Abstract
Autophagy is a finely orchestrated cellular catabolic process that requires multiple autophagy-related gene products (ATG). The ULK1 complex functions to integrate upstream signals to downstream ATG proteins through an unknown mechanism. Here, we identified an interaction between mammalian FIP200 and ATG16L1, essential components of the ULK1 and ATG5 complexes, respectively. Further analyses demonstrate that this is a direct interaction mediated by a short domain of ATG16L1 which we term the FIP200-Binding Domain (FBD). The FBD is not required for ATG16L1 self-dimerization or interaction with ATG5. Importantly, FBD-deleted ATG16L1 mutant is defective in mediating amino acid starvation-induced autophagy, which requires the ULK1 complex. Intriguingly, this mutant retains its function in supporting glucose deprivation-induced autophagy, a ULK1 complex-independent process. Our study has therefore identified a novel interaction between the ULK1 and ATG5 complexes that can distinguish ULK1-dependent and -independent autophagy processes.
Degradation of cellular contents can occur via the proteasome or lysosome systems1. A number of pathways exist that deliver substrates for degradation to either systems. Macroautophagy (commonly referred to as autophagy) is one pathway that facilitates the degradation of long lived proteins, damaged organelles and infectious pathogens, resulting in the clearance of toxic materials and increased nutrients availability in the cell2. Because of these substrates, autophagy has been implicated in a number of physiological and pathological processes including development, pathogen infection, neurodegeneration and cancer3.
During autophagy, a number of protein complexes orchestrate the formation of a lipid bilayer (termed phagophore or pre-autophagosome) which upon maturation, engulfs cytoplasmic materials and forms the autophagosome. The eventual fusion of autophagosomes with lysosomes results in the degradation of the autophagosome content and the recycling of nutrients back into the cytoplasm. Key players in these events include a family of ubiquitin-like proteins, such as LC3, which are involved in the phagophore maturation and possibly cargo selection4–7. Membrane targeting of LC3 is essential for autophagy and requires a series of ubiquitin-like conjugation events that lead to the conjugation of cytosolic LC3 (LC3–I) to membrane bound, phosphatidylethanolamine (PE)-conjugated form (LC3–II)4. These events are catalyzed by autophagy-related proteins (or ATG proteins) with E1, E2 and E3-like enzymatic activities termed ATG7, ATG3 and ATG5–ATG12, respectively. The ATG5–ATG12 conjugate (herein referred to as ATG5–12) formation is also catalyzed by similar conjugation events but requires a distinct E2-like enzyme termed ATG108. In addition to its possible role as an E3-like enzyme during LC3–II formation, the ATG5–12 conjugate was shown to form a large protein complex with ATG16L1 which is thought to be required to specify the site of LC3 conjugation during autophagy9. Depletion of any of ATG5, ATG16L1 or ATG7 completely abolishes autophagosome formation10–12. Meanwhile, depletion of ATG3 results in defective maturation of the phagophore structure13. LC3 can also be recruited to single membrane structures, for example during phagocytosis or entotic cell clearance14–16. In these cases, the core machinery utilized during LC3–II formation is also required.
A number of upstream signaling complexes can regulate autophagy including the Vps34 and ULK1 complexes. The Vps34 complex is comprised of a number of proteins, including the Vps34 lipid kinase, p150, Beclin and ATG14, and is essential for phagophore formation and proper recruitment of ATG proteins to the phagophore17,18. On the other hand, the ULK1 complex, comprised of the protein kinase ULK1 and several regulatory components including ATG13 and FIP200, is suppressed by the mTORC1 kinase activity19,20. Once mTORC1 activity is inhibited, for example by amino acid deprivation or cytotoxic response, the ULK1 complex becomes activated and stimulates autophagy. Mechanistically, how the ULK1 complex coordinates with other ATG complexes during autophagy is not clear, and the protein substrate(s) of its kinase activity required for autophagy induction have not yet been identified.
In this study we sought to identify novel players in autophagy that are particularly required during the initial phagophore formation stage. To do so, we developed a system to isolate membrane-localized ATG proteins. Using this system, we conducted tandem affinity purification and identified FIP200 as a direct binding partner of ATG16L1. We further provide evidence to demonstrate that this novel interaction between the ULK1 complex and the ATG5 complex is required for ULK1 complex-dependent but not ULK1 complex-independent autophagy processes.
Results
Pre-autophagosomes accumulate in the absence of ATG3
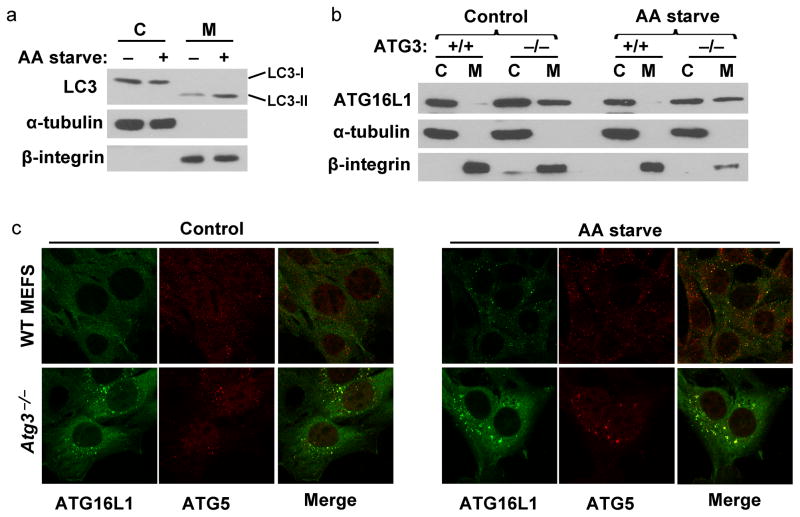
In order to study the molecular events during pre-autophagosome structures formation, we separated membrane-bound proteins from cytosolic proteins by subcellular fractionation. As can be seen in figure 1a, cytosolic LC3 (LC3–I) was separated from membrane-bound LC3 (LC3–II) in wild type mouse embryonic fibroblasts (MEFs). However, using this method, we hardly detected the accumulation of upstream ATG proteins, such as ATG16L1, in the membrane fraction of wild type MEFs even when autophagy was induced by amino acid deprivation (Fig. 1b). This suggests that the recruitment of upstream ATG proteins to pre-autophagosome structures may occur in a transient manner. Previous studies suggest that in the absence of ATG3, pre-autophagosome structures are unable to mature into autophagosomes and therefore accumulate in the cell likely leading to stabilization of upstream autophagy events which are transient in ATG3-expressing cells13,21. Therefore, we tested the subcellular localization of ATG16L1 in Atg3−/−. Interestingly, ATG16L1 accumulated on the membrane fraction of these cells (Fig. 1b). We observed no detectable increase of ATG16L1 recruitment to the membrane fraction after 2 hrs of amino acid starvation in Atg3−/−, in agreement with previous studies21. Similarly, we show by immunofluorescence analyses that ATG16L1 positive structures, which co-localize with endogenous ATG5, were detectable even under nutrient rich conditions in the absence of ATG3 (Fig. 1c).
