RNA unwinding (Table I).
We have shown previously that DNA unwinding catalyzed by Hel-His is not particularly sensitive to the presence of a DNA trap (16). Not surprisingly, DNA unwinding catalyzed by the other proteins is likewise insensitive to a trap (data not shown), suggesting that they are all processive DNA helicases.
DNA Translocation Assays
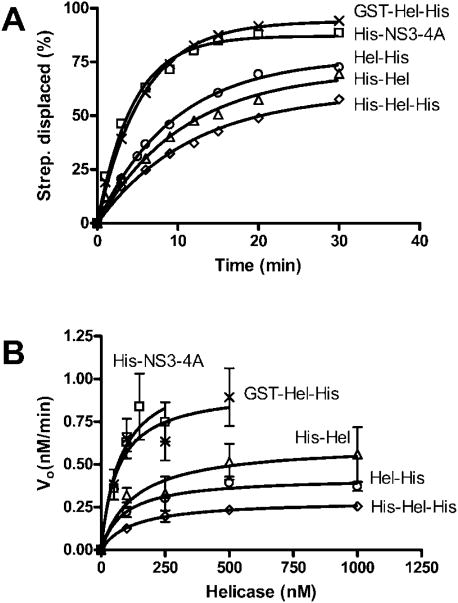
Morris et al. (21) have shown that in addition to unwinding RNA and DNA, the full-length HCV genotype 1b NS3 protein is able to strip streptavidin from biotin-labeled DNA. Because the HCV helicase can only strip streptavidin from the 5′-end of DNA, these assays provide evidence that the protein translocates only in a 3′–5′ direction. We recently confirmed these experiments using Hel-His, but because our studies used a protein derived from genotype 1a, no firm conclusions were reached regarding the role of the protease in DNA translocation (16). We have therefore repeated these studies using all the proteins described here (Fig. 6). The new data show that the protease domain accelerates the translocation rate of the protein on ssDNA but that this effect can be substituted entirely by a GST fusion peptide (GST-Hel-His). This correlates with faster DNA unwinding seen with His-NS3-4A and GST-Hel-His. An N-terminal His tag does not appear to aid translocation because His-Hel moves only some what faster than Hel-His, whereas His-Hel-His moves slightly slower. The initial translocation rates of each protein seem to obey Michaelis-Menten kinetics (Fig. 6B), unlike initial rates of RNA unwinding (Fig. 3E), or DNA unwinding (Fig. 5A). This may simply be because the enzyme and substrate are in a state of rapid equilibrium or because the reactions are not proces-sive. The addition of a DNA trap to these reactions abolishes any detectable dissociation of ssDNA from streptavidin (data not shown).
Fig. 6. Displacement of streptavidin from a 5′-biotinylated oligonucleotide.
HCV helicase was incubated at 37 °C with a 32P-la-beled oligonucleotide with biotin at its 5′-end bound to streptavidin. When ATP is added to the reaction, streptavidin is displaced from the oligonucleotide leading to a shift in the electrophoretic mobility of the [32P]DNA. A, amounts of streptavidin displaced from 5′-biotinylated ssDNA versus time in reactions containing 100 nM of His-NS3-4A (□), His-Hel (Δ), His-Hel-His (◊), Hel-His (○), and GST-Hel-His (×). B, reactions were repeated at various protein concentrations at which initial rates of streptavidin displacement were determined for reactions containing His-NS3-4A (□), His-Hel (Δ), His-Hel-His (◊), Hel-His (○), and GST-Hel-His (×). Data are fit to the Michaelis-Menten equation.
Binding Assays
The issue of whether or not helicases unwind nucleic acid duplexes as monomers, dimers, or higher order oligomers is still hotly debated. Numerous studies have concluded that HCV helicase functions as a monomer (10, 34, 35), whereas many others have presented clear evidence for oligomerization (17, 36, 37). It has been proposed that these differences might be because the protease region is required for stable oligomerization (17, 37), but this has not yet been directly demonstrated. Although all of the proteins here migrate mainly as monomers on both gel filtration columns and native polyacrylamide gels, the sigmoidal dependence of unwinding on protein concentration suggests that interprotein interactions might be critical for RNA unwinding (Fig. 3E). To address this issue further, binding assays were performed to quantify the interaction of each helicase with ssDNA.
