Abstract
We assessed tissue macrophage gene expression in different mouse organs. Diversity in gene expression among different populations of macrophages was remarkable. Only a few hundred mRNA transcripts stood out as selectively expressed by macrophages over DCs and many of these were not present in all macrophages. Nonetheless, well-characterized surface markers, including MerTK and FcγR1 (CD64), along with a cluster of novel transcripts were distinctly and universally associated with mature tissue macrophages. TCEF3, C/EBPα, BACH1, and CREG-1 were among the top transcriptional regulators predicted to regulate these core macrophage-associated genes. Other transcription factor mRNAs were strongly associated with single macrophage populations. We further illustrate how these transcripts and the proteins they encode facilitate distinguishing macrophage versus DC identity of less characterized populations of mononuclear phagocytes.
Introduction
The team of immunologists and computational biologists that comprise the Immunological Genome Project (ImmGen) share the goal of generating an exhaustive definition of gene expression and regulatory networks of the mouse immune system using shared resources and rigorously controlled data generation pipelines1. Here, we have turned attention to gene expression and regulatory networks in tissue resident macrophages.
Macrophages are professional phagocytic cells, often long-lived, that reside in all organs to maintain tissue integrity, clear debris, and respond rapidly to initiate repair upon injury or innate immunity following infection2,3. Accordingly, macrophages are specialized for degrading and detoxifying engulfed cargo and they are potent secretagogues with the capacity to display an array of phenotypes4. Macrophages can also present antigens, but lack the potency in stimulating T cells observed in dendritic cells, and usually fail to mobilize to lymphoid tissues where naïve T cells are abundant. Partially overlapping functions between macrophages and dendritic cells, reflected by overlapping molecular profiles, have for decades fueled some debate over the origins and overall distinction between macrophages and dendritic cells (DCs)5.
In the last several years, significant progress has been made in identifying precursors specific to DCs6–8. Moreover, transcription factors have been identified, such as Batf3, which are essential for the development of some DCs but not required for macrophage specification9. Recent advances have also been made in deciphering the development of tissue macrophages. Counter to the prevalent idea that monocytes were precursors for tissue macrophages, some earlier work contended that tissue macrophages arise from primitive hematopoietic progenitors present in the yolk sac during embryonic development independently of the monocyte lineage10 and strong support for this contention has very recently emerged through fate-mapping and genetic models11,12. Thus, in the adult, maintenance of tissue macrophage involves local proliferation, again independently of monocytes and definitive hematopoiesis10,12. In this context, MAFB/cMAF has been shown to regulate macrophage self-renewal13. Some transcription factors that drive development of given macrophage types such as osteoclasts14 or red pulp macrophages15 have also been reported. However, much remains to be revealed regarding the transcriptional regulatory pathways that control other types of macrophages or global regulatory pathways that govern macrophages as a group of related cells3. The database generated by the Immunological Genome Project creates a unique resource to compare gene expression profiles and to identify regulatory pathways that specify or unify macrophage populations from different organs. Our analysis of the macrophage transcriptome in this context enables the analysis of networks of genes and their regulators that can be used to better distinguish different types of macrophages and pinpoint the differences between macrophages and DCs.
Results
Tissue macrophage diversity
As part of ImmGen, we sorted several tissue macrophages populations from C57BL/6J mice according to strict, standardized procedures and analyzed these populations using whole-mouse genome Affymetrix Mouse Gene 1.0 ST Arrays. Sorting strategies for these populations can be found on the ImmGen website (http://immgen.org), and gene expression data are deposited in the Gene Expression Omnibus (GEO, accession # GSE15907).
We began our analysis by examining the gene expression profiles of resting macrophage populations that have historically been highly characterized and accepted as bona fide resident tissue macrophages16. Though some classical macrophages, such as Kupffer cells of the liver and metallophilic as well marginal zone macrophages of the spleen proved elusive for definitive identification and/or isolation by flow cytometric cell sorting, four resting macrophage populations submitted to Immgen met the criteria of bona fide macrophage populations: peritoneal macrophages, red pulp splenic macrophages, lung macrophages, and microglia (brain macrophages). We thus focused our initial analysis on these four key macrophage populations. Principal component analysis (PCA) of all genes expressed by the four sorted populations and several DC populations revealed a relatively greater distance between the different macrophages compared with DCs (Fig. 1a). Pearson correlation values were high for replicates within a given DC or macrophage population as per the quality control standards of Immgen; variability within replicates for a single population varied from 0.908 ± 0.048 for microglia to 0.995 ± 0.001 for peritoneal macrophages. Pearson correlations in gene expression profiles between different populations of DCs yielded coefficients ranging from 0.877 (liver CD11b+ versus spleen CD8+ DCs) to 0.966 (spleen CD4+CD11b+ versus spleen CD8+ DCs) (mean of all DC populations 0.931), whereas the correlation coefficients between different tissue macrophages ranged from 0.784 (peritoneal versus splenic red pulp) to 0.863 (peritoneal versus lung) with a mean of 0.812 (Fig. 1b). Several thousand mRNA transcripts were differentially expressed by at least 2-fold when, for example, lung macrophages were compared to red pulp splenic macrophages (Fig. 1c). This degree of diversity was greater than that observed when DCs from different subsets (CD103+ versus CD11b+) were compared from different organs (Fig. 1c). Finally, a dendrogram applied to the various populations showed that DCs clustered more closely than did macrophages (Fig. 1d), and this was true whether we considered all gene transcripts in the array (data not shown) or only the top 15% ranked by cross-population max/min ratio or coefficient of variation (Fig. 1d). Overall, these comparisons indicate a pronounced diversity among tissue macrophage populations.
Figure 1. Analysis of macrophage diversity.
(a) Relative distance between different types of macrophages and DCs was assessed using principal component analysis. (b) Correlation matrix of macrophages and dendritic cells based on all genes probes. (c) Examples of the relatively greater diversity between macrophage populations than DCs were plotted. The number of probes increased by a minimum of 2-fold for each population is indicated. (d) Hierarchical clustering of macrophages and dendritic cells based on the top 15% most variable genes.
