Abstract
Despite enormous body plan variation, genes regulating embryonic development are highly conserved. Here, we probe the mechanisms that predispose ancient regulatory genes to reutilization and diversification rather than evolutionary loss. The Hox gene fushi tarazu (ftz) arose as a homeotic gene but functions as a pair-rule segmentation gene in Drosophila. ftz shows extensive variation in expression and protein coding regions but has managed to elude loss from arthropod genomes. We asked what properties prevent this loss by testing the importance of different protein motifs and partners in the developing CNS, where ftz expression is conserved. Drosophila Ftz proteins with mutated protein motifs were expressed under the control of a neurogenic-specific ftz cis-regulatory element (CRE) in a ftz mutant background rescued for segmentation defects. Ftz CNS function did not require the variable motifs that mediate differential cofactor interactions involved in homeosis or segmentation, which vary in arthropods. Rather, CNS function did require the shared DNA-binding homeodomain, which plays less of a role in Ftz segmentation activity. The Antennapedia homeodomain substituted for Ftz homeodomain function in the Drosophila CNS, but full-length Antennapedia did not rescue CNS defects. These results suggest that a core CNS function retains ftz in arthropod genomes. Acquisition of a neurogenic CRE led to ftz expression in unique CNS cells, differentiating its role from neighboring Hox genes, rendering it nonredundant. The inherent flexibility of modular CREs and protein domains allows for stepwise acquisition of new functions, explaining broad retention of regulatory genes during animal evolution.
Keywords: molecular evolution, fushi tarazu, protein evolution
The sets of genes regulating embryonic development are highly conserved throughout the animal kingdom despite the enormous diversity of body plans they control (1). How heterogeneity in body patterning is achieved through the action of these conserved regulatory genes is still largely unknown. Much of our current understanding rests on the observation that genes diversify through changes in cis-regulatory elements (CREs), whereas the transcription factors they encode are thought to be constrained, presumably because changes in protein coding regions of key developmental regulators would be highly detrimental (2, 3). This notion of static regulatory proteins is challenged by the finding that several Hox genes have acquired new biological roles during evolution (4–8). These genes were able to take on new roles because of redundancy, yet their ability to change function raises additional questions. Why were these genes not simply lost due to redundancy? Did these genes take on new required functions in a single step, with the new function imposing positive selection? Did changes occur stepwise, with retention at intermediate stages due to drift?
The fushi tarazu (ftz) gene arose as a duplication of an Antennapedia (Antp)-like Hox gene, presumably early in protostome lineages (9, 10). Overlap in expression and function with neighboring Hox genes Antp and/or Sex-combs reduced (Scr) allowed diversification: ftz neofunctionalized to take on a role in segmentation in higher insects, whereas Antp and Scr retained ancestral functions in determining segment identity (4, 5, 10–12). The segmentation function of ftz, studied in depth in Drosophila, required a change in expression pattern from a single Hox-like domain to seven stripes in the primoridia of alternate body segments (10, 13, 14). Ftz segmentation function also requires an LXXLL motif that mediates interaction with an obligate cofactor, the orphan nuclear receptor Ftz-F1 (15–20). Interestingly, ectopic expression of a Ftz protein lacking its homeodomain caused an anti-ftz phenotype, and this protein was able to rescue segmentation defects in ftz mutants (21, 22), suggesting relaxed selection on the Ftz homeodomain for its role in segmentation. In contrast, the LXXLL motif was strictly required for Ftz segmentation function (18–20).
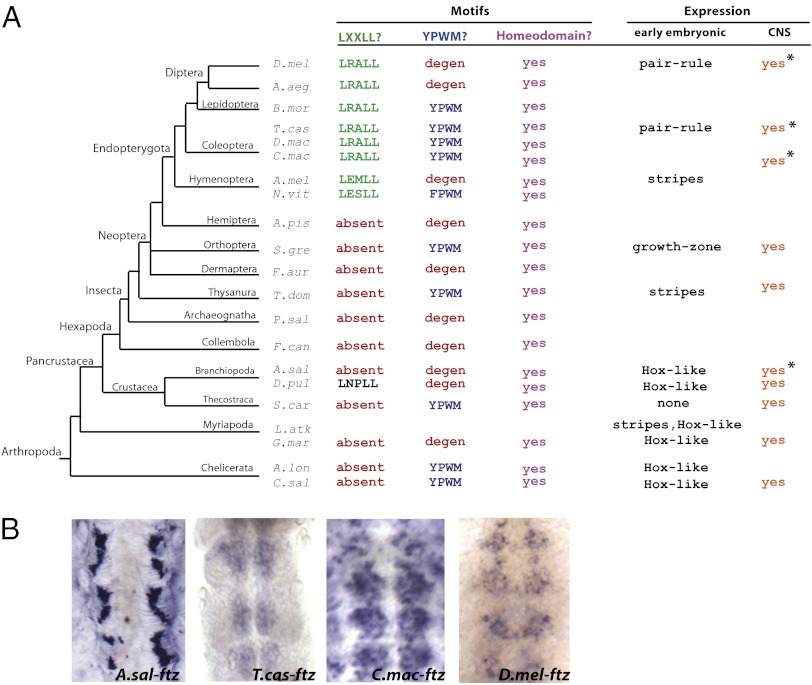
Tracking changes within an established arthropod phylogeny revealed unexpected lability in ftz expression and Ftz protein domains (4): Hox-like expression is retained in several myriapods and chelicerates but is virtually lost in a crustacean (Fig. 1A). Striped expression was reported in the basal insect Thermobia (23) and in several holometabolous insects (13, 24, 25). ftz likely lost striped expression in at least one lineage, represented by extant grasshoppers, where ftz is expressed in the growth zone but not in stripes (26). In addition to these changes in expression, Ftz stably acquired an LXXLL motif at the base of holometabolous insects, suggesting these Ftz proteins could interact with Ftz-F1. Interestingly, an ancestral YPWM motif that mediates interaction of Hox proteins with cofactor Extradenticle (Exd) (27) independently degenerated at least six times in arthropods.