Figure 1. ATG16L1 stably localizes to membrane compartments in Atg3−/−.
(A) Subcellular fractionation of membrane (M) and cytoplasmic (C) cell fractions of wild type MEFs. Cells were either left untreated (control) or incubated in amino acid-free media for 2 hrs before harvesting (AA starve). Samples were analyzed by Western blot using antibodies against α-tubulin and β-integrin as markers for membrane and cytosolic fractions, respectively. Antibodies against LC3 were used to verify localization of autophagosome structures to the membrane compartment. (B) Western blot analysis of wild type (+/+) or Atg3 knockout (−/−) MEFs treated as in (A). Note that ATG16L1 can stably localize to the membrane fraction only in Atg3−/−. (C) Immunofluorescence analysis of wild type or Atg3−/− stably expressing S-tagged ATG16L1. Control or AA starved cells, as in (A), were fixed and processed for immunofluorescence staining using anti-S tag antibodies to stain for ATG16L1 or anti-ATG5 antibodies. C: Cytosolic fraction; M: Membrane fraction; AA starve: Amino Acid starvation; WT: Wild Type.
Identification of FIP200 as an ATG16L1-interacting partner
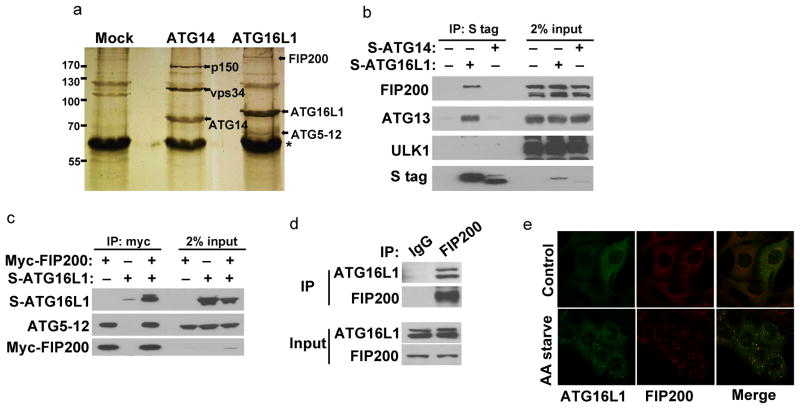
We further sought to identify novel protein-protein interactions of membrane bound ATG proteins in Atg3−/− using ATG14 and ATG16L1 as baits. To do so, we used tandem affinity purification to purify ATG14 and ATG16L1 complexes followed by SDS–PAGE analysis and silver staining (Fig. 2a). We observed prominent bands in the ATG14 sample that correspond to known ATG14 interacting partners, including Vps34 and p150, thereby confirming that this method is able to purify functional autophagy complexes. In the case of ATG16L1 purification, we detected a prominent distinct band of a molecular weight greater than 170 kDa in addition to the ATG5–12 conjugate, the known binding partner of ATG16L1. Mass spectrometric analysis revealed the identity of this band as FIP200, an essential component of the ULK1 complex. Previously, the ULK1 complex was proposed to be functionally linked to the ATG5 complex, but how these two complexes cross-talk is unclear.
Figure 2. Identification of FIP200 as a binding partner of ATG16L1.
(A) Silver staining was used to analyze protein complexes after tandem affinity purification using Atg3−/− stably expressing FLAG-S-tagged ATG14 or ATG16L1. (B) ATG16L1 but not ATG14 can interact with FIP200. Samples are whole cell lysates from 293T cells expressing S-tagged ATG proteins and subjected to pull down using S protein agarose. Bound proteins were analyzed by Western blot using antibodies against endogenous FIP200, ATG13 or ULK1. (C) Interaction between FIP200 and the ATG5–12–ATG16L1 complex in 293T whole cell lysate. Cell lysates expressing myc-FIP200 were subjected to anti-myc antibodies immunoprecipitation followed by Western blot analysis to detect S-tagged ATG16L1 and endogenous ATG5–12 conjugate. (D) Endogenous FIP200 and ATG16L1 can co-immunoprecipitate. Endogenous FIP200 was immunoprecipitated from Atg3−/− followed by Western blot analysis to detect endogenous ATG16L1 binding. (E) ATG16L1 and FIP200 co-localize. Wild type MEFs stably expressing GFP-ATG16L1 were left untreated (control) or incubated in the absence of amino acid (AA starve) for 2 hrs before fixation and processing for immunofluorescence staining. Anti-FIP200 antibodies were used to stain for endogenous FIP200. (*) denotes IgG heavy chain; AA starve: Amino Acid starvation.