Distinct molecular signatures among tissue macrophages
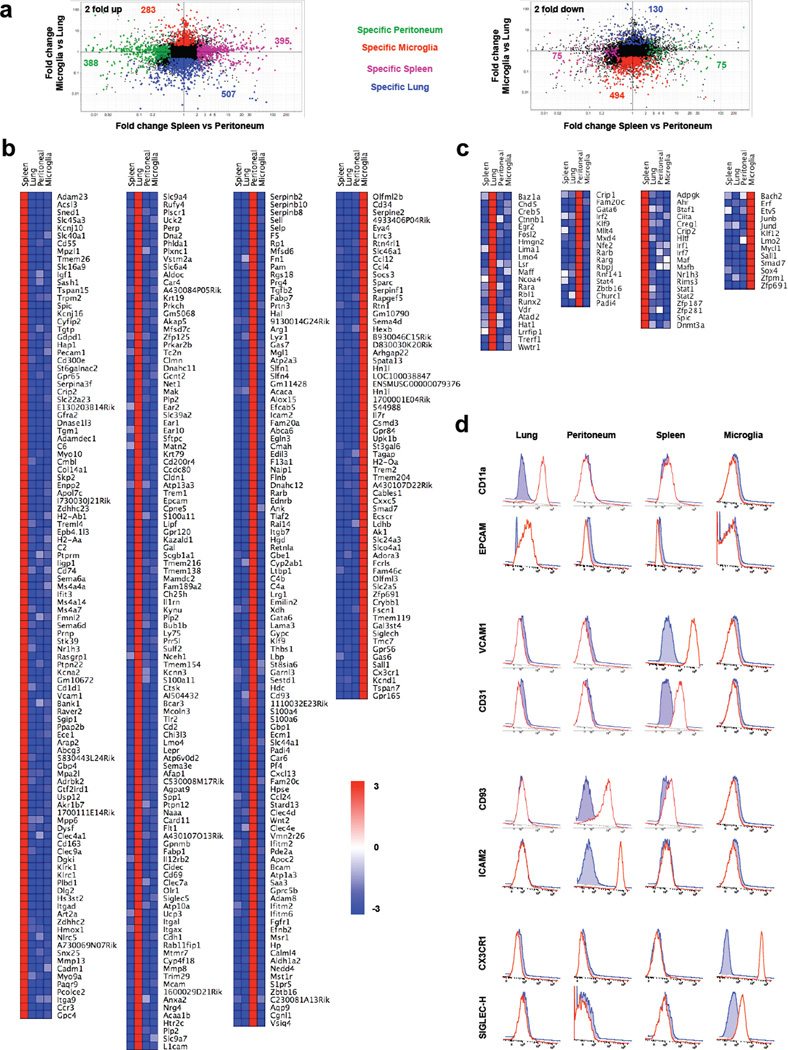
The diversity among these four classical macrophage populations extended to gene families previously associated with macrophage function - chemokine receptors, Toll-like receptors, C-type lectins, and efferocytic receptors. For example, at least one distinct chemokine receptor observed in each population was prominently expressed above the others (Supplementary Fig. 1a). Diversity among Toll-like receptors, C-type lectin domain members and efferocytic receptors was also remarkable (Supplementary Fig. 1b–d). Indeed, only a few of the mRNA transcripts profiled in these categories, including mRNA encoding the Mer tyrosine kinase receptor (MerTK) involved in phagocytosis of apoptotic cells17, and toll-like receptors Tlr4, Tlr7, Tlr8, and Tlr13, showed relatively uniform expression across all macrophages compared. Hundreds of mRNA transcripts were selectively increased or decreased by at least 2-fold in only one of the macrophage populations (Fig. 2a), and microglia in particular displayed low expression of hundreds of transcripts that were expressed in other macrophage populations (Fig. 2a). Using Ingenuity pathway analysis tools, we found each specific signature to be enriched in groups of transcripts with predicted specific functions, including those with oxidative metabolism in brain macrophages, lipid metabolism in lung macrophages, eicosanoid signaling in peritoneal macrophages, and readiness for interferon responsiveness in red pulp macrophages (Supplementary Table 1). Considering that the gene expression profiles of four macrophage populations were simultaneously compared, the number of transcripts that were increased or decreased ≥ 5-fold in only one macrophage population relative to all three of the others was striking (Fig. 2b). We also noted that many transcripts were especially strongly reduced in only one population compared to the others (Supplementary Fig. 2). Several transcription factors were markedly increased in just one of the four macrophage populations (Fig. 2c). For example, Spic was restricted to splenic red pulp macrophages, fitting with previous work revealing that this transcription factor plays a critical role in controlling the development of these cells15. Diversity at the gene expression level translated to the protein level. For example, CD11a and EPCAM were detected on lung macrophages but not microglia, spleen or peritoneal macrophages; VCAM-1 and CD31 were selectively displayed by spleen macrophages; CD93 and ICAM-2 were expressed by peritoneal macrophages but not the others macrophages; and CX3CR1 and SiglecH were selectively found in microglia (Fig. 2d). All together, these data indicate that macrophage populations in different organs express many unique mRNA transcripts that equip them for specialized local functions.
Figure 2. Unique gene expression profiles of macrophages from different organs.
(a) Scatter plots depict in distinct colors the mRNA transcripts that are ≥ 2-fold increased (left) or decreased (right) in one macrophage population compared to the remaining three populations. (b) Heat map and gene lists reveal mRNA transcripts uniquely expressed by single macrophage populations by ≥ 5 fold. (c) Transcription factor mRNA transcripts increased in only one of the four macrophage populations by ≥ 2 fold. (d) Specific cell surface markers for each macrophage populations, identified from the gene expression profiling data were validated by flow cytometry. Macrophages reacting with the antibodies tested matched the pattern of gene expression observed in (b). Shaded blue line shows isotype control and red line specific antibody.
Identification of core transcripts expressed in tissue macrophages but low to absent in classical DCs
In the midst of the rather vast diversity among macrophages from different organs, we next wondered if we could identify a core gene expression profile that generally unified macrophages over other types of immune cells. Among all hematopoietic cells, the cells anticipated to be most similar to macrophages are DCs5. To search for mRNA transcripts that distinguished macrophages from DCs, we compared the four selected prototypical macrophage populations to the most well-defined classical DC populations, including resting CD8+ and CD4+CD11b+ splenic DCs, CD103+ tissue DCs and various populations of lymph node MHC-IIhi CD11c+ migratory DCs18. Because tissue CD11b+ DCs may be contaminated with macrophages19, tissue CD11b+ DCs were initially excluded from the comparison. This comparison revealed only 14 transcripts that were expressed in all 4 macrophages but not expressed in DCs (Table 1, upper left column, bolded gene names). These included few of those anticipated to be highly expressed in macrophages, including Fcgr1 (CD64) and Tlr4. Two of these molecules, G-CSF receptor (Csf3r) and the MHC-I-related gene Mr1 involved in activation of mucosal-associated invariant T (MAIT) cells20, function at least partly at the cell surface. In agreement with the pattern of mRNA expression, we found MR1 protein on spleen and lung macrophages but not classical DCs (Supplementary Fig. 3), suggesting that MR1 on macrophages rather than DCs may drive MAIT cell activation. Other transcripts encode proteins involved in signal transduction, such as Fert2 encoding the fms/pfs-related protein kinase, or in metabolism and lipid homeostasis such as peroxisomal trans-2-enoyl-CoA reductase (Pecr) and alkyl glycerol monooxygenase (Tmem195), the latter being the only enzyme that cleaves the O-alkyl bond of ether lipids like platelet-activating factor that has been shown to be actively catabolized in association with macrophage differentiation in vitro21.
Table 1.
Gene induced in tissue macrophages relative to classical and migratory DCs
| All 4 Mϕ populations | All 4 Mϕ populations | |||
|---|---|---|---|---|
| Except Peritoneal Mϕ | Except Lung Mϕ | Except Microglia | Except Splenic red pulp Mϕ |
|
| Pecr | Xrcc5 | Mafb | Hgf | Cd151 |
| Tmem195 | Gm4878 | Itga9 | Pilrb2 | Lonrf3 |
| Ptplad2 | Slco2b1 | Cmklr1 | Mgst1 | Acy1 |
| 1810011H11Rik | Gpr77 | Fez2 | Klra2 | |
| Fert2 | Gpr160 | Tspan4 | Rnasel | C5ar1 |
| Tlr4 | P2ry13 | Abcc3 | Fcgr4 | Pld1 |
| Pon3 | Tanc2 | Nr1d1 | Rhoq | Gpr177 |
| Mr1 | Sepn1 | Ptprm | Fpr1 | Arsk |
| Arsg | Ctsf | Cd302 | Plod3 | |
| Fcgr1 | Il1a | Tfpi | Slc7a2 | Cd33 |
| Camk1 | Asph | Slc16a7 | Cebpb | |
| Fgd4 | Dnase2a | Ptgs1 | Slc16a10 | Atp6ap1 |
| Sqrdl | Slc38a7 | C1qa | Slpi | Pros1 |
| Csf3r | Siglece | Engase | Mitf | Dhrs3 |
| Itgb5 | C1qb | Snx24 | Rnf13 | |
| Plod1 | Rhob | C1qc | Lyplal1 | Man2b2 |
| Tom1 | Mavs | Timp2 | St7 | Ltc4s |
| Myo7a | Atp13a2 | Slc11a1 | ||
| A930039A15Rik | Slc29a1 | 4632428N05Rik | Tlr8 | |
| Pld3 | Slc15a3 | Sesn1 | Gbp6 | |
| Tpp1 | Tmem86a | Plxnb2 | 6430548M08Rik | |
| Ctsd | Tgfbr2 | Apoe | C130050O18Rik | |
| Pla2g15 | Tnfrsf21 | Pilra | ||
| Lamp2 | Pilrb1 | |||
| Pla2g4a | Lpl | |||
| MerTK | Pstpip2 | |||
| Tlr7 | Serpinb6a | |||
| Cd14 | Slc38a6 | |||
| Tbxas1 | Abcc5 | |||
| Fcgr3 | Lrp1 | |||
| Sepp1 | Pcyox1 | |||
| Glul | Hmox1 | |||
| Cd164 | Slc17a5 | |||
| Tcn2 | Emr1 | |||
| Dok3 | Hgsnat | |||
| Ctsl | ||||
| Tspan14 | ||||
| Comt1 | ||||
| Tmem77 | ||||
| Abca1 | ||||
Bolded genes depict those whose signal intensities indicated that DC populations did not express them. Nonbolded genes were expressed in DCs, but more highly expressed in macrophages (Mϕ). See methods for full description of how gene expression comparisons were executed and cut-offs were generated.