Fig. 1.
ftz is expressed in the developing CNS throughout arthropods, despite diversity in Ftz cofactor interaction motifs and early expression patterns. (A) An arthropod phylogeny showing the presence/absence of functional Ftz motifs and expression patterns during embryogenesis. The LXXLL motif (green) is required for pair-rule function in Drosophila and mediates interaction with the Ftz cofactor Ftz-F1. LXXLL was stably acquired at the base of Endopterygota. The YPWM mediates interaction with the homeotic cofactor Exd. This motif is present in Ftz in some arthropods (blue), but has degenerated in many lineages (red). All Ftz sequences have a homeodomain (purple). The early embryonic expression pattern of ftz has been reported as Hox-like (Crustacea, Myriapoda, Chelicerata), in the growth zone (Orthoptera), and in stripes (Thysanura, Hymenoptera, Coleoptera, Diptera). ftz CNS expression has been reported in many arthropods (orange). Asterisks indicate expression patterns examined in this study. (B) Neuronal expression of ftz was analyzed by in situ hybridization in embryos using probes to Artemia (A.sal), Tribolium (T.cas), Callosobruchus (C.mac), and Drosophila (D.mel) ftz sequences, as indicated.
Despite these dynamic changes in sequence and expression, it is striking that the ftz gene is retained in all arthropod genomes examined to date. Here we show that extensive functional variation in ftz in arthropods is balanced by constraints of a core function in the developing central nervous system (CNS). The LXXLL segmentation motif and degenerate homeotic motif (FNWS) in Drosophila Ftz are dispensable for CNS function, but a homeodomain is required for activation of Eve expression in RP2 neurons in the CNS. Interestingly, the Antp homeodomain can substitute for the Ftz homeodomain in this core function, suggesting that acquisition of a neurogenic ftz CRE led to ftz expression in a unique group of cells, differentiating its role from neighboring Hox genes. However, even here, changes in protein sequence contribute to functional specificity, as full-length Antp cannot substitute for Ftz. Here we provide strong evidence to support the hypothesis that constraint on one protein domain for one tissue-specific function has led to long-term retention of a gene during evolution while enabling extensive diversification in other protein domains that play critical roles in other tissues. Together, these results suggest that evolutionary diversification in gene function can occur through differential selection for individual subfunctions of a single protein, which play different roles in different cell types.
Results
ftz CNS Expression Is Conserved over 550 My of Arthropod Evolution.
Given the diversity in ftz expression and protein motifs, it is surprising that the gene is retained in all arthropod genomes examined (Fig. 1A). ftz is expressed in the embryonic CNS in a broad range of arthropods, including myriapods (28–30), crustaceans (4, 31), insects (14, 23, 24, 26), and a distant lophotrochozoa, where the ftz ortholog Lox5 is expressed in the CNS (10, 32). This CNS expression is conserved in arthropods with diverse Ftz sequences and early expression patterns (Fig. 1B). Artemia Ftz is 201 amino acids long, lacks LXXLL and YPWM motifs, and has weak Hox-like expression in early nauplii (4). ftz from Tribolium and Callosobruchus beetles encode proteins that are 290 and 368 residues, respectively; both sequences have LXXLL and YPWM motifs and are expressed in stripes (24, 33). Drosophila Ftz is 410 amino acids long, includes an LXXLL but no YPWM motif, and is expressed in stripes (14). Thus, despite diversity in sequence and expression, conservation of ftz expression in the developing CNS appears to be a constant feature of extant ftz genes.
Candidate Ftz Cofactors Are Not Coexpressed with Ftz in the CNS.
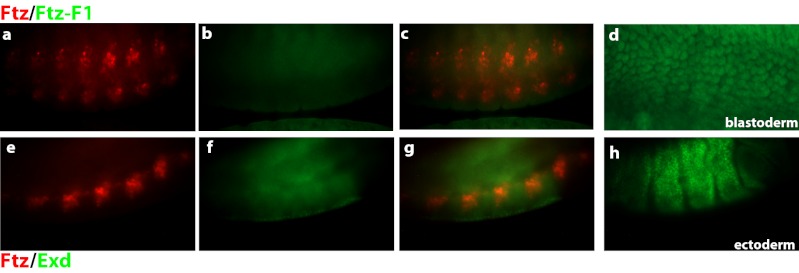
During the blastoderm stage of Drosophila development, Ftz interacts with Ftz-F1, and together they bind composite sites in regulatory regions of segmentation target genes, activating their expression (15, 17, 19, 34). If Ftz regulates CNS target genes by this mechanism, a minimal requirement is coexpression of Ftz and Ftz-F1 in this tissue. However, Ftz-F1 expression was not detectable in Ftz+ neurons (Fig. 2 A–C; Ftz, red; Ftz-F1, green). Nuclear Ftz-F1 expression was readily detected using the same antibody at the blastoderm stage (Fig. 2D). Additionally, ftz-f1 RNA was not detected at this stage of development (35). Because Ftz retains the “W” residue in the YPWM motif critical for interaction with Exd, we asked whether Ftz could use Exd as a cofactor in regulating gene expression in the CNS. Although Exd is expressed at this time, expression was localized to the ectoderm (Fig. 2H), and expression did not overlap with Ftz+ neurons (Fig. 2 E–G). Thus, neither Ftz-F1 nor Exd colocalizes with Ftz in the developing CNS.
Fig. 2.
Ftz is not coexpressed with known cofactors in the CNS. (A) Drosphila Ftz colocalizes with cofactor Ftz-F1 during the blastoderm stage of development but not in the CNS. Ftz (red) expression in a cluster of cells in every segment of the developing CNS. (B) Only faint background staining was observed with anti–Ftz-F1 antibody (green). (C) Merge of images in A and B shows Ftz and Ftz-F1 do not colocalize in the CNS. (D) Nuclear Ftz-F1 expression is detected using this same antibody at the blastoderm stage of development. (E) Ftz-expressing neurons (red) do not overlap with (F) Exd (green), which was expressed in the nuclei of ectodermal cells. (G) Merge of images from D and E shows expression of Ftz and Exd in different cell layers (Exd out of focus in this figure; in focus in H).
Cofactor Interaction Motifs in Dm-Ftz Are Dispensable for CNS Function.
