SUMMARY
Mammary epithelial stem cells are vital to tissue expansion and remodeling during various phases of postnatal mammary development. Basal mammary epithelial cells are enriched in Wnt-responsive cells and can reconstitute cleared mammary fat pads upon transplantation into mice. Lgr5 is a Wnt-regulated target gene and was identified as a major stem cell marker in the small intestine, colon, stomach, hair follicle and also in kidney nephrons. Here we demonstrate the outstanding regenerative potential of a rare population of Lgr5-expressing (Lgr5+) mammary epithelial cells (MECs). We found that Lgr5+ cells reside within the basal population, are superior to other basal cells in regenerating functional mammary glands (MGs), are exceptionally efficient in reconstituting MGs from single cells and exhibit regenerative capacity in serial transplantations. Loss-of-function, depletion experiments of Lgr5+ cells from transplanted MECs or from pubertal MGs revealed that these cells are not only sufficient but also necessary for postnatal mammary organogenesis.
Keywords: Lgr5, stem cell, mammary gland, regenerative potential
INTRODUCTION
Adult stem cells are characterized by their ability to both self renew and to differentiate into specialized cells. Unraveling the hierarchy of mammary stem and progenitor cells has been of great interest since the mammary gland (MG) undergoes extensive tissue expansion and remodeling at various phases throughout adult life. Moreover, deciphering the stem cell players contributing to normal mammary development is key to understand subsequent pathologies such as cancer transformation. During pubertal development, which happens between 3–8 weeks of age in mice, the mammary epithelium undergoes glandular expansion. This yields a branching network of ducts composed of two primary epithelial cell lineages: myoepithelial/basal and luminal. During pregnancy, the epithelium goes through additional lobuloalveolar differentiation to allow lactation (Deome et al., 1959; Shackleton et al., 2006; Stingl and Caldas, 2007; Stingl et al., 2006; Visvader and Lindeman, 2006; Welm et al., 2003; Woodward et al., 2005). The MG can be regenerated efficiently by transplanting mammary epithelial cells (MECs) into cleared mammary fat pads. Serial transplantation and limiting dilution assays of primary cultures derived from clonal outgrowths have pointed to the existence of a rare subset of mammary cells that function as stem cells and reconstitute functional MGs (Kordon and Smith, 1998). This basal population, which includes mammary stem cells, is characterized by the surface antigen profile Lin−CD24+CD29high or Lin−CD24lowCD49fhigh (Shackleton et al., 2006; Stingl et al., 2006) and is enriched in Wnt-responsive cells (Zeng and Nusse, 2010).
Wnt signaling has been implicated in different stages of mammary development as well as in mammary oncogenesis (Boras-Granic et al., 2006; Brisken et al., 2000; Chu et al., 2004; Lindvall et al., 2006; Lindvall et al., 2009; Nusse and Varmus, 1982; Roelink et al., 1990). The Wnt co-receptor Lrp5 has been described as a marker of mammary stem cells (Badders et al., 2009) and secreted Wnt proteins are proposed as important self-renewal factors for MG stem cells (Zeng and Nusse, 2010). Lgr5, a downstream target of Wnt, was identified as a marker of adult stem cell populations in the small intestine, colon (Barker et al., 2007), stomach (Barker et al., 2010) and hair follicle (Barker et al., 2008), organs which undergo extensive postnatal regeneration. Recently, lineage tracing experiments revealed that Lgr5+ stem/progenitor cells also contribute to nephron formation during kidney development (Barker et al., 2012).
Here, we unmask the regenerative potential of a rare Lgr5-expressing (Lgr5+) mammary cell population and their indispensible contribution to pubertal mammary development.
RESULTS
Lgr5 Expression is Restricted to a Rare Subpopulation of Cytokeratin 14+, Lin−CD24+CD49fhigh Basal Cells
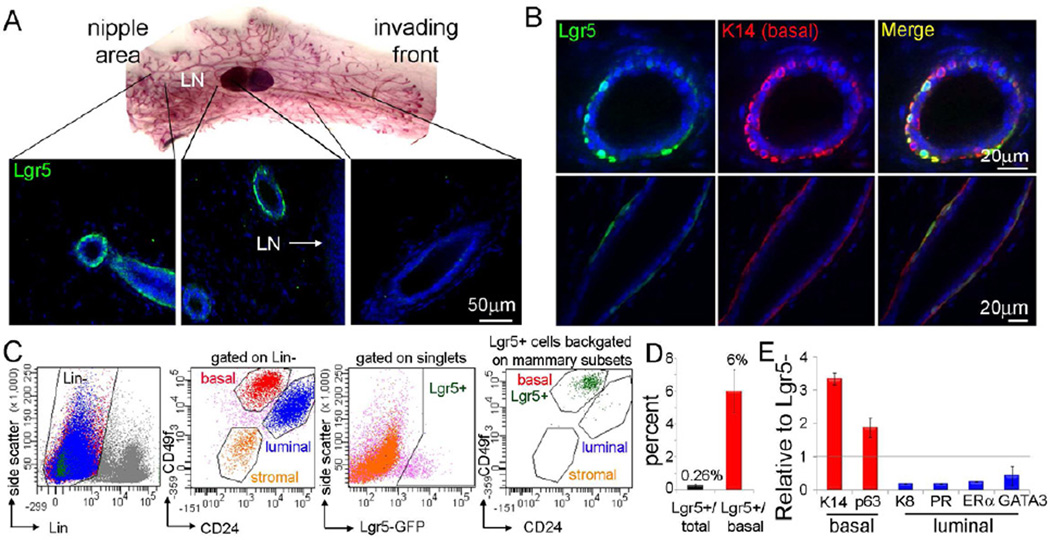
To identify cells that express Lgr5 in the MG, we used the Lgr5 knock-in mouse model in which EGFP reporter gene expression is driven by the endogenous Lgr5 regulatory sequences (Barker et al., 2007). In adult pubertal MGs, only 14% [± 2% standard error (SE)] of ducts had Lgr5+ cells that were localized to the nipple side (taking the lymph node as a point of reference), as previously illustrated (Van Keymeulen et al., 2011). The nipple is where the fetal epidermis initially invaginates into the mammary fat pad and is the origin growth point of the mammary epithelium (Figure 1A). Lgr5+ cells were a subset of cytokeratin 14 positive (K14+) cells and were localized to the supra-basal position (Figure 1B), similar to that previously described for mammary stem cells (Sleeman et al., 2007). In MGs, adult stem cells have been defined by flow cytometry as a rare subset of Lin−CD24+CD29high (Shackleton et al., 2006) or Lin−CD24lowCD49fhigh basal cells (Stingl et al., 2006), and a subpopulation of such cells exhibit the capacity to regenerate an entire MG in vivo. The vast majority of Lgr5+ cells were basal Lin−CD24+CD49fhigh (Figure 1C and S1) and were quite rare, comprising 0.26% (1 Lgr5+ cell per 386 cells) of total dissociated cells in pubertal MGs (Figure 1D). Previous studies have estimated the frequency of mammary stem cells or MRUs from adult virgin mouse MG to be 1 per 1,400 dissociated cells [for FVB background (Stingl et al., 2006)]; in contrast, 3–7% of cells in intestinal crypts express Lgr5 (Barker et al., 2007). In pubertal glands, among the mammary basal cells, only 6% were Lgr5+ (Figure 1D); this was corroborated by the expression profile of Lgr5+ cells, which showed high levels of basal but low levels of luminal epithelial markers (Figure 1E).
