Abstract
Small-cell lung cancer (SCLC) is an exceptionally aggressive disease with poor prognosis. Here, we obtained exome, transcriptome and copy-number alteration data from approximately 53 samples consisting of 36 primary human SCLC and normal tissue pairs and 17 matched SCLC and lymphoblastoid cell lines. We also obtained data for 4 primary tumors and 23 SCLC cell lines. We identified 22 significantly mutated genes in SCLC, including genes encoding kinases, G protein–coupled receptors and chromatin-modifying proteins. We found that several members of the SOX family of genes were mutated in SCLC. We also found SOX2 amplification in ~27% of the samples. Suppression of SOX2 using shRNAs blocked proliferation of SOX2-amplified SCLC lines. RNA sequencing identified multiple fusion transcripts and a recurrent RLF-MYCL1 fusion. Silencing of MYCL1 in SCLC cell lines that had the RLF-MYCL1 fusion decreased cell proliferation. These data provide an in-depth view of the spectrum of genomic alterations in SCLC and identify several potential targets for therapeutic intervention.
Lung cancer is the leading cause of cancer mortality in the United States, where it is responsible for over 160,000 deaths annually1. Approximately 10–15% of the new lung cancer cases diagnosed each year are SCLC2. The genomic landscape of SCLC is of particular interest compared to those of other solid tumors, given the unique biological characteristics of this tumor type3. SCLC is an exceptionally aggressive malignancy with a high proliferative index and an unusually strong predilection for early metastasis.
Previous efforts to characterize the genetic alterations present in SCLC tumors identified high prevalence of inactivating mutations in TP53 (75–90%)4, RB1 (60–90%)5,6 and PTEN (2–4%)7, rare activating mutations in PIK3CA, EGFR and KRAS8–10, amplification of MYC family members, EGFR and BCL2, and loss of RASSF1A, PTEN and FHIT6,11.
A better understanding of the genomic changes in this cancer will be essential to developing new therapeutics. To this end, we have applied next-generation sequencing technologies to characterize multiple exomes and a single genome of primary SCLC, as well as exomes of SCLC cell lines, together with genome-wide copy-number analysis and whole-transcriptome sequencing.
Specifically, we characterized 80 human SCLCs, including 36 primary SCLC human tumor and adjacent normal sample pairs and 17 paired SCLC cell lines and their patient-matched lymphoblastoid cell lines, as well as 4 primary SCLC tumors and 23 SCLC cell lines without matched normal controls (Supplementary Table 1). We sequenced and analyzed the complete genome of one SCLC tumor–normal tissue pair.
Exome capture, sequencing and analysis of 42 SCLC tumor–normal tissue pairs identified 26,406 somatic mutations. Approximately 30% (7,977) of these mutations were protein altering (Fig. 1a and Supplementary Table 2). The somatic mutations identified included 7,154 missense, 536 nonsense, 12 stop loss, 243 essential splice site, 32 protein-altering insertion and/or deletion (indel), 2,674 synonymous, 11,460 intronic and 4,295 other types (Fig. 1a and Supplementary Tables 3 and 4). Comparison of the protein-altering changes identified in this study with those reported in the Catalogue of Somatic Mutations in Cancer (COSMIC)12 showed that 98% (7,824/7,977) of these variations are newly identified somatic changes. Nineteen percent of the protein-altering somatic mutations reported were validated using RNA sequencing (RNA-seq) data or mass spectrometry genotyping, with a validation rate of 91% (Supplementary Table 3). We confirmed the effect of several splice-site mutations using RNA-seq data (Supplementary Table 3). We validated all of the indels reported using Sanger sequencing (Supplementary Table 4). One sample represented a distinct profile, with 2,953 mutations (757 validated protein-altering variants; Fig. 1a and Supplementary Table 3). Given the exceptionally high number of mutations in this sample, we excluded it from our calculations of the background mutation rate. Excluding the hypermutated sample, the SCLC tumors had an average of 175 protein-altering single-nucleotide variants (range 31–388) with a mean nonsynonymous mutation rate of 5.5 mutations per megabase (Fig. 1a). This is comparable to the 92 protein-altering variants observed in the previously sequenced genome of a single SCLC cell line13.
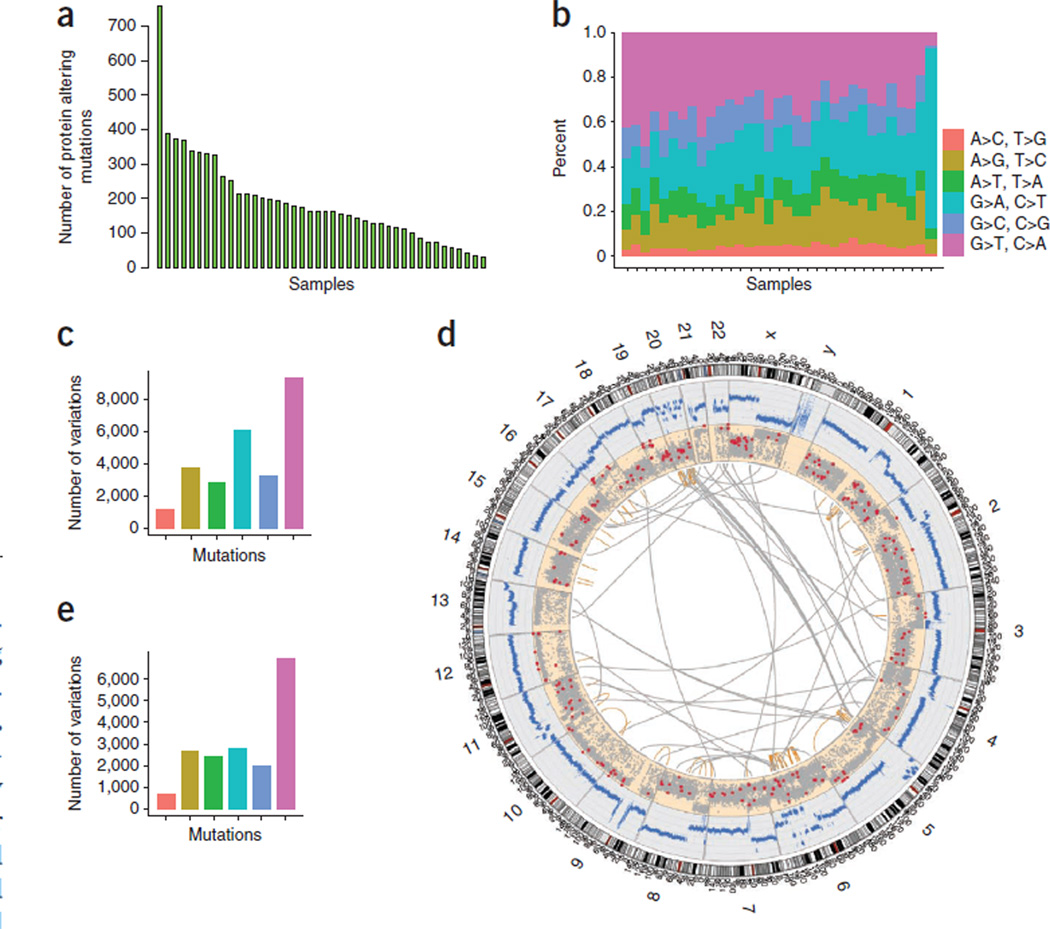
Figure 1. SCLC somatic mutations.