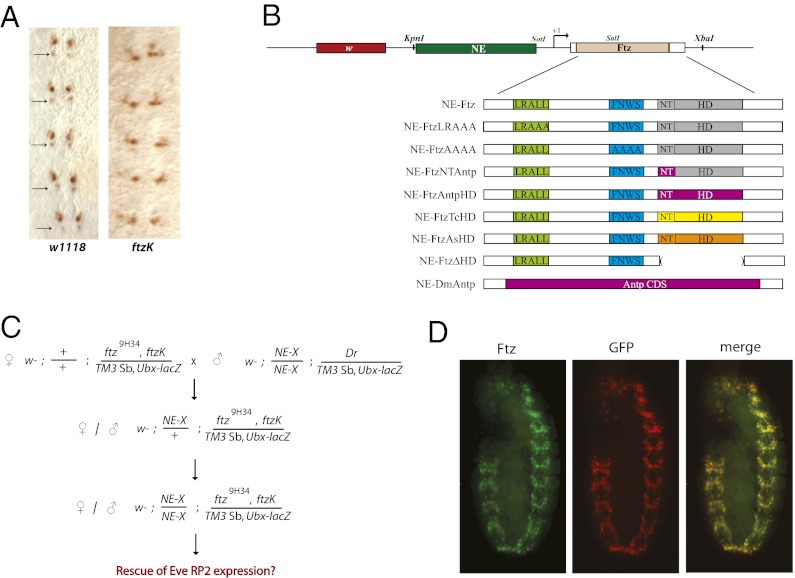
If the evolutionary constant function of Ftz is its role in the CNS, the LXXLL and YPWM motifs should be dispensable for this function, because many Ftz proteins lack one or both motifs (Fig. 1A). To test this, we made use of a Drosophila line carrying a rescue transgene that lacks the ftz neurogenic element (ftzK) (36, 37). Expression of ftzK in a ftz9H34 background rescues segmentation defects but not CNS function; RP2 neurons fail to develop, as evidenced by lack of Even-skipped (Eve) expression (Fig. 3A) (36). To test whether characterized Ftz motifs are necessary for CNS function, we generated a series of transgenes containing the ftz neurogenic element (NE), basal promoter, and WT coding sequence (NE-Ftz) or with mutations in protein motifs (NE-X; Fig. 3B). LRALL was changed to LRAAA, which abolishes interaction with Ftz-F1 (NE-FtzLRAAA; ref. 11); the FNWS motif was changed to AAAA (NE-FtzAAAA) to abolish potential interaction with Exd; and several mutations were made in the homeodomain: (i) the N-terminal arm of the homeodomain (SKRTRQTY) was changed to that of Antp (RKRGRQTY; NE-FtzNTAntp); (ii) the Ftz homeodomain was swapped with the homeodomain from Antp (NE-FtzAntpHD), Tribolium (NE-FtzTcHD), or Artemia (NE-FtzAsHD); and (iii) the Ftz homeodomain was deleted (NE-FtzΔHD). Last, the entire Ftz coding region was replaced with that of Antennapedia (NE-DmAntp). For each NE-X, flies carrying two copies of the transgene on chromosome II were crossed with ftzK, ftz9H34/TM3 Ubx-lacZ (Fig. 3C). Rescue efficiency was calculated as the percentage of ftzK, ftz9H34 embryos that showed Eve expression in RP2 neurons. To confirm that the ftz CRE used here was sufficient to drive transgene expression in a ftz-like CNS pattern, the Ftz coding sequence was replaced with GFP (NE-GFP). Indeed, NE-GFP expression was detected only in the CNS and overlapped with all native Ftz+ neurons (Fig. 3D).
Fig. 3.
Strategy to test the role of Ftz protein motifs in Drosophila CNS function. (A) Eve expression is detected in RP2 neurons of stage 10–12 embryos (Left, arrows), but missing in ftzK mutants (Right) (36). (B) Schematic of constructs designed to test motif function in the CNS. All transgenes included the 2.2-kb ftz NE (37), basal promoter, coding region, and ∼200 bp downstream of the 3′UTR. Transgenes: the segmentation LRALL motif (bright green) was mutated to LRAAA in NE-FtzLRAAA; the degenerate homeotic FNWS motif (blue) changed to AAAA in NE-FtzAAAA; the N-terminal arm of Ftz (gray, NT) changed to that of Dm-Antp (purple) in NE-FtzNTAntp; the Ftz homeodomain (gray) swapped with that of Dm-Antp (purple) in NE-FtzAntpHD; the Ftz homeodomain (gray) swapped with that of Tribolium (yellow) in NE-FtzTcHD; the Ftz homeodomain (gray) swapped with that of Artemia (orange) in NE-FtzAsHD; the homeodomain deleted from NE-FtzΔHD (shown with empty parentheses); and the entire Ftz coding region replaced with Antennapedia (purple) in NE-DmAntp. (C) Crossing scheme used to test CNS functional rescue. All crosses were carried out with multiple independent lines for each of the transgenes shown in B, indicated as NE-X. After establishing flies homozygous for NE-X and carrying ftzK on a ftz9H34 chromosome, these lines were self-crossed, and embryos were tested for rescue of Eve RP2 neuron expression. (D) NE-GFP lines were double-stained with antibodies against Ftz (green) and GFP (red). Expression of these proteins overlapped (merge, yellow), showing that the ftz NE used to drive expression was sufficient to test ftz function in the CNS.