Figure 1. Lgr5 Expression is Restricted to a Rare Subpopulation of Cytokeratin 14+, Lin−CD24+CD49fhigh Mammary Basal Cells.
(A) The expression of Lgr5 was examined in cryosections from 7-week-old Lgr5-EGFP MGs with an anti-GFP antibody (green). Carmine stain of a representative MG whole mount demonstrates that Lgr5+ ducts are located to the nipple area, but not to the invading front. Around the lymph node there are some positive and negative ducts. LN, lymph node.
(B) Cryosections co-stained with anti-GFP and anti-K14. Lgr5+ cells (green) are a located to the supra-basal layer of the ducts and are a subpopulation of the myoepithelial K14+ cells (red).
(C) MGs isolated from Lgr5-EGFP mice and analyzed by flow cytometry, for the expression of the cell surface markers Ter119, CD45, CD31 (Lin), CD24 and CD49f. All Lgr5+ cells (GFP+) were part of the Lin−CD24+CD49fhigh cells (stem cell-enriched population). Lgr5+ cells are 0.3% of total mammary cells and 2.5% of Lin−CD24+CD49fhighbasal cells. GFP+ cells within the luminal population are 0.009% of total.
(D) Summary of flow cytometry data in Figure 1C, Lgr5+ cells in 7.5-week-old pubertal female mice.% of Lgr5+ cells of total (n=14) and of Lin−CD24+CD49fhighbasal cells (n=7). Also see Figure S1.
(E) Real-time, quantitative PCR analysis of the Lgr5+ cell population (relative to Lgr5− mammary cells) revealed they are high for basal but not luminal markers. PR, progesterone receptor; ERα, estrogen receptor α. See also Table S1.
Error bars represent SE.
Within the Lin−CD24+CD49fhigh Basal Population, Lgr5+ Cells are Highly Potent in Generating Functional Mammary Outgrowths
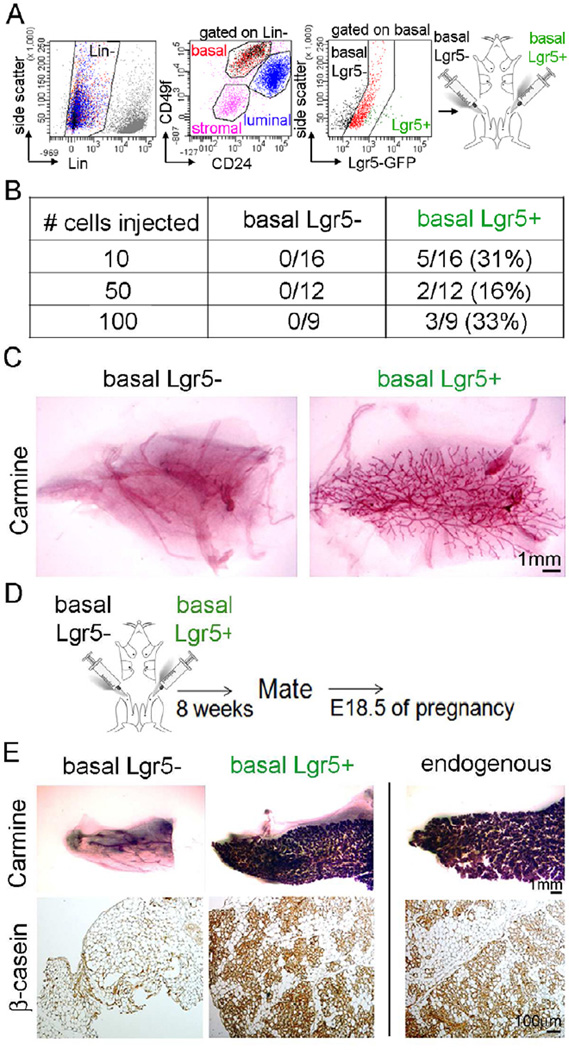
The analysis described above revealed that Lgr5+ cells are a subset of the Lin−CD24+CD49fhigh basal cells previously reported to include stem cells (Shackleton et al., 2006). To assess MG reconstitution competence, we challenged the Lgr5+ cells for mammary regeneration and compared them to Lgr5-negative (Lgr5−) basal cells in limiting dilution experiments (Figure 2A). In these experiments, we transplanted 10, 50 and 100 Lgr5+ versus Lgr5− basal cells into cleared fat pads. The number of cells transplanted was chosen on the lower range, to increase the stringency of the assay, focus on a small subset of basal cells, and avoid false negatives owing to Lgr5+ cells that express low levels of GFP and could therefore be sorted into the Lgr5− group. We found that within the basal population, Lgr5+ cells generated MGs far more efficiently than did basal Lgr5− cells. On average, 27% (± 5% SE) of Lgr5+ cells were able to regenerate a full MG, within the 10–100 cell range, or 1 MRU per 3.7 Lgr5+ cells (Figure 2B and 2C). We then tested functionality upon pregnancy (Figure 2D) and found that these outgrowths were able to undergo full lactational lobuloalveolar differentiation and express the milk protein, β-casein (Figure 2E). Characterization of single basal Lgr5+ cells vs. basal Lgr5− cells revealed that the different functional mammary reconstitution abilities of the two subsets are based on differences in gene expression of lineage differentiation, stem cell and pluripotency markers, demonstrating that these populations are distinct (Figure S2).
Figure 2. Within the Lin−CD24+CD49fhigh Basal Population, Lgr5+ Cells are Highly Potent in Generating Functional Mammary Outgrowths.
(A) Lgr5+ (GFP+) and non-expressing (GFP−) cells from Lgr5-EGFP were isolated by flow cytometry from the Lin−CD24+CD49fhigh basal population and injected (10, 50 or 100 cells) into cleared mammary fat pads. Outgrowths were analyzed 6 weeks post-transplantation.
(B) Transplanted basal Lgr5+ cells have higher numbers of outgrowths compared to the basal Lgr5-cells. Data pooled from 3 different experiments.
(C) Whole mount carmine-stained representative outgrowths show that 10 basal Lgr5+ cells are able to reconstitute a full MG vs. no outgrowth for basal Lgr5-transplanted cells.
