Abstract
Topoisomerases are ubiquitous enzymes that control DNA supercoiling and entanglements. They are essential during transcription and replication and topoisomerase inhibitors are among the most effective and most commonly used anticancer and antibacterial drugs. This review consists in two parts. In the first part (“Lessons”), it gives background information on the catalytic mechanisms of the different enzyme families (6 different genes in humans and 4 in most bacteria), describes the “interfacial inhibition” by which topoisomerase-targeted drugs act as topoisomerase poisons and describes clinically relevant topoisomerase inhibitors. It generalizes the interfacial inhibition principle, which was discovered from the mechanism of action of topoisomerase inhibitors, and discusses how topoisomerase inhibitors kill cells by trapping topoisomerases on DNA rather than by classical enzymatic inhibition. Trapping protein-DNA complexes extends to a novel mechanism of action of PARP inhibitors and could be applied to the targeting of transcription factors. The second part of the review focuses on the challenges for discovery and precise use of topoisomerase inhibitors, including targeting topoisomerase inhibitors using chemical coupling and encapsulation for selective tumor delivery, use of pharmacodynamic biomarkers to follow drug activity, complexity of the response determinants for anticancer activity and patient selection, prospects of rational combinations with DNA repair inhibitors targeting tyrosyl-DNA-phosphodiesterases 1 and 2 (TDP1 and TDP2) and PARP, and the unmeet need to develop inhibitors for type IA enzymes.
1. Foreword
The first part of this assay summarizes the known mechanisms by which drugs target topoisomerases, complementing and updating more detailed reviews.1–12 The relatively unknown mechanism of action of topoisomerase inhibitors can be traced to the complexity of the topic with 6 different genes in humans cells and bacteria, drugs acting as interfacial inhibitors that trap ternary complexes, and drug cytotoxic mechanisms mediated by the trapping of topoisomerases on DNA rather than by classical enzymatic inhibition. The second part of the review addresses the remaining challenges for the development and precise use of topoisomerase inhibitors for the treatment of cancers and infections.
2. DNA topoisomerases
Topoisomerases are universal and present in eukaryotes, archaebacteria and eubacteria.13–19 Human cells encode six topoisomerases whereas bacteria generally contain only 4 topoisomerases and lack the type IB enzymes (Table 1 and Fig. 1).5 The ubiquity of topoisomerases stems from DNA’s double-helical (duplex) structure and length, which promote DNA entanglement in the compacted nucleus of eukaryotic cells or the nucleoid of bacteria. The opening of duplex DNA and separation of its two strands during transcription and replication generate supercoiling (torsional tension) on both sides of the open DNA segment. Excessive positive supercoiling tightens the DNA and prevents further strand separation thereby stalling the polymerases. Negative supercoiling behind the polymerases, on the other hand, tends to extend DNA strand separation and facilitates the formation of abnormal nucleic acid structures such as R-loops, which can stall RNA polymerase when the transcripts remain bound to the unwound DNA template. Negative supercoiling also promotes the formation of non-canonical DNA structures such as z-DNA, intramolecular hairpins and guanosine quartets (G4’s). Topoisomerases prevent the formation of such potentially deleterious structures by removing free supercoiling.
Table 1.