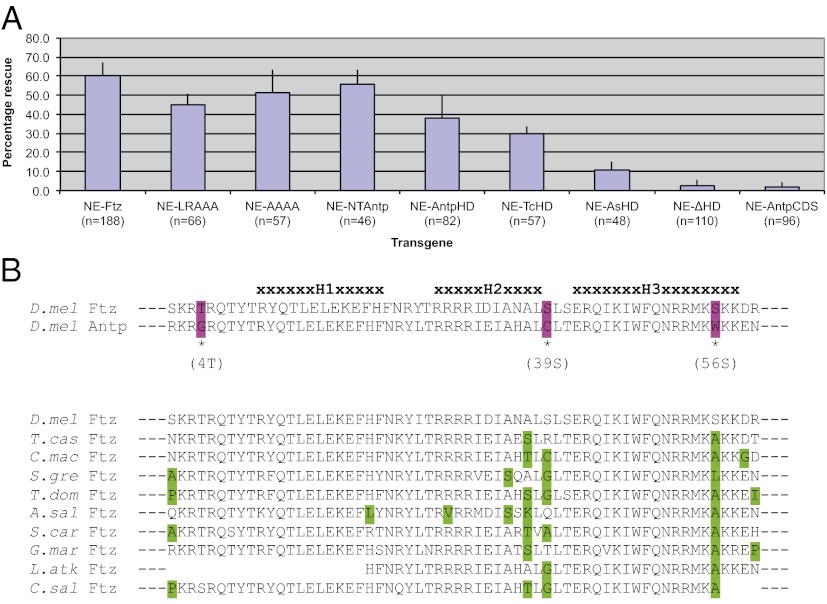
As shown in Fig. 4A, Eve RP2 expression was rescued by expression of NE-Ftz. NE-FtzLRAAA, in which the LXXLL motif is inactivated, also rescued Eve RP2 expression. Note that because the observed level of rescue of NE-FtzLRAAA was slightly lower than other transgenes, we cannot rule out the possibility that the LXXLL motif or nearby residues are important for the structure or function of Ftz in the Drosophila CNS.
Fig. 4.
The homeodomain is required for Ftz function in the CNS, whereas cofactor interaction motifs are dispensable. (A) Rescue of Eve expression in RP2 neurons by different transgenes, indicated by the percentage of ftz mutant embryos that displayed Eve RP2 neuronal staining, is shown. Bars indicate 1 SD. Ftz proteins lacking the cofactor interaction motifs LXXLL and YPWM effectively rescued ftz CNS function, as did transgenes with the Antp homeodomain. However, deletion of the homeodomain abolished rescue. Full-length Antp did not substitute for Ftz in the CNS. (B) Homeodomain alignments highlighting differences in sequence. Dm-Ftz and Dm-Antp homeodomain sequences are 95% similar and 83% identical, with only three nonconservative substitutions at the amino acid level (Upper, purple). Alignment of Ftz homeodomains from species where CNS expression has been reported (Lower: green = nonconservative substitutions).
NE-FtzAAAA, in which the degenerate YPWM motif is mutated, rescued Eve RP2 expression. This finding suggests that the degenerate YPWM motif in Drosophila Ftz, including the conserved W in the FNWS motif, is not necessary for Ftz CNS function. Taken together with the expression data above, these results indicate that Ftz function in the CNS is independent of Ftz-F1 and Exd.
Homeodomain Is Required for Ftz CNS Function.
In contrast to the motifs that vary in Ftz from different species, the homeodomain was absolutely required for CNS function, as NE-FtzΔHD showed virtually no rescue of Eve RP2 expression (Fig. 4A). The N-terminal arm of the homeodomain confers specificity and is used to classify Hox paralog groups (reviewed in ref. 38). To test whether a Ftz group homeodomain was required for CNS function, its N-terminal arm was substituted with that of Antp. This protein, NE-FtzNTAntp, rescued Eve RP2 expression, suggesting that N-terminal homeodomain specificity is not necessary for Ftz CNS function. To test the extent of entire homeodomain specificity, the Ftz homeodomain was replaced with that of Antp (NE-FtzAntpHD), which also effectively rescued Eve RP2 expression. Substituting the Ftz homeodomain with that of Tribolium Ftz also supported rescue of CNS function, whereas the Artemia Ftz HD substitution only weakly rescued it. We suggest that these Ftz homeodomains from distant taxa have diverged in specific or general manners that decrease their activity when expressed in Drosophila, similar to the full-length Sg-Ftz, which was less active in Drosophila than more closely related Ftz proteins (12).
The results presented above suggest that residues that differentiate Ftz and Antp family homeodomains are not involved in Ftz CNS function. Although these homeodomains are similar, there are 10 amino acid differences, 3 of which are nonconservative (Fig. 4B, Upper, purple). Interestingly, these three nonconservative substitutions are residues that could be phosphorylated in Ftz but not Antp. However, because NE-FtzAntpHD was able to rescue Eve RP2 expression, phosphorylation at these sites must not be crucial for Ftz homeodomain function in regulating Eve RP2 expression in the CNS, although we cannot rule out specificity in other CNS functions of Ftz. Also, an alignment of Ftz homeodomain sequences from arthropods with documented ftz CNS expression suggests that these three nonconservative residues are not highly constrained, especially at position 39 of the homeodomain, which exhibits great diversity (Fig. 4B, Lower).
Ftz Function in the CNS Is Not Dependent on Antp and Cannot Be Substituted by Antp.
Antp is also expressed in the developing CNS, raising the possibility that NE-FtzAntpHD rescued Ftz CNS function because Antp is activated by Ftz in the CNS. However, in WT embryos, Antp expression in the CNS does not overlap with Ftz (Fig. S1 A and B), suggesting separate roles of each; also, Antp expression persists longer than Ftz. Further, Antp expression was not altered in ftzK embryos (Fig. S1 C–F). Thus, Ftz has a distinct role in the developing CNS, and Ftz does not regulate Eve RP2 indirectly via activation of Antp.
Because Antp was not responsible for the observed rescue by ftz transgenes, but the Antp HD fully substituted for the Ftz HD in CNS function, we asked whether the full-length Antp protein, if expressed in Ftz+ cells under the control of the ftz NE, was capable of rescuing ftz CNS defects. No rescue of Eve expression in RP2 neurons was observed (Fig. 4A), suggesting that Ftz and Antp have diverged in residues other than the homeodomain or LXXLL or YPWM motifs, which are necessary for Ftz CNS activity.