(D) Mice transplanted with 10 Lgr5+ cells were mated with males and their MGs analyzed on day 18.5 (E18.5) of pregnancy.
(E) Whole mount Carmine-stained mammary epithelial outgrowths from E18.5 pregnant female mice transplanted with 10 basal Lgr5+ cells that underwent full lobuloalveolar differentiation (basal Lgr5+), comparable to the endogenous epithelium in MG #3 of the recipient mouse (upper panels). MG sections from the same mice stained positive for the milk protein, β-casein (lower panels; brown).
See also Figure S2 and Table S2.
Lgr5+ cells can Regenerate a Mammary Gland from a Single Cell and Maintain a Regenerative Potential Through Serial Transplantations
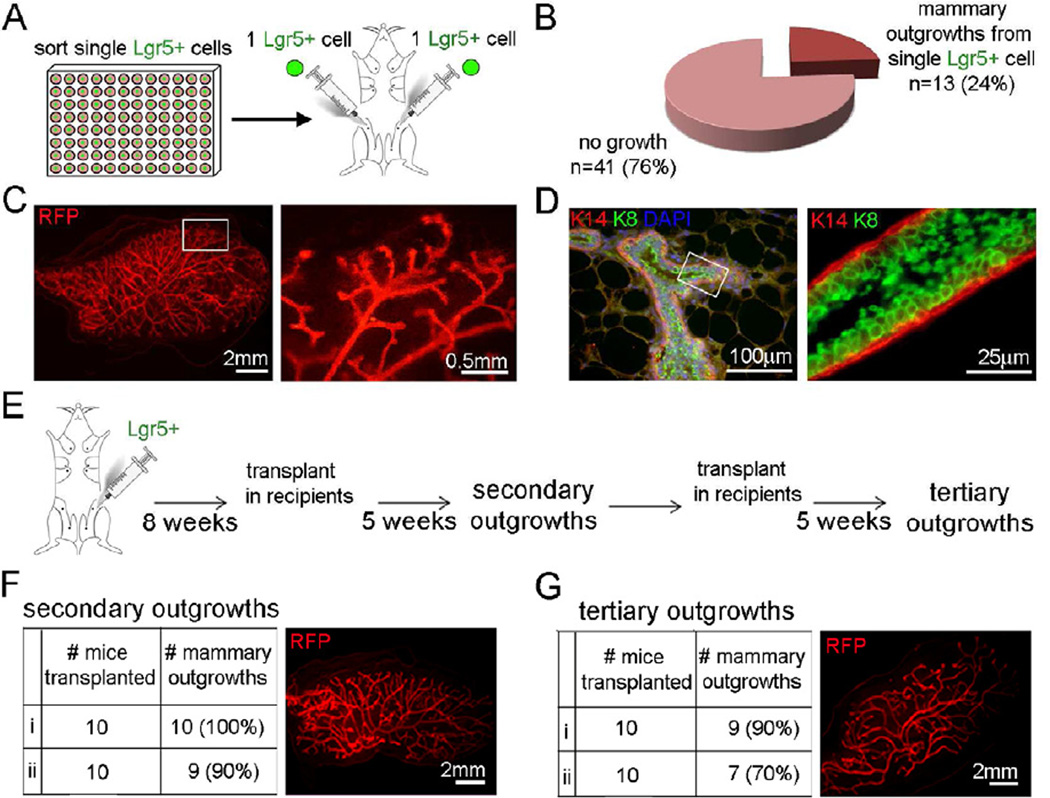
Since Lgr5+ cells within the basal cell population were highly efficient in regenerating a full MG in limiting dilution experiments, we tested them for classical stem cell characteristics of multipotency and self-renewal. First, we assessed their ability to regenerate fully differentiated MGs from single cells (Figure 3A). We observed that 13 outgrowths were generated from 54 single Lgr5+ transplanted cells (Figure 3B), demonstrating that 24% of Lgr5+ single cells were able to regenerate a full MG equivalent to 1 MRU per 4.2 Lgr5+ cells. These results are similar to those of the limiting dilution experiments (Figure 2). On close examination, we observed substantial epithelial outgrowth in the mammary fat pads (Figure 3C, Figure S3) and demonstrated that these single transplanted Lgr5+ cells were multipotent, as they were able to differentiate into both mammary epithelial lineages (myoepithelial/basal K14+ and luminal K8+ cells) (Figure 3D). In addition, when we serially transplanted epithelial outgrowths from primary transplants of Lgr5+ cells (Figure 3E), the Lgr5+ outgrowths retained their regenerative potential through secondary and tertiary transplants, demonstrating a long-term, regenerative potential (Figure 3F, 3G).
Figure 3. Lgr5+ cells can Regenerate a Mammary Gland from a Single Cell and Maintain Regenerative Potential through Serial Transplantations.
(A) Single mammary Lgr5+ (GFP+) cells from Lgr5-EGFP crossed into the Life Act-RFP mice were isolated by flow cytometry into 96-well plates and transplanted into cleared mammary fat pads. Outgrowths were analyzed at 8 weeks post-transplantation.
(B) From transplants of single adult mammary Lgr5+ cells in 54 mammary glands, 13 mammary outgrowths were observed.
(C) A representative RFP+ mammary outgrowth from a single Lgr5+ cell, exhibiting a full epithelial tree (left) with ductal structures at higher magnification of boxed area (right). (D) Outgrowths from single Lgr5+ cells differentiate into the myoepithelial (K14+ in red) and luminal (K8+ in green) lineages (left). Boxed area magnified (right). See also Figure S3.
(E) Mammary outgrowth from 2 mice transplanted with 100 Lgr5+ cells (isolated from Lgr5-EGFP crossed into the LifeAct-RFP mice) were collected and transplanted into 10 mice each for secondary and the same for tertiary outgrowths.
(F,G) Lgr5+ outgrowths retain their regenerative potential through secondary (F) and tertiary (G) transplants. RFP images are representative of the mammary outgrowths.