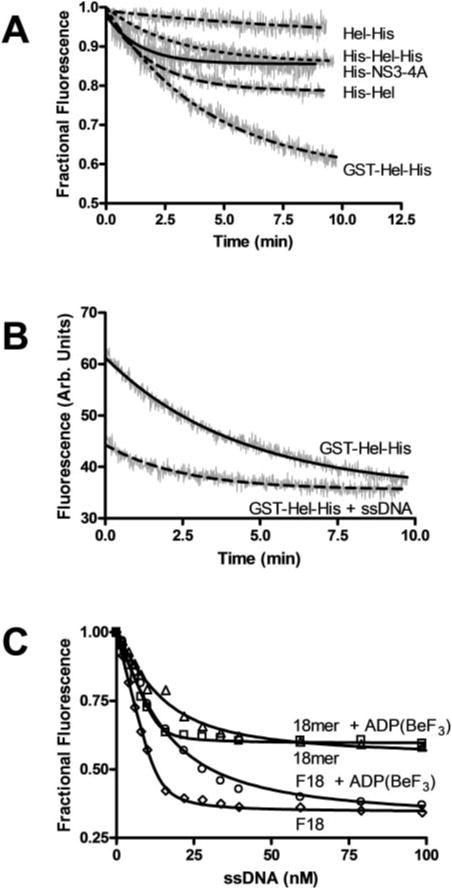
Previously, intrinsic protein fluorescence has been used to examine ssDNA interaction with HCV helicase (15, 16, 22, 35, 38). All of these studies used recombinant proteins lacking the protease domain and exploit the fact that ssDNA binds near a solvent exposed tryptophan to quench intrinsic protein fluorescence (7). Previously, we have performed these studies using Hel-His (see Fig. 8 of Ref. 16). However, it was extremely difficult to repeat these experiments with the other proteins examined here because the intrinsic protein fluorescence of each changed more rapidly in the absence of added oligonucleo-tide. As shown in Fig. 7A the intrinsic protein fluorescence of Hel-His decreases only about 5% over 10 min. Others have reported similar drifts (35, 38), which were corrected for by performing “blank” titrations where buffer was added at the same intervals as the additions of ssDNA. The data were then corrected for this minor drift.
Fig. 7. Helicase-ssDNA binding monitored by changes in intrinsic protein fluorescence.
Intrinsic protein fluorescence was measured by exciting the sample at 280 nm and reading the emission at 340 nm. A, change in fractional fluorescence with time in samples incubated at 25 °C containing 50 nM His-NS3-4A, His-Hel, His-Hel-His, Hel-His, or GST-Hel-His. B, protein fluorescence of the GST-Hel-His protein monitored in the presence and absence of 100 nM 18-nucleotide-long oligonucleotide with the same sequence as the shorter strand used in the DNA helicase assays (18-mer). C, effect of the 18-mer on intrinsic protein fluorescence (expressed as fractional fluorescence) in the absence (□) and presence (Δ) of the nonhydrolyzable ATP analog ADP-(BeF3). The titrations were repeated with the same oligonucleotide with a fluorescein moiety attached to its 3′-end (F18) both in the absence (◊) and presence of ADP(BeF3) (○). Data are fit to Equation 1 by nonlinear regression.
This drifting was more obvious with proteins containing peptides attached to the N terminus of the helicase (Fig. 7A). The intrinsic protein fluorescence of the full-length protein, His-NS3-4A, and the N-terminal His-tagged proteins decreased almost 20%, whereas that of GST-Hel-His decreased almost 40%. To investigate the physical basis for this fluorescence drift, the experimental conditions were varied systematically. The drift was independent of pH, salt concentration, buffer composition, temperature, or divalent metal ion concentration. Only the concentration of Tween 20 affected drift rates, with faster changes in intrinsic protein fluorescence occurring at lower concentrations of detergent. The data in Fig. 7 were obtaining in the presence of 0.2% Tween 20. Higher detergent concentrations were not used because background fluorescence from higher concentrations of Tween 20 made observations of intrinsic protein fluorescence difficult.
There are two possible explanations for the drift in intrinsic protein fluorescence: either the protein is slowly changing the conformation (denaturing) or aggregating. Two lines of evidence support the idea that the changes in intrinsic protein fluorescence reflect protein oligomerization rather than denaturation. First, no changes in specific activity in either helicase or ATPase assays were measured when any of the helicase proteins were incubated for several hours at temperatures as high as 37 °C. Second, oligomerization of the HCV helicase has been observed by others using cross-linking (35), yeast two-hybrid assays (17, 39), and gel filtration chromatography (9, 17). Although the vast majority of the proteins studied here migrate as monomers (or in the case of His-NS3-4A, a het-erodimer), a small amount of the HCV proteins can be detected in the void volume of the columns, as was first reported by Sali et al. (see Fig. 3 of Ref. 9).
The previously reported oligomerization of HCV helicase has been reported to be greatly enhanced by the presence of oligonucleotides. The presence of ssDNA oligonucleotides allows for more apparent dimers, trimers, tetramers, and pentamers to be detected by cross-linking (36) and gel filtration chromatography (17). Thus, if the changes in protein fluorescence reflect a slow oligomerization, then the rates of fluorescence change should be accelerated in the presence of a ssDNA oligonucleotide. To test this hypothesis, the experiments in Fig. 7A were repeated in the presence of DNA. Indeed, added oligonucleotides accelerated the rate of intrinsic protein fluorescence decrease for all proteins. The data obtained with GST-Hel-His are shown in Fig. 7B. When ssDNA is present, it not only quenches fluorescence, but it also accelerates drift. The intrinsic protein fluorescence stabilizes earlier in the presence of DNA than in the absence of DNA (Fig. 7B). Although this observation supports the idea that fluorescence changes reflect oligomerization, it greatly complicates the analysis of DNA binding to HCV helicase when intrinsic protein fluorescence is monitored.