Classification of topoisomerases
| Type | Polarity | Mechanism | Humans | Bacteria | ||||
|---|---|---|---|---|---|---|---|---|
| Genes | Proteins | Drugs | Genes | Proteins | Drugs | |||
| IA | 5'-PY | Strand passage | TOP3A | Top3α | none | TOPA | Topo I | none |
| TOP3B | Top3β | none | TOPB | Topo III | none | |||
| IB | 3'-PY | Rotation | TOP1 | Top1 | anticancer | usually | ||
| TOP1MT | Top1mt | none (?) | none | |||||
| GYRA | Gyrase | |||||||
| TOP2A | Top2α | GYRB | ||||||
| IIA | 5'-PY | Strand passage | anticancer | Antibiotics | ||||
| ATPase | TOP2B | Top2β | PARC | Topo IV | ||||
| PARE | ||||||||
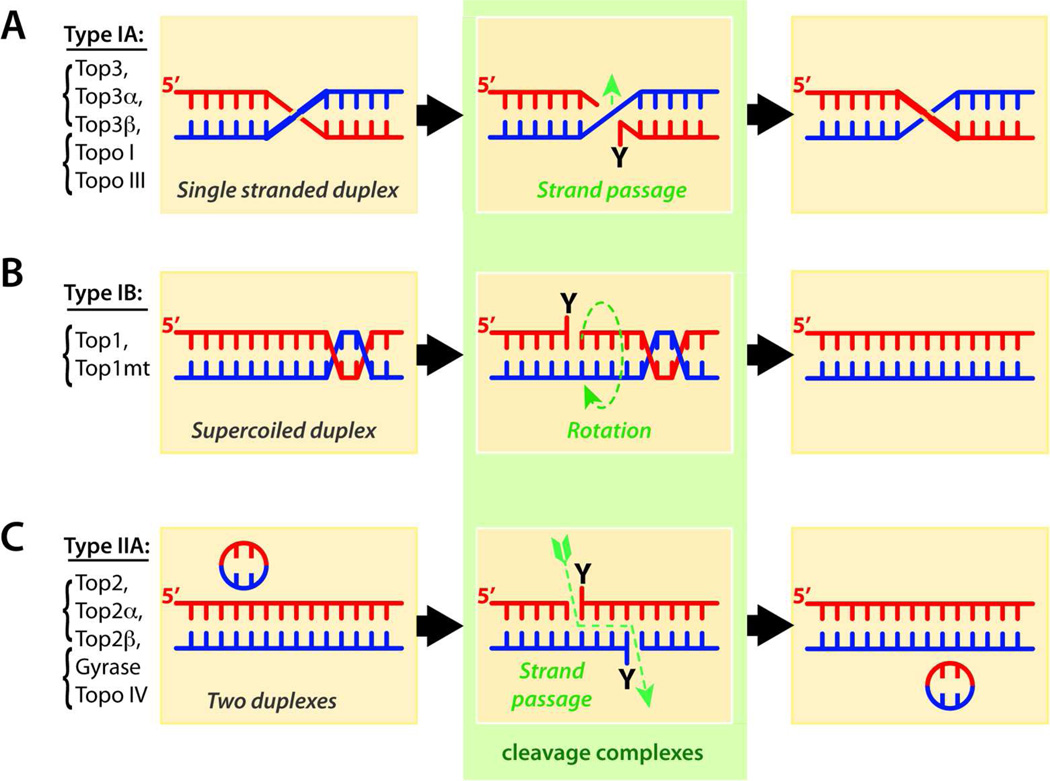
Type I enzymes are monomeric and cleave one strand of DNA for catalysis. Type II enzymes are homodimeric (humans) or heterotetrameric (bacteria), and cleave both strands of duplex DNA with a 5’-four-base overhang (see Fig. 2).
Figure 1.
Differential catalytic mechanisms of topoisomerases. Reactions are represented from left to right. Type I enzymes cleave one strand to process DNA entanglements whereas type II cleave both strands by concerted action of each Top2 monomer (see Table 1). Type IA and IIA enzymes (panels A and C) cleave DNA by covalently attaching their catalytic tyrosine to the DNA 5’-end. Type IA enzymes cleave a single-stranded segment and let another single-strand pass through the break, whereas type IIA let a duplex pass through the concerted breakage of both strands. For both type IA and IIA enzymes, the 3’-ends are tightly bound during strand passage, which keeps the passing DNA in an enzyme cavity before resealing of the ends (not shown; for details see 15,24,25). By contrast to type IA and IIA enzymes, type IB topoisomerases (panel B) form 3’-phosphotyrosine bonds and relax DNA supercoiling by controlled rotation of the broken 5’-end around the intact strand.23,155