Depletion Experiments Demonstrate that Lgr5+ Cells are Necessary for Postnatal Mammary Gland Organogenesis
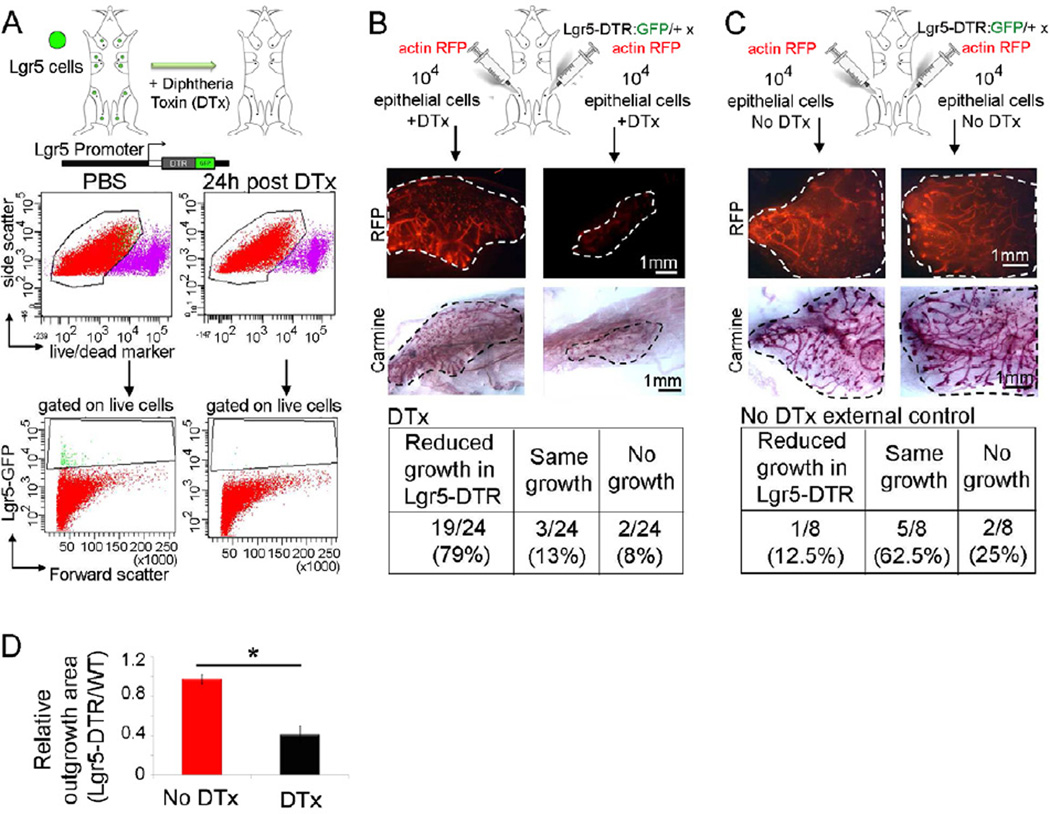
To determine whether Lgr5+ cells are not only sufficient but also necessary for postnatal MG organogenesis, we used the Lgr5-DTR:GFP mice to deplete Lgr5+ cells following administration of diphtheria toxin (DTx) (Figure 4A). This mouse model was used previously to demonstrate the dispensability of intestinal Lgr5+ cells under steady state conditions (Tian et al., 2011). However, depletion of Lgr5+ cells from transplanted MECs immediately post-transplantation, impaired the outgrowth of Lgr5-DTR:GFP donor epithelium, compared to the contralateral MG transplanted with WT MECs (Figure 4B). As an additional control, we found that the majority of MECs from Lgr5-DTR:GFP and WT mice not treated with DTx (i.e. in the presence of Lgr5+ cells) were able to reconstitute mammary outgrowth (Figure 4C). Uncleared, endogenous mammary tissue from the WT recipient mice was not affected by DTx administration (Figure S4B). The total outgrowth area for Lgr5-DTR:GFP epithelial transplants (including impaired ducts, as shown in Figure S4A) was also significantly reduced in DTx-treated mice relative to the contralateral WT transplants (Figure 4D). These experiments indicate that, although all other epithelial cells were not depleted, the absence of Lgr5+ cells was detrimental to adequate MG reconstitution. This protocol allowed targeted MG Lgr5+ cell depletion, since the recipient mice do not carry the Lgr5-DTR:GFP transgene. Mammosphere-forming assays in culture confirmed the indispensability of Lgr5+ cells (Figure S4C).
Figure 4. Depletion Experiments Demonstrate that Lgr5+ Cells are Necessary for Mammary Gland Epithelial Reconstitution.
(A) Depletion of Lgr5+ cells was achieved utilizing Lgr5-DTR:GFP crossed into actin-RFP mice, injected with 50ng/g BW DTx, analyzed 24h post DTx i.p. (Lgr5+ cells are 0.1% of total dissociated mammary cells versus 0% in DTx- injected mice).
(B) Isolated primary MECs of Lgr5-DTR:GFP mice or WT littermates transplanted into contralateral pre-cleared mammary fat pads with or without DTx administration. MGs collected 3 weeks post-transplantation had significantly impaired outgrowths in the Lgr5-DTR:GFP transplants vs. the WT controls.
(C) To assess the growth potential of the Lgr5-DTR:GFP and control littermate, mice transplanted with the same cells as in (B) but not treated with DTx reveal no difference between the two contralateral sides.
(D) Outgrowth area for Lgr5-DTR:GFP epithelial transplants (including impaired ducts) relative to the contralateral WT transplants is significantly reduced in DTx- treated mice (*p= 0.006). Bars represent SE.
See also Figure S4.