(a) Histogram of the number of mutations in each primary tumor sample. (b) Base-level transitions and transversions in each SCLC sample shown in a. (c) Average number of transitions and transversions in the SCLC samples based on the exome sequencing data. (d) Whole genome of an SCLC sample shown as a Circos plot. Copy-number changes measured using sequencing reads are shown in blue. Somatic nonsynonymous, splice-site and stop-gain mutations are shown as red dots. Other somatic mutations are depicted as gray dots. Intra- (orange lines) and interchromosomal (gray lines) rearrangements are also shown. (e) Average number of transitions and transversions in the whole-genome sequence of an SCLC sample. Colors in c and e correspond to those defined in b.
Analysis of the base-level transitions and transversions showed that G-to-T transversions were predominant, followed in prevalence by G-to-A and A-to-G transitions (Fig. 1b), both at the exome (Fig. 1c) and whole-genome (Fig. 1d,e and Supplementary Fig. 1) levels. This pattern is consistent with demonstrated effects of tobacco smoke carcinogens on DNA13.
In assessing the whole-genome data for an SCLC tumor–normal tissue pair, we found 59,784 somatic mutations, of which 286 were protein-altering changes (256 missense, 19 nonsense, 11 essential splice site, 77 synonymous, 13,924 intronic and 45,497 others). The average whole-genome mutation rate was 21.34 mutations per mega-base (Fig. 1d). Previously, 22,910 somatic variants were reported for the NCI-H209 SCLC cell line13.
Our mutation analysis identified protein-altering somatic single-nucleotide variants in 5,179 genes, including 4,775 genes that were mutated in the non-hypermutated SCLC sample set. Frequently mutated classes included genes encoding kinases, G protein–coupled receptors and chromatin-modifying proteins. To further understand the impact of the mutations on gene function, we applied SIFT14, Polyphen15 and Condel16 and found that ~53% of the somatic mutations identified are likely to have functional consequences according to at least two of the three methods (Supplementary Table 3). In contrast, only approximately 17% of germline variants identified in the normal samples are predicted by these methods to have a functional impact (Supplementary Fig. 2).
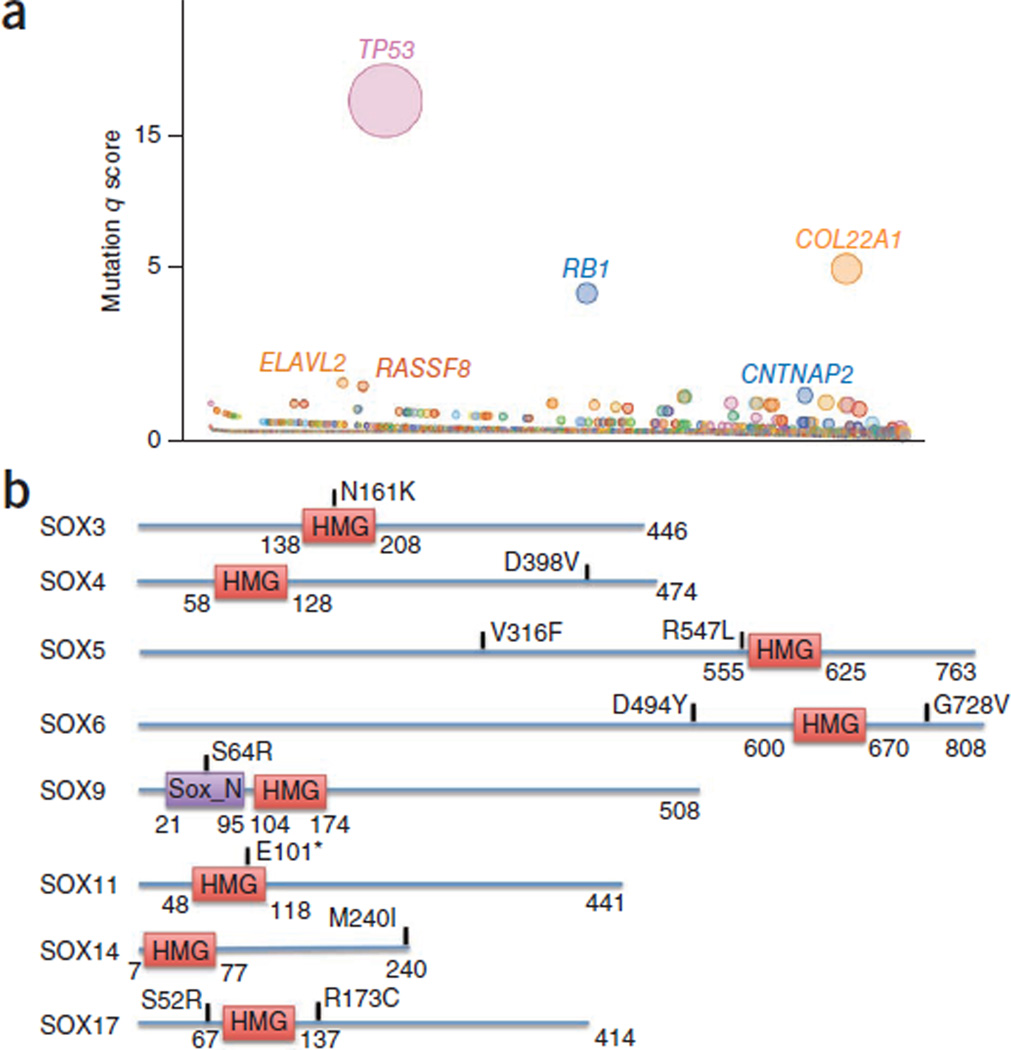
To further assess the relevance of mutated genes, we applied a q-score metric17 to rank significantly mutated cancer-associated genes. We identified 22 significantly mutated genes in SCLC (q score ≥ 1; false discovery rate ≤ 10%; Supplementary Table 5). These genes included TP53 and RB1 and several genes that have not previously been reported as mutated in SCLC (Fig. 2a and Supplementary Table 5). To further confirm the relevance of the 22 genes, we assessed the mutation frequency for these genes using exome data from a set of 21 additional samples (Supplementary Table 6). We found a significant correlation between the mutation frequencies of the 22 genes in the initial sample set and the validation cohort (P = 1.16 × 10−5, r2 = 0.63; Supplementary Table 7). In addition, we found that 42 genes that were mutated in our primary tumor samples (Supplementary Table 8) were also previously reported to be mutated in the genome of the NCI-H209 SCLC cell line13.