Topoisomerase remove supercoiling by different mechanisms. Type IB enzymes work by letting the broken strand rotate around the intact strand (Fig. 1B),20–23 whereas type IA and type IIA enzymes work by passing one strand or one duplex, respectively, from the same DNA molecule through the single- or double-strand break generated by the topoisomerase in another duplex (Fig. 1 and Table 1).15,24,25
Replication of circular DNA molecules and chromatin loops produces interlinked DNA products (catenanes)5 that need to be removed by the strand passing activities of topoisomerases. While type IIA enzymes (Fig. 1C and Table 1) act as full decatenases, passing one duplex through another, the strand passing activity of type IA enzymes is restricted to single-stranded DNA segments adjacent to duplex regions (Fig. 1A), which enables Top3 to pass a DNA single-strand and resolve hemicatenanes and double-holiday junctions following replication of supercoiled DNA.26
Topoisomerases always break DNA by transesterification reactions using an active site tyrosine as the nucleophile that attacks the DNA phosphodiester backbone. Type IA and IIA enzymes break the DNA by attacking and bonding to the 5’-phosphate whereas type IB enzymes break DNA by covalent attachment to the 3’-phosphate (Table 1 and Figs. 1 and 2). The resulting 3’-hydroxyl ends in the case of type IA and IIA enzymes and 5’-hydroxyl ends in the case of the type IB enzymes reverse the phosphotyrosyl bonds, thereby enabling the release of the topoisomerase and religation of the DNA (Figs. 1 & 2). The nicking-closing activities of topoisomerases are remarkably fast (up to 6000 cycles per minute for Top1 and 250 for Top2);6,23 yet the enzymes are susceptible to the drugs selectively when the DNA is in the cleaved state (Fig. 2).23
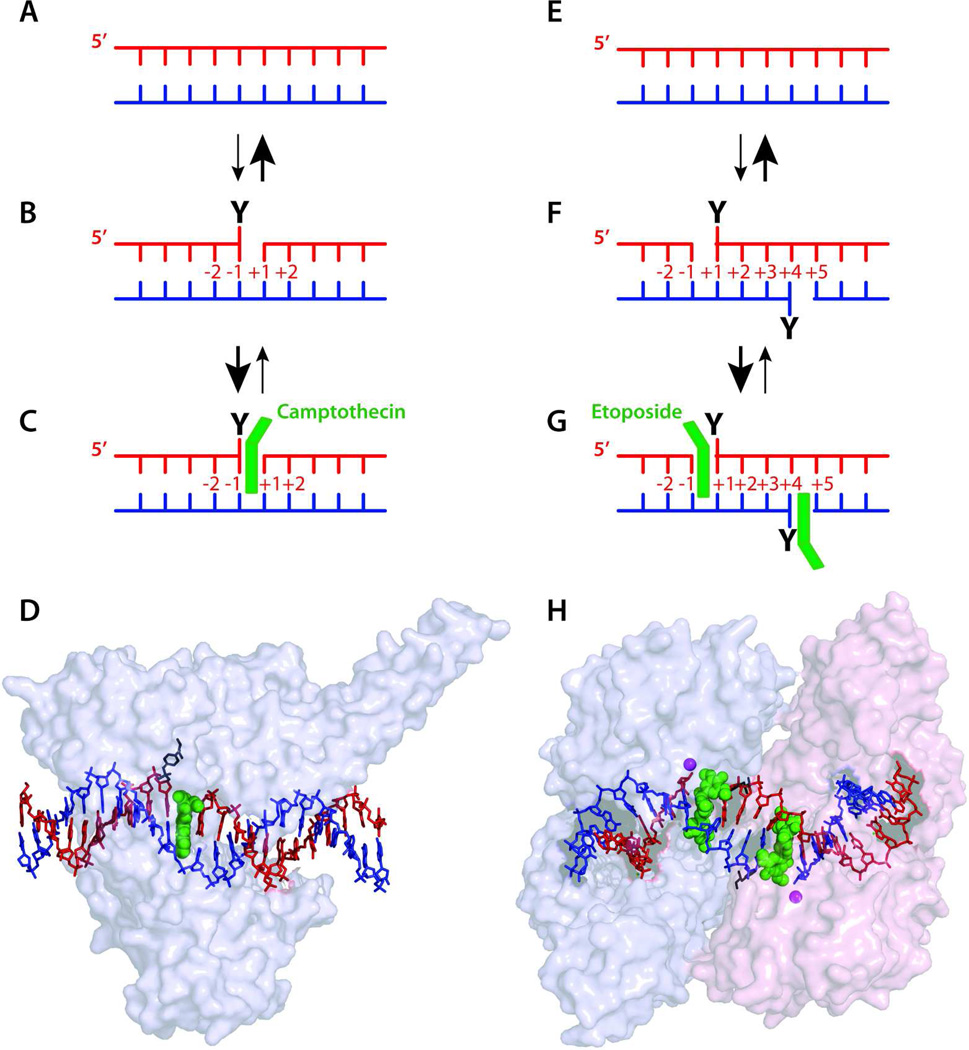
Figure 2.