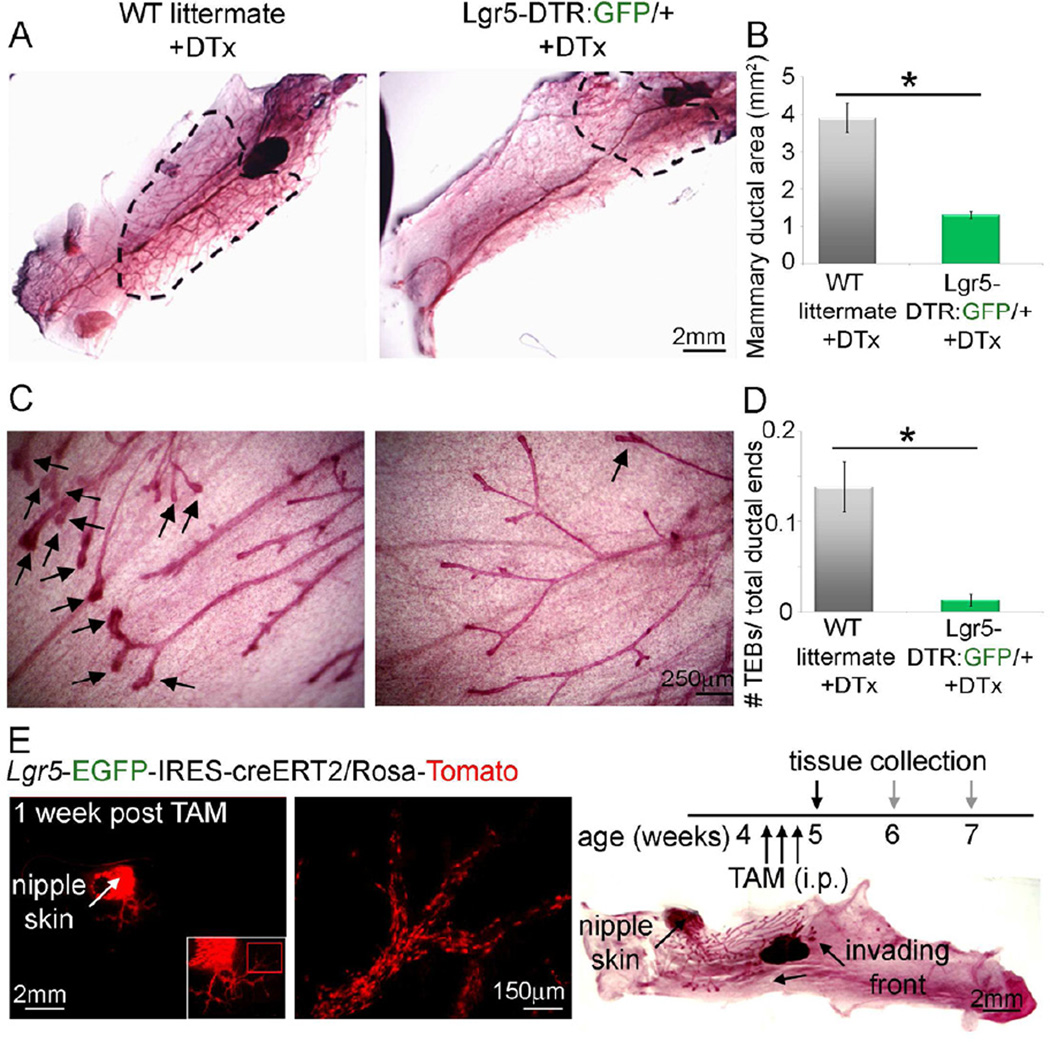
To complement the results above, we examined the role of Lgr5+ cells in postnatal MG organogenesis, in a more physiological setting, by injecting DTx to pubertal mice that were either Lgr5-DTR:GFP or WT littermates, (Figure 5). Depletion of Lgr5+ cells during pubertal MG development resulted in impaired ductal invasion (Fig. 5A, B) and interestingly, also in a significant reduction in the number of terminal end buds (TEBs) at the epithelial invading front (Fig. 5C, D), even though Lgr5+ cells (Fig. 1A) and their lineage-specific progeny (Fig. 5E, Fig. S5) are absent from the TEBs. In this context, although Lgr4 has been shown to play a minor role in MG development (Oyama et al., 2011), Lgr4+ cells were not interchangeable with Lgr5+ cells, since a significant phenotype was observed upon Lgr5+ cell depletion. These data show that under normal physiology, although all other cells (including additional progenitor cells) were not depleted, the presence of Lgr5+ cells is necessary for MG pubertal development and reinforce the depletion results in the transplantation setting
Figure 5. Depletion of Lgr5+ Cells During Pubertal Development Results in Impaired Ductal Invasion and Terminal End Bud Formation.
(A) Carmine-stained MG of 4.5-week-old Lgr5-DTR:GFP mice (n=6) or WT littermates (n=4) that were i.p. injected with DTx demonstrate significantly reduced ductal invasion in the Lgr5-DTR:GFP mice.
(B) Quantification of data presented in (A).
(C) Depletion of Lgr5+ cells from Lgr5-DTR:GFP mice resulted in significant reduction in the number of TEBs per MG versus WT littermates.
(D) Quantification of data presented in (C).
(E) Whole mounts of 5-week-old Lgr5-EGFP-IRES-creERT2/Rosa-Tomato mice one week past start of Tamoxifen (TAM) induction, indicated that Lgr5+ cell progeny are close to the nipple area (left) and, according to their localization and shape, mark myoepithelial cells (middle, enlargement of red boxed area in left) and not TEBs in the invading front (Carmine-stained tissue on right).
Error bars represent SE.
See also Figure S5.
DISCUSSION
Classically, stem cells are characterized by their ability to self-renew as well as to differentiate into specialized cells. According to these criteria, Lgr5+ cells have been identified as adult stem cells in the small intestine, colon (Barker et al., 2007), stomach (Barker et al., 2010), and hair follicle (Barker et al., 2008). Our study now shows that Lgr5+ cells are also adult stem cells in the MG. By transplantation assays, we demonstrated that most Lgr5+ cells are a subset of the basal population previously shown to include the mammary stem cells, exhibiting superior reconstitution capabilities as compared to other cells within that population and are also extremely efficient in regenerating a MG from a single cell. The reconstituted MG epithelial tree was also functional, as it was able to undergo adequate differentiation during pregnancy and produce a milk protein. Lgr5+ cells were multipotent and maintain regenerative potential in serial transplantations and therefore sufficient for postnatal MG organogenesis. They were also necessary for MG organogenesis as shown in depletion assays in both transplantation and physiological settings.
The frequency of MRUs was previously estimated to be between 1 MRU per 8–17 cells using transplants of single cells or 1 per 64 cells within the Lin−CD24+CD29high population of mammary cells (Shackleton et al., 2006) or 1 per 60 cells (for FVB background) and 1 per 90 cells (for C57BL/6 background) within the Lin-CD24+CD49fhigh cells (Stingl et al., 2006) in limiting dilution experiments. More recently, the stem cell frequency within the adult Lin−CD24+CD49fhigh population was estimated as 1 per 50 cells when co-injected with Matrigel (Spike et al., 2012). The reconstitution capabilities of 1 per 4 cells that we observed are remarkable, bringing us closer to obtaining a homogeneous population of MRUs.
Although previous transplant experiments suggested a common progenitor for both major mammary epithelial lineages (myoepithelial/basal and luminal) (Shackleton et al., 2006; Stingl et al., 2006), a recent study that utilized lineage tracing assays pointed to two different progenitors for these lineages as early as birth (Van Keymeulen et al., 2011), and therefore suggested a more restricted fate for the Lgr5+cells, which was reinforced in a recent study (de Visser et al., 2012) and also in our study. These data point to important differences between lineage-tracing and transplantation techniques. Indeed, individual stem cells can have different roles under physiological, homeostatic conditions visualized by lineage tracing (van Amerongen et al., 2012), compared to when they are challenged to regenerate an entire organ in the transplant assays (Keller et al., 2011). Thus, lineage-tracing experiments using an Lgr5-CreER line show that Lgr5+ cells give rise only to myoepithelial cells in pubertal MGs (Van Keymeulen et al., 2011), whereas our transplant experiments demonstrated that a single Lgr5+ cell is sufficient to regenerate a complete mammary epithelium and differentiates into both myoepithelial and luminal cells. The transplant assays might therefore uncover a regenerative potential of Lgr5+ cells that would be inhibited during MG pubertal development. However, in all the previous studies, depletion of a specific cell population in the presence of all the other cells was never attempted. We now have demonstrated that, in the MG, the unique approach of specific Lgr5+ cell depletion resulted in significantly impaired organogenesis, indicating that Lgr5+ cells are required during both regeneration from transplanted MECs, but also, and more importantly, during physiological pubertal development.