To this small number of mRNA transcripts, we added probe sets that were not absent in expression by DCs, but were at least 2-fold lower in signal intensity in all single DC populations than the lowest intensity of that same probe set in each macrophage population. Thus, we were able to add 25 more transcripts to this “macrophage core” list (Table 1, lower left column, non-bolded gene names), including those known to be associated with macrophages like Cd14, Mertk, Fcrg3 (CD16) and Ctsd (Cathepsin D).
F4/80, encoded by the gene Emr1, has served as the most definitive marker of macrophages to date16,22. However, in order to identify additional mRNA transcripts widely associated with macrophages with the core list of macrophage-associated genes, including Emr1 (F4/80), Mafb, and Cebpb, we found it necessary to adjust the criteria of the above approach to include transcripts expressed in only 3 of 4 macrophage populations because, for instance, Emr1 mRNA was low in microglia. Making this adjustment expanded the list of mRNA transcripts associated with macrophages, adding another 93 genes (Table 1). Additional macrophage-associated genes like Mrc1 (CD206, mannose receptor), Marco and Pparg were not identified until we loosened the criteria such that only 2 out of 4 prototypic macrophages needed to express a given transcript that was otherwise absent or low on DCs (Table 2). The mRNA encoding CD68 (Cd68), widely used to identify tissue macrophages, was expressed at similar levels in DCs and macrophages and so excluded from the list. However, at the protein level, it was still several orders of magnitude higher in macrophages than in DCs of the spleen (Supplemental Fig. 4), an organ where mRNA levels were scarcely different. In summary, numerous transcripts, 366 altogether (Tables 1 and 2), were absent or markedly reduced in classical DCs relative to macrophages. However, because of the great diversity among macrophages, only 39 of these transcripts are shared by all tissue macrophages we compared.
Table 2.
Macrophage-induced genes present in 2 out of 4 tissue macrophage populations
| Peritoneal + Splenic red pulp |
Peritoneal + Lung | Lung + Splenic red pulp |
Peritoneal + Microglia |
Lung + Microglia | Microglia + Splenic red pulp |
|---|---|---|---|---|---|
| Ccl24 | Marco | Dmxl2 | Hnmt | Scamp5 | Lhfpl2 |
| Gstk1 | P2ry2 | Dip2c | Mtus1 | Ppp1r9a | Osm |
| Aspa | Aifm2 | Galnt3 | C3ar1 | Tppp | Mgll |
| 2810405K02Rik | Clec4e | Niacr1 | Dagla | Abcb4 | Bhlhe41 |
| B430306N03Rik | Plcb1 | Bckdhb | Wrb | Kcnj2 | Ang |
| Fcna | Kcnn3 | Angptl4 | Gab1 | P2ry12 | D8ertd82e |
| Gm5970 | Arhgap24 | Lrp4 | Fkbp9 | ||
| Aoah | Cd93 | Sh3bgrl2 | Slc37a2 | X99384 | |
| Cd5l | Fundc2 | Gm5150 | Rab11fip5 | Adrb1 | Serpine1 |
| Gm4951 | Tspan32 | Tcfec | 6230427j02rik | Slc16a6 | Abhd12 |
| Nr1d1 | Lmbr1 | Sh2d1b1 | Scn1b | Rab3il1 | Ms4a6d |
| Mlkl | Adarb1 | Galnt6 | Scamp1 | Mfsd11 | Cebpa |
| Vnn3 | Fzd4 | Pdgfc | Msrb2 | Flcn | Lpcat3 |
| Igf1 | F7 | Abca9 | Tmem63a | Manea | |
| Ccr1 | 6720489N17Rik | Plxdc2 | P2rx7 | Ctss | |
| Ptgis | Hspa12a | Pparg | Adam15 | Hpgds | Ccl3 |
| Pitpnc1 | Cav1 | Megf9 | Itgam | Hpgd | Cryl1 |
| Fam43a | Nt5e | Adcy3 | Itga6 | Lpcat2 | Man1c1 |
| Itsn1 | 1190002a17rik | Enpp1 | Vkorc1 | Slc7a8 | Ctns |
| Ifi27l1 | Il18 | 1700017b05rik | Maf | Sgk1 | |
| Rasgrp2 | Cav2 | Siglec1 | Smad3 | Tmem86a | Pag1 |
| Aldh6a1 | Gda | Clec4n | Smpd1 | Slc36a1 | Tgfbr1 |
| Epb4.1l1 | Frrs1 | Lgals8 | Naglu | Gna12 | Clec5a |
| Cryzl1 | Tspan5 | Nceh1 | Pmp22 | Adap2 | |
| Lrp12 | Pdk4 | Lipa | Man2b2 | Lgmn | |
| Cd300ld | Slc36a4 | 4931406c07rik | Tnfrsf1a | Hist1h1c | |
| Pla2g7 | Fam3c | Sirpa | Lifr | Lair1 | |
| Cfp | Ms4a8a | Rasgef1b | Tlr13 | Slc40a1 | |
| Sdc3 | Atoh1 | Wdfy3 | Slc25a37 | Csf1r | |
| Dusp7 | Alox5 | Ermp1 | Grn | P4ha1 | |
| Tbc1d2b | Thbd | Asah1 | Iffo1 | ||
| Igsf6 | Gstm1 | Ear1 | Dusp6 | ||
| Man2a1 | Cxcl2 | Ear10 | |||
| Zswim6 | Nhlrc3 | Ano6 | |||
| Ifnar2 | Fry | Mrc1 | |||
| Trf | F10 | Camk2d | |||
| Blvrb | Sord | Gab3 | |||
| Cd38 | Ncf2 | Syne2 | |||
| Ctsb | Hexa | Axl | |||
| Tmem87b | Dram1 | Tcf7l2 | |||
| Itfg3 | Plaur | Ctsc | |||
| Ninj1 | G6pdx | D730040f13rik | |||
| Fn1 | Slc15a3 | ||||
| Cybb | Plk3 | ||||
| Dennd4c | Hebp1 | ||||
| Mpp1 | Dst | ||||
| S100a1 | Blvra | ||||
| Gsr | Sort1 | ||||
| Abcd2 | Slc12a7 | ||||
| Dab2 | Clec4a3 | ||||
| Ccl6 | |||||
| Sepx1 | |||||
| Prdx5 | |||||
| Dusp3 | |||||
| Pgd | |||||
| Gp49a | |||||
| Capg | |||||
| Cndp2 | |||||
| Vps13c | |||||
| Adipor2 | |||||
| App | |||||
| Atg7 | |||||
| Cebpb |
Bolded genes depict those whose signal intensities indicated that they were not expressed by DC populations used in comparison to spleen, brain, peritoneal, and lung macrophages. Nonbolded genes were expressed in DCs, but more highly expressed in macrophages.