Interfacial inhibition for Top1 (left) and Top2 inhibitors (right). Under normal conditions, Top1 and Top2 cleave and religate DNA very rapidly (A–B and E–F). Religation is faster than cleavage and cleavage complexes are transient. Drugs (green) (C–D and G–H) bind reversibly (C and G) at the interface of the cleaved DNA and the enzyme by forming a ternary complex (see text for details). The pdb coordinates for D and H are 1T8I and 3QX3, respectively.6
3. Topoisomerase inhibitors and the interfacial inhibition principle
The molecular mechanism of action of topoisomerase inhibitors, i.e. their specific binding at the interface of topoisomerase-DNA complexes, led to the interfacial inhibition concept.6,27,28 We proposed this hypothesis initially for Top2 inhibitors to explain the sequence selective trapping of Top2cc by different drugs; namely the preference for an adenine at position −1 in the case of doxorubicin and other anthracyclines (Fig. 3C),29 and for cytosine −1 and adenine +1 for etoposide and m-AMSA, respectively.30,31 Similarly for Top1, camptothecin preferentially traps a subset of Top1cc; those with a guanine +1.32 A unifying model emerged by which one drug molecule stacks against the base pairs flanking the topoisomerase-induced cleavage site (Fig. 2C & G).29,30
Figure 3.
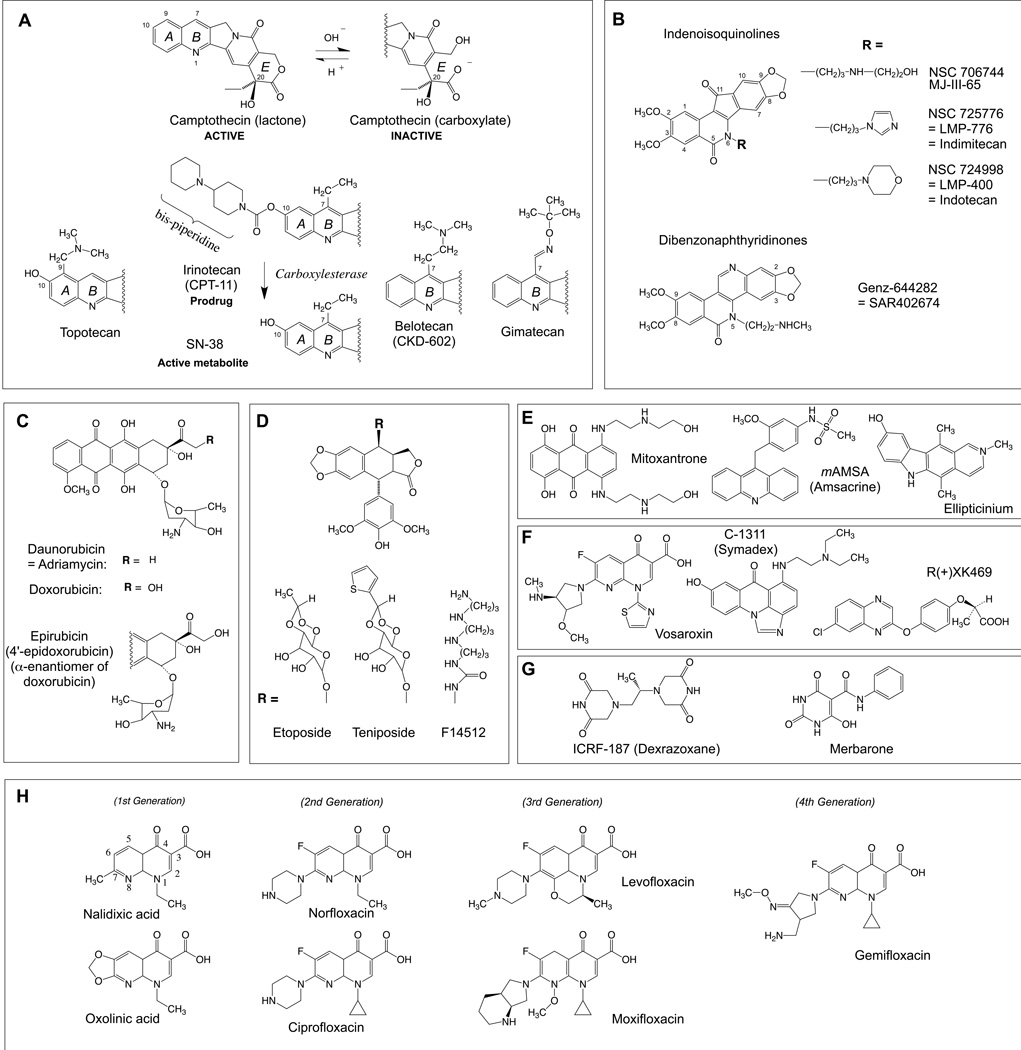
Structure of anticancer and antibacterial topoisomerase inhibitors. A: Camptothecins. B: Non-camptothecin Top1 inhibitors in clinical trials. C: Anthracyclines. D: Demethylepipodophyllotoxin derivatives, including the clinical trial drug F14512 with its spermine side chain. E: Other Top2cc-targeted intercalative drugs. F: Three Top2cc-targeted drugs in clinical trials in addition to F14512 shown in panel D. G: Top2 catalytic inhibitors. G: Quinolone antibacterials.