Because quenching of intrinsic protein fluorescence provided a poor method to examine helicase DNA interactions (for proteins other than Hel-His) several fluorescently labeled DNA molecules were screened to find one that displayed a large fluorescence change upon protein binding. We found that the molecule that gave the best signal was an 18-nucleotide-long oligonucleotide with a fluorescein moiety attached to its 3′-end (F18). To show that the fluorescein itself did not influence the interaction between helicase and the oligonucleotide, its binding to Hel-His was analyzed by monitoring changes of intrinsic protein fluorescence (Fig. 7C). F18 and the same oligonucleotide lacking the fluorescein (18-mer) bound similarly both in the presence and absence of the nonhydrolyzable ATP analog ADP(BeF3). As was seen previously, DNA binds HCV helicase about 10-fold weaker in the presence of ADP(BeF3) (16, 22). This has been interpreted to mean that ATP binding allows the protein to slide along DNA (22). Using this assay, the apparent KD for the 18-mer in the absence of ADP(BeF3) is 1.9 ± 0.3 nM, whereas the KD with F18 is 2.5 ± 0.3. These values are not significantly different. Likewise, the KD of the 18-mer with ADP(BeF3) is 26 ± 2, and the KD for F18 is similar at 30 ± 2.
The shapes of the binding isotherms in Fig. 7C clearly indicate tight binding, but interestingly, the inflection points do not match the total amount of protein monomers in solution (50 nM, as measured from the protein extinction coefficient). To fit a standard equation describing the binding of a ligand to a protein, the ligand concentration must be adjusted by a factor, n (see Equation 1, which was derived from Equations 2, 3, and 4 of Ref. Ref. 35). The factor, n, can be interpreted as the number of protein monomers that bind a single oligonucleotide. Although a value of n = 3.7 was determined from the data in Fig. 7C by nonlinear regression analysis, a similar value can be determined qualitatively simply by examining the graph. All curves inflect around 13.5 nM DNA, which is about the amount of DNA necessary to bind all 50 nM protein. Hence, three or four proteins monomers bind a single template, or they each cover about 5 nucleotides of the 18-mer.
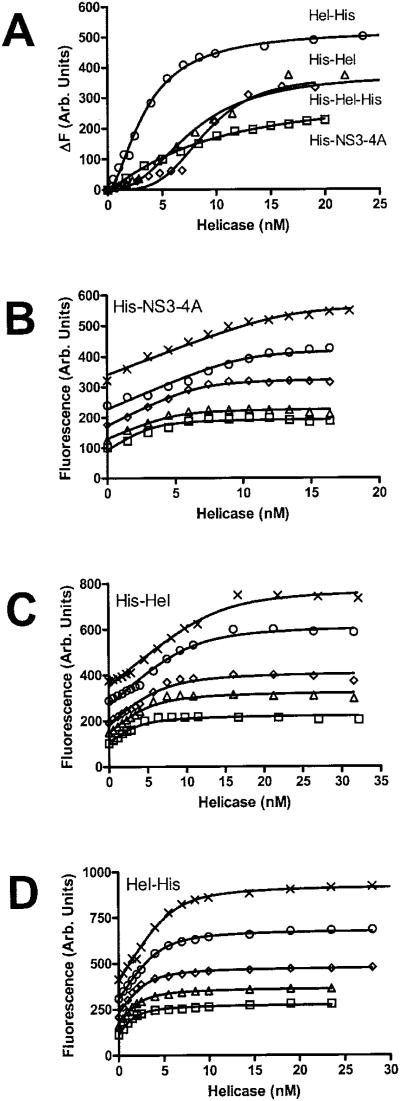
Changes in F18 fluorescence upon addition of all the heli-cases studied here except GST-Hel-His is shown in Fig. 8A. In control experiments, neither addition of buffer nor a Hel-His mutant (F444A) lacking detectable ATPase, helicase, or binding activity (16) produced any changes in F18 fluorescence. GST-Hel-His did not yield analyzable data in this assay because after fluorescence increased upon protein addition, fluorescence began to decrease slowly. Fluorescence of the F18 complex made with the other proteins was much more stable.
Fig. 8. Analysis of ssDNA binding using a fluorescein-labeled oligonucleotide.
The fluorescence of F18 was monitored at an excitation wavelength of 492 nm and an emission of 518 nm in the absence and the presence of various concentrations of various HCV helicase constructs. A, change in fluorescence (ΔF) when 2 nM F18 is titrated with His-NS3-4A (□), His-Hel (Δ), His-Hel-His (◊), or Hel-His (○). The other three panels show how raw fluorescence of 0.5 nM (□), 0.75 nM (Δ), 1 nM (◊), 1.5 nM (○) or 2 nM (×) F18 changes upon addition of His-NS3-4A (B), His-Hel (C), or Hel-His (D). Data are fit to Equation 2 using global nonlinear least squares analysis to yield the values listed in Table II.