To verify the interaction between ATG16L1 and FIP200, we expressed ATG16L1 in 293T cells and found that ATG16L1, but not ATG14, can pull down FIP200 as well as ATG13 (Fig. 2b). In addition, stably expressed ATG16L1 in Atg3−/− can also pull down endogenous FIP200, ATG13 and to a lower extent ULK1 (Supplementary Fig. 1a). Reciprocal pull down of FIP200 expressed in 293T cells can co-precipitate ATG16L1 as well as the ATG5–12 conjugate (Fig. 2c). Furthermore, we tested the potential interaction of endogenous FIP200 with ATG16L1. Because their interaction is most likely transient and stabilized by ATG3 deletion, we used Atg3−/− for this purpose. As shown in figure 2d, immunoprecipitation of endogenous FIP200 can co-precipitate endogenous ATG16L1 in these cells. Using immunofluorescence analysis we show that ATG16L1 punctate structures almost completely co-localized with FIP200 in both wild type MEFs (Fig. 2e) and Atg3−/− (Supplementary Fig. 1b) as was described previously21,22.
Interaction between FIP200 and ATG16L1 is direct
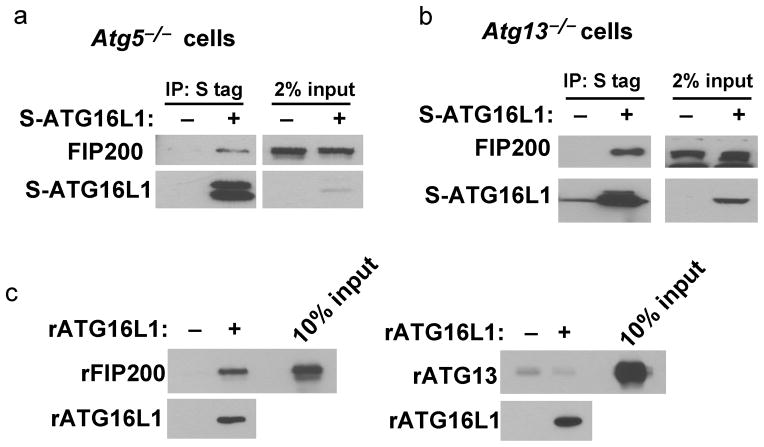
Having shown that the ATG16L1-containing ATG5 complex and the FIP200-containing ULK1 complex can co-precipitate, we further sought to determine which proteins directly mediate the interaction between these two complexes. First, we found that ATG16L1 and FIP200 can interact in MEF cells lacking the expression of ATG5 (Fig. 3a and Supplementary Fig. 2a) or ATG13 (Fig. 3b), indicating that ATG5 and ATG13 are dispensable for the interaction between ATG16L1 and FIP200. Furthermore, ATG5 and its ATG12 conjugation mutant (ATG5 K130R) were unable to pull down FIP200 (Supplementary Fig. 2b). In addition, we show that purified recombinant ATG16L1 can interact with recombinant FIP200 (Fig. 3c, left panel) but not with recombinant ATG13 (Fig. 3c, right panel), ATG3 or ATG7 (Supplementary Fig. 2c). Overall, these results indicate that ATG16L1 and FIP200 directly interact with each other independently of other complex components.
Figure 3. Interaction between ATG16L1 and FIP200 is direct.
(A) ATG5 is not required for interaction between ATG16L1 and FIP200. S protein agarose pull down of Atg5−/− lysates expressing S-tagged ATG16L1 (S-ATG16L1) followed by Western blot analysis using antibodies against FIP200 to detect binding to endogenous FIP200. (B) ATG13 is not required for binding between ATG16L1 and FIP200. Atg13−/− stably expressing S-tagged ATG16L1 were subjected to S protein agarose pull down as in (B). (C) Recombinant ATG16L1 (rATG16L1) was incubated with either recombinant FIP200 (rFIP200, left panel) or recombinant ATG13 (rATG13, right panel). As negative controls, rFIP200 or rATG13 were incubated in the absence of rATG16L1 under identical conditions.
Identification of ATG16L1 domain required for FIP200 binding
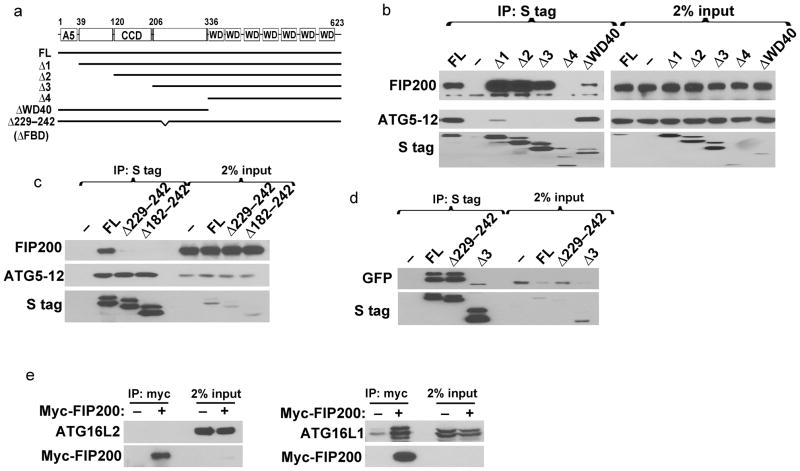
In order to determine the function of the ATG16L1–FIP200 interaction in autophagy, we first sought to map the ATG16L1 domain required to mediate its binding to FIP200. To do so, we generated a series of ATG16L1 truncation fragments (depicted in Fig. 4a) based on previous structural analysis of yeast ATG1623–25. These include truncations of ATG16L1 that lack ATG5-binding region (mediated through the N-terminal region), coiled-coil domain (CCD, required for self-dimerization), and seven WD40 repeats (located at the C-terminal half). We expressed the truncation fragments of ATG16L1 in 293T cells and tested their interaction with endogenous FIP200 and ATG5. As shown in figure 4b, ATG16L1 fragments that were defective in ATG5 binding can still bind to FIP200, indicating that FIP200 and ATG5 interact with ATG16L1 through distinct domains. The ATG16L1 fragment lacking a region between the WD40 and coiled-coil domains (Δ4 fragment, residues 1–335 deleted) were defective in binding to FIP200. These results suggest that the interaction between ATG16L1 and FIP200 is mediated through residues 206–335 of ATG16L1. This region of ATG16L1 was previously shown to be required for Rab33B binding26.