Co-expressed gene modules in macrophages and predicted transcriptional regulators
The computational biology groups of the ImmGen project analyzed the transcriptional program of the entire large database generated in the ImmGen project (Jojic et al, in preparation; Supplementary Note 1). First, mRNA transcripts were clustered into 334 fine modules based on patterns of co-expression. Then a novel algorithm termed Ontogenet, developed for the ImmGen dataset, was applied in order to find a regulatory program for each fine module, based on its expression pattern, the expression pattern of regulators and the position of the cells on the hematopoietic lineage tree. ImmGen modules, including the gene lists in each module, and regulatory program metadata are available online (http://www.immgen.org/ModsRegs/modules.html), and the numbering of the modules reported on the website is used herein.
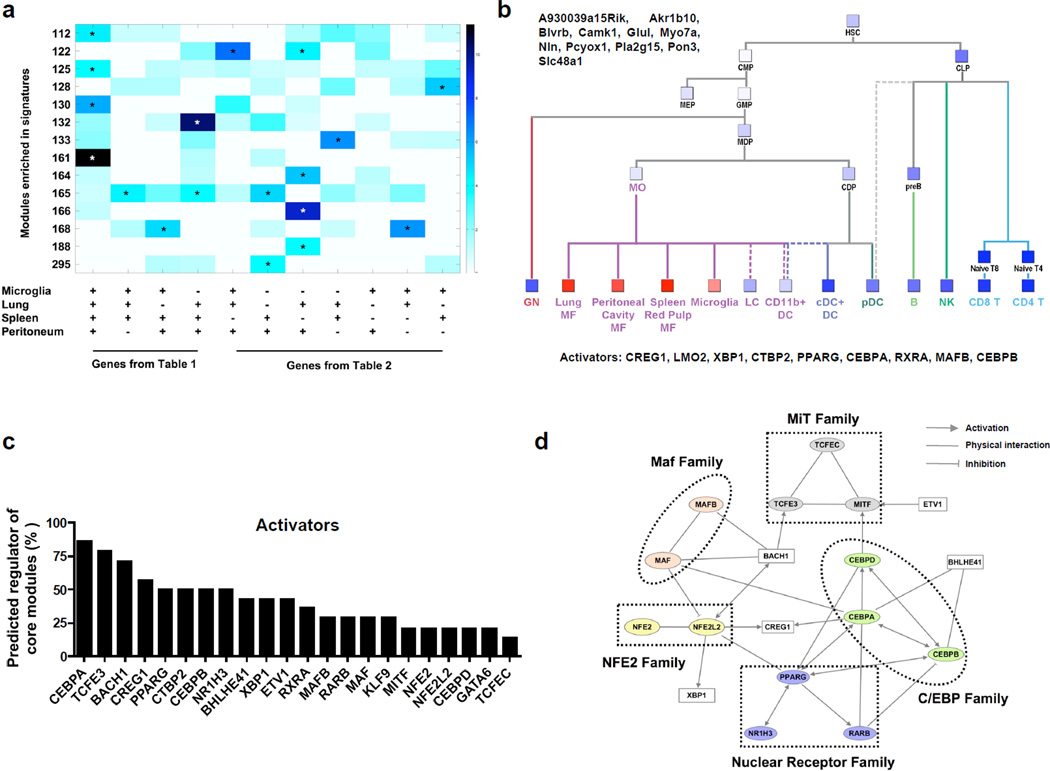
When the list of the 366 mRNA transcripts associated with macrophages was mapped according to the their placement into various fine modules, 14 modules were significantly enriched for the macrophage-associated gene signature we identified (Fig. 3a). In particular, the 11 genes that comprise module 161 were significantly induced in all 4 macrophages used to generate the list of macrophage-associated genes (Fig. 3a). Other modules, such as module 165, contained genes significantly induced in several specific groups of macrophages, but not in all (Fig. 3a). The 11 genes that comprise module 161 (A930039a15Rik, Akr1b10, Blvrb, Camk1, Glul, Myo7a, Nln, Pcyox1, Pla2g15, Pon3, Slc48a) are involved in redox regulation, heme biology, lipid metabolism, and vesicular trafficking (Supplementary Table 3). Beyond the comparison to DCs, genes in module 161, expressed in all macrophages, were not expressed by any other hematopoietic cell types including granulocytes (GN) or any of the blood monocyte (MO) subsets (Fig. 3b), importantly indicating that this list of genes is selectively associated with mature macrophage differentiation in the hematopoietic system.
Figure 3. Identification of gene modules enriched for macrophage-related gene signatures and their predicted regulators.
(a) The overlap size of ImmGen modules of co-expressed genes with all macrophage-associated genes signatures (Table 1 and 2) is depicted graphically as a heat map. Only modules significantly enriched for at least one signature are shown. Stars mark significant overlap size by hypergeometric test (Methods). (b) Simplified hematopoietic tree showing mean expression of genes in module 161 (red – high expression; blue – low expression). Listed are genes that constitute module 161 (top) and the predicted positive regulators of the module (bottom). (c) A bar graph listing the positive regulators (activators) predicted by the Ontogenet algorithm to regulate two or more modules listed in a. The frequency that each factor was associated with the 14 modules is depicted. (d). Physical and regulatory interactions between the 18 most frequently represented regulators across the 14 macrophage-associated modules were interrogated using Ingenuity analysis tools. The scheme uses arrows to depict links where there are established physical interactions, or known pathways of co-activation or inhibition.
As a framework for future studies on the transcriptional control of macrophage development, maintenance and function, we examined the predicted activators Ontogenet assigned to the modules associated with the macrophage core genes. As a specific example, the activators predicted by Ontogenet algorithm to control the expression of the 11 gene transcripts that form module 161 are listed in Fig. 3b. Overall, a highly overlapping set of 22 regulators emerged in the 14 macrophage-associated modules (Fig. 3c). In particular, TCFE3, C/EBPα and BACH1 were predicted activators in a majority of these modules (>75%) and especially novel regulators like CREG-1 also came up prominently. Among the 22 regulators associated with the 14 modules, 18 of them are predicted using Ingenuity pathway tools to interact in a regulatory network based on known protein-protein interactions or mutual transcriptional regulation (Fig. 3d). These regulators represent 5 main families of transcriptional factors as depicted in Fig. 3d. The statistical evaluation score generated for this network revealed a p-value ≤ 10−35.
Beyond modules of genes that unified the 4 tissue macrophage populations we studied, several modules were selectively associated with a single macrophage population (Supplementary Table 4). In these specific modules, predicted regulators included Spi-C for red pulp macrophages, confirming a regulation that is already known15 and thus supporting the predictive power of the algorithm, and GATA6 as a regulator of peritoneal macrophages (Supplementary Table 4).