Figure 2. Significantly mutated genes in SCLC.
(a) Genes evaluated for significance on the basis of q score are shown. Each gene is represented as a circle, where the size of the circle is proportional to the observed frequency of mutation in that gene. Genes are arranged on the x axis in order of increasing number of expected mutations from left to right. Genes with significant q scores are labeled. (b) Alterations affecting the SOX family. *, nonsense change; HMG, high-mobility group; Sox_N, Sox developmental protein N terminal.
Mutational hotspots are indicative of genes that are relevant to cancer. In this study, we have identified 17 genes with 18 hotspot mutations (Supplementary Table 9). By comparing our mutations with those reported in COSMIC12 and a large-scale colon cancer mutation screen18, we identified an additional 150 hotspot mutations in 116 genes (Supplementary Table 9). Besides known hotspots in TP53, RB1, PIK3CA, CDKN2A and PTEN, several new hotspot mutations were identified. These included genes encoding Ras family regulators (RAB37, RASGRF1 and RASGRF2), chromatin-modifying enzymes or transcriptional regulators (EP300, DMBX1, MLL2, MED12L, TRRAP and RUNX1T1), ionotropic glutamate receptor (GRID1), kinases (STK38, LRRK2, PRKD3 and CDK14), protein phosphatases (PTPRD and PPEF2) and G protein–coupled receptors (GPR55, GPR113 and GPR133). Further, three of the genes with the top q scores—RUNX1T1, CDYL and RIMS2—contained a hotspot mutation.
In addition to the hotspots, we found mutations clustering in particular gene families and pathways (Supplementary Table 10). Evidence of clustering was found in genes in the phosphatidylinositol 3-kinase (PI3K) pathway (PIK3CA, AKT1–3, MTOR, RPS6KA2 and RPS6KA6), the mediator complex (MED12, MED12L, MED13, MED13L, MED15, MED24, MED25, MED27 and MED29), Notch and Hedgehog family members (NOTCH1, NOTCH2, NOTCH3 and SMO), glutamate receptor family members (GRIA1, GRIA2, GRIA3, GRIA4, GRIND1, GRID2 and GRM1–3, GRM 5, GRM 7 and GRM 8), SOX family members (SOX3, SOX4, SOX5, SOX6, SOX9, SOX11, SOX14 and SOX17; Fig. 2b) and DNA repair and/or checkpoint pathway genes (ATM, ATR, CHEK1 and CHEK2). The mutations in SOX family members were mutually exclusive (Supplementary Fig. 3). In contrast to non–small-cell lung cancer (NSCLC)12, we did not observe any SCLC samples with a KRAS mutation. Among the receptor tyrosine kinase genes, we identified mutations in FLT1, FLT4, KDR and KIT and members of the Ephrin family (EPHA1–7 and EPHB4). Notably, the KIT mutation affecting codon 761 has previously been reported in mast cell activation disorder and is likely an activating change19 (Supplementary Fig. 4).
Chromosomal copy-number analysis of 56 SCLC samples identified recurrent copy gains and losses (Supplementary Tables 11 and 12). Genes with copy-number loss included the previously reported RB1, RASSF1 and FHIT (Fig. 3) and several genes not previously known to be altered in SCLC, including KIF2A and CNTN3 (refs. 6,20). Among the genes with recurrent copy-number gain, we confirmed previously reported amplifications involving MYC, SOX4 and KIT (Fig. 3a, Supplementary Fig. 4b and Supplementary Table 11)6,20,21.
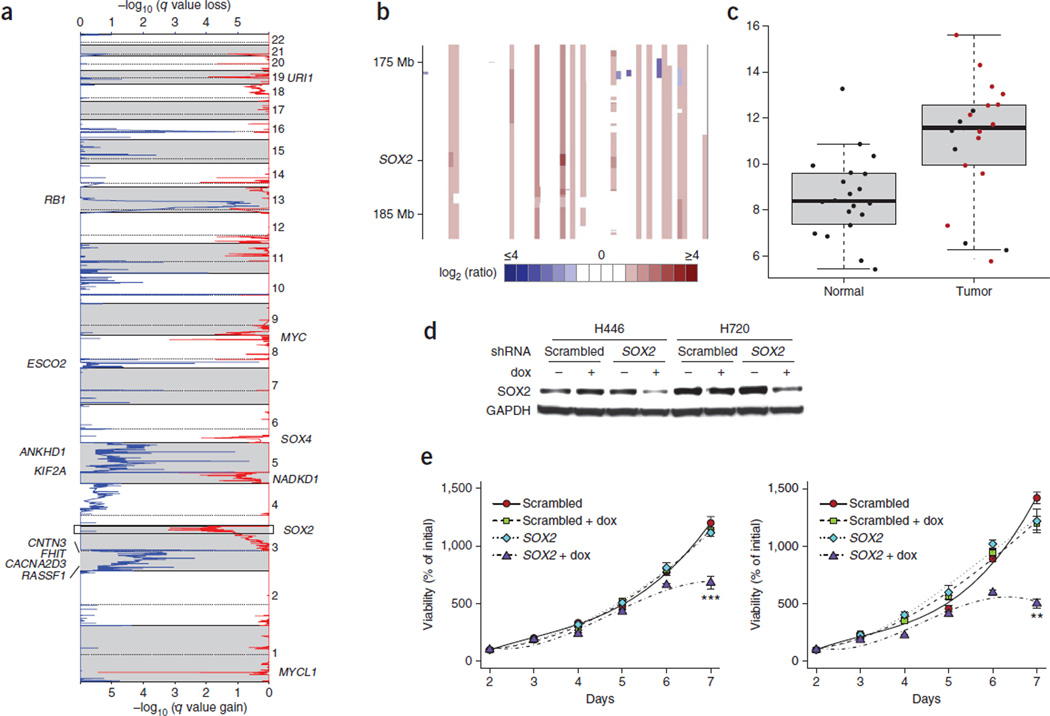
Figure 3. SOX2 is amplified in SCLC and drives proliferation.
(a) GISTIC plot depicting recurrent amplifications in SCLC samples (n = 56) with copy-number data. (b) Heatmap of segmented copy-number log2 (ratio) values from the 3q chromosomal region containing the SOX2 locus. (c) Box plot of SOX2 expression in SCLC and adjacent normal samples measured by RNA-seq. Samples with SOX2 amplification are highlighted in red. Error bars at the top indicate the maximum values excluding outliers, and error bars at the bottom indicate the minimum values excluding outliers. Outliers are defined as values more than the third quartile +1.5 × IQR or less than the first quartile −1.5 × IQR, where IQR is the innerquartile range. (d,e) Doxycycline-inducible shRNA targeting of SOX2 suppresses SOX2 protein levels (d) and inhibits cell proliferation (e) in H460 and H720 SCLC lines compared to scrambled control shRNA. Error bars in e, s.e.m. **P < 0.01; ***P < 0.001.