Discussion
The ftz gene shows extensive variation throughout arthropods (Fig. 1A) (reviewed in refs. 5, 6, and 33). These variations comprise both loss and gain of expression patterns (e.g., loss of Hox-like expression in Artemia and gain of stripes in Thermobia) and loss and gain of cofactor interaction motifs (e.g., multiple losses of the YPWM motif and gain of the LXXLL motif in holometabolous insects). The loss of Hox-like expression combined with the absence of known cofactor-interaction motifs in species such as Artemia are suggestive of nonfunctionalization events that would be expected to lead to loss of the ftz gene entirely (39). However, ftz has been retained in all arthropod genomes examined to date. In contrast to the dynamic changes in ftz described above, ftz expression in the developing CNS appears to be a constant feature, as observed by Akam and colleagues several years ago, when even a smaller number of ftz genes had been examined (40). This suggests that the selective pressure stabilizing ftz in the genome is its role in the CNS. This hypothesis predicts that ftz CNS function would not require cofactor interactions and protein domains mediating them that are present in Ftz from some taxa but absent in others. Indeed, we found that rescue of ftz CNS function was achieved by proteins lacking the YPWM or LXXLL motifs. Only the homeodomain, which is present in all Ftz proteins, was indispensable. Thus, ftz expression and protein variation are balanced by constraints of a core CNS function that is broadly conserved in protostomes.
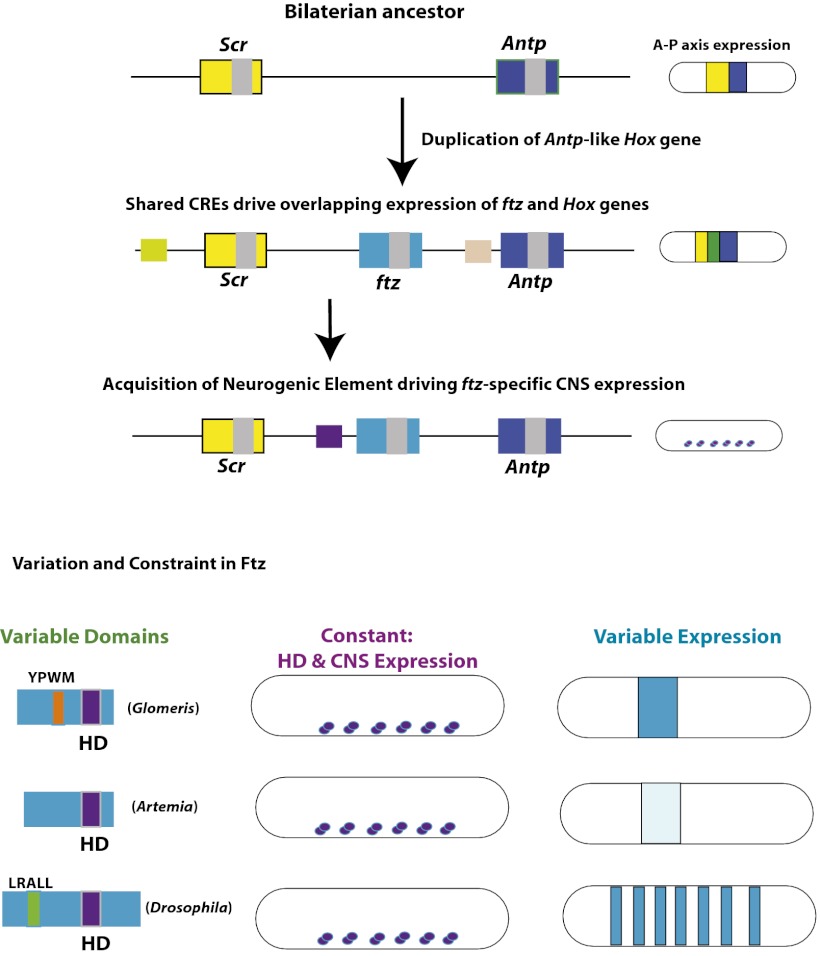
We propose that the modular structure of both CREs and protein domains permitted lability of ftz in nature, with the nonredundant neurogenic expression serving as a stabilizer for ftz in early protostome genomes (Fig. 5). Following duplication from an Antp-like ancestral gene, ftz remained under the control of neighboring CREs directing Hox-like expression of Scr and/or Antp (Fig. 5, yellow-green and tan squares). Remnants of this traditional collinear Hox expression remain in the mite and centipede, where ftz expression overlaps with Scr (10, 29); the millipede, where ftz expression overlaps with several Hox genes (30); and in Drosophila, where CREs that border the Dm-ftz gene direct expression that overlaps with Scr (41) and Antp (42, 43). A unique role for Ftz in the CNS resulted from a cis-regulatory change, specifically the acquisition of a NE directing ftz expression in specific cells of the developing CNS, different from those where the Antp-like ancestor was expressed (Fig. 5, purple square). This was likely a neofunctionalization event, although we cannot rule out the possibility that the ftz/Antp ancestor was expressed in these CNS cells. The observation that the Antp homeodomain can substitute for the Ftz homeodomain in CNS function suggested that Antp itself could have taken on the essential role in the CNS, had it come under control of the NE. However, the fact that the entire Antp coding region under control of the ftz NE did not rescue Eve RP2 expression suggests otherwise. Thus, either protein differences in a bilaterian ancestor prevented Antp from taking on this particular CNS role or the Drosophila Ftz and Antp proteins diverged in function after an initial cis-regulatory change that imparted different roles on them.
Fig. 5.
Acquisition of a ftz NE constrains the homeodomain, preventing its loss while permitting flexibility. (Upper) A bilaterian ancestor had a Hox cluster in which Scr (yellow) and Antp (dark blue) expression along the A-P axis were in regions adjacent to one another. After the Hox duplication that produced ftz (teal), expression overlapped (green) with neighboring Hox genes because shared CREs drove expression in the same regions along the anterior-posterior axis. Due to redundancy, ftz was allowed to diverge. At some later point, a unique ftz neurogenic CRE arose (purple), which drove ftz expression in a subset of neurons. (Lower) Because CNS function is dependent on the homeodomain (HD), this part of the protein has been maintained in all extant arthropod Ftz sequences. Other regions of the protein sequence have diversified, such as LXXLL acquisition (green) and YPWM (orange) degeneration, which vary in different species, exemplified by Glomeris, Artemia, and Drosophila Ftz. Additionally, unique CREs have arisen, driving ftz expression in stripes (lower right), allowing for cooption into alternate developmental pathways in different arthropod lineages.