Previous studies indicate that mammary stem cells are likely to be present in any portion of the epithelial branches (Kordon and Smith, 1998). Our study showed that Lgr5+ cells, although able to regenerate a full MG, are clustered towards the nipple area in pubertal MGs, where the branching of the epithelium originates and they or their progeny are not found at the invading front of the ductal tree. However, Lgr5+ cell depletion in the transplants resulted in significantly impaired reconstitution, although all other epithelial cells were not targeted for depletion. Moreover, Lgr5+ cell depletion during physiological MG organogenesis also resulted in impaired ductal invasion and specifically was characterized by diminished TEBs. TEBs are essential to pubertal MG development and contain additional progenitor populations [as Axin 2+ cells (van Amerongen et al., 2012)]. Our data indicate that even if there are additional stem/progenitor cells that contribute to MG organogenesis, Lgr5+ cells are not only sufficient, but also essential for this process and suggest a crosstalk between various stem/progenitor cells during normal MG development.
Stem cells are key for understanding both normal development as well as associated pathologies. In fact, Lgr5 was first described as a gene expressed in colon cancer cells (van de Wetering et al., 2002). Moreover, it has since been postulated that transformation of Lgr5+ stem cells drives malignant progression in the small intestine and colon (Barker et al., 2009) and stem cell activity has been demonstrated in Lgr5+ cells in mouse intestinal adenoma (Schepers et al., 2012). Lgr5 is also over-expressed in other cancers (McClanahan et al., 2006; Oskarsson et al., 2011; Yamamoto et al., 2003), including breast cancer (Oskarsson et al., 2011). The fact that Lgr5+ cells are particularly efficient in regenerating a full mammary gland suggests they could also effectively play an active role in breast cancer once they are transformed. Since Wnt signaling has been implicated in different stages of mammary oncogenesis, future studies should explore the role of Lgr5+ cells as breast cancer stem cells. Moreover, R-spondins were recently shown to potentiate Wnt/β-catenin signaling through Lgr5 (Carmon et al., 2011; de Lau et al., 2011; Gong et al., 2012). Since local epithelial R-spondin 1 signaling is required for normal development of the mammary gland (Chadi et al., 2009), future studies evaluating the role of Lgr5 as a receptor for R-spondin during mammary development and cancer are worth pursuing.
EXPERIMENTAL PROCEDURES
Mouse Strains
C57BL/6J (Jackson Laboratories), β-actin-RFP (Long et al., 2005), LifeAct-RFP (Riedl et al., 2010), Lgr5-EGFP-IRES-creERT2 (Lgr5-EGFP) (Barker et al., 2007), Lgr5-DTR:GFP (Tian et al., 2011) and Ai14 Rosa-Tomato (Madisen et al., 2010) mice were bred and maintained in the UCSF animal facility according to IACUC guidelines. All mice were maintained in C57BL6J background. β-actin-RFP and LifeAct-RFP reporter mice were used interchangeably to specifically identify and visualize mammary outgrowths from the donor mice.
Mammary Cell Preparations
MGs were dissected from pubertal (7–9 week-old) female mice. For flow cytometry and limiting dilution experiments, after mechanical dissociation with a scalpel, the tissue was placed in culture medium (DMEM/F12 with 5 ng/ml insulin (UCSF Cell Culture Facility), 50 ng/ml gentamycin (UCSF Cell Culture Facility) containing 2 mg/ml collagenase-1 (Sigma) and digested for 30 min at 37°C. The resulting suspension was sequentially resuspended in 2 U/µl DNAse for 3 min at RT, washed and dissociated with 2 ml 0.05% trypsin/EDTA (UCSF Cell Culture Facility) for 10 min at 37°C and filtered through a 70 µm filter. Erythrocytes were lysed with Red Blood Cell Lysis Buffer [protocol ID PS00000002 (Gilman et al., 2002)] for 1 min at room temperature. For the diphtheria toxin depletion experiments, epithelium-enriched organoids were prepared as described previously (Ewald et al., 2008) then dissociated with 2 ml 0.05% trypsin/EDTA and filtered as described above.
Cell Labeling, Flow Cytometry and Sorting
Antibodies against the mouse antigens CD45, CD31, TER119, CD49f, CD24 were purchased from eBioscience. For the single cell transplants, single Lgr5-GFP+ cells were sorted into 96-well plates in minimal medium + 2.5 nM FGF2 (Ewald et al., 2008). Flow cytometry was performed using LSRII, for data analysis and FACS ARIA for cell sorting (BD Biosciences).
Mammary Fat Pad Transplantation
Cleared fat pads from 3-week-old female nude mice (Simenson) were transplanted with 1–100 MECs in 50/50 Matrigel/minimal medium+2.5 nM FGF2 (Ewald et al., 2008). The tissues cleared from the MGs were Carmine-stained as described below, to validate adequate clearing of the native epithelium (to ensure that the native epithelium had not yet reached the lymph node). The transplanted mammary epithelium was allowed to grow from 3 to 8 weeks and mammary outgrowths were analyzed by whole mount staining with Carmine, whole mount fluorescence or flow cytometry. For the secondary and tertiary transplants, pieces of mammary fat pad containing epithelium were transplanted into cleared fat pads from 3-week-old female nude mice. Mammary outgrowths were analyzed 5 weeks after transplants. Outgrowths were considered positive when the epithelium invaded at least half of the fat pad. For single cell transplants and serial transplantation experiments, Lgr5-EGFP-IRES-creERT2 were crossed into the LifeAct-RFP reporter mice, and for Lgr5 depletion experiments Lgr5-DTR:GFP mice were crossed into the β-actin-RFP reporter mice, to allow easier and reliable detection of outgrowths.
Histochemistry, Immunohistochemistry and Immunofluorescence
Mammary whole mounts were stained with Carmine Alum (Sigma). Cryo- or paraffin sections from the inguinal (#4) MGs of Lgr5-EGFP mice or from mammary outgrowths were labeled using the following primary antibodies: GFP (Abcam, ab5450, 1:200), cytokeratin 14 (Covance, PRB-155P, 1:500), cytokeratin 8 (Troma 1, Developmental Studies Hybridoma Bank, Iowa, 1:50) and β-casein (ABBIOTEC, #250558, 1:200).
Real Time PCR
Sorted cell populations were lysed and RNA was extracted using a Qiagen mini kit (74104). cDNA synthesis was performed using the Invitrogen SuperScript III system (18080-051), and quantitative reverse transcription-PCR was done via the Sybrgreen (Applied Biosystems, 4309155) method and an Eppendorf Realplex Mastercycler. Primer sequences are listed in Table S1. Primers were purchased from SABiosciences. Relative quantification of gene expression was calculated according the Pfaffl method. Target gene expression in each cell subpopulation was normalized to HPRT and GAPDH reference gene expression. The data reported are one representative experiment of three independent sorting and qRT-PCR experiments.