Use of the macrophage core signature to evaluate identity and heterogeneity of less characterized macrophage and DC populations
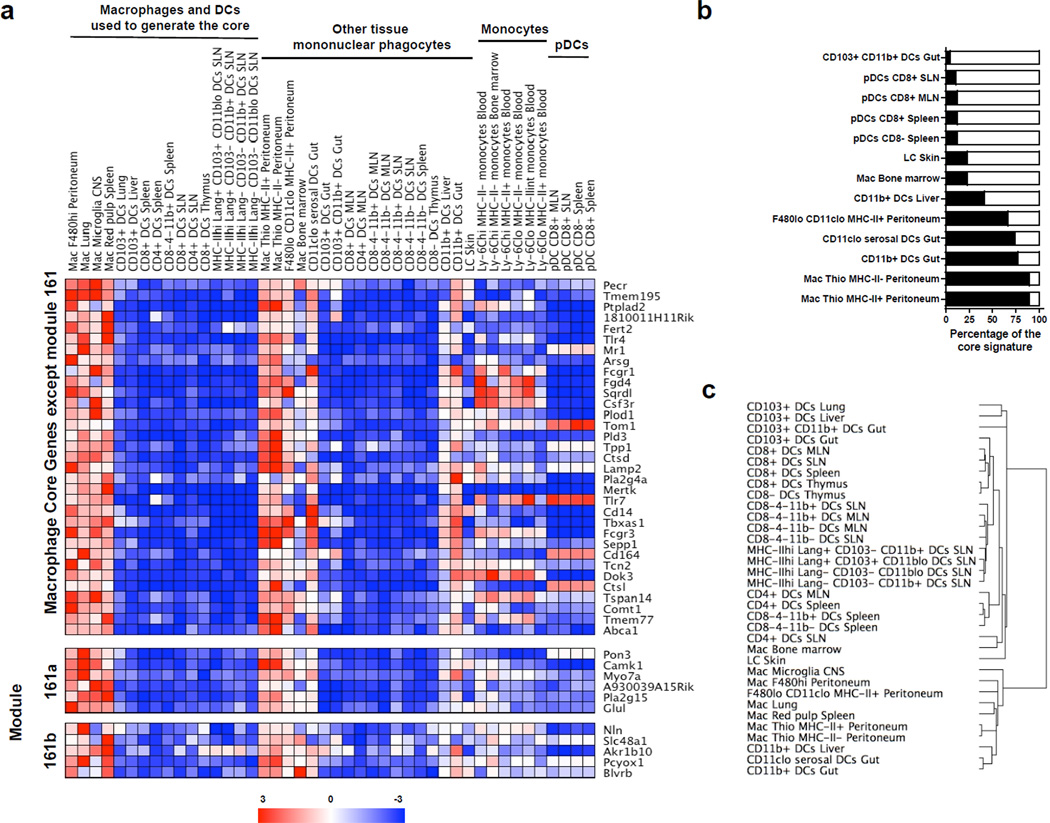
Finally, we utilized the resting macrophage core signature defined above to assess mononuclear phagocyte populations that we earlier excluded from our core analysis due to low levels of information on a given population or controversial discussions in the literature about origins or functional properties, including whether they should be classified as DCs or macrophages. In the ImmGen database (http://www.immgen.org), each population was assigned a classification as DCs or macrophage (Mac) a priori. The names of these populations will be used below and in Figure 4 for clarity and consistency with the database. These populations included resting and thioglycollate-elicited (Thio) mononuclear phagocytes that express CD11c and MHC II (Supplementary Fig. 5), skin Langerhans cells, bone marrow macrophages23, and putative CD11b+ tissue DCs including those in the liver and gut. All thioglycollate-elicited cells from the peritoneal cavity, even those co-expressing CD11c and MHC II, strongly expressed genes in the macrophage core, including module 161 itself, similar to the prototypic macrophage populations used to generate the core (Fig. 4a and 4b), indicating that these cells are indeed macrophages despite co-expression of CD11c and MHC II. On the other hand, Langerhans cells, and CD11c+ MHC-II+ CD11b+ cells from the liver (CD11b+ liver DCs in the ImmGen database), did not robustly express the macrophage core signature or module 161 alone, nor did bone marrow macrophages (Fig. 4a and 4b). CD11c+ MHC-II+ CD11b+CD103− cells from the intestinal lamina propria and CD11clo MHC-II+ CD11b+ cells from the serosa, though previously called DCs in many studies, expressed macrophage core genes including those from module 161, suggesting a strong relationship to macrophages (Fig. 4a and 4b). Accordingly, these cells are now called CD11b+ gut macrophages and CD11clo serosal macrophages herein and on the Immgen website. We clustered these mononuclear phagocytes based on their expression of the 39-gene macrophage core to model their relatedness to each other (Fig. 4c). Langerhans cells of the skin and bone marrow macrophages were positioned at the interface between DCs and macrophages, with a distal relationship to classical DCs but failing to cluster with macrophages (Fig. 4c).
Figure 4. Expression of macrophage core genes by other populations of mononuclear phagocytes.
(a) Heat map depicts the 39 gene transcripts increased in spleen, brain, peritoneal, and lung macrophages compared to classical and migratory DCs. Genes from module 161 that were included among these 39 genes are segregated and labeled “161a.” Other members of module 161 that did not meet the criteria for inclusion on Table 1 are labeled “161b.” Populations of tissue-derived mononuclear phagocytes that were not included in the generation of this list of genes are shown in the middle of the heat map. Various subsets of blood monocytes and plasmacytoid DCs are depicted further to the right on the heat map. (b) The frequency that the 39 genes in a were expressed in these populations at a signal intensity at least 2-fold higher than the highest expressing DC in the original comparison is depicted. (c) A dendrogram depicting the relationship between a wide variety of mononuclear phagocytes based on their expression of the list of 39 common macrophage-enriched genes.
As mentioned earlier, non-lymphoid tissue CD11b+ DCs have been argued to be heterogeneous19. Thus, we reasoned that the use of antibodies to cell surface proteins identified as macrophage-specific from our gene expression analysis may discriminate macrophage “contaminants” in a heterogeneous population. Furthermore, we aimed to determine if the same cell surface markers may also prove valuable in identifying macrophages universally, including in organs beyond those we initially analyzed and/or where F4/80 has not proved sufficiently definitive. We homed in CD14, FcγRI, and MerTK as cell surface proteins in the group of 39 mRNA transcripts deemed to be low or absent in DCs but present in all macrophages, and to which quality mAbs have been generated. Indeed, all of these proteins were expressed on all of the 4 resident macrophage populations used in our primary analysis (Fig. 5a), with lower CD14 levels compared with FcγRI and MerTK (Fig. 5a). Two of these tissues, spleen and lung, have significant DC populations. In the spleen, MerTK, FcγRI, and CD14 did not stain CD8+ or CD11b+ DCs (Fig. 5a). However, in the lung where interstitial pulmonary macrophages are CD11b−, there may still be an underlying heterogeneity in lung CD11b+ DCs that includes a subset of CD11b+ macrophages19,24{Satpathy, 2012 #3482}. Indeed, CD14, FCγR1 and MerTK were expressed by a portion of lung CD11b+ DCs, but not by CD103+ DCs (Fig. 5a). Gating on MerTK+ FCγR1+ cells revealed the vast majority of such cells were SiglecF+ lung macrophages, but a small proportion of MerTK+FCγR1+ cells in the lung were SiglecF− cells expressing high levels of MHC II (Fig. 5b). Gating on DCs (Fig. 5c) revealed that CD11b+ DCs could be divided into CD11b+ CD24+ FCγR1lo MerTK− CD14int and CD11b+ CD24lo FCγR1+ MerTK+ CD14hi cells (Fig. 5d). Thus, the latter likely comprises a population of macrophages that cosegregates with DCs using many markers, but are not DCs. Indeed, the CD11b+ DCs were segregated within Immgen on the basis of CD24 expression based on the likelihood that those expressing CD24 were true DCs, but those without CD24 were not. Our findings suggest that this possibility is highly likely and points to the utility of using markers like MerTK and FCγR1 as a panel to facilitate the identification of macrophages from DCs.
Figure 5. Examination of macrophage core transcripts at the protein level in multiple tissues.