To account for the stereospecificity of camptothecins (i.e. only the natural 20-S-isomer is active against Top1; see Fig. 3A) 33 and for the high drug resistance of specific Top1 mutants,34 we also proposed that the enzyme forms specific amino acid contacts with the drug as it stacks against the bases flanking the cleaved DNA. This led to the ternary complex hypothesis with the drug simultaneously interacting with the DNA and the enzyme (Fig. 2C & G).
It took over 10 years to confirm by x-ray crystallography the Top1-DNA-camptothecin model (Fig. 2D) with camptothecin stacking against the +1 guanine and forming a hydrogen bond network with the enzyme.35–38 The confirmation of the Top2cc trapping interfacial model (Fig. 2C) for antibiotics and anticancer drugs was obtained more recently (Fig. 2G).39–42 The interfacial inhibition principle extends beyond topoisomerase inhibitors. A large number of natural products act as interfacial inhibitors not only by binding at the interface of nucleic acids and proteins (α-amanitin, aminoglycoside antibiotics), but also at the interface of polypeptides that form multi-proteins complexes and move around each other to perform their biological function (vinblastine, colchicine, rapamycin, brefeldine A, benzodiazepines, anesthetics).6
4. Mechanism of action of topoisomerase inhibitors: trapping of topoisomerase-DNA complexes versus catalytic inhibition
Topoisomerase inhibitors are exquisitely selective and without ambiguity eligible as “targeted therapies”. Clinically relevant Top1 inhibitors (Fig. 3) do not affect Top2, and, conversely, Top2 inhibitors do not trap Top1 enzymes. Furthermore, the inhibitors of bacterial topoisomerases (gyrase and Topo IV) are inactive against host cell topoisomerases (Top2 and Top1), which accounts for their antibacterial potency without impact on the host genome.