The data obtained with Hel-His at multiple oligonucleotide concentrations (Fig. 8D) fit an equation similar to the one used to analyze changes in intrinsic protein fluorescence to yield values for KD, and n (Table II). However, when the same experiments are done with His-NS3-4A (Fig. 8B), His-Hel (Fig. 8C) or His-Hel-His (not shown), the resulting isotherms look very different from those obtained with Hel-His (Fig. 8D). Binding isotherms obtained with His-Hel-His (not shown) were virtually identical to those obtained with His-Hel. The isotherms obtained with His-Hel and His-Hel-His are S-shaped at low protein concentrations, very steep, and have clearly defined inflection points. The inflection points of His-Hel are at twice the protein concentration seen with Hel-His (compare Fig. 8C and D), indicating that twice the amount of protein is required to saturate the binding sites on the DNA. Isotherms obtained with His-NS3-4A inflect in the same region as those obtained with His-Hel (Fig. 8C). The data are fit to the simple binding model described in Equation 2 to yield the parameters described in Table II.
Table II. ssDNA binding.
Fluorescence data obtained when oligonucleotide F18 (Fig. 8) is titrated with HCV helicase in the presence and absence of the nonhydrolysable ATP analog ADP(BeF3) was fit to Equation 2 to yield an apparent dissociation constant and a factor n, which describes the number of helicase monomers bound to a single oligonucleotide.
| Protein | No nucleotide | ADP(BeF3) | ||
|---|---|---|---|---|
|
|
|
|||
| KD | n protein/DNA | KD | n protein/DNA | |
| nM | nM | |||
| His-NS3–4A | 0.059 ± 0.029 | 6.7 ± 0.3 | 3.4 ± 0.5 | 6.6 ± 1.0 |
| His-Hel | 0.16 ± 0.06 | 7.0 ± 0.5 | NDa | ND |
| His-Hel-His | 0.11 ± 0.08 | 8.2 ± 0.8 | ND | ND |
| Hel-His | 0.23 ± 0.3 | 2.9 ± 0.2 | 3.8 ± 0.25 | 3.0 ± 0.1 |
ND, not determined.
Table II also summarizes the results of the same set of experiments when they were conducted in the presence of the nonhydrolyzable ATP analog ADP(BeF3). Importantly, both Hel-His and His-NS3-4A appear to release ssDNA upon ATP binding, indicating that the full-length enzyme could function using the same bind-slide mechanism proposed by Levin et al. (22).
If one closely examines the data in Fig. 8, one might conclude that the protease domain not only allows more monomers to assemble on ssDNA, but also that the full-length and the N-terminally tagged proteins appear to bind more cooperatively. In support of a cooperative model, when a Hill coefficient is added to Equation 2 (as an exponential modifying protein concentration), somewhat better fits were obtained for all the data. Such an analysis is not presented here because the molecular meaning of such an arbitrary factor is unclear. Nevertheless, the assembly of HCV helicase on DNA is evidently more complicated than that described previously because individual monomers appear to interact cooperatively. Although an analysis of cooperativity would clearly be informative, it is beyond the scope of this study.
ATP Hydrolysis
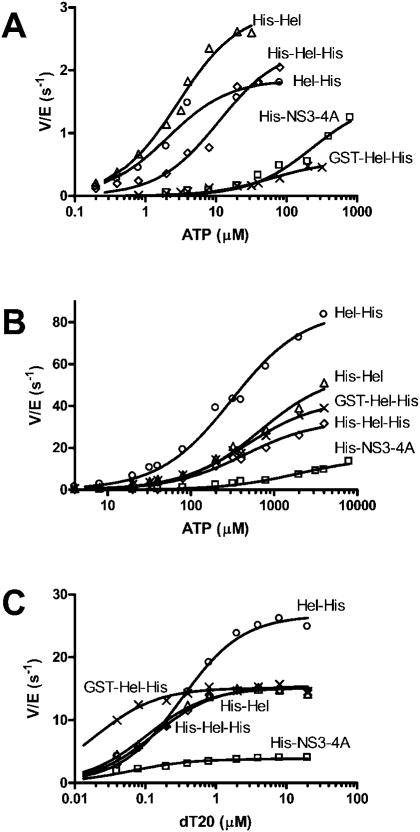
Both RNA and DNA unwinding by HCV helicase are fueled by the hydrolysis of ATP. In all the helicase assays described above, no reactions were detected in the absence of ATP. Because the various proteins studied here unwound at different rates, it is conceivable that these differences are caused by different rates of ATP hydrolysis. Because HCV helicase hydrolyzes ATP in the absence of nucleic acids, and the ATPase rate is stimulated by DNA (and RNA), five different kinetic constants can be defined: the Km and Vmax in the presence and absence of nucleic acid, and the concentration of nucleic acid that yields 50% Vmax (KDNA). These factors were determined by titrating each helicase with ATP in the absence (Fig. 9A) and presence (Fig. 9B) of saturating amounts of poly(U) RNA (the polymer that most efficiently stimulates ATP hydrolysis (40)), and by measuring ATP hydrolysis at saturating ATP concentrations at different concentrations of a DNA oligonucleotide (Fig. 9C). The resulting constants are summarized in Table III.
Fig. 9. ATP hydrolysis.
Initial rates of ATP hydrolysis were measured at various ATP concentrations in the absence (A) and presence of 1 mg/ml poly(U) RNA (B). Rates (nM/s) were divided by the total amount of enzyme in the assay (nM) to compare the activities of His-NS3-4A (□), His-Hel (Δ), His-Hel-His (◊), Hel-His (○), and GST-Hel-His (×). Data are fit to the Michaelis-Menten equation to yield the parameters summarized in Table III. C, specific activity of ATP hydrolysis was measured at 4 mM ATP at various concentrations of a 20-nucleotide ho-mopolymer of deoxythymidine. Data are fit to Equation 3 to determine KDNA, which is reported in Table III.