(a) Histograms of brain, peritoneum, spleen, and lung stained for CD14, CD64, and MerTK to examine expression in macrophages and DCs from these organs. Shaded blue line shows isotype control and red line specific antibody. (b) MerTK+CD64+ cells were more than 95% Siglec F+ macrophages, but some MHC II+Siglec F− cells were also found in this population. c) Lung DC gating strategy is shown, with DCs being CD45+ cells lacking Siglec F, expressing CD11c and MHC II. d) Gating on lung DCs DCs revealed a significant reactivity for CD14, CD64, and MerTK in the CD11b+ CD24− putative DCs, but lack of MerTK and CD64 in CD24+ CD11b+ DCs. (e) Liver CD45+ cells were plotted to show F4/80 and CD11c staining. Eosinophils were gated (Siglec F+ high SSC+) to reveal that they overlay with another population in gate 1. Replotting gates without (w/o) eosinophils revealed 4 low SSC subsets of cells that differentially express F4/80 and CD11c. These 4 gates were examined with regard to expression of MerTK, CD64, and MHC II. Finally, reverse gating on MerTK+ CD64+ cells was carried these gated cells were plotted based on F4/80 and CD11c. f) A similar approach was as in “e” was carried out here in adipose tissue. Each analysis in this figure was based on studies from at least two replicative experiments with 3 mice per group.
We next turned to two tissues—liver and adipose tissue--that were not analyzed in Immgen with respect to gene expression profiling in macrophages to determine if the use of MerTK and FcγRI staining would facilitate the identification of macrophages in those organs and distinguish them from DCs. In liver, we started with a classical approach of plotting F4/80 versus CD11c. Eosinophils are now recognized as high side-scatter, F4/80+ cells that express Siglec F universally25. Indeed, among macrophages, Siglec F is observed only on macrophages in the lung26,27 (as used to identify lung macrophages here). In the liver, the level of F4/80 on eosinophils overlaid with that of another population of low side-scatter, F4/80+ cells that were CD11clo in liver (Fig. 5e, left plot), so that even after excluding Siglec F+, high side-scatter eosinophils, four gates of cells expressing varying levels of F4/80 and CD11c were found (Fig. 5e). MerTK and FcγRI was highly expressed in two of these gates, suggesting that the cells with the highest level of F4/80 (gate 2) and many that expressed lower F4/80 (in gate 3) were two populations of F4/80hi and F4/80lo liver macrophages, corresponding to the two types of macrophages believed to be present in many organs16. The liver CD45+ cells with highest CD11c were MerTK−FcγRI−, suggesting they were liver DCs (Fig. 5e). Reverse gating revealed that all MerTK+FcγRI+ cells fell into one of the two putative macrophage gates (Fig. 5e). A relatively similar picture was seen in adipose tissue (Fig. 5f), where the cells with highest F4/80 were MerTK+FcγRI+ and those with higher CD11c and lower F4/80 were MerTK−FcγRI−. In both liver and adipose tissue, MHC II was high on macrophages and DCs (Fig. 5e, f). Because F4/80 and CD11c are both expressed by many tissue macrophages and DCs, albeit at levels that are somewhat different, distinguishing macrophages and DCs based on these traditional markers can be difficult. MerTK and FcγRI staining offers the advantage of sharp differences in the magnitude of expression between macrophages and DCs. Thus, we propose that MerTK and FcγRI costaining provides a powerful approach to identifying macrophages universally and selectively in mouse tissues.
Discussion
The large and unique database and accompanying bioinformatic analysis of the Immunological Genome Project provide novel insight into macrophage populations isolated from various organs of mice. A striking initial revelation was that macrophage populations from different organs are considerably diverse, and it is likely that further profiling in macrophages will expand upon this diversity. Only a very small group of mRNA transcripts were associated with all macrophages but not DCs. Proteins previously predicted to distinguish macrophages from other cell types, such as F4/80, CD68 and CD115 (C-fms/Csf1r), did not emerge as the most powerful markers of macrophages. However, many canonical genes did, including those encoding CD14, the high-affinity FcγR FcγR1 (CD64), the Mer tyrosine kinase involving in efferocytosis MerTK, cathespin D, and a fms/fps protein kinase FERT2 that may strongly impact CD115 signaling (but which has not yet been studied in macrophages). The identification of these genes as selectively macrophage-associated reinforce the key role of macrophages in innate immunity, efferocytosis, and clearance of debris, whereas genes associated with antigen presentation and migration to lymphoid tissues were more associated with DCs (Miller at el., In Press). However, our data do suggest that macrophages may have a greater role in activation of MAIT cells than DCs. Based upon follow-up protein expression analysis of MerTK and FcγRI in macrophages from six different tissues, we propose that analysis of MerTK and FcγRI should serve as a starting point for identifying macrophages in tissues, as staining for these markers appears to identify F4/80hi macrophages and other macrophages that express somewhat lower levels of F4/8016 in all tissues. We believe staining for MerTK and FcγRI has advantage over, but can also powerfully be used in addition to, traditional staining for F4/80, CD11c, and MHC II. The levels of F4/80 and CD11c are often overlapping between macrophages and DCs in nonlymphoid tissues, but it appears that DCs do not co-express MerTK and FcγRI.
Beyond these cell surface markers closely associated with macrophage identity, we uncover other transcripts associated only with macrophages among hematopoietic cells. In particular, immGen module 161 identified a group of genes (A930039a15Rik, Akr1b10, Blvrb, Camk1, Glul, Myo7a, Nln, Pcyox1, Pla2g15, Pon3, Slc48a) that are co-expressed across all the ImmGen dataset and whose functions with the established broad roles of macrophages, but none of them have previously been considered macrophage markers. Both the genes from this module and their predicted regulators would deserve considerable attention in the future.
The Ontogenet algorithm makes it possible to extend the macrophage-associated genes we identified to regulatory programs that may control them. Induced expression of a single module (#330) in red pulp macrophages over all other macrophages and the predictions generated by the algorithm that this module is regulated by SPI-C support the reliability of the algorithm predicted regulatory programs, as SPI-C is already known to be required selectively for red pulp macrophage development or maintenance15. Exciting new information also emerged, such as strong association of modules unique to peritoneal macrophages that are predicted to be regulated by GATA6.
Gene transcripts that were highly expressed in multiple macrophage populations but not highly expressed in DCs were associated with predicted transcriptional regulatory programs that strongly differed from those uncovered in DCs (Miller et al., Submitted). The predicted regulatory programs of modules enriched for macrophage-associated genes include several members of the MiT family of transcription factors that has been recognized to be specifically expressed in macrophages3 as well as transcription factors not previously associated with macrophages, such as BACH1 and CREG-1. BACH1 has been little studied in macrophages but has recently been linked to osteoclastogenesis28 and is a regulator of heme oxygenase 129. CREG1 (cellular repressor of E1A-stimulated genes) is a secreted regulator30,31 associated broadly with differentiation32 and cellular senescence33 that was strongly associated with macrophage-enriched gene modules, though it has never been studied in the context of macrophage biology. The Ontogenet algorithm predicts RXRα as the most prominent key activator of the highly specific and universal macrophage module genes, module # 161. Future analysis of these predictions is expected to be highly fruitful in revealing how macrophage identity and function is controlled.
To date, the Immunological Genome Project has mainly focused on cells recovered from resting, uninfected mice, where macrophages mainly derive from the yolk sac12. Macrophage polarization in the context of infection and inflammation is a topic of great interest that this study has scarcely been able to address. beyond finding that monocytes recruited to the peritoneum in response to thioglycollate upregulate mRNA transcripts observed in resting tissue macrophages, even though monocytes are not precursors for resting tissue macrophages as they are for inflammatory macrophages. The foundations laid herein suggest that future additions to the ImmGen database of macrophages recovered during disease states will add enormously to our understanding of how to manipulate these crucial cells to favor desired outcomes in disease. Based on the great diversity of macrophages in different organs, which we anticipate will hold up even in inflamed organs, such studies may be expected to ultimately generate therapeutic approaches to selectively target macrophages in diseased organs without affecting others cell types.