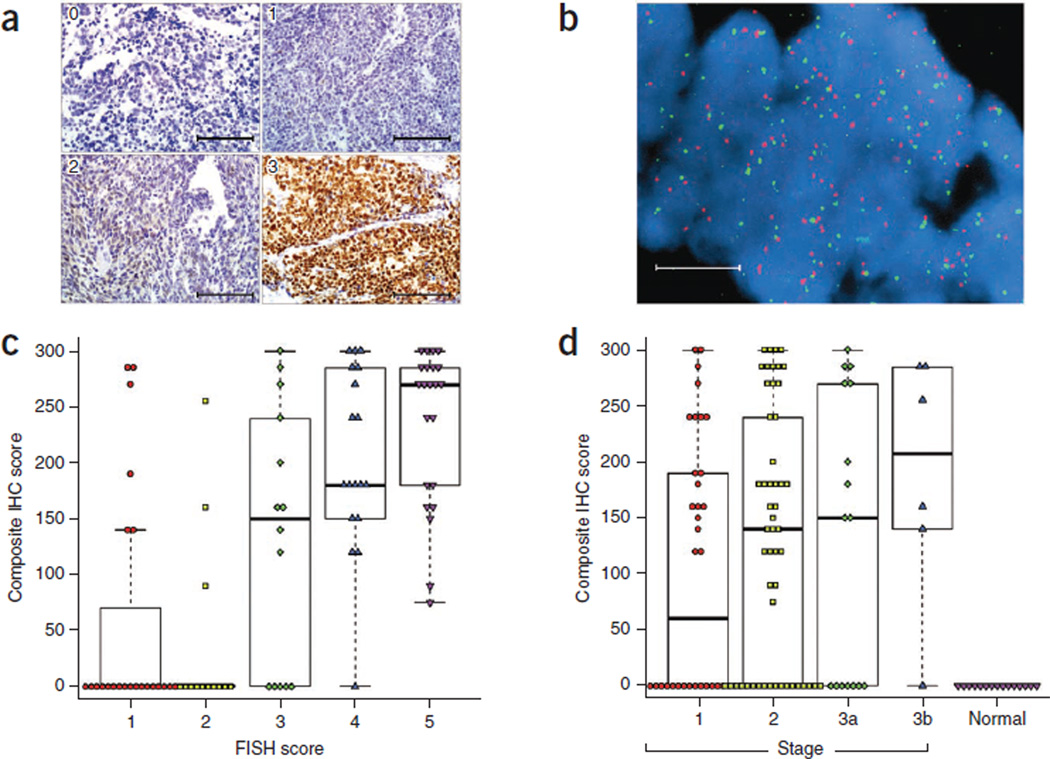
In addition, we identified high levels of amplification (copy number of ≥4) of SOX2 in ~27% (15/56) of the SCLC samples (Fig. 3b). RNA-seq data showed that the majority of the SCLC samples, including those with SOX2 amplification, had higher SOX2 expression compared to adjacent normal samples (Fig. 3c). We further examined the expression of SOX2 by immunohistochemistry (IHC) and copy-number change by FISH in an independent cohort of 110 primary SCLC tumor samples (Fig. 4a,b). Expression of SOX2 was strongly correlated with increased gene copy number and with clinical stage (Fig. 4c,d).
Figure 4. SOX2 gene amplification and protein expression in SCLC.
SOX2 protein expression was assessed by IHC, and SOX2 gene copy number was assessed by FISH on a set of 110 SCLC cases and 15 normal lung controls. (a) Representative images showing variability of staining intensity by IHC, from 0 to 3. Scale bars, 100 µm. (b) Representative image showing very high SOX2 copy number by FISH. Red, SOX2 probe; green, centromeric probe. Scale bar, 10 µm. (c) Correlation between SOX2 IHC score (staining intensity × percent with positively stained nuclei) and SOX2 FISH score (1–6). (d) Composite SOX2 IHC score of SCLC samples by stage and normal lung controls. Plots in c and d are box plots where the box encloses the first to third quartiles, the bar inside the box represents the median, the whisker at the top indicates the maximum value excluding outliers and the whisker at the bottom indicates the minimum value excluding outliers. Outliers are defined as values more than the third quartile +1.5 × IQR or less than the first quartile −1.5 × IQR, where IQR is the interquartile range.
To further assess the relevance of SOX2 in SCLC, we analyzed a panel of SCLC cell lines for SOX2 protein expression and gene copy number (Supplementary Fig. 5). Among these cell lines, H446 and H720 both had strong SOX2 protein expression, and H720 was found to have elevated gene copy number. SOX2 has previously been implicated in the maintenance of proliferative potential and stem cell function22–25. To test whether H446 and H720 were dependent on SOX2 for continued growth and proliferation, we stably transduced them with lentiviruses carrying either a doxycycline-inducible SOX2-targeting short hairpin RNA (shRNA) or a scrambled control shRNA. Induction of SOX2 shRNA in both H446 and H720 resulted in lower amounts of SOX2 protein and reduced cell proliferation (Fig. 3d,e). Previously, amplification of SOX2 and its role as an oncogene have been reported in lung and esophageal squamous cell carcinoma26. Our findings further support the idea of SOX2 as a genuine SCLC driver gene.
Analysis of RNA-seq data obtained from SCLC samples for fusion transcripts identified 41 gene fusions, including 4 recurrent fusions (Supplementary Table 13). A majority of the predicted gene fusions were intrachromosomal (83%, 34/41). All of the gene fusions reported were verified and confirmed to be somatic by RT-PCR Supplementary Table 13). A fusion involving RLF and MYCL1 (Supplementary Fig. 6a) was found in one primary SCLC tumor and four SCLC cell lines (H889, HCC33, H1092 and COR-L47). RLF and MYCL1 are ~259 kb apart and are encoded by opposing strands. The observed fusion requires an inversion event that brings exon 1 of RLF in frame with MYCL1, leading to the expression of a fusion protein composed of the first 79 amino acids of RLF and a MYCL1 protein lacking its first 27 amino acids, thereby generating a 446-residue fusion protein. The clinical sample that had the RLF-MYCL1 fusion also overexpressed MYCL1. This fusion has previously been noted27, but its role as an oncogene in SCLC has not been established. We found that small interfering RNA (siRNA)-mediated targeting of MYCL1 in H1092 and CORL47 fusion-positive cells effectively reduced the proliferation of these cells, strongly suggesting a functional role for MYCL1 in SCLC (Supplementary Fig. 6).