Diphtheria Toxin-mediated Cell Depletion
Mammary fat pads from 3-week-old female nude mice (Simenson) were cleared to remove all endogenous epithelium, and the recipient mice were allowed to grow bigger before transplantation and therefore become more resilient to DTx toxicity. Four to five weeks later, 104 MECs from Lgr5-DTR:GFP or WT littermates were contra-laterally transplanted into pre-cleared fat pads in Matrigel/minimal medium+2.5nM FGF2, 1:1 (Ewald et al., 2008) containing 1µg/ml DTx (Sigma), or no DTx in external controls, to achieve immediate but local Lgr5+ cell depletion. After 6 days, mice were injected intraperitoneally (i.p.) with 50ng/g body weight (BW) DTx, 3 times/week for 1.5 weeks to maintain Lgr5+ cell depletion throughout the experiment. Mammary tissue was collected 3 weeks post-transplantation, which is sufficient time to yield mammary outgrowths. Due to possible DTx toxicity at the concentration of 50ng/g BW, which allows full Lgr5+ depletion in the mammary, the treatment regimen above could not be prolonged further to allow outgrowths to fully progress so the internal controls of outgrowths from WT cells, which are also subjected to DTx, serve as a reference to the Lgr5-DTR:GFP outgrowths. The external control group was i.p. injected with PBS, under a similar regimen.
In a separate set of experiments, 4.5-week-old Lgr5-DTR or WT littermates were injected i.p. with 50ng/g BW DTx, 3 times per week for 1.5 weeks. Inguinal MGs were retrieved, Carmine-stained and the ductal-invaded area was calculated. Calculation was done using ImageJ software- the ductal area calculated is demarcated (the lymph node is the point of reference for ductal invasion). Additionally, terminal end buds were manually counted directly from MG whole mounts. Due to the possible effect of DTx depletion on additional organs in Lgr5-DTR:GFP mice and DTx toxicity, these experiments could not be prolonged beyond the current endpoint.
In vivo Tamoxifen induction
Four-week-old Lgr5-EGFP-IRES-creERT2/Rosa-Tomato female mice were i.p. injected with 5mg of Tamoxifen (Sigma) diluted in sunflower oil (Sigma) every other day for a total of 3 days (15 mg total), as indicated in (Van Keymeulen et al., 2011). Mammary glands were collected at 5, 6 and 7 weeks of age and Cre induction was assessed by whole mount fluorescence while epithelial outgrowths were visualized by Carmine staining.
Supplementary Material
HIGHLIGHTS.
Lgr5+ cells regenerate mammary epithelium more effectively than other basal cells
Single Lgr5+ cells are exceptionally efficient in reconstituting mammary glands
Depletion of Lgr5+ cells leads to impaired mammary gland pubertal development
Lgr5+ cells are sufficient and necessary for postnatal mammary gland organogenesis
ACKNOWLEDGMENTS
We thank Dr. Nirav Bhakta for help with the single-cell PCR analysis, Dr. Brian Biehs and Dr. Hua Tian for initial help with the Lgr5-DTR:GFP breeding, Dr. Roland Wedlich-Soeldner for providing the LifeAct-RFP mice, Michael Kissner for assistance with flow cytometry, Ying Yu and Elena Atamaniuc for mice husbandry and genotyping and Joanne Dai for technical help. This work was supported by grants from the National Cancer Institute and the National Institute of Environmental Health Sciences (R01 CA057621 and U01 ES019458 to ZW) and by Department of Defense postdoctoral fellowships to VP (W81XWH-11-01-0139) and to DAL (W81XWH-11-1-0742) and the Weizmann Institute of Science-National Postdoctoral Award Program for Advancing Women in Science (to VP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes 5 Figures and 2 Tables, Supplemental Experimental Procedures and References.
REFERENCES
- Badders NM, Goel S, Clark RJ, Klos KS, Kim S, Bafico A, Lindvall C, Williams BO, Alexander CM. The Wnt receptor, Lrp5, is expressed by mouse mammary stem cells and is required to maintain the basal lineage. PLoS One. 2009;4:e6594. doi: 10.1371/journal.pone.0006594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, Huch M, Kujala P, van de Wetering M, Snippert HJ, van Es JH, Sato T, Stange DE, Begthel H, van den Born M, et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- Barker N, Rookmaaker MB, Kujala P, Ng A, Leushacke M, Snippert H, van de Wetering M, Tan S, Van Es JH, Huch M, et al. Lgr5(+ve) Stem/Progenitor Cells Contribute to Nephron Formation during Kidney Development. Cell Rep. 2012;2:540–552. doi: 10.1016/j.celrep.2012.08.018. [DOI] [PubMed] [Google Scholar]
- Barker N, van Es JH, Jaks V, Kasper M, Snippert H, Toftgard R, Clevers H. Very long-term self-renewal of small intestine, colon, and hair follicles from cycling Lgr5+ve stem cells. Cold Spring Harb Symp Quant Biol. 2008;73:351–356. doi: 10.1101/sqb.2008.72.003. [DOI] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Boras-Granic K, Chang H, Grosschedl R, Hamel PA. Lef1 is required for the transition of Wnt signaling from mesenchymal to epithelial cells in the mouse embryonic mammary gland. Dev Biol. 2006;295:219–231. doi: 10.1016/j.ydbio.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Brisken C, Heineman A, Chavarria T, Elenbaas B, Tan J, Dey SK, McMahon JA, McMahon AP, Weinberg RA. Essential function of Wnt-4 in mammary gland development downstream of progesterone signaling. Genes Dev. 2000;14:650–654. [PMC free article] [PubMed] [Google Scholar]
- Carmon KS, Gong X, Lin Q, Thomas A, Liu Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc Natl Acad Sci USA. 2011;108:11452–11457. doi: 10.1073/pnas.1106083108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadi S, Buscara L, Pechoux C, Costa J, Laubier J, Chaboissier MC, Pailhoux E, Vilotte JL, Chanat E, Le Provost F. R-spondin1 is required for normal epithelial morphogenesis during mammary gland development. Biochem Biophys Res Commun. 2009;390:1040–1043. doi: 10.1016/j.bbrc.2009.10.104. [DOI] [PubMed] [Google Scholar]
- Chu EY, Hens J, Andl T, Kairo A, Yamaguchi TP, Brisken C, Glick A, Wysolmerski JJ, Millar SE. Canonical WNT signaling promotes mammary placode development and is essential for initiation of mammary gland morphogenesis. Development. 2004;131:4819–4829. doi: 10.1242/dev.01347. [DOI] [PubMed] [Google Scholar]