Table III. Kinetic analysis of ATP hydrolysis.
Initial rates of ATP hydrolysis (Fig. 9) in the presence and absence of 1 mg/ml poly(U) RNA were fit to ATP concentration using the Michaelis-Menten equation to yield Vmax and Km values. Also shown is the KDNA, defined as the DNA concentration needed to reach half of the Vmax seen at saturating DNA levels.
| Protein | Basal | RNA stimulated | KDNA | ||
|---|---|---|---|---|---|
|
|
|
||||
| Vmax/ET | Km | Vmax/ET | Km | ||
| s−1 | μM | s−1 | μM | nM | |
| His-NS3–4A | 1.5 ± 0.2 | 230 ± 72 | 15 ± 1.3 | 1,900 ± 410 | 64 ± 25 |
| His-Hel | 3.0 ± 0.1 | 2.8 ± 0.37 | 56 ± 3.0 | 670 ± 96 | 105 ± 18 |
| His-Hel-His | 2.4 ± 0.2 | 11 ± 2.4 | 33 ± 1.2 | 440 ± 47 | 134 ± 19 |
| Hel-His | 1.8 ± 0.1 | 2.1 ± 0.42 | 86 ± 3.0 | 307 ± 34 | 324 ± 32 |
| GST-Hel-His | 0.56 ± 0.05 | 61 ± 15 | 43 ± 1.4 | 470 ± 45 | 20 ± 4 |
Because ATP hydrolysis fuels helicase action, one might reasonably assume that those proteins that efficiently unwind RNA also hydrolyze ATP the most rapidly. Surprisingly, this is not the case. Even in the absence of RNA (Fig. 9A), His-NS3-4A and GST-Hel-His hydrolyze ATP more slowly than the other proteins. Although the Vmax for His-NS3-4A almost reaches that obtained with Hel-His, dramatically more ATP is required to reach these rates. The Km with His-NS3-4A is 82-fold higher than that seen with Hel-His. Likewise, when the protein is tagged with an N-terminal GST peptide (GST-Hel-His), the Km for ATP is 10–21-fold higher than it is with the same protein with simply a His tag at the N terminus (His-Hel, His-Hel-His). If ATP is in a rapid equilibrium with each of these helicases, the data would suggest that the bulky N-terminal additions somehow prevent ATP from binding to the helicase.
Similar, but less striking Km differences are seen in the presence of RNA (Fig. 9B and Table III). In this case the Km for ATP in the reaction catalyzed by the full-length protein is 63 times higher than it is in reactions catalyzed by Hel-His. Un like in the absence of RNA, however, the GST-Hel-His has a Km that more resembles the other proteins. Also noteworthy is the fact that His-NS3-4A again hydrolyzes ATP more slowly than the fragments lacking a protease domain. The Vmax catalyzed by His-NS3-4A is only 17% of that catalyzed by Hel-His. Because the Vmax values were more similar in the absence of RNA, these data suggest that although ATP hydrolysis by the full-length protein is stimulated 10-fold, the protein lacking a protease is stimulated up to 54-fold.
As was seen with saturating RNA (Fig. 9B), a saturating amount of ssDNA (Fig. 9C) stimulates ATP hydrolysis catalyzed by Hel-His better than that catalyzed by the other proteins. Again, His-Hel, His-Hel-His, and GST-Hel-His have similar Vmax values, but all have a significantly higher Vmax than His-NS3-4A. The KDNA values listed in Table III reveal that less ssDNA is required to stimulate GST-Hel-His and His-NS3-4A than the other proteins, possibly indicating tighter binding. His-Hel-His and His-Hel have similar weaker KDNA values, and Hel-His interacts with DNA in this assay the weakest with a KDNA 5-fold higher than His-NS3-4A.
Discussion
Until now it has been generally assumed that the HCV NS3 protease and helicase are two independent units that are simply combined into a single multifunctional protein. Most previous studies have not noted differences in helicase (11, 23, 41), RNA binding (42), or RNA replication assays (24) when NS3 was truncated to delete the protease. In early studies Heilek and Peterson (11), concluded that the protease domain had little or no effect on helicase activity. In fact, in their assays, the full-length protein seemed to unwind RNA more slowly. Likewise, Gallinari et al. (23) concluded that there were no significant differences between the full-length protein and the isolated helicase domain in either ATPase or RNA unwinding assays. Both full-length NS3 and a truncated helicase fragment have been shown specifically to bind sequences in the HCV genome, and the protease is apparently not required for this interaction (42). In another interesting study that showed an ability of NS3 to stimulate RNA synthesis catalyzed by the HCV NS5B RNA-dependent RNA polymerase, both full-length and truncated NS3 behaved similarly (24). In contrast to these studies, Howe et al. (43) reported that a single chain recombi-nant protein where NS4A is attached to the N terminus of NS3 was more active in helicase assays than either NS3 or a truncated protein. That single chain NS4-NS3 protein was later crystallized and its structure is shown in Fig. 1B. When a molecular model of the single chain NS4-NS3 protein (5) is compared with one of a helicase fragment with a bound DNA oligonucleotide (7), it is clear that the protease active site and its NS4A cofactor are each more than 30 Å from the known DNA binding site and putative ATP binding site (Fig. 1B). The protease and helicase are connected via a flexible linker, and only a few residues in the protease domain contact residues in the helicase domain.