Figure 4. Amino acids 229–242 of ATG16L1 are required for its interaction with FIP200.
(A) Schematic presentation of ATG16L1 fragments used encompassed the following amino acids: FL: 1–623; Δ1: 39–623; Δ2: 120–623; Δ3: 206–623; Δ4: 336–623; ΔWD40: 1–335. The Δ229–242 lacks amino acids 229–242 and is termed ΔFBD (for FIP200-Binding Domain) in subsequent figures. (B) S-tagged fragments depicted in (A) were expressed in 293T, followed by S protein agarose pull down. Interaction with endogenous FIP200 and ATG5–12 was analyzed by Western blot. Note that residual binding of Δ1 fragment to ATG5–12 is most likely due to dimerization with endogenous ATG16L1. (C) S-tagged deletion mutants of ATG16L1 lacking residues 229–242 or 182–242 expressed as in (B). (D) FL GFP-tagged ATG16L1 was co-expressed with S-tagged FL, Δ229–242 or Δ3 mutants of ATG16L1 followed by S protein agarose pull down. Note, the Δ3 mutant lacking the CCD is unable to self-dimerize. (E) 293T whole cell lysate expressing myc-tagged FIP200 was subjected to anti-myc immunoprecipitation and Western blot analysis using antibodies against endogenous ATG16L2 (left panel) or ATG16L1 (right panel). WD: WD40 repeats; FL: Full Length; CCD: Coiled-Coil Domain.
The Δ4 fragment includes a large deletion of the N-terminal half of ATG16L1 and is therefore unable to bind ATG5 or self-dimerize in addition to its inability to bind FIP200. Therefore, we sought to identify a smaller region of ATG16L1 that is required for FIP200 binding. By further mutational analyses we identified a shorter region of ATG16L1 within residues 206–335 to be required for FIP200 binding. As shown in figure 4c, ATG16L1 mutant deleted of residues number 229–242 (denoted as Δ229–242) as well as an additional mutant harboring a larger deletion (Δ182–242) were defective in binding to FIP200 when compared to the full length protein. Importantly, Δ229–242 mutant of ATG16L1 still retained its ATG5 binding ability (Fig. 4c) as well as its ability to dimerize with full length protein (Fig. 4d). Interestingly, residues 229–242 were previously shown dispensable for Rab33B binding26. In addition, these residues are not conserved in ATG16L2, which does not support autophagy despite its ability to self-dimerize and bind to ATG526, but is conserved in ATG16L1 from various vertebrate species (Supplementary Fig. 3). Consistently, we found that FIP200 was unable to bind ATG16L2 (Fig. 4e). These results demonstrate that residues 229–242 can differentiate the ability of ATG16L1 to bind to FIP200 from binding to ATG5 or self-dimerization. We name this region as the FIP200-binding domain (FBD) of ATG16L1.
ΔFBD mutant is defective in ULK1-dependent autophagy
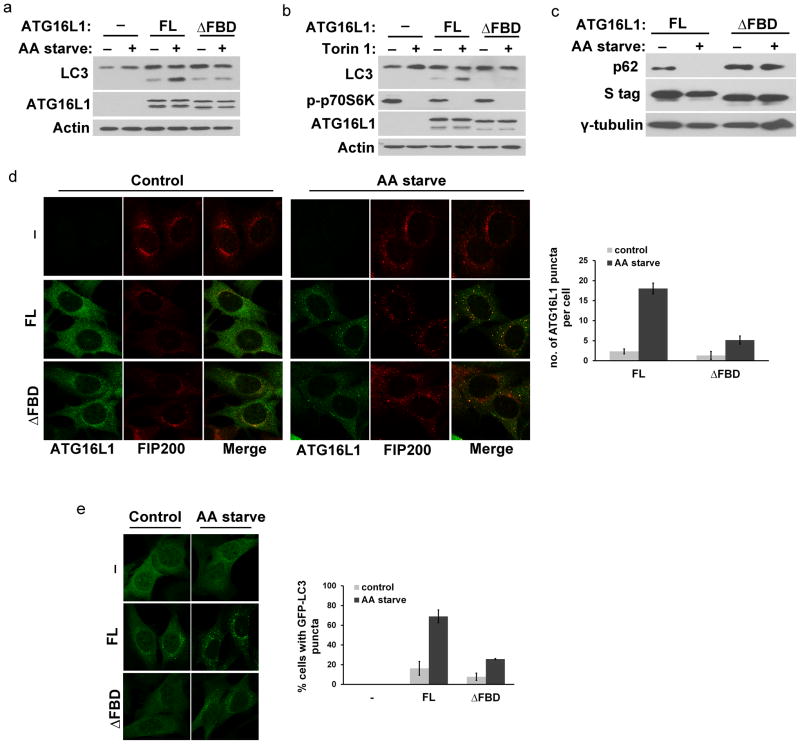
To test whether the FIP200–ATG16L1 interaction is required for the function of ATG16L1 in autophagy, we reconstituted Atg16l1−/− with either full length or FIP200-binding domain truncated mutant (ΔFBD). In the absence of ATG16L1 expression, autophagy was completely disrupted10. Meanwhile, when we expressed full length ATG16L1 or ATG16L1 FBD basal levels of autophagy was restored, as measured by LC3–I to LC3–II conversion (Fig. 5a, compare lanes 3 and 5). However, when we induced autophagy by depriving cells of amino acids, ATG16L1ΔFBD-expressing cells showed greatly reduced autophagy compared to cells expressing full length ATG16L1 (Fig. 5a, compare lanes 4 and 6). We obtained similar results when we induced autophagy by treating cells with the selective mTOR inhibitor, Torin 1 (Fig. 5b). The defect of ATG16L1ΔFBD in mediating amino acid starvation-induced autophagy is further supported by impeded p62 degradation (a well establish autophagosome substrate) in the ATG16L1ΔFBD-reconstituted cells (Fig. 5c). On the other hand, we found that the ΔWD40 mutant, which retains its ability to bind to FIP200 (Fig. 4b), can restore amino acid starvation-induced autophagy to similar levels to full length protein (Supplementary Fig. 4). Consistently, we show that ATG16L1ΔFBD exhibits reduced ability to localize to punctate structures corresponding to pre-autophagosomes (Fig. 5d). The formation of GFP–LC3 punctate structures, a marker of autophagosomes, was also markedly reduced in cells reconstituted with ATG16L1ΔFBD compared to cells reconstituted with full length ATG16L1 (Fig. 5e). Therefore, the FIP200–ATG16L1 interaction is crucial for the function of ATG16L1 during amino acid starvation-induced autophagy, a process which requires the FIP200-containing ULK1 complex.