- de Lau W, Barker N, Low TY, Koo BK, Li VS, Teunissen H, Kujala P, Haegebarth A, Peters PJ, van de Wetering M, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- de Visser KE, Ciampricotti M, Michalak EM, Tan DW, Speksnijder EN, Hau CS, Clevers H, Barker N, Jonkers J. Developmental stage-specific contribution of LGR5(+) cells to basal and luminal epithelial lineages in the postnatal mammary gland. J Pathol. 2012;228:300–309. doi: 10.1002/path.4096. [DOI] [PubMed] [Google Scholar]
- Deome KB, Faulkin LJ, Jr, Bern HA, Blair PB. Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer Res. 1959;19:515–520. [PubMed] [Google Scholar]
- Ewald AJ, Brenot A, Duong M, Chan BS, Werb Z. Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev Cell. 2008;14:570–581. doi: 10.1016/j.devcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman AG, Simon MI, Bourne HR, Harris BA, Long R, Ross EM, Stull JT, Taussig R, Bourne HR, Arkin AP, et al. Overview of the Alliance for Cellular Signaling. Nature. 2002;420:703–706. doi: 10.1038/nature01304. [DOI] [PubMed] [Google Scholar]
- Gong X, Carmon KS, Lin Q, Thomas A, Yi J, Liu Q. LGR6 is a high affinity receptor of R-spondins and potentially functions as a tumor suppressor. PLoS One. 2012;7:e37137. doi: 10.1371/journal.pone.0037137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller PJ, Arendt LM, Kuperwasser C. Stem cell maintenance of the mammary gland: it takes two. Cell Stem Cell. 2011;9:496–497. doi: 10.1016/j.stem.2011.11.008. [DOI] [PubMed] [Google Scholar]
- Kordon EC, Smith GH. An entire functional mammary gland may comprise the progeny from a single cell. Development. 1998;125:1921–1930. doi: 10.1242/dev.125.10.1921. [DOI] [PubMed] [Google Scholar]
- Lindvall C, Evans NC, Zylstra CR, Li Y, Alexander CM, Williams BO. The Wnt signaling receptor Lrp5 is required for mammary ductal stem cell activity and Wnt1-induced tumorigenesis. J Biol Chem. 2006;281:35081–35087. doi: 10.1074/jbc.M607571200. [DOI] [PubMed] [Google Scholar]
- Lindvall C, Zylstra CR, Evans N, West RA, Dykema K, Furge KA, Williams BO. The Wnt co-receptor Lrp6 is required for normal mouse mammary gland development. PLoS One. 2009;4:e5813. doi: 10.1371/journal.pone.0005813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JZ, Lackan CS, Hadjantonakis AK. Genetic and spectrally distinct in vivo imaging: embryonic stem cells and mice with widespread expression of a monomeric red fluorescent protein. BMC Biotechnol. 2005;5:20. doi: 10.1186/1472-6750-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClanahan T, Koseoglu S, Smith K, Grein J, Gustafson E, Black S, Kirschmeier P, Samatar AA. Identification of overexpression of orphan G protein-coupled receptor GPR49 in human colon and ovarian primary tumors. Cancer Biol Ther. 2006;5:419–426. doi: 10.4161/cbt.5.4.2521. [DOI] [PubMed] [Google Scholar]
- Nusse R, Varmus HE. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982;31:99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- Oskarsson T, Acharyya S, Zhang XH, Vanharanta S, Tavazoie SF, Morris PG, Downey RJ, Manova-Todorova K, Brogi E, Massague J. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med. 2011;17:867–874. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama K, Mohri Y, Sone M, Nawa A, Nishimori K. Conditional knockout of Lgr4 leads to impaired ductal elongation and branching morphogenesis in mouse mammary glands. Sex Dev. 2011;5:205–212. doi: 10.1159/000329476. [DOI] [PubMed] [Google Scholar]
- Riedl J, Flynn KC, Raducanu A, Gartner F, Beck G, Bosl M, Bradke F, Massberg S, Aszodi A, Sixt M, et al. Lifeact mice for studying F-actin dynamics. Nat Methods. 2010;7:168–169. doi: 10.1038/nmeth0310-168. [DOI] [PubMed] [Google Scholar]
- Roelink H, Wagenaar E, Lopes da Silva S, Nusse R. Wnt-3, a gene activated by proviral insertion in mouse mammary tumors, is homologous to int-1/Wnt-1 and is normally expressed in mouse embryos and adult brain. Proc Natl Acad Sci USA. 1990;87:4519–4523. doi: 10.1073/pnas.87.12.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M, Clevers H. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–735. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Sleeman KE, Kendrick H, Robertson D, Isacke CM, Ashworth A, Smalley MJ. Dissociation of estrogen receptor expression and in vivo stem cell activity in the mammary gland. J Cell Biol. 2007;176:19–26. doi: 10.1083/jcb.200604065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike BT, Engle DD, Lin JC, Cheung SK, La J, Wahl GM. A mammary stem cell population identified and characterized in late embryogenesis reveals similarities to human breast cancer. Cell Stem Cell. 2012;10:183–197. doi: 10.1016/j.stem.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl J, Caldas C. Molecular heterogeneity of breast carcinomas and the cancer stem cell hypothesis. Nat Rev Cancer. 2007;7:791–799. doi: 10.1038/nrc2212. [DOI] [PubMed] [Google Scholar]
- Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, de Sauvage FJ. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Amerongen R, Bowman AN, Nusse R. Developmental Stage and Time Dictate the Fate of Wnt/beta-Catenin-Responsive Stem Cells in the Mammary Gland. Cell Stem Cell. 2012;11:387–400. doi: 10.1016/j.stem.2012.05.023. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, Sharma N, Dekoninck S, Blanpain C. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Lindeman GJ. Mammary stem cells and mammopoiesis. Cancer Res. 2006;66:9798–9801. doi: 10.1158/0008-5472.CAN-06-2254. [DOI] [PubMed] [Google Scholar]
- Welm B, Behbod F, Goodell MA, Rosen JM. Isolation and characterization of functional mammary gland stem cells. Cell Prolif. 2003;36(Suppl 1):17–32. doi: 10.1046/j.1365-2184.36.s.1.3.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward WA, Chen MS, Behbod F, Rosen JM. On mammary stem cells. J Cell Sci. 2005;118:3585–3594. doi: 10.1242/jcs.02532. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Sakamoto M, Fujii G, Tsuiji H, Kenetaka K, Asaka M, Hirohashi S. Overexpression of orphan G-protein-coupled receptor, Gpr49, in human hepatocellular carcinomas with beta-catenin mutations. Hepatology. 2003;37:528–533. doi: 10.1053/jhep.2003.50029. [DOI] [PubMed] [Google Scholar]
- Zeng YA, Nusse R. Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell Stem Cell. 2010;6:568–577. doi: 10.1016/j.stem.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.