The therapeutic mechanism of action of topoisomerase inhibitors revealed another new paradigm for drug action (in addition to interfacial inhibition detailed above): enzyme poisoning rather than catalytic inhibition drives drug activity. This concept first emerged for the antibacterial topoisomerase inhibitors and was demonstrated for the anticancer topoisomerase inhibitors soon after Top1 was discovered as the target of camptothecin.43 Indeed, yeast cells lacking Top1 are immune to camptothecin.44,45 Similarly, human cancer cells depleted for Top1 become resistant to camptothecin,46 implying that Top1 is required for the cytotoxicity of camptothecin, whereas lack of Top1 catalytic activity (as in cells lacking Top1) is tolerated. Biochemical evidence for the requirement of Top1 for the cytotoxicity of camptothecins and non-camptothecin Top1 inhibitors is supported by the formation of Top1cc in cells treated with Top1 inhibitors.46–49 Induction of Top1cc in biochemical system is actually routinely used to discover and evaluate Top1 inhibitors.50,51
Genetic evidence for Top2 requirement for the anticancer activity of Top2 inhibitors (Table 1) or for the requirement of gyrase or/and topo IV for the antibiotics (Table 1) has been more difficult to obtain than for Top1 inhibitors because cells lacking type IIA topoisomerases are not viable. Nevertheless, several independent studies established that reducing Top2 (and Top1) levels in tumors minimizes drug activity.34,46,52–54 Conversely, the therapeutic activity of doxorubicin is correlated with Top2 overexpression in the case of amplification of the TOP2A locus together with the HER2 locus on chromosome 17 in a subset of breast cancers.55 Biochemical evidence for the trapping of Top2-DNA complexes (Top2cc) by anticancer drugs is relatively straightforward and multiple assays can be used to detect the Top2cc not only with recombinant Top256 but also in cells.57–59
Because the cytotoxic effect of topoisomerase inhibitors requires and is positively correlated with the levels and activity of topoisomerases, assays are being developed to measure the enzymes in patient samples to monitor drug anticancer activity (see Table 2 and section 8).60,61
Table 2.
Challenges for the discovery and use of topoisomerase inhibitors
| Challenges | Possible answers (new approaches) |
|---|---|
| 1. New topoisomerase targets |
|
| 2. New topoisomerase inhibitors (in addition to #1 above) |
|
| 3. Pharmacodynamic (PD) biomarkers to rapidly evaluate tumor drug response |
|
| 4. Cancer patient selection |
|
| 5. Optimize drug combinations |
|
The enzyme poisoning mechanism of action first identified for topoisomerase inhibitors has recently been extended to poly(adenosine diphosphoribose) polymerase (PARP) inhibitors. Indeed, as in the case of Top1 inhibitors, knocking out PARP renders cells immune to PARP inhibitors, and treating cells with PARP inhibitors such as olaparib produces PARP1 and PARP2 DNA complexes.62 These findings imply that the remarkable activity of PARP inhibitors in breast and ovarian cancers and in Ewing sarcoma can be related, at least in part to the trapping of PARP-DNA complexes when the cancer cells are deficient in homologous recombination repair (BRCA and Fanconi anemia genetic deficiencies).62 The fact that three very important classes of anticancer drugs, the Top1, Top2 and PARP inhibitors act by poisoning protein-DNA complexes suggests the possibility that other DNA processing protein complexes, such as transcription factors including the Myc-Max heterodimer could be targeted by interfacial inhibitors. Interfacial inhibitors would lock them on the DNA and thereby initiate a cytotoxic cascade to kill cancer cells.
5. Anticancer Top1-targeted drugs
Camptothecin derivatives are the only FDA-approved Top1-targeted anticancer drugs (Fig. 3A). They are water-soluble semi-synthetic derivatives of the plant alkaloid, camptothecin. The potent anticancer activity of camptothecin was known for ≈ 20 years33 before the identification of Top1 as its molecular target.43,47
Topotecan (Hycamtin®) is routinely prescribed for ovarian cancer, and recurrent small cell lung cancer (SCL).63 Irinotecan (Camptosar®, Campto®) is widely used in gastrointestinal (colorectal and gastro esophageal) malignancies.63 Topotecan and irinotecan are also used in primary brain malignancies (glioblastomas), sarcomas, and cancers of the cervix. Irinotecan is a prodrug, which is readily hydrolyzed to its active metabolite, SN-38 by carboxyl esterase (Fig. 3A).
Dose-limiting toxicities are myelosuppression for both topotecan and irinotecan and diarrhea for irinotecan. Bone marrow toxicity is common to all other classical cytotoxics and is probably related to the high proliferative index of bone marrow cells and to cell death priming.64 Severe diarrheas are only observed with irinotecan and their mechanism is not fully understood. It has been related to the hepatic elimination of SN-38 and its glucuronated metabolite that produce high intestinal concentrations of SN-38.63
In spite of their potent anticancer activity, all the camptothecins (Fig. 3A) suffer from well-defined limitations.3 In addition to their dose-limiting toxicity, which prevents the use of curative doses,65 camptothecins are rapidly inactivated by E-ring opening (Fig. 3A). Indeed, the E-ring α-hydroxylactone spontaneously hydrolyzes within minutes at physiological pH to camptothecin carboxylate, which is sequestered by its high affinity binding to serum albumin. Irinotecan and topotecan are also substrates for the drug efflux transporters, especially ABCG2.66 Finally, both irinotecan and topotecan are formulated for IV administration and oral formulations are not been pursued because of the intestinal toxicity of irinotecan.