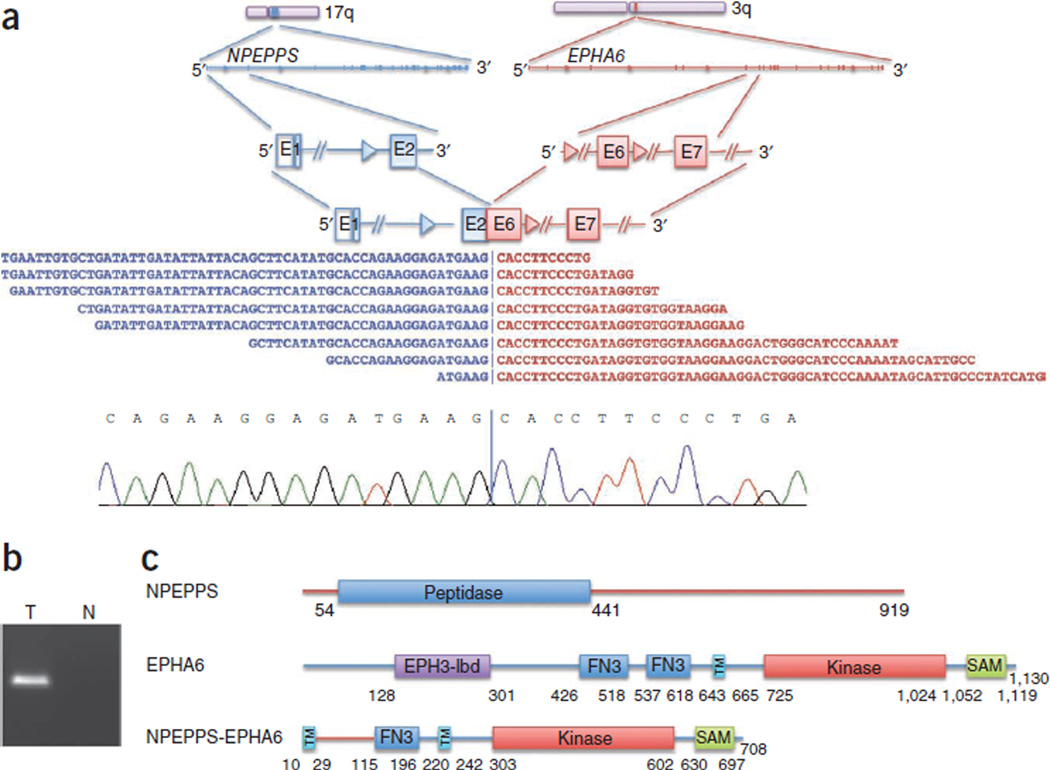
Multiple gene fusions involving kinase genes have recently been shown to be activating28. We identified four such fusions—NPEPPS-EPHA6, SKP1-CDKL3, NEK4-SFMBT1 and ZAK-RAPGEF4—that are predicted by sequence to result in functional fusion proteins (Fig. 5 and Supplementary Figs. 7–9). The roles of these fusion products in cancer remain to be elucidated.
Figure 5. Kinase fusions.
(a) NPEPPS-EPHA6 fusion identified using RNA-seq (top) along with a representative Sanger sequencing chromatogram derived from this fusion product (bottom). E, exon. (b) Independent product derived by RT-PCR confirming the NPEPPS-EPHA6 somatic fusion resolved on an agarose gel. RT-PCR was performed on a tumor (T) and normal (N) sample. (c) Schematic of the NPEPPS-EPHA6 fusion protein. EPH3-lbd, Ephrin receptor ligand–binding domain; FN3, fibronectin type 3 domain; TM, transmembrane domain; SAM, sterile α motif.
In this study, we have identified multiple new recurrent somatic mutations in SCLC, including multiple mutations and copy-number alterations in SOX gene family members. The potential role of SOX family members in SCLC is further emphasized here by the identification of SOX2 amplification and overexpression in approximately a quarter of the SCLC samples analyzed. SOX proteins have an important role in diverse biological processes, including cell type specification. Among the SOX family members, SOX2 in particular is a key factor in the maintenance of pluripotency and self-renewal of stem cells23. Aberrant SOX2 expression has also been implicated in reprogramming mature cells to acquired pluripotency24. Its expression in mouse fibroblasts, together with FoxG1, has been shown to generate self-renewing neural precursor cells25. Conditional deletion of Sox2 in mice indicates its critical role in lung development22. Conversely, overexpression of SOX2 in lung epithelial cells has been shown to promote tumorigenesis29.
Notably, conditional induction of SOX2 in lung epithelial cells is also known to increase the number of neural progenitor cells30. SCLCs are tumors with neuroendocrine features. SOX2 protein overexpression has previously been noted in high-grade SCLC31, and immunoreactive antibodies against SOX2 have been detected in sera from SCLC patients32. These observations, together with the frequent amplifications identified here, imply that SOX2 has an important role as a putative lineage-survival oncogene in SCLC. This suggestion is further supported by the correlation of SOX2 expression with SCLC stage and the role of SOX2 expression in maintaining SCLC proliferation.
The recurrent nature of the RLF-MYCL1 fusion and its functional relevance provide additional opportunities for therapeutic intervention in SCLC. Recently, oncogenic kinase gene fusions have become a major focus of interest in the therapeutic targeting of NSCLC33–35. Understanding the role of tumor-specific in-frame kinase fusion transcripts identified in SCLC in this study may provide promising opportunities for targeted therapy development.
ONLINE METHODS
Samples, DNA and RNA preparations
In this study, we have characterized 80 human SCLCs, including 36 primary SCLC human tumor and adjacent normal sample pairs and 17 paired SCLC cell lines and their patient-matched lymphoblastoid lines, as well as 4 primary SCLC tumors and 23 SCLC cell lines without matched normal controls (Supplementary Table 1).
Patient-matched fresh-frozen primary SCLC tumors and normal tissue samples were obtained from commercial sources or the Johns Hopkins tissue repository (Supplementary Table 1). All samples used in the study had appropriate IRB approval and informed consent from study participants. All tumor and normal tissues were subjected to review by a pathologist to confirm diagnosis and tumor content. The Qiagen AllPrep DNA/RNA kit was used to prepare DNA and RNA.
Exome capture and sequencing
We analyzed the exomes of 42 SCLC samples (30 primary tumor–normal tissue pairs and 12 paired cell lines) and their patient-matched normal samples to assess their mutational burden. We also obtained exome data for an additional 21 SCLC samples that included 5 primary SCLC tumors and 16 SCLC cell lines (Supplementary Table 1). Exome capture was performed using the Agilent SureSelect Human All Exome kit (38 Mb or 50 Mb). The SureSelect 50 Mb kit includes all of the capture probes from the 38 Mb kit plus some additional content derived from CCDS, GENCODE and RefSeq. Exome capture libraries were sequenced by HiSeq 2000 (Illumina) to generate 2 × 75-bp paired-end data (Supplementary Table 1). Targeted mean coverage of 80× and 162× with 96% and 92% of bases covered at ≥10× was achieved for 38 Mb and 50 Mb exome libraries, respectively (Supplementary Table 2).