Although no role for the protease in helicase action has been identified previously, recently the NS4A protein has been suggested to help load helicase on RNA (44). This model is based mainly on the report by Pang et al. (25) that the presence of NS4A reduces the amount of preincubation time required for maximal rates of unwinding in the presence of a DNA trap. Also supporting this idea is the observation that when NS4A is covalently tethered to NS3, the helicase activity appears to be enhanced (43). It is therefore possible that the specific effects seen here are not caused by the protease, but instead NS4A. However, we are skeptical that the effects seen here are the result of NS4A for a couple of reasons. First, we did not find that the preincubation time influenced the RNA (or DNA) helicase activity of any of the proteins studied here (data not shown). Second, all assays were performed at low ionic strength, where NS4A might be dissociated from NS3. NS3 and NS4A bind via hydrophobic interaction stabilized by the presence of salt. When salt is absent, NS4A no longer stimulates HCV protease, indicating that it may be dissociated (45). Further examination of the role of NS4A will require purified NS4A which can be quantitatively added back to helicase assays. Unfortunately, this will be a very difficult experiment because of the insoluble nature of the NS4A protein.
The experiments described here clearly delineated effects of the NS3 protease domain on its helicase function which were discussed as either specific or nonspecific effects. For example, an NS3 protein lacking the protease domain unwinds duplex RNA slower than the full-length protein, but this rate can be accelerated by either the presence of an N-terminal His tag or GST tag (a nonspecific effect). On the other hand, no fusion protein can enable the helicase fragment to unwind RNA efficiently in the presence of a DNA trap (a specific effect). Of course, it is recognized these supposedly specific effects might in fact be nonspecific because non-HCV peptides that were not tested might duplicate these effects.
One can speculate about how the protease-NS4A complex might nonspecifically influence the helicase based on the molecular model shown in Fig. 1B. All helicase fragments used here begin with a loop that extends along the back of the helicase connecting to domain 1 (red in Fig. 1). It is possible that this region that is not resolved in the structures of the helicase fragments (6–8), when not anchored to the back of the helicase, allows the protein to assume conformations that do not bind RNA tightly during helicase movements. The protease or fusion peptides might simply act like an anchor. Specific effects of the protease-NS4A complex are more difficult to explain. Perhaps a groove between the protease and helicase domain 2 provides a binding site for the strand of RNA not yet seen in the published molecular models. We plan to test some of these ideas using structure-based site-directed mutagenesis.
The results presented here also explain why many prior studies have only noted monomers of the HCV helicase, whereas others have detected dimers or higher order oligomers. As shown in Fig. 8 and Table II, the numbers of helicase monomers binding to a single oligonucleotide depend on the type of protein utilized. Furthermore, although the precise mechanism has not yet been elucidated, the proteins appear to act cooperatively in unwinding (Figs. 3 and 5) and binding assays (Fig. 8). Although prior studies with the fragment alone suggest that the HCV helicase moves along RNA as monomers like an inchworm (7, 10, 16, 22, 34), the mechanism of action of the full-length protein is clearly more complex.
Such oligomerization could be indicative of a rolling model like the ones proposed to explain the mechanism of action of other helicases such as Rep (46) and UvrD (33, 47). Based on an analysis of crystal packing interactions, Cho et al. (8) proposed that HCV helicase could form a dimer by forming contacts between helicase domains 1 and 2 of two separate monomers. Other evidence for helicase dimer formation has come from yeast two-hybrid assays that suggest that NS3 protein forms a dimer via domain 1 of the helicase (17, 39). Using a reverse two-hybrid screen, Khu et al. (17) have isolated three mutations in domain 1 which are unable to form dimers: T266A, Y267S, and M288T. It is possible that without the protease tethered to the back of the helicase, these residues in domain 1 might not be positioned properly to form a dimer. Again this could be tested using mutagenesis and the assays described here.
Both the helicase and protease portion of NS3 are evolutionarily conserved with cellular proteins, namely the DExD/H box helicase family and the chymotrypsin-like ser-ine protease family. It would appear that two proteins had simply combined into a single protein. This would be of a clear advantage to the virus because by having added the protease to the helicase, the virus essentially converted a DNA helicase into an RNA helicase. Why a primordial RNA virus would possess a helicase that only unwinds DNA is not known, but nevertheless, the current protein provides an interesting possibility for replication regulation. Based on our data, we propose that the helicase could disengage from RNA simply by rotating the protease away from the back of the helicase domain. Such a rotation has already been proposed by Yao et al. (5), who observed that such a rotation would be necessary for the NS3 protease to cleave the rest of the viral polyprotein. If the protease swings away, then the helicase might behave more like the isolated domain fragments that appear to fall from RNA rapidly during translocation. It is also possible that inhibitors of the protease could lock the protein in a conformation that unwinds RNA poorly and therefore inhibit viral replication by both inhibiting polyprotein processing and RNA replication.