Materials and Methods
Mice
Six-week-old male C57BL/6J mice purchased from Jackson Laboratory were used for sorting and validation. CX3CR1-GFP knock in mice were from Jackson Laboratories, and Mr1 knockout mice20 were generated, bred, maintained at the Washington University School of Medicine. Mice were housed in specific pathogen–free facilities at the Mount Sinai School of Medicine or Washington University School of Medicine and experimental procedures were performed in accordance with the animal use oversight committees at these respective institutions. Most of the populations in the study were sorted from resting mice. However, for thioglycollate-elicited peritoneal macrophages, macrophages were harvested from the peritoneal cavity 5 days after instilling 1 ml of 3% thioglycollate.
Cell identification and isolation
All cells were purified using the sorting protocol and antibodies listed on http://www.immgen.org. Cells were directly sorted from mouse tissues and were processed from tissue procurement to a second round of sorting into Trizol within 4 h using a Beckton-Dickinson Aria II instrument. Resting red pulp macrophages from the spleen were sorted after nonenzymatic disaggregation of the spleen and were identified as F4/80hi cells that lacked B220 and high levels of CD11c and MHC II34,35; macrophages from the resting peritoneum were collected in a peritoneal lavage and stained to identify CD115hi cells that were F4/80hi MHC II−; resting pulmonary macrophages were isolated from Liberase III-digested lungs (15 min. digest) and macrophages were identified as SiglecF+ CD11c+ cells with low levels of MHC II26,27; and resting brain microglial macrophages were sorted from Liberase III-digested, Percoll-gradient separated cells that were CD11b+ CD45lo F4/80lo11. The Data Browser in the Immgen website is a resource for pdf files showing FACS dot plots that depict the purification strategies and purity after isolation of these and all other populations. A list of abbreviations used in the Immgen database relevant to macrophages and DCs can be found in Supplementary Note 1.
Microarray analysis, normalization, and dataset analysis
RNA was amplified and hybridized on the Affymetrix Mouse Gene 1.0 ST array by the Immgen consortium using double-sorted cell populations sorted directly into TRIzol. These procedures followed a highly standardized protocol for data generation and QC documentation (pdf documents found under “Protocols,” available on http://www.immgen.org; Supplementary Notes 2–4). A table listing QC data, replicate information, and batch information for each sample is also available on the website. All datasets have been deposited at National Center for Biotechnology Information/Gene Expression Omnibus under accession number GSE15907. Data analysis utilized GenePattern analysis software. Raw data were normalized using the robust multi-array algorithm, returning linear values between 10 to 20,000. A common threshold for positive expression at 95% confidence across the dataset was determined to be 120 (Supplementary Note 4). Differential gene expression signatures were identified and visualized using the “Multiplot” module of GenePattern (http://www.broadinstitute.org/cancer/software/genepattern/). Differentially expressed probesets were considered as those with a coefficient of variation less than 0.5 and a p value ≥ 0.05 (Student’s T-test). Signature transcripts were clustered (mean centered) using the “Hierarchical Clustering” module of GenePattern, employing Pearson’s correlation as a metric, and data were visualized using the “Hierarchical Clustering Viewer” heat map module. Clustering analyses performed across ImmGen centered on the most variable gene sets (objectively defined as the top 15% genes ranked by cross-population max/min ratio), to avoid noise from genes at background variation. Pearson correlation plots of gene expression profiles between different cell populations were generated using Express Matrix software. Pathway analysis as well as transcription factor network construction were performed using Ingenuity Systems Pathway Analysis (IPA) software. This software calculates a significance score for each network. The score is generated using a p-value indicative of the likelihood that the assembly of a set of focus genes in a network could be explained by random chance alone.
Principle component analysis (PCA) analysis
Only the 4417 genes whose variance of expression across all samples from the ten cell types was at least within the 80th percentile of variance were considered for the PCA analysis. The RMA normalized and log2 transformed expression levels were used. PCA was conducted using MATLAB.
Generation of the core macrophage signature
A macrophage core signature was generated by comparing brain, lung, peritoneal, and red pulp macrophages to populations deemed not to be macrophages, but authentic DCs. These DCs included CD103+ DCs from lung and liver, CD8+ DCs from spleen and thymus, CD4+CD11b+ DCs from spleen, CD4−CD8−CD11b+ DCs from spleen, and all DC populations (resident and migratory) isolated from skin-draining lymph nodes. A first list of possible genes defining macrophages was generated using the median value in the group of macrophages or DCs for each probe set in order to generate a list of probe sets with a differential median expression that was ≥ 2-fold higher in the group of macrophages with a statistical significance of p < 0.05 (Student’s T-test) and a coefficient of variation less than 0.5. Then this list of probe sets was filtered to remove any probe sets that did not show ≥ 120 (the threshold for positive expression) normalized intensity value in at least 2 macrophage populations. From the remaining probe sets, we compared the mean expression values of each macrophage and DC population, filtering to identify the lowest mean value in any single macrophage population to the highest mean value in any of the DCs. The probe sets that were at least 2.0-fold higher in expression in the lowest expressing macrophage compared with the highest expressing DC comprised the core genes were retained (Table 1, left column). To account for genes observed in only some macrophages, but still not expressed in DCs, we also generated lists wherein one or two macrophages were allowed to be excluded from consideration, but the criteria for comparing the remaining macrophages to the DCs was otherwise carried forward as described.
Generation of gene modules and prediction of module regulators
The gene modules, Ontogenet algorithm and regulatory programs are described in (Jojic et al, in preparation; Method found in Supplementary Note 5). Briefly, the expression data normalization was done as part of the ImmGen pipeline, March 2011 release. Data was log2 transformed. For gene symbols represented on the array with more than one probeset, only the probeset with the highest mean expression was retained. Of those, only the 7996 probesets displaying a standard deviation higher than 0.5 across the entire dataset were used for the clustering. Clustering was performed by Super Paramagnetic Clustering36 with default parameters, resulting in 80 stable coarse modules of co-expressed genes. Each coarse module was further clustered by hierarchical clustering into more fine modules, resulting in 334 fine modules.
A novel algorithm termed Ontogenet was developed for the ImmGen dataset (Jojic et al, in preparation, Supplementary Note 5). Ontogenet finds a regulatory program for each coarse and fine module, based on regulators expression and the structure of the lineage tree. The regulatory program uses a form of regularized linear regression, in which each cell type can have its own regulatory program, but regulatory programs of related cells are encouraged to be similar. This allows switching in the regulatory program but still allows robust fitting given the available data. To visualize the expression of a module on the lineage tree, the expression of each gene was standardized by subtraction of the mean and division by its standard deviation across all dataset. Replicates were averaged. Mean expression of each module was projected on the tree. Expression values are color coded from minimal (blue) to maximal (red).
Association between the macrophage core signature and gene modules
Hypergeometric test for two groups was used to estimate the enrichment of all ImmGen fine modules for the 11 gene signatures listed in Tables 1 and 2. Benjamini Hochberg FDR <= 0.05 was applied to the p-value table of all 11 signatures across all 334 fine modules.
Antibodies used for validation studies
Anti-mouse CD11c (N418), CD11b (M1/70), CD24a (30-F1), CD45 (30-F11), CD14 (Sa2-8) MHC-II (M5/114.15.2), F4/80 (BM8), CD8a (53-6.7), CD103 (2E7), CD11a (M17/4), EPCAM (G8.8), VCAM1 (429), CD31 (390), CD93 (AA4.1), ICAM2 (3C4 mIC2/4), CD68 (FA-11), and isotype control mAbs were from Ebioscience or Biolegend. Anti-mouse MERTK (BAF591) was from R&D Systems. Anti-mouse FCGR1 (X54-5/7.1) and SiglecF (E50-2440) were from BD Bioscience. Anti-mouse Mr1 antibody was previously described20. Anti-SiglecH antibody was a king gift from Marco Colonna (Washington University School of Medicine).