RNA-seq
We obtained RNA-seq data for 55 samples (24 primary tumor–normal tissue pairs, 7 primary tumors, 2 adjacent normal samples and 22 SCLC cell lines) using the TruSeq RNA Sample Preparation kit (Illumina). Libraries were multiplexed two per lane and sequenced on HiSeq 2000 to obtain at least ~30 million paired-end (2 × 75-bp) reads per sample.
Sequence data processing
All sequencing reads were evaluated for quality using the Bioconductor ShortRead package36. Sample identity was confirmed by comparing data derived from exome sequencing and RNA-seq against Illumina 2.5 M array data as described18.
Variant calling and validation
Sequencing reads were mapped to the UCSC human reference genome (GRCh37/hg19) using Burrows-Wheeler Aligner (BWA) software37 set to default parameters. Local realignment, duplicate marking and raw variant calling were performed as described previously38. Known germline variations represented in dbSNP Build 131 (ref. 39) but not represented in COSMIC v56 (ref. 12) were filtered out. Variations present in the tumor sample but absent in matched normal tissue were predicted to be somatic. Predicted somatic variations were additionally filtered to include only positions with a minimum of 10× coverage in both the tumor and matched normal tissue, as well as an observed variant allele frequency of <3% in the matched normal tissue and a significant difference in variant allele counts, as determined using Fisher’s exact test. To control for possible low-level tumor contamination in adjacent normal tissue, the allele frequency cutoff was expanded to 5% if a gene was significantly mutated, allowing for an additional 11 variants to be included. We performed whole-genome sequencing of the 1 hypermutated sample and only report the 755 protein-altering variants that were found in both the exome and whole-genome data for this sample. This sample was excluded from background mutation rate calculations. For unpaired samples, in addition to dbSNP, variants were filtered against normal variants from this data set, as well as normal variants from a published colon data set18. In addition, data from 2,500 normal exomes in the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project was used to filter out variants and hotspot mutations. To evaluate the performance of the variant calling algorithm, we randomly selected 594 protein-altering variants and validated them using Sequenom, as described previously17. Of these variants, 91% (539) were validated as somatic. All variants that were invalidated were removed from the final set. Variants that were also validated by RNA-seq are labeled as VALIDATED: RNA-Seq to show confirmed expression of the variant (Supplementary Table 3). Indels were called using the GATK Indel Genotyper Version 2 (ref. 28). Indel validation was performed as described in a recent study18. The effects of all nonsynonymous somatic mutations on gene function were predicted using SIFT14, PolyPhen15 and Condel16. All variants were annotated using Ensembl (release 59).
Mutational significance
We evaluated the mutational significance of genes using a previously described method17, with the addition of an expression filter, as mutation rates are known to vary with expression level13,40. The hypermutated sample was excluded from analysis so that it did not affect the background mutation rate. Because of the variability in background mutation, the uniform background mutation rate used to assess the significance of mutation in cancer-associated genes is at times lower than the actual mutation rate in some regions, resulting in false positive candidates, such as the olfactory genes, seeming to be significantly mutated cancer-associated genes. To address this, a recent study used an RNA-seq–based expression filter to focus on expressed genes, thereby potentially filtering out genes that are expressed at very low levels or are not expressed at all41. In this study, we classified average gene expression on the basis of RNA-seq data into tertiles (high, medium and low) and used this information to remove low expressors that would otherwise be identified as significantly mutated cancer-associated genes.
Whole-genome, RNA-seq and pathway analysis
Whole-genome analysis, RNA-seq–based expression assessment and pathway-level analysis were performed as described previously18.
SNP array data generation and analysis
Illumina HumanOmni2.5_4v1 arrays were used to assay 56 samples (36 primary tumor–normal pairs, 15 SCLC cell line–normal pairs, 1 SCLC cell line and 4 unpaired primary tumors) for genotype, DNA copy number and loss of heterozygosity (LOH) at ~2.5 million SNP positions. These samples all passed our quality control metrics for sample identity and data quality. A subset of 2,295,239 high-quality SNPs was selected for all analyses.
After making modifications to permit use with Illumina array data, we applied the PICNIC42 algorithm to estimate total copy number, allele-specific copy number and LOH, as described recently18. Recurrent genomic regions with DNA copy gain and loss were identified using GISTIC, version 2.0 (ref. 43).
Fusion detection and validation
Fusion identification and validation were performed as has been recently described18.
Cell lines and culture conditions
All cell lines used in the study, except where noted, were cultured in RPMI 1640 supplemented with 10% FBS. H446 and H720 were cultured in RPMI 1640 with 10% tetracycline-free FBS (Hyclone, R10). Cell line identity for lines used to assess SOX2 copy number was confirmed by short tandem repeat (STR) profiling using the StemElite ID System (Promega). HCC33, HCC2433, H289, H2141, H2107, H209, H1963, H1672, H1607, H1450, H1339, H1184, H2171, HCC1772, HCC970, H128 and H2195 SCLC cell lines, their patient-matched lymphoblastoid lines and their culture conditions have been described previously44–46 (Supplementary Table 1). Additional SCLC cell lines were obtained from the American Type Culture Collection (ATCC).
Doxycycline-inducible shRNA-expressing cell lines and protein blotting
Scrambled or SOX2-targeting (TRC Clone TRCN0000003253) shRNAs were cloned as annealed oligonucleotides (Sigma) into Tet-pLKO-puro (Addgene plasmid 21915) digested with AgeI and EcoRI according to published protocols47,48. Sequence-verified clones were used to produce lentiviral particles according to TRC protocols. Lentiviral supernatants were used to infect cultured H446 or H720 cells in R10 medium at low multiplicity of infection in the presence of 8 µg/ml polybrene for 16 h. After incubation, medium was replaced with fresh R10, and cells were cultured for an additional 24 h before being selected and maintained in 500 ng/ml puromycin. The optimal doxycycline dose for inducible knockdown was determined to be 2 µg/ml, which was the minimum dose that resulted in maximal knockdown of SOX2 after 96 h. The effect of SOX2 knockdown on the amount of SOX2 protein was assessed by protein blot using antibody to SOX2 (Cell Signaling Technology 27485) or GAPDH (Santa Cruz Biotechnology, sc-25778) horseradish peroxidase (HRP)-conjugated secondary antibodies, followed by signal detection with chemiluminescence (GE Healthcare Life Sciences).