Further changes in ftz expression, including degeneration of CREs driving overlapping expression with Scr and/or Antp, were permitted by their redundancy, as predicted by classical models (39, 44, 45). The escape from collinearity permitted further changes in ftz expression, exemplified by striped expression in insects, and sequence changes that resulted in a switch in cofactor interaction (5). However, despite this rather extreme lability, loss of ftz was constrained by a unique and required developmental role in the CNS. In contrast to the strict requirement for a homeodomain in the CNS, Ftz function in segmentation could be rescued by a Ftz protein lacking the homeodomain (21, 22). This segmentation function strictly required the LXXLL motif (18–20), which is not necessary for CNS function (Fig. 4A). Thus, there seem to be balancing constraints on different portions of Ftz for different functions: the homeodomain is constrained by CNS function, shared broadly across diverse taxa, whereas the LXXLL motif is constrained in holometabolous insects, presumably because of its role in segmentation (Fig. 1A).
The extreme flexibility of ftz challenges notions that embryonic regulatory genes are highly static and changes in gene function occur by a single mechanism. This finding is a striking example of mosaic pleiotropy enabling regulatory protein evolution, whose prevalence in other embryonic transcription factors is likely more widespread than previously realized. We propose that stepwise changes in function—both loss of function and neofunctionalization—build on each other to allow the sequential acquisition of new functions during evolution. This process is active, even for an embryonically active transcription factor predicted to be highly constrained, because changes in function (either loss or gain) would be highly detrimental to development, as evidenced by mutations made in the laboratory. Constraints on one protein domain for one tissue-specific function—in this case, the homeodomain required for CNS activity—have led to long-term retention of ftz during evolution while enabling extensive diversification in other domains dispensable for this core function. Thus, protein modularity permits differential selection for functions carried out by individual domains, thereby expanding the repertoire of material available for functional variation without changes in gene or isoform number. Cis-regulatory changes and protein multitasking build on each other, achieving a balance between constraint and variation. This inherent flexibility of an ancient set of regulatory genes allows for functional diversification and may explain their long-term retention during animal evolution.
Materials and Methods
Arthropod Embryo Collection and Fixation.
One-week-old Artemia nauplii were fixed according to ref. 4. Callosobruchus embryos that were 2 d old were collected by first soaking mung beans with eggs in a dilute bleach solution, scraping the eggs off the beans with a paintbrush, and then fixing according to standard Drosophila protocols. Drosophila embryos were collected over 2 h, aged for 5–6 h on apple juice plates at 25 °C, and then fixed according to standard protocols.
Ftz Rescue Transgenes.
The ∼2.2-kb fragment containing the NE extending from the XbaI to BalI restriction sites in the 10-kb genomic region sufficient for rescue of ftz mutants (37) was inserted into pCasper4, followed by insertion of the ftz basal promoter (∼40 bp upstream of the TSS) and 5′ UTR, coding region, 3′UTR, and ∼200 bp downstream of the polyadenylation signal using standard techniques. Mutations to the ftz coding region were made using site-directed mutagenesis (primer sequences available on request). Homeodomain deletion and swaps were done by fusion PCR. DNA sequences were verified for all fragments generated by PCR. Transgenic flies were generated by Rainbow Transgenic Flies, Inc. (Camarillo, CA). Due to lethality issues when expressing Hox-like genes with attB lines, traditional P-element integration techniques were used, such that transgenes were inserted randomly into the Drosophila genome (46). For each construct, three to seven independent lines were established that were homozygous viable on the second chromosome. Males homozygous for NE-Ftz constructs (NE-X), carrying Dr/TM3SbUbx-lacZ on chromosome III, were crossed with ftz9H34, ftzK/TM3Sb, Ubx-lacZ virgin females, and males and females carrying one copy of NE-Ftz and ftz9H34, ftzK/ TM3SbUbx-lacZ were selected and self-crossed (Fig. 3C). Rescue efficiency was measured by calculating the percentage of embryos homozygous for ftz9H34, ftzK (β-galactosidase negative embryos) that showed Eve antibody staining in any number of RP2 neurons in stage 10–12 embryos. Rescue percentages from several independent transgene lines were averaged together. To confirm that the ftz cis-regulatory elements present in the transgene were sufficient to drive transgene expression in the Ftz+ cells of the CNS, a transgene in which a GFP-coding sequence was placed downstream of the NE, ftz basal promoter, and first 169 amino acids of the Ftz coding region was generated. GFP was detected in an identical pattern to native Ftz protein, as visualized by double antibody staining of GFP and Ftz (Fig. 3D).
Analysis of Gene Expression Patterns.
In situ hybridizations were performed according to established protocols in Drosophila, Tribolium (47), and Artemia (4, 48, 49). Callosobruchus embryos were first dissected from their thick vitelline membrane and then stained according to Drosophila protocols. Digoxygenin-labeled probes were made with T7/T3 polymerase using embryonic cDNA, detected with a sheep anti-digoxigenin antibody (1:2,000; Roche), and stained with nitro-blue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolyl-phosphate (BCIP) according to the manufacturer’s instructions. Drosophila antibody stainings were performed according to established antibody protocols (50). Primary antibodies used were as follows: mouse anti-Ftz (1:1,000) (51); guinea pig anti-Eve (1:1,000), and rabbit anti-GFP (1:1,000; Invitrogen). Secondary antibodies used were as follows: anti-mouse Alexa488 (1:500; Molecular Probes), anti-rabbit Alexa568 (1:500; Molecular Probes), and biotinylated anti-guinea pig (1:1,000; KPL). Embryos were mounted in Vectashield mounting solution with DAPI (Vector Laboratories) and scored for rescue and photographed by Leica DMRB microscopy.
Supplementary Material
Acknowledgments
We thank Urs Kloter and Walter Gehring for maintaining and providing the ftzK line, Diane Duncan and John Reinitz for antibodies, and the Bloomington Stock Center for fly lines. This work was improved by comments on the manuscript from Steve Mount, Jeff Shultz, and Eric Haag. This work was supported by National Science Foundation Grant IOS-0950765.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1210847110/-/DCSupplemental.