Figure 5. The FIP200-binding mutant of ATG16L1 is defective in amino acid starvation-induced autophagy.
(A) Amino acid starvation-induced autophagy is reduced in cells reconstituted with the ATG16L1ΔFBD mutant. Atg16l1−/− reconstituted with FL or ATG16L1ΔFBD were either left untreated or amino acid-starved for 2 hrs. Bafilomycin A1 was included for 2 hrs in all samples. (B) Cells as in (A) were treated with Torin 1 (1 μM) for 4 hrs prior to harvesting. (C) p62 degradation induced by amino acid starvation is impeded in Atg16l1−/− reconstituted with ATG16L1ΔFBD. Cells were left untreated or starved of amino acids for 2 hrs. (D) Atg16l1−/− expressing the indicated constructs were subjected to immunofluorescence analysis using antibodies against ATG16L1 and FIP200. Quantification of ATG16L1 punctate structures per cell is shown in the right panel with standard deviation error bars included of 3 independent experiments of approximately 16 cells counted per condition. (E) GFP-LC3 expressed in Atg16l1−/− reconstituted with FL or ATG16L1ΔFBD and treated as in (D). Right panel shows quantification of GFP-LC3 punctate structures with standard deviation error bars included of 3 independent experiments. AA starve: Amino Acid starvation; FL: Full Length; FBD: FIP200-Binding Domain.
ΔFBD mutant is fully active during ULK1-independent autophagy
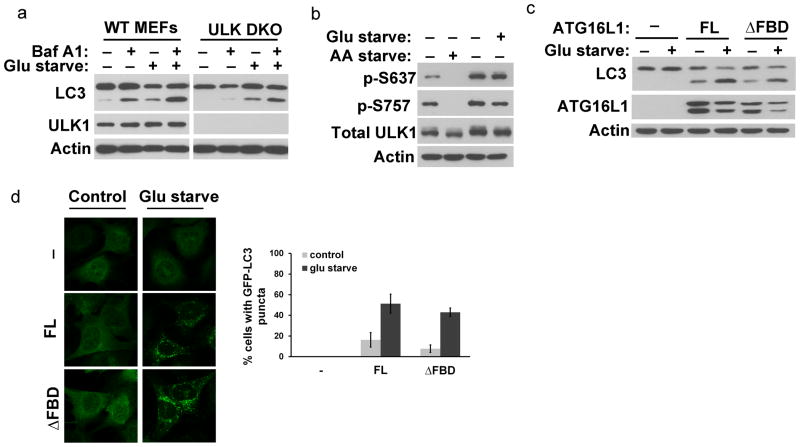
The inability of ATG16L1ΔFBD to fully restore autophagy can be potentially due to an overall misfolding of the protein caused by the introduced truncation rather than its deficiency in binding FIP200. If so, our results in figure 5 would not be able to support a functional relevance of the FIP200–ATG16L1 interaction. Therefore, we sought to assess whether ATG16L1ΔFBD possesses any biological function that is independent of FIP200 and thereby the ULK1 complex. Recently, glucose starvation was suggested to induce autophagy in a ULK1 complex-independent manner which correlates with an increase in cellular ammonia levels and lack of mTORC1 inhibition27,28. We confirmed this by using ULK1 and ULK2 double knockout MEFs (ULK DKO) where glucose starvation induced LC3–II formation which is susceptible to lysosomal degradation (Fig. 6a). Meanwhile, we observed that glucose starvation did not induce ULK1 dephosphorylation on residues that are dephosphorylated during amino acid starvation29 (Fig. 6b). Interestingly, we when reconstituted Atg16l1−/− with full length ATG16L1 or ATG16L1ΔFBD comparable LC3–I to LC3–II conversion upon glucose starvation were exhibited whereas the absence of ATG16L1 expression completely abolished LC3 conjugation (Fig. 6c) indicating that ATG16L1ΔFBD is fully functional during such ULK1 complex-independent autophagy. Similarly, we observed comparable GFP–LC3 punctate structure formation between full length ATG16L1 and ATG16L1ΔFBD expressing cells (Fig. 6d). Overall, these analyses suggest that, unlike ULK1 complex-dependent autophagy induced by amino acid starvation, ULK1 complex-independent autophagy induced by glucose starvation does not require FIP200 binding to ATG16L1.
Figure 6. ATG16L1ΔFBD mutant retains full activity during ULK1 complex-independent, glucose starvation-induced autophagy.