Cell viability and proliferation assays
Stable cell lines were plated in quad-ruplicate at a density of 1 × 103 cells per well in opaque 96-well plates in the presence or absence of 2 µg/ml doxycycline. Cells were plated in replicate plates for each time point tested. ATP content was measured as an indicator of metabolically active cells using the CellTiter-Glo Luminescent Cell Viability Assay (Promega) read on a SpectraMax M2e plate reader in luminescence mode (Molecular Devices). Viability was normalized between cell lines at 48 h to correct for differences in the initial number of cells plated in each group. All experiments were repeated a minimum of three times with similar results, and one representative experiment is shown.
Analysis of copy-number variation in SCLC cell lines
SOX2 copy number was assessed by quantitative RT-PCR using TaqMan Copy Number Assays (Hs02719379_cn) on a StepOnePlus Real-Time PCR System (Applied Biosystems). RPPH1 served as the reference gene (Applied Biosystems). Copy-number calls relative to normal human genomic DNA (Promega) were made with CopyCaller v2.0 (Applied Biosystems).
Tissue microarrays
SCLC tissue microarrays were obtained from US Biomax (LC703, LC802a, LC1009 and LC10010a) for IHC and FISH as fresh-cut slides. The four tissue microarrays contain replicate cores and a small set of overlapping cases. For analysis, missing or inconclusive cores were removed, and the replicate or overlapping case core with the highest percentage of tumor area was used for analysis, yielding 110 unique SCLC cases and 15 normal lung cases. Histological diagnosis with SCLC was confirmed by an attending pathologist.
Immunohistochemistry
IHC for SOX2 was performed on the tissue microarrays using a Leica Bond-III automated slide stainer (Leica Microsystems). The 4-µm sections were deparaffinized and subjected to antigen retrieval with Cell Conditioning Solution (high pH CC1 standard, Ventana Medical Systems) for 60 min. Sections were then incubated for 44 min with rabbit monoclonal antibody to SOX2 (1:100 dilution; clone SP76, Cellmarque). Reactions were developed through biotin-free, polymer detection (Ultra-view, Ventana Medical Systems) according to the manufacturer’s instructions.
Scoring was performed on each sample. Nuclear labeling was scored by intensity (no (0), weak (1), moderate (2) or strong (3)) and for extent (expressed as the percentage of nuclei that were positive). Results were expressed by assigning a composite IHC score that was calculated by multiplying the intensity score by the percentage of nuclei with positive staining, with a maximum value of 300.
FISH analysis
FISH was performed on the tissue microarrays. The BAC clone RP11-459K6 containing a human DNA insert from the genomic region of SOX2 (previously validated by PCR) was used for preparation of the SOX2 FISH probe. The SOX2 probe was validated for chromosome mapping and quality of hybridization in the human lymphoblastoid cell line AG09391 (Coriell Institute).
One slide of each tissue microarray was subjected to a two-color FISH assay using a mixture of the SOX2 probe (red) and a commercially available probe for the chromosome 3 centromere (Kreatech) (green). The steps before hybridization were performed using the Zymed Spot-Light Tissue Pretreatment kit (Invitrogen) according to the manufacturer’s instructions.
Analysis was performed on an epifluorescence microscope using single interference filter sets for blue (DAPI), green (FITC) and red (Texas red). For each interference filter, monochromatic images were acquired and merged using CytoVision (Leica Microsystems). Tumor cells were scored for copy-number signals of SOX2 in 30–50 cells. In this analysis, a scoring system was proposed to identify increased levels of copy number per cell. Scores were assigned on a scale from 1–6 (according to pattern of copy-number gain, median per-cell change): 1 (no, 1–2), 2 (low, 2–3), 3 (moderate, 3–4), 4 (high, 4–5), 5 (very high, >5), 6 (gene amplification, gene clusters).
MYCL1 knockdown studies
The SCLC cell lines, NCI-H1092, CORL47 and NCI-H2171 were transfected with siRNA pools targeting MYCL1 (Dharmacon) or with a non-targeting control siRNA (Dharmacon) following a reverse transfection protocol. The cells were incubated at 37 °C for 5 d after transfection and were subjected to a cell viability assay using the CellTiter-Glo kit (Promega).
MYCL1 (Hs00420495_m1) and GAPDH (Hs00266705_g1) TaqMan probes and primers were obtained from Life Technologies and were used to assess knockdown according to the manufacturer’s instructions. Data were analyzed using the ΔΔCT method by normalizing to GAPDH and mock-transfected controls. TaqMan reactions were performed in duplicate to obtain a mean value and s.d. P values were calculated by t test.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Genentech DNA Sequencing and Oligo groups for their help with the project. We thank M.A. Huntley and J. Degenhardt for bioinformatics support and the Pathology Core Labs for providing histology, IHC and tissue management support. This work was also supported by grants from the Burroughs Wellcome Fund, the Flight Attendant Medical Research Institute, the Johns Hopkins Specialized Programs of Research Excellence (SPORE) NCI P50CA058184 (M.V.B. and C.M.R.), the Colorado SPORE NCI P50 CA058187 (M.V.-G) and the University of Texas SPORE NCI P50CA70907 (J.D.M., A.F.G. and K.E.H.). D.D.P. is supported by the Coordenacao de Aperfeicoamento de Passoal de Nivel Superior (CAPES) Foundation and the Ministry of Education of Brazil.
Footnotes
Accession codes. Sequencing and genotype data have been deposited at the European Genome-phenome Archive, which is hosted by the European Bioinformatics Institute (EBI), under accession EGAS00001000334.