References
- 1.Carroll SB, Grenier JK, Weatherbee SD. From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design. 2nd Ed. Malden, MA: Blackwell; 2005. [Google Scholar]
- 2.Carroll SB. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell. 2008;134(1):25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 3.Xue L, Noll M. The functional conservation of proteins in evolutionary alleles and the dominant role of enhancers in evolution. EMBO J. 1996;15(14):3722–3731. [PMC free article] [PubMed] [Google Scholar]
- 4.Heffer A, Shultz JW, Pick L. Surprising flexibility in a conserved Hox transcription factor over 550 million years of evolution. Proc Natl Acad Sci USA. 2010;107(42):18040–18045. doi: 10.1073/pnas.1010746107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pick L, Heffer A. Hox gene evolution: Multiple mechanisms contributing to evolutionary novelties. Ann N Y Acad Sci. 2012;1256:15–32. doi: 10.1111/j.1749-6632.2011.06385.x. [DOI] [PubMed] [Google Scholar]
- 6.Mann RS, Carroll SB. Molecular mechanisms of selector gene function and evolution. Curr Opin Genet Dev. 2002;12(5):592–600. doi: 10.1016/s0959-437x(02)00344-1. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt-Ott U, Rafiqi AM, Lemke S. Hox3/zen and the evolution of extraembryonic epithelia in insects. Adv Exp Med Biol. 2010;689:133–144. doi: 10.1007/978-1-4419-6673-5_10. [DOI] [PubMed] [Google Scholar]
- 8.McGregor AP. How to get ahead: The origin, evolution and function of bicoid. Bioessays. 2005;27(9):904–913. doi: 10.1002/bies.20285. [DOI] [PubMed] [Google Scholar]
- 9.Grenier JK, Garber TL, Warren R, Whitington PM, Carroll S. Evolution of the entire arthropod Hox gene set predated the origin and radiation of the onychophoran/arthropod clade. Curr Biol. 1997;7(8):547–553. doi: 10.1016/s0960-9822(06)00253-3. [DOI] [PubMed] [Google Scholar]
- 10.Telford MJ. Evidence for the derivation of the Drosophila fushi tarazu gene from a Hox gene orthologous to lophotrochozoan Lox5. Curr Biol. 2000;10(6):349–352. doi: 10.1016/s0960-9822(00)00387-0. [DOI] [PubMed] [Google Scholar]
- 11.Löhr U, Pick L. Cofactor-interaction motifs and the cooption of a homeotic Hox protein into the segmentation pathway of Drosophila melanogaster. Curr Biol. 2005;15(7):643–649. doi: 10.1016/j.cub.2005.02.048. [DOI] [PubMed] [Google Scholar]
- 12.Löhr U, Yussa M, Pick L. Drosophila fushi tarazu. a gene on the border of homeotic function. Curr Biol. 2001;11(18):1403–1412. doi: 10.1016/s0960-9822(01)00443-2. [DOI] [PubMed] [Google Scholar]
- 13.Hafen E, Kuroiwa A, Gehring WJ. Spatial distribution of transcripts from the segmentation gene fushi tarazu during Drosophila embryonic development. Cell. 1984;37(3):833–841. doi: 10.1016/0092-8674(84)90418-5. [DOI] [PubMed] [Google Scholar]
- 14.Carroll SB, Scott MP. Localization of the fushi tarazu protein during Drosophila embryogenesis. Cell. 1985;43(1):47–57. doi: 10.1016/0092-8674(85)90011-x. [DOI] [PubMed] [Google Scholar]
- 15.Yu Y, et al. The nuclear hormone receptor Ftz-F1 is a cofactor for the Drosophila homeodomain protein Ftz. Nature. 1997;385(6616):552–555. doi: 10.1038/385552a0. [DOI] [PubMed] [Google Scholar]
- 16.Guichet A, et al. The nuclear receptor homologue Ftz-F1 and the homeodomain protein Ftz are mutually dependent cofactors. Nature. 1997;385(6616):548–552. doi: 10.1038/385548a0. [DOI] [PubMed] [Google Scholar]
- 17.Florence B, Guichet A, Ephrussi A, Laughon A. Ftz-F1 is a cofactor in Ftz activation of the Drosophila engrailed gene. Development. 1997;124(4):839–847. doi: 10.1242/dev.124.4.839. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz CJE, et al. FTZ-Factor1 and Fushi tarazu interact via conserved nuclear receptor and coactivator motifs. EMBO J. 2001;20(3):510–519. doi: 10.1093/emboj/20.3.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yussa M, Löhr U, Su K, Pick L. The nuclear receptor Ftz-F1 and homeodomain protein Ftz interact through evolutionarily conserved protein domains. Mech Dev. 2001;107(1–2):39–53. doi: 10.1016/s0925-4773(01)00448-8. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki T, Kawasaki H, Yu RT, Ueda H, Umesono K. Segmentation gene product Fushi tarazu is an LXXLL motif-dependent coactivator for orphan receptor FTZ-F1. Proc Natl Acad Sci USA. 2001;98(22):12403–12408. doi: 10.1073/pnas.221552998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Copeland JW, Nasiadka A, Dietrich BH, Krause HM. Patterning of the Drosophila embryo by a homeodomain-deleted Ftz polypeptide. Nature. 1996;379(6561):162–165. doi: 10.1038/379162a0. [DOI] [PubMed] [Google Scholar]
- 22.Fitzpatrick VD, Percival-Smith A, Ingles CJ, Krause HM. Homeodomain-independent activity of the fushi tarazu polypeptide in Drosophila embryos. Nature. 1992;356(6370):610–612. doi: 10.1038/356610a0. [DOI] [PubMed] [Google Scholar]
- 23.Hughes CL, Liu PZ, Kaufman TC. Expression patterns of the rogue Hox genes Hox3/zen and fushi tarazu in the apterygote insect Thermobia domestica. Evol Dev. 2004;6(6):393–401. doi: 10.1111/j.1525-142X.2004.04048.x. [DOI] [PubMed] [Google Scholar]