Clearly, the full-length and truncated versions unwind RNA very differently, and the activity of the isolated helicase fragments can be greatly influenced by the nature of the fusion protein attached to its N terminus. This means that many of the general conclusions of numerous studies that utilized recombinant proteins encoding only fragments of the NS3 protein will need to be reconsidered, and many of the more rigorous studies will need to be repeated with the full-length NS3-protein.
Acknowledgments
We thank Fred Jaffe and Ruth Gallagher for valuable technical assistance
Footnotes
The abbreviations used are: HCV, hepatitis C virus; GST, glutathione S-transferase; MOPS, 4-morpholinepropanesulfonic acid; NS3, nonstructural protein 3; ss, single-stranded; F18, 5′-GCC TCG CTG CCG TCG CCA-fluorescein.
This work was supported by the American Association for the Study of Liver Diseases Liver Scholar Award from the American Liver Foundation and National Institutes of Health Grant AI052395 (both to D.N.F.).
References
- 1.Tan SL, Pause A, Shi Y, Sonenberg N. Nat Rev Drug Discov. 2002;1:867–881. doi: 10.1038/nrd937. [DOI] [PubMed] [Google Scholar]
- 2.Kolykhalov AA, Mihalik K, Feinstone SM, Rice CM. J Virol. 2000;74:2046–2051. doi: 10.1128/jvi.74.4.2046-2051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frick DN. Curr Org Chem. 2003;7 in press. [Google Scholar]
- 4.Yanagi M, Purcell RH, Emerson SU, Bukh J. Proc Natl Acad Sci U S A. 1997;94:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao N, Reichert P, Taremi SS, Prosise WW, Weber PC. Struct Fold Des. 1999;7:1353–1363. doi: 10.1016/s0969-2126(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 6.Yao N, Hesson T, Cable M, Hong Z, Kwong AD, Le HV, Weber PC. Nat Struct Biol. 1997;4:463–467. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]
- 7.Kim JL, Morgenstern KA, Griffith JP, Dwyer MD, Thomson JA, Murcko MA, Lin C, Caron PR. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 8.Cho HS, Ha NC, Kang LW, Chung KM, Back SH, Jang SK, Oh BH. J Biol Chem. 1998;273:15045–15052. doi: 10.1074/jbc.273.24.15045. [DOI] [PubMed] [Google Scholar]
- 9.Sali DL, Ingram R, Wendel M, Gupta D, McNemar C, Tsarbopoulos A, Chen JW, Hong Z, Chase R, Risano C, Zhang R, Yao N, Kwong AD, Ramanathan L, Le HV, Weber PC. Biochemistry. 1998;37:3392–3401. doi: 10.1021/bi972010r. [DOI] [PubMed] [Google Scholar]
- 10.Preugschat F, Danger DP, Carter LH, 3rd, Davis RG, Porter DJ. Biochemistry. 2000;39:5174–5183. doi: 10.1021/bi9923860. [DOI] [PubMed] [Google Scholar]
- 11.Heilek GM, Peterson MG. J Virol. 1997;71:6264–6266. doi: 10.1128/jvi.71.8.6264-6266.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin L, Peterson DL. Arch Biochem Biophys. 1995;323:47–53. doi: 10.1006/abbi.1995.0008. [DOI] [PubMed] [Google Scholar]
- 13.Lin C, Kim JL. J Virol. 1999;73:8798–8807. doi: 10.1128/jvi.73.10.8798-8807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du MX, Johnson RB, Sun XL, Staschke KA, Colacino J, Wang QM. Biochem J. 2002;363:147–155. doi: 10.1042/0264-6021:3630147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lam AMI, Keeney D, Eckert PQ, Frick DN. J Virol. 2003;77:3950–3961. doi: 10.1128/JVI.77.7.3950-3961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lam AMI, Keeney D, Frick DN. J Biol Chem. 2003;278:44514–44524. doi: 10.1074/jbc.M306444200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khu YL, Koh E, Lim SP, Tan YH, Brenner S, Lim SG, Hong WJ, Goh PY. J Virol. 2001;75:205–214. doi: 10.1128/JVI.75.1.205-214.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubendorff JW, Studier FW. J Mol Biol. 1991;219:45–59. doi: 10.1016/0022-2836(91)90856-2. [DOI] [PubMed] [Google Scholar]
- 20.Morris PD, Raney KD. Biochemistry. 1999;38:5164–5171. doi: 10.1021/bi9822269. [DOI] [PubMed] [Google Scholar]
- 21.Morris PD, Byrd AK, Tackett AJ, Cameron CE, Tanega P, Ott R, Fanning E, Raney KD. Biochemistry. 2002;41:2372–2378. doi: 10.1021/bi012058b. [DOI] [PubMed] [Google Scholar]
- 22.Levin MK, Gurjar MM, Patel SS. J Biol Chem. 2003;278:23311–23316. doi: 10.1074/jbc.M301283200. [DOI] [PubMed] [Google Scholar]