To avoid the dose-limiting toxicity of camptothecins and their short half lives and to reduce normal tissue toxicity while increasing drug delivery to tumors, camptothecins have been conjugated to a macromolecular core as in etirinotecan pegols (NKTR-102). NK-102 is in advanced clinical development (Phase III) for ovarian, breast and colon cancers. The macromolecular conjugation allows slow drug release with lower peak concentration, extended half-life (up to 15 days), and enhanced tumor penetration through leaky tumor vasculature. Another comparable approach is liposomal formulation (see section 8.3).
The chemical instability of camptothecins has been impossible to overcome by simple semi-synthetic derivations of any of the camptothecins including topotecan and irinotecan derivatives.3,33 The intact α-hydroxylactone E-ring is indeed critical for the binding of camptothecins to the Top1-DNA cleavage complex.35–38
Two families of non-camptothecin Top1 inhibitors that overcome the E-ring instability of camptothecins are in clinical development (Fig. 3B).4,67 The indenoisoquinolines were discovered by screening the NCI Developmental Program drug database for compounds producing cytotoxicity profiles highly correlated with camptothecin across the 60 diverse cancer cell lines of the NCI drug screen [the NCI-6068].3,69,70 Two derivatives are currently in clinical trials indotecan (LMP400) and indimitecan (LMP776). They are developed by the NCI and Purdue University, and licensed to Linus Oncology. In addition to their chemical stability, the indenoisoquinolines offer several advantages over the camptothecins. They target additional genomic sites, their cleavage complexes are markedly more persistent than for camptothecins,51,71 they overcome multidrug resistance drug efflux pumps51 and they produce less bone marrow suppression at equal antitumor activity 72.
The other non-camptothecin in clinical trials is 8,9-dimethoxy-5-(2-N-methylaminoethyl)-2,3-methylenedioxy-5H-dibenzo[c,h][1,6]naphthyridin-6-one (Genz-644282; SAR402674) (Fig. 3B), was developed from a structure-activity relationship conducted around the dibenzo[c,h][1,6]naphthyridin-6-one compound family.80 Genz-644282 has equivalent or superior activity in xenograft models compared with standard drugs and has a favorable cytotoxic profile in vitro in bone marrow and tumor cell colony forming assays.81,82 The compound was licensed to Genzyme by Rutgers University and is now in the drug development portfolio of Sanofi (SAN).67 Genz-644282 has comparable Top1-targeting activity as the indenoisoquinolines.73 Both the indenoisoquinolines (LMP776 and LMP400, indimitecan and indotecan) and Genz-644282 appear active and relatively well tolerated in Phase I clinical trials. 83
6. Anticancer Top2-targeted drugs
All the drugs shown in Figure 3 (panel C–F) inhibit Top2 by targeting Top2cc and inhibiting their religation,1,5,7,11,74 most likely through interfacial inhibition (see section 3 and Fig. 2). They offer a broad spectrum of chemical diversity,11 potency,75,76 sequence selectivity,31 and ability to trap concerted Top2cc.5,29,75–77 We refer to “concerted Top2cc” as those where both strands of the DNA are cleaved simultaneously with a canonical 4 base pair overhang stagger (Figs. 1 & 2).
The chemical diversity of Top2-targeted drugs was recently reviewed, and we invite the reader to consult the excellent overview of C. Bailly.11 We will focus on the clinical use and chemical biology of prototypical Top2-targeted drugs and the drugs in clinical trials.