(A) Glucose starvation-induced autophagy occurs independently of the ULK1 complex. WT and ULK1–ULK2 double knockout MEFs (ULK DKO) were cultured in full growth media or in glucose free media for 20 hrs followed by Bafilomycin A1 treatment for additional 3 hrs. (B) ULK1 phosphorylation was examined in wild type MEFs starved of amino acids for 2 hrs or glucose for 20 hrs. ULK1 phospho-specific antibodies against phospho-serines 637 and 757 were used as indicated. (C) ΔFBD mutant of ATG16L1 can fully reconstitute autophagy induced by glucose starvation. Atg16l1−/− reconstituted with full length or ΔFBD mutant of ATG16L1 were either left untreated or glucose-starved for 20 hrs and samples were analyzed by Western blot using the indicated antibodies. (D) Cells as in (C) expressing GFP-LC3 were subjected to fluorescence analysis. The quantification of cells with GFP-LC3 punctate structures is shown in the right panel with standard deviation error bars included of 3 independent experiments. Approximately 400 cells were counted per condition. AA starve: Amino Acid starvation; Glu starve: Glucose starvation; FL: Full Length; FBD: FIP200-Binding Domain; WT: Wild Type.
DISCUSSION
In this study we provide evidence for a direct interaction between the ULK1 complex component, FIP200, and the ATG5 complex component, ATG16L1. This interaction is specifically required for ULK1 complex-dependent autophagy, thus providing mechanistic insights into how the ULK1 complex communicates with other ATG complexes, such as the ATG5 complex. Meanwhile the ATG5 complex (including its component ATG16L1) belongs to the essential core autophagy machinery, the ULK1 complex appears to only mediate autophagy induced by certain specific triggers. The biochemical LC3 conjugation reaction is intact in cells with genetic deletion of ULK130,31 and LC3–II formation takes place at basal levels in cells depleted of the ULK1 complex components22,32. Indeed, the ULK1 complex, but not the ATG5 complex, is dispensable for glucose starvation-induced autophagy27.
Could ATG16L1 be a converging point mediating other upstream autophagy signals in addition to those through ULK1? This would be an interesting hypothesis to consider. It is clear, however, that the FBD domain of ATG16L1 is not necessarily responsible for all other signals, because ATG16L1ΔFBD is functionally intact during glucose starvation-induced autophagy. Another recognizable structure in ATG16L1 is its C-terminal WD40 repeats, which is a versatile protein-protein interaction domain and is not present in yeast ATG16. Although not required for autophagy triggered by amino acid starvation, it is possible that this region mediates certain other autophagy signals. If so, then the C-terminal region of ATG16L1 enables the mammalian autophagy pathway to sense more diverse and complex signals compared to its yeast counterpart. On the other hand, it is also possible that the WD40 repeats in mammalian ATG16L1 might only be relevant to non-autophagy processes10,33. In this sense, the potential pathological role of the Crohn’s disease-associated ATG16L1 mutation (T300A), which lies within the WD40 repeats, during both autophagy-related and non-autophagy-related processes should be explored34,35.
A detailed comparison of ATG16L1 with its homolog ATG16L2 should also shed light on the function of ATG16L1 during autophagy. Unlike ATG16L1, ATG16L2 is unable to support autophagy or localize to the phagophore structures despite its ability to bind ATG5, self-oligomerize and form a large protein complex with the ATG5–12 complex26. ATG16L1 from various vertebrate species all possess highly conserved FBD regions, whereas ATG16L2 lacks this domain. Consistently, we found ATG16L2 failed to interact with FIP200. However, whether the lack of FBD renders ATG16L2 inactive in autophagy is unclear, as ATG16L1 with its FBD deleted can still mediate ULK1 complex-independent autophagy. In addition, neither yeast nor C. elegans ATG16 have this domain (there is no true FIP200 homolog in these organisms either). Furthermore, previous in vitro biochemical studies suggest that yeast ATG16 is not required for the E3-like activity of the ATG5–12 during the conjugation of ATG8 (the yeast homologue of LC3) to PE36. Thus in mammalian cells, the difference between ATG16L1 and ATG16L2 in autophagy does not appear to be due to their differential influence on the E3-like enzymatic activity of the ATG5 complex. As such, the exact structural and biochemical basis that renders ATG16L1 but not ATG16L2 to be an essential functional component in autophagy has yet to be defined.
In conclusion, this study has uncovered a novel functional interaction between two upstream ATG complexes, and demonstrated that ATG16L1 is not only an essential structural component of the ATG5 complex but also a signaling protein that can mediate specific upstream signals during autophagy such as those transduced by the ULK1 complex.
ONLINE METHODS
Cell culture and treatments
Wild type Mouse Embryonic Fibroblasts (MEFs), Atg3 knockout MEFs (Atg3−/−), Atg16l1−/−, Atg5−/−, Atg13−/−, ULK1 and ULK2 Double knockout MEFs (ULK DKO) and 293T cells were cultured in DMEM supplemented with 10% FBS, L-Glutamine (2 mM), Pencillin (10 Units/mL) and Streptomycin (0.1 mg/ml). Stable overexpression of GFP-LC3 or F-S-ATG proteins was obtained by retroviral infection using pBabe expression plasmid followed by blasticidin or puromycin selection. For transient expression, Lipofectamine 2000 (Invitrogen) was used according to the manufacturer’s instructions.
For amino acid starvation experiments, cells were grown in DMEM lacking amino acids and serum typically for 2 hrs prior to harvesting. Glucose starvation was performed by culturing cells in DMEM lacking glucose and sodium pyruvate, supplemented with 10% dialyzed FBS and glutamine for 20 hrs. Control cells were also grown in 10% dialyzed FBS and glutamine but in the presence of glucose and sodium pyruvate.
Antibodies and reagents
For Western blot analysis, the following antibodies were used: anti-LC3 (1:2000, Sigma, #L7543); anti-Actin (1:5000, Sigma, #A5316); anti-ULK1 (1:2000, Sigma, #A7481); anti-ATG13 (1:3000, Sigma, SAB4200100); anti-ATG7 (1:2000, Santa Cruz, clone H300); anti-p62/SQSTM1 (1:10,000, MBL, #PM045); anti-ATG16L1 (1:3000, MBL, clone 1F12); anti-α-tubulin (1:1000, Calbiochem, #CP06); anti-S tag (1:1000, Novagen); anti-ATG5 (1:1000, Sigma, #A0731); anti-FIP200 (1:1000, ProteinTech, #10043-2-AP); anti-myc tag (1:1000, Santa Cruz, clone 9E10); anti-T7 tag HRP (1:5000, Novagen); anti-ATG16L2 (1:1000, Abgent, #K292); anti-phospho-p70 S6 Kinase (1:1000, p-p70S6K, Cell Signaling, #9205); control rabbit IgG (Millipore, 12–370). The anti-β-integrin antibody (1:1000) is a generous gift from Dr Filippo Giancotti; phospho-specific antibodies against ULK1 (p-757 and p-637, 1:1000) were described previously29.