- 24.Brown SJ, Hilgenfeld RB, Denell RE. The beetle Tribolium castaneum has a fushi tarazu homolog expressed in stripes during segmentation. Proc Natl Acad Sci USA. 1994;91(26):12922–12926. doi: 10.1073/pnas.91.26.12922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dearden PK, et al. Patterns of conservation and change in honey bee developmental genes. Genome Res. 2006;16(11):1376–1384. doi: 10.1101/gr.5108606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dawes R, Dawson I, Falciani F, Tear G, Akam M. Dax, a locust Hox gene related to fushi-tarazu but showing no pair-rule expression. Development. 1994;120(6):1561–1572. doi: 10.1242/dev.120.6.1561. [DOI] [PubMed] [Google Scholar]
- 27.Mann RS, Chan S-K. Extra specificity from extradenticle: The partnership between HOX and PBX/EXD homeodomain proteins. Trends Genet. 1996;12(7):258–262. doi: 10.1016/0168-9525(96)10026-3. [DOI] [PubMed] [Google Scholar]
- 28.Damen WG. Parasegmental organization of the spider embryo implies that the parasegment is an evolutionary conserved entity in arthropod embryogenesis. Development. 2002;129(5):1239–1250. doi: 10.1242/dev.129.5.1239. [DOI] [PubMed] [Google Scholar]
- 29.Hughes CL, Kaufman TC. Exploring the myriapod body plan: Expression patterns of the ten Hox genes in a centipede. Development. 2002;129(5):1225–1238. doi: 10.1242/dev.129.5.1225. [DOI] [PubMed] [Google Scholar]
- 30.Janssen R, Damen WG. The ten Hox genes of the millipede Glomeris marginata. Dev Genes Evol. 2006;216(7–8):451–465. doi: 10.1007/s00427-006-0092-5. [DOI] [PubMed] [Google Scholar]
- 31.Mouchel-Vielh E, Blin M, Rigolot C, Deutsch JS. Expression of a homologue of the fushi tarazu (ftz) gene in a cirripede crustacean. Evol Dev. 2002;4(2):76–85. doi: 10.1046/j.1525-142x.2002.01063.x. [DOI] [PubMed] [Google Scholar]
- 32.Kourakis MJ, et al. Conserved anterior boundaries of Hox gene expression in the central nervous system of the leech Helobdella. Dev Biol. 1997;190(2):284–300. doi: 10.1006/dbio.1997.8689. [DOI] [PubMed] [Google Scholar]
- 33.Heffer A, Lohr U, Pick L. 2011. ftz evolution: Findings, hypotheses and speculations (response). BioEssays 33(12):910–918.
- 34.Hou HY, et al. Stripy Ftz target genes are coordinately regulated by Ftz-F1. Dev Biol. 2009;335(2):442–453. doi: 10.1016/j.ydbio.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomancak P, et al. 2002. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol 3(12):RESEARCH0088.
- 36.Doe CQ, Hiromi Y, Gehring WJ, Goodman CS. Expression and function of the segmentation gene fushi tarazu during Drosophila neurogenesis. Science. 1988;239(4836):170–175. doi: 10.1126/science.2892267. [DOI] [PubMed] [Google Scholar]
- 37.Hiromi Y, Kuroiwa A, Gehring WJ. Control elements of the Drosophila segmentation gene fushi tarazu. Cell. 1985;43(3 Pt 2):603–613. doi: 10.1016/0092-8674(85)90232-6. [DOI] [PubMed] [Google Scholar]
- 38.Mann RS, Lelli KM, Joshi R. Hox specificity unique roles for cofactors and collaborators. Curr Top Dev Biol. 2009;88:63–101. doi: 10.1016/S0070-2153(09)88003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Force A, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151(4):1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alonso CR, Maxton-Kuechenmeister J, Akam M. Evolution of Ftz protein function in insects. Curr Biol. 2001;11(18):1473–1478. doi: 10.1016/s0960-9822(01)00425-0. [DOI] [PubMed] [Google Scholar]
- 41.Calhoun VC, Levine M. Long-range enhancer-promoter interactions in the Scr-Antp interval of the Drosophila Antennapedia complex. Proc Natl Acad Sci USA. 2003;100(17):9878–9883. doi: 10.1073/pnas.1233791100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hiromi Y, Gehring WJ. Regulation and function of the Drosophila segmentation gene fushi tarazu. Cell. 1987;50(6):963–974. doi: 10.1016/0092-8674(87)90523-x. [DOI] [PubMed] [Google Scholar]
- 43.Pick L, Schier A, Affolter M, Schmidt-Glenewinkel T, Gehring WJ. Analysis of the ftz upstream element: Germ layer-specific enhancers are independently autoregulated. Genes Dev. 1990;4(7):1224–1239. doi: 10.1101/gad.4.7.1224. [DOI] [PubMed] [Google Scholar]
- 44.Ohno S. Evolution by Gene Duplication. Berlin: Springer-Verlag; 1970. [Google Scholar]
- 45.Lynch M, O’Hely M, Walsh B, Force A. The probability of preservation of a newly arisen gene duplicate. Genetics. 2001;159(4):1789–1804. doi: 10.1093/genetics/159.4.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218(4570):348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 47.Schinko J, Posnien N, Kittelmann S, Koniszewski N, Bucher G. 2009. Single and double whole-mount in situ hybridization in red flour beetle (Tribolium) embryos. Cold Spring Harb Protoc 2009(8):pdb prot5258. [DOI] [PubMed]
- 48.Manzanares M, Marco R, Garesse R. Genomic organization and developmental pattern of expression of the engrailed gene from the brine shrimp Artemia. Development. 1993;118(4):1209–1219. doi: 10.1242/dev.118.4.1209. [DOI] [PubMed] [Google Scholar]
- 49.Copf T, Schröder R, Averof M. Ancestral role of caudal genes in axis elongation and segmentation. Proc Natl Acad Sci USA. 2004;101(51):17711–17715. doi: 10.1073/pnas.0407327102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gutjahr T, Vanario-Alonso CE, Pick L, Noll M. Multiple regulatory elements direct the complex expression pattern of the Drosophila segmentation gene paired. Mech Dev. 1994;48(2):119–128. doi: 10.1016/0925-4773(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 51.Kellerman KA, Mattson DM, Duncan I. Mutations affecting the stability of the fushi tarazu protein of Drosophila. Genes Dev. 1990;4(11):1936–1950. doi: 10.1101/gad.4.11.1936. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.