Supplementary Material
Acknowledgements
We are extremely grateful to all of our colleagues in the ImmGen Consortium and wish to extend special thanks to Vladimir Jojic, Jeff Ericson, Scott Davis and Christophe Benoist for their critical contributions. We also thank eBioscience and Affymetrix for material support of the ImmGen Project. We are additionally grateful to Marco Colonna for provision of mAbs and other reagents used during this study. ImmGen is funded by R24 AI072073 from NIH/NIAID, spearheaded by Christophe Benoist. Additional support for the present body of work was funded by NIH grants R01AI049653 and R01AI061741 to GJR and NIH grants P50GM071558-03 and R01DK08854 to AM. EG was supported by a postdoctoral fellowship from the American Heart Association (10POST4160140). ARM was supported by an NIH postdoctoral fellowship 5T32DA007135-27.
Footnotes
Conflict of interest: The authors have declared no conflict of interest related to this work
References
- 1.Heng TS, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 2.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 3.Hume DA. Differentiation and heterogeneity in the mononuclear phagocyte system. Mucosal Immunol. 2008;1:432–441. doi: 10.1038/mi.2008.36. [DOI] [PubMed] [Google Scholar]
- 4.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geissmann F, Gordon S, Hume DA, Mowat AM, Randolph GJ. Unravelling mononuclear phagocyte heterogeneity. Nat Rev Immunol. 2010;10:453–460. doi: 10.1038/nri2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fogg DK, et al. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science. 2006;311:83–87. doi: 10.1126/science.1117729. [DOI] [PubMed] [Google Scholar]
- 7.Onai N, et al. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat Immunol. 2007;8:1207–1216. doi: 10.1038/ni1518. [DOI] [PubMed] [Google Scholar]
- 8.Liu K, et al. In vivo analysis of dendritic cell development and homeostasis. Science. 2009;324:392–397. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hildner K, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi K. Development and differentiation of macrophages and related cells: Historical review and current concepts. Journal of Clinical and Experimental Hematopathology. 2001;41:1–33. [Google Scholar]
- 11.Ginhoux F, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schulz C, et al. A Lineage of Myeloid Cells Independent of Myb and Hematopoietic Stem Cells. Science. 2012 doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 13.Aziz A, Soucie E, Sarrazin S, Sieweke MH. MafB/c-Maf deficiency enables self-renewal of differentiated functional macrophages. Science. 2009;326:867–871. doi: 10.1126/science.1176056. [DOI] [PubMed] [Google Scholar]
- 14.Teitelbaum SL, Ross FP. Genetic regulation of osteoclast development and function. Nat Rev Genet. 2003;4:638–649. doi: 10.1038/nrg1122. [DOI] [PubMed] [Google Scholar]
- 15.Kohyama M, et al. Role for Spi-C in the development of red pulp macrophages and splenic iron homeostasis. Nature. 2009;457:318–321. doi: 10.1038/nature07472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schulz C, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 17.Lemke G, Rothlin CV. Immunobiology of the TAM receptors. Nat Rev Immunol. 2008;8:327–336. doi: 10.1038/nri2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohl L, et al. CCR7 governs skin dendritic cell migration under inflammatory and steady-state conditions. Immunity. 2004;21:279–288. doi: 10.1016/j.immuni.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 19.Hashimoto D, Miller J, Merad M. Dendritic cell and macrophage heterogeneity in vivo. Immunity. 2011;35:323–335. doi: 10.1016/j.immuni.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chua WJ, et al. Endogenous MHC-related protein 1 is transiently expressed on the plasma membrane in a conformation that activates mucosal-associated invariant T cells. J Immunol. 2011;186:4744–4750. doi: 10.4049/jimmunol.1003254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elstad MR, Stafforini DM, McIntyre TM, Prescott SM, Zimmerman GA. Platelet-activating factor acetylhydrolase increases during macrophage differentiation. A novel mechanism that regulates accumulation of platelet-activating factor. J Biol Chem. 1989;264:8467–8470. [PubMed] [Google Scholar]
- 22.Geissmann F, Gordon S, Hume DA, Mowat AM, Randolph GJ. Unraveling mononuclear phagocyte heterogeneity. Nat Rev Immunol. 2010 doi: 10.1038/nri2784. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chow A, et al. Bone marrow CD169+ macrophages promote the retention of hematopoietic stem and progenitor cells in the mesenchymal stem cell niche. J Exp Med. 2011;208:261–271. doi: 10.1084/jem.20101688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hashimoto D, et al. Pretransplant CSF-1 therapy expands recipient macrophages and ameliorates GVHD after allogeneic hematopoietic cell transplantation. J Exp Med. 2011;208:1069–1082. doi: 10.1084/jem.20101709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim HJ, Alonzo ES, Dorothee G, Pollard JW, Sant'Angelo DB. Selective depletion of eosinophils or neutrophils in mice impacts the efficiency of apoptotic cell clearance in the thymus. PLoS One. 2010;5:e11439. doi: 10.1371/journal.pone.0011439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sung SJ, et al. A major lung CD103 (alphaE)-beta 7 integrin-positive epithelial dendritic cell population expressing langerin and tight junction proteins. J. Immunol. 2006;176:2161–2172. doi: 10.4049/jimmunol.176.4.2161. [DOI] [PubMed] [Google Scholar]
- 27.Desch AN, et al. CD103+ pulmonary dendritic cells preferentially acquire and present apoptotic cell-associated antigen. J Exp Med. 2011;208:1789–1797. doi: 10.1084/jem.20110538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hama M, et al. Bach1 regulates osteoclastogenesis via both heme oxygenase-1 dependent and independent pathways. Arthritis Rheum. 2011 doi: 10.1002/art.33497. [DOI] [PubMed] [Google Scholar]
- 29.Sun J, et al. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J. 2002;21:5216–5224. doi: 10.1093/emboj/cdf516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veal E, Eisenstein M, Tseng ZH, Gill G. A cellular repressor of E1A-stimulated genes that inhibits activation by E2F. Mol Cell Biol. 1998;18:5032–5041. doi: 10.1128/mcb.18.9.5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sacher M, et al. The crystal structure of CREG, a secreted glycoprotein involved in cellular growth and differentiation. Proc Natl Acad Sci U S A. 2005;102:18326–18331. doi: 10.1073/pnas.0505071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veal E, Groisman R, Eisenstein M, Gill G. The secreted glycoprotein CREG enhances differentiation of NTERA-2 human embryonal carcinoma cells. Oncogene. 2000;19:2120–2128. doi: 10.1038/sj.onc.1203529. [DOI] [PubMed] [Google Scholar]
- 33.Moolmuang B, Tainsky MA. CREG1 enhances p16(INK4a) -induced cellular senescence. Cell Cycle. 2011;10:518–530. doi: 10.4161/cc.10.3.14756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nahrendorf M, et al. The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J Exp Med. 2007;204:3037–3047. doi: 10.1084/jem.20070885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Idoyaga J, Suda N, Suda K, Park CG, Steinman RM. Antibody to Langerin/CD207 localizes large numbers of CD8alpha+ dendritic cells to the marginal zone of mouse spleen. Proc Natl Acad Sci U S A. 2009;106:1524–1529. doi: 10.1073/pnas.0812247106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blatt M, Wiseman S, Domany E. Superparamagnetic clustering of data. Phys Rev Lett. 1996;76:3251–3254. doi: 10.1103/PhysRevLett.76.3251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.