Bafilomycin A1 (Baf A1, inhibitor of lysosome degradation) was purchased from Sigma and used at a final concentration of 20 nM. Torin 1 (an mTOR specific inhibitor) was purchase from Tocris Bioscience and used at a final concentration of 1 μM.
Plasmids
Flag-S-tagged proteins (F-S-) were cloned into pBabe-F-S retroviral vector using SalI cloning sites. ATG16L1 full length, fragments and mutants were generated based on mouse ATG16L1 using the following forward primers: full length (amino acids 1–623): 5′ GCAGCAGTCGACATGTCGTCGGGCCTGCGCGC; Δ1 (amino acids 39–623): 5′ GCAGCAGTCGACATGTATACCAAGTTGCTGGAAAAG; Δ2 (amino acids 120–623): 5′ GCAGCAGTCGACATGAAGGACAAGGAGATACAGATG; Δ3 (amino acids 206–623): 5′ GCAGCAGTCGACATGAATGCAGAGAATGAGAAGGAC; Δ4 (amino acids 336–623): 5′ GCAGCAGTCGACATGGCGCATGACGGAGAGGTCAAC. The above constructs used the following common reverse primer: 5′ CTTAAGTCGACTCAAGGCTGTGCCCACAGCAC. The ΔWD40 (amino acids 1–335): reverse primer: 5 ′ CCGAAGTCGACTCAATCGAAGACATACGAGGCAGTAG. The following deletion mutants were obtained by two-step PCR using the following forward primers: Δ229–242: 5′ AAGGAGCTTGCAGAAGCAGCAATTGTGGATGAGACCTCA; Δ182–242: GCCCTAGAAGAGAAACTGAGGATTGTGGATGAGACCTCA. C-terminally tagged GFP-ATG16L1 was cloned into pBabe vector using MfeI and SalI sites. The myc-tagged FIP200 was expressed from pCMV vector as described previously19. ATG14 was cloned into pBabe-F-S vector using SalI/MfeI restriction sites. ATG5 and the ATG5 K130R mutant (unable to conjugate to ATG12) were expressed from pCDNA3.1 with HA-FLAG-S-tags cloned using BamHI/XbaI restriction sites.
Tandem affinity purification
Cytosolic and membrane fractions were obtained using the subcellular fractionation kit (Thermo Scientific). Membrane fractions were obtained from 18 X 15 cm plates of Atg3−/− stably expressing Flag-S-tagged ATG16L1 or ATG14 (F-S-ATG16L1 or F-S-ATG14, respectively) starved for 2 hrs in amino acid deficient media. Membrane fractions were then spun at 60,000g for 2 hrs. Pre-cleared lysates were incubated overnight with Flag-beads (Sigma) at 4°C followed 3 washes in NP-40 buffer (100mM NaCl, 25mM Hepes pH7.5, 1.5 mM MgCl2, 1 mM EDTA, 1.5 mM β-ME and 0.5% NP-40, supplemented with protease inhibitor cocktail and proteasome inhibitor MG132, Sigma). Bound complexes were then eluted in NP-40 buffer containing 200 μg/ml 3XFLAG peptide (Sigma) for 30 min at 4 °C with rotation. Eluates were further incubated with S protein agarose (Novagen) for 6 hrs at 4 °C with rotation. Following four washes in NP-40 buffer, samples were eluted in SDS-sample buffer and protein complexes were analyzed by SDS-PAGE, silver staining and mass spectrometry.
Binding assays
Cell lysates were obtained from 293T or MEF cells grown in 10 cm plates and harvested by direct lysing in NP-40 buffer (except that NaCl concentration was increased to 150 mM). Cell lysates were cleared by spinning at 12,000g for 5 min at 4 °C, and incubated with either S protein agarose (Novagen) for 5 hrs at 4 °C or with anti-myc antibodies (Santa Cruz, clone 9E10) for 3 hrs followed by protein G agarose for an additional 2 hrs. Beads were then washed 3 times with NP-40 buffer and bound proteins were analyzed by SDS-PAGE and Western blot analyses. For endogenous co-immunoprecipitation assays, Atg3−/− were harvest as above and cleared cell lysates were incubated with 0.3 μg FIP200 antibodies or control rabbit IgG overnight at 4 °C followed by protein G agarose for an additional 3 hrs. Beads were washed 4 times with NP-40 buffer and protein complexes were analyzed by Western blot analysis.
Recombinant proteins purification was performed as described previously19,37,38. Briefly, T7-tagged recombinant ATG16L1 and ATG3 were purified from BL21 E. coli cells, whereas, recombinant FIP200, ATG13 and ATG7 were purified from SF-9 cells19,37,38. For recombinant protein binding, recombinant ATG16L1 was incubated with anti-ATG16L1 antibodies and protein G for 3 hrs at 4 °C with rotation followed by 2 washes in NP-40 buffer. Beads were then incubated with recombinant ATG13, ATG7 or ATG3 for 1 hr at 4 °C followed by 3 washes in NP-40 buffer. As negative controls, recombinant proteins were incubated with protein G pre-incubated with anti-ATG16L1 antibody but in the absence of recombinant ATG16L1. Bound proteins were analyzed by SDS-PAGE and Western blot analysis using anti-T7-HRP (to detect ATG3), anti-ATG16L1, anti-ATG13 or anti-ATG7 antibodies. Alternatively, S-tagged ATG16L1 expressed in wild type or Atg5−/− was precipitated using S protein agarose, followed by extensive washings and incubation with recombinant FIP200 as above.