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
C.M.R. and S.S. conceived the study and designed the experiments. E.W.S. and S.D. performed the exome and whole-genome sequencing, RNA-seq and copy-number analysis. Z.M. and Y.G. performed validation of the fusions. Z.M. managed exome capture. J.T.P., E.A.B., S.C., V.J., B.S.J., W.Y. and C.P. performed biological validated studies. J. Shin, D.D.P., P.B.I. and M.V.-G. performed SOX2 IHC and FISH studies. K.E.H., A.F.G. and J.D.M. provided reagents and analysis support. J. Stinson, C.K.F., D.B., C.S.R. and J.G. collected sequencing data and performed mutation validation. F.G. and Z.Z. predicted the functional effects of mutations. E.W.S., P.M.H., R.B., T.D.W. and R.G. provided bioinformatics support, including the algorithm for variant calling, fusion detection and copy-number calling. R.B. and P.M.H. analyzed SNP array data. H.K., H.M.S., P.B.I., M.V.B. and A.F.G. provided pathology support. F.J.d.S., D.S.S., R.L.Y. and J.D.M. provided critical analysis and organizational support. D.S.S., E.W.S., S.D., Z.M., C.M.R. and J.T.P. wrote the manuscript, which was reviewed and edited by the other coauthors.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J. Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J. Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 3.Hann CL, Rudin CM. Fast, hungry and unstable: finding the Achilles’ heel of small-cell lung cancer. Trends Mol. Med. 2007;13:150–157. doi: 10.1016/j.molmed.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wistuba II, Gazdar AF, Minna JD. Molecular genetics of small cell lung carcinoma. Semin. Oncol. 2001;28:3–13. [PubMed] [Google Scholar]
- 5.Mori N, et al. Variable mutations of the RB gene in small-cell lung carcinoma. Oncogene. 1990;5:1713–1717. [PubMed] [Google Scholar]
- 6.Arriola E, et al. Genetic changes in small cell lung carcinoma. Clin. Transl. Oncol. 2008;10:189–197. doi: 10.1007/s12094-008-0181-1. [DOI] [PubMed] [Google Scholar]
- 7.Yokomizo A, et al. PTEN/MMAC1 mutations identified in small cell, but not in non-small cell lung cancers. Oncogene. 1998;17:475–479. doi: 10.1038/sj.onc.1201956. [DOI] [PubMed] [Google Scholar]
- 8.Tatematsu A, et al. Epidermal growth factor receptor mutations in small cell lung cancer. Clin. Cancer Res. 2008;14:6092–6096. doi: 10.1158/1078-0432.CCR-08-0332. [DOI] [PubMed] [Google Scholar]
- 9.Shibata T, Kokubu A, Tsuta K, Hirohashi S. Oncogenic mutation of PIK3CA in small cell lung carcinoma: a potential therapeutic target pathway for chemotherapy-resistant lung cancer. Cancer Lett. 2009;283:203–211. doi: 10.1016/j.canlet.2009.03.038. [DOI] [PubMed] [Google Scholar]
- 10.Onuki N, et al. Genetic changes in the spectrum of neuroendocrine lung tumors. Cancer. 1999;85:600–607. doi: 10.1002/(sici)1097-0142(19990201)85:3<600::aid-cncr10>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 11.Sher T, Dy GK, Adjei AA. Small cell lung cancer. Mayo Clin. Proc. 2008;83:355–367. doi: 10.4065/83.3.355. [DOI] [PubMed] [Google Scholar]
- 12.Forbes SA, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38:D652–D657. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pleasance ED, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng PC, Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002;12:436–446. doi: 10.1101/gr.212802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.González-Pérez A, Lopez-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am. J. Hum. Genet. 2011;88:440–449. doi: 10.1016/j.ajhg.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kan Z, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 18.Seshagiri S, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012 Aug 15; doi: 10.1038/nature11282. published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molderings GJ, et al. Multiple novel alterations in Kit tyrosine kinase in patients with gastrointestinally pronounced systemic mast cell activation disorder. Scand. J. Gastroenterol. 2007;42:1045–1053. doi: 10.1080/00365520701245744. [DOI] [PubMed] [Google Scholar]
- 20.D’Angelo SP, Pietanza MC. The molecular pathogenesis of small cell lung cancer. Cancer Biol. Ther. 2010;10:1–10. doi: 10.4161/cbt.10.1.12045. [DOI] [PubMed] [Google Scholar]
- 21.Medina PP, et al. The SRY–HMG box gene, SOX4, is a target of gene amplification at chromosome 6p in lung cancer. Hum. Mol. Genet. 2009;18:1343–1352. doi: 10.1093/hmg/ddp034. [DOI] [PubMed] [Google Scholar]
- 22.Tompkins DH, et al. Sox2 is required for maintenance and differentiation of bronchiolar Clara, ciliated, and goblet cells. PLoS ONE. 2009;4:e8248. doi: 10.1371/journal.pone.0008248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wegner M. SOX after SOX: SOXession regulates neurogenesis. Genes Dev. 2011;25:2423–2428. doi: 10.1101/gad.181487.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 25.Lujan E, Chanda S, Ahlenius H, Sudhof TC, Wernig M. Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc. Natl. Acad. Sci. USA. 2012;109:2527–2532. doi: 10.1073/pnas.1121003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bass AJ, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat. Genet. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mäkelä TP, Saksela K, Evan G, Alitalo K. A fusion protein formed by L-myc and a novel gene in SCLC. EMBO J. 1991;10:1331–1335. doi: 10.1002/j.1460-2075.1991.tb07652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson DR, et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat. Med. 2011;17:1646–1651. doi: 10.1038/nm.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu Y, et al. Evidence that SOX2 overexpression is oncogenic in the lung. PLoS ONE. 2010;5:e11022. doi: 10.1371/journal.pone.0011022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gontan C, et al. Sox2 is important for two crucial processes in lung development: branching morphogenesis and epithelial cell differentiation. Dev. Biol. 2008;317:296–309. doi: 10.1016/j.ydbio.2008.02.035. [DOI] [PubMed] [Google Scholar]
- 31.Sholl LM, Long KB, Hornick JL. Sox2 expression in pulmonary non-small cell and neuroendocrine carcinomas. Appl. Immunohistochem. Mol. Morphol. 2010;18:55–61. doi: 10.1097/PAI.0b013e3181b16b88. [DOI] [PubMed] [Google Scholar]
- 32.Güre AO, et al. Serological identification of embryonic neural proteins as highly immunogenic tumor antigens in small cell lung cancer. Proc. Natl. Acad. Sci. USA. 2000;97:4198–4203. doi: 10.1073/pnas.97.8.4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwak EL, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl. J. Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bergethon K, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J. Clin. Oncol. 2012;30:863–870. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takeuchi K, et al. RET, ROS1 and ALK fusions in lung cancer. Nat. Med. 2012;18:378–381. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 36.Morgan M, et al. ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics. 2009;25:2607–2608. doi: 10.1093/bioinformatics/btp450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martincorena I, Seshasayee AS, Luscombe NM. Evidence of non-random mutation rates suggests an evolutionary risk management strategy. Nature. 2012;485:95–98. doi: 10.1038/nature10995. [DOI] [PubMed] [Google Scholar]
- 41.Barbieri CE, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greenman CD, et al. PICNIC: an algorithm to predict absolute allelic copy number variation with microarray cancer data. Biostatistics. 2010;11:164–175. doi: 10.1093/biostatistics/kxp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mermel CH, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simms E, Gazdar AF, Abrams PG, Minna JD. Growth of human small cell (oat cell) carcinoma of the lung in serum-free growth factor–supplemented medium. Cancer Res. 1980;40:4356–4363. [PubMed] [Google Scholar]
- 45.Phelps RM, et al. NCI–Navy Medical Oncology Branch cell line data base. J. Cell. Biochem. Suppl. 1996;24:32–91. doi: 10.1002/jcb.240630505. [DOI] [PubMed] [Google Scholar]
- 46.Carney DN, Bepler G, Gazdar AF. The serum-free establishment and in vitro growth properties of classic and variant small cell lung cancer cell lines. Recent Results Cancer Res. 1985;99:157–166. doi: 10.1007/978-3-642-82533-0_16. [DOI] [PubMed] [Google Scholar]
- 47.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 48.Wiederschain D, et al. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle. 2009;8:498–504. doi: 10.4161/cc.8.3.7701. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.