- 23.Gallinari P, Brennan D, Nardi C, Brunetti M, Tomei L, Steinkuhler C, De Francesco R. J Virol. 1998;72:6758–6769. doi: 10.1128/jvi.72.8.6758-6769.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piccininni S, Varaklioti A, Nardelli M, Dave B, Raney KD, Mc-Carthy JE. J Biol Chem. 2002;277:45670–45679. doi: 10.1074/jbc.M204124200. [DOI] [PubMed] [Google Scholar]
- 25.Pang PS, Jankowsky E, Planet PJ, Pyle AM. EMBO J. 2002;21:1168–1176. doi: 10.1093/emboj/21.5.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lohmann V, Korner F, Dobierzewska A, Bartenschlager R. J Virol. 2001;75:1437–1449. doi: 10.1128/JVI.75.3.1437-1449.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blight KJ, McKeating JA, Rice CM. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu B, Gates AT, Isken O, Behrens SE, Sarisky RT. J Virol. 2003;77:5352–5359. doi: 10.1128/JVI.77.9.5352-5359.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grobler JA, Markel EJ, Fay JF, Graham DJ, Simcoe AL, Lud-merer SW, Murray EM, Migliaccio G, Flores OA. J Biol Chem. 2003;278:16741–16746. doi: 10.1074/jbc.M212602200. [DOI] [PubMed] [Google Scholar]
- 30.Bukh J, Pietschmann T, Lohmann V, Krieger N, Faulk K, Engle RE, Govindarajan S, Shapiro M, St Claire M, Bartenschlager R. Proc Natl Acad Sci U S A. 2002;99:14416–14421. doi: 10.1073/pnas.212532699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paolini C, De Francesco R, Gallinari P. J Gen Virol. 2000;81:1335–1345. doi: 10.1099/0022-1317-81-5-1335. [DOI] [PubMed] [Google Scholar]
- 32.Gwack Y, Kim DW, Han JH, Choe J. Eur J Biochem. 1997;250:47–54. doi: 10.1111/j.1432-1033.1997.00047.x. [DOI] [PubMed] [Google Scholar]
- 33.Ali JA, Lohman TM. Science. 1997;275:377–380. doi: 10.1126/science.275.5298.377. [DOI] [PubMed] [Google Scholar]
- 34.Porter DJ, Short SA, Hanlon MH, Preugschat F, Wilson JE, Willard DH, Jr, Consler TG. J Biol Chem. 1998;273:18906–18914. doi: 10.1074/jbc.273.30.18906. [DOI] [PubMed] [Google Scholar]
- 35.Levin MK, Patel SS. J Biol Chem. 2002;277:29377–29385. doi: 10.1074/jbc.M112315200. [DOI] [PubMed] [Google Scholar]
- 36.Levin MK, Patel SS. J Biol Chem. 1999;274:31839–31846. doi: 10.1074/jbc.274.45.31839. [DOI] [PubMed] [Google Scholar]
- 37.Locatelli GA, Spadari S, Maga G. Biochemistry. 2002;41:10332–10342. doi: 10.1021/bi026082g. [DOI] [PubMed] [Google Scholar]
- 38.Preugschat F, Averett DR, Clarke BE, Porter DJT. J Biol Chem. 1996;271:24449–24457. doi: 10.1074/jbc.271.40.24449. [DOI] [PubMed] [Google Scholar]
- 39.Flajolet M, Rotondo G, Daviet L, Bergametti F, Inchauspe G, Tiollais P, Transy C, Legrain P. Gene (Amst) 2000;242:369–379. doi: 10.1016/s0378-1119(99)00511-9. [DOI] [PubMed] [Google Scholar]
- 40.Suzich JA, Tamura JK, Palmer-Hill F, Warrener P, Grakoui A, Rice CM, Feinstone SM, Collett MS. J Virol. 1993;67:6152–6158. doi: 10.1128/jvi.67.10.6152-6158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim DW, Gwack Y, Han JH, Choe J. Virus Res. 1997;49:17–25. doi: 10.1016/s0168-1702(97)01452-4. [DOI] [PubMed] [Google Scholar]
- 42.Banerjee R, Dasgupta A. J Virol. 2001;75:1708–1721. doi: 10.1128/JVI.75.4.1708-1721.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Howe AY, Chase R, Taremi SS, Risano C, Beyer B, Malcolm B, Lau JY. Protein Sci. 1999;8:1332–1341. doi: 10.1110/ps.8.6.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silverman E, Edwalds-Gilbert G, Lin RJ. Gene (Amst) 2003;312:1–16. doi: 10.1016/s0378-1119(03)00626-7. [DOI] [PubMed] [Google Scholar]
- 45.Gallinari P, Paolini C, Brennan D, Nardi C, Steinkuhler C, De Francesco R. Biochemistry. 1999;38:5620–5632. doi: 10.1021/bi982892+. [DOI] [PubMed] [Google Scholar]
- 46.Wong I, Lohman TM. Science. 1992;256:350–355. doi: 10.1126/science.256.5055.350. [DOI] [PubMed] [Google Scholar]
- 47.Maluf NK, Fischer CJ, Lohman TM. J Mol Biol. 2003;325:913–935. doi: 10.1016/s0022-2836(02)01277-9. [DOI] [PubMed] [Google Scholar]
- 48.Gibrat JF, Madej T, Bryant SH. Curr Opin Struct Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]