Microscopy
For fluorescence analysis, cells stably were grown on glass coverslips in a 6-well plate. 24 hrs later, cells were either left untreated or treated as indicated. Coverslips were then fixed with 3.7% Paraformaldehyde in 20 mM HEPES pH7.5 for 30 min at room temperature followed by permeabilization in 0.1% triton in PBS for 5 min. Slides were then incubated in primary antibodies in blocking buffer (PBS supplemented with 1% BSA) at 37 °C for 2 hrs followed by incubation with Alexa Fluor secondary antibodies (Invitrogen) for 30 min at room temperature. Following extensive washings, coverslips were then mounted on microscope slides and visualized under Nikon Eclipse Ti-U Confocal Microscope using a 60X magnification objective. Images were acquired using Nikon EZ-C1 image acquisition software and edited using Photoshop.
Supplementary Material
Acknowledgments
We thank S. Akira (Osaka University, Osaka, Japan) for Atg16l1−/−, M. Komatsu (Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan) for Atg3−/−, N. Mizushima (Tokyo Medical and Dental University, Tokyo, Japan) for Atg5−/−, C. Thompson (Memorial Sloan Kettering Cancer Center, MSKCC, New York, NY, USA) for ULK1 and ULK2 Double Knockout MEFs and H. Erdjument-Bromage (MSKCC, New York, NY, USA) for mass spectrometric analyses. We thank members of the X.J. laboratory for critical reading of the manuscript and discussion. This work was supported in part by NIH U54CA137788/U54CA132378 and R01CA113890 funds (to X.J.); National Cancer Institute RO1CA154649, the Benjamin Friedman Research Fund and the Louis V. Gerstner, Jr. Young Investigators Fund (to M.O.); as well as an NIH 1F32CA162691 fellowship (to N.G.).
Footnotes
AUTHOR CONTRIBUTIONS N.G. and X.J. designed the study and wrote the paper; N.G. and O.F. performed the experiments; N.G., O.F., M.O. and X.J. analyzed the data.
COMPETING FINANCIAL INTERESTS The authors declare no competing interests.
References
- 1.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 2.Wirawan E, Vanden Berghe T, Lippens S, Agostinis P, Vandenabeele P. Autophagy: for better or for worse. Cell Res. 2012;22:43–61. doi: 10.1038/cr.2011.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:859–64. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjorkoy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 7.Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol. 2010;12:836–41. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 8.Nemoto T, et al. The mouse APG10 homologue, an E2-like enzyme for Apg12p conjugation, facilitates MAP-LC3 modification. J Biol Chem. 2003;278:39517–26. doi: 10.1074/jbc.m300550200. [DOI] [PubMed] [Google Scholar]
- 9.Fujita N, et al. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saitoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–8. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 11.Mizushima N, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–68. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komatsu M, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–34. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sou YS, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell. 2008;19:4762–75. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Florey O, Kim SE, Sandoval CP, Haynes CM, Overholtzer M. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat Cell Biol. 2011;13:1335–43. doi: 10.1038/ncb2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez J, et al. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci U S A. 2011;108:17396–401. doi: 10.1073/pnas.1113421108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Florey O, Overholtzer M. Autophagy proteins in macroendocytic engulfment. Trends Cell Biol. 2012;22:374–80. doi: 10.1016/j.tcb.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–72. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong Y, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–76. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganley IG, et al. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung CH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6:764–76. doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hara T, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizushima N, et al. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679–88. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- 24.Fujioka Y, Noda NN, Nakatogawa H, Ohsumi Y, Inagaki F. Dimeric coiled-coil structure of Saccharomyces cerevisiae Atg16 and its functional significance in autophagy. J Biol Chem. 2010;285:1508–15. doi: 10.1074/jbc.M109.053520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsushita M, et al. Structure of Atg5.Atg16, a complex essential for autophagy. J Biol Chem. 2007;282:6763–72. doi: 10.1074/jbc.M609876200. [DOI] [PubMed] [Google Scholar]
- 26.Ishibashi K, et al. Atg16L2, a novel isoform of mammalian Atg16L that is not essential for canonical autophagy despite forming an Atg12-5-16L2 complex. Autophagy. 2011;7:1500–13. doi: 10.4161/auto.7.12.18025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheong H, Lindsten T, Wu J, Lu C, Thompson CB. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc Natl Acad Sci U S A. 2011;108:11121–6. doi: 10.1073/pnas.1107969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eng CH, Yu K, Lucas J, White E, Abraham RT. Ammonia derived from glutaminolysis is a diffusible regulator of autophagy. Sci Signal. 2010;3:ra31. doi: 10.1126/scisignal.2000911. [DOI] [PubMed] [Google Scholar]
- 29.Shang L, et al. Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc Natl Acad Sci U S A. 2011;108:4788–93. doi: 10.1073/pnas.1100844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alers S, et al. Atg13 and FIP200 act independently of Ulk1 and Ulk2 in autophagy induction. Autophagy. 2011;7:1423–33. doi: 10.4161/auto.7.12.18027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kundu M, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei H, et al. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev. 2011;25:1510–27. doi: 10.1101/gad.2051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujita N, et al. Differential involvement of Atg16L1 in Crohn disease and canonical autophagy: analysis of the organization of the Atg16L1 complex in fibroblasts. J Biol Chem. 2009;284:32602–9. doi: 10.1074/jbc.M109.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rioux JD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hampe J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–11. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 36.Hanada T, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 37.Gao Z, et al. Processing of autophagic protein LC3 by the 20S proteasome. Autophagy. 2010;6:126–37. doi: 10.4161/auto.6.1.10928. [DOI] [PubMed] [Google Scholar]
- 38.Shao Y, Gao Z, Feldman T, Jiang X. Stimulation of ATG12-ATG5 conjugation by ribonucleic acid. Autophagy. 2007;3:10–6. doi: 10.4161/auto.3270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.