The anthracycline daunorubicin (daunomycin) was discovered in the 1950’s from Streptomyces soil bacteria as an extremely potent anticancer drug. It remains used today primarily for the treatment of acute leukemia.78 Doxorubicin (adriamycin), another bacterial toxin, was discovered soon after daunorubicin and is more widely used.78 It is active in first line therapy for breast cancers, bone and soft tissue sarcomas, bladder cancers, anaplastic thyroid cancer, Hodgkin’s and non-Hodgkin’s lymphomas and multiple myeloma.79 Epirubicin (4’-epi-doxorubicin), an active isomer of doxorubicin (Fig. 3C) was developed later (FDA approval in 1999) to limit the side effects of doxorubicin, possibly due to its faster elimination. Epirubicin is used in breast, esophageal and gastric cancers.79 The molecular pharmacology and mechanism of action of anthracyclines are complex. In addition to their anti-Top2 activity, anthracyclines are potent DNA intercalators and generate reactive oxygen intermediates. Their effects on Top280 first proposed by Kohn and coworkers81 was demonstrated well after their approval by the FDA as anticancer agents.
The Top2 inhibitory effect of anthracyclines exhibits notable peculiarities. First, because of the very effective DNA intercalation of anthracyclines, the trapping of Top2 cleavage complexes, which is achieved at submicromolar drug concentration decreases as drug concentration increases, resulting in a bell-shape concentration response with lack of trapping of Top2cc at or above 10 µM concentration.56,82 Second, compared to etoposide, anthracyclines trap Top2cc with high selectivity at limited genomic sites with an adenine at the −1 position (see Fig. 2).29 Third, most of the Top2cc are concerted and correspond to DNA double-strand breaks.75 Finally, the reversal to Top2cc is slow upon drug removal, explaining the potent effects of the anthracyclines on Top2.
Besides bone marrow suppression, which is common to all topoisomerase-targeted anticancer drugs, anthracyclines are cardiotoxic. This dose-limiting and cumulative cardiotoxicity was until recently primarily attributed to the generation of reactive oxygen species.78 However, a recent study has linked cardiotoxicity to top2β targeting in the nucleus and subsequent mitochondrial damage.83 Although interesting, this possibility will probably require further studies to elucidate whether doxorubicin could damage mitochondria more directly by targeting mitochondrial Top2β.84
Etoposide (VP-16; Vepesid®) (Fig. 3D) is widely used in oncology for a broad range of solid tumors including small cell lung cancers, testicular and germ cell tumors, endocrine tumors, osteosarcomas and Ewing’s sarcomas, neuroblastomas and Kaposi sarcoma. Like doxorubicin, etoposide was developed clinically and approved by the FDA (in 1983) without knowing that Top2 was its molecular target. It is a semi-synthetic demethylepipodophyllotoxin derivative without activity on tubulin (by contrast to the podophyllotoxins). Etoposide stands apart from other Top2cc-targeted drugs for the following reasons. First, it is the most selective Top2cc-targeted drug currently in the clinic. As it does not act as a DNA intercalating agent, the Top2cc produced by etoposide form in a monotonic manner without decrease at high drug concentration.85–87 Second, the Top2cc trapped by etoposide are frequently uncoupled (“non-concerted”) with the majority being single-strand breaks instead of the expected double-strand breaks during concerted inhibition86,87 (see beginning of this section), suggesting that etoposide traps Top2 homodimers asymmetrically with a single drug molecule bound into one of the two breaks (see Fig. 2G but with a single drug molecule and only one of the homodimers trapped in the cleavage complex). Third, as mentioned in section 3, the base sequence preference of etoposide is determined by the presence of cytosine at position −1.30 Fourth, etoposide produces a very high frequency of Top2cc compared to the intercalating Top2 inhibitors.88,89 Fifth, the Top2cc trapped by etoposide are readily reversible upon drug wash out, which is different from the anthracyclines. Finally, etoposide traps both Top2α and β very effectively,59,90 whereas doxorubicin tends to target more selectively cellular Top2α over Top2β.91 The trapping of Top2β has been related to the induction of secondary leukemia in patient previously treated with etoposide,92 and linked to Top2β-mediated DNA translocations (see section 8.2).93,94
The anthracenedione mitoxantrone (Novantrone®) (Fig. 3E) was developed as a synthetic analog of anthracyclines at the American Cyanamid Laboratories in the late 1970s78 and approved by the FDA in 1996 for prostate cancer. Like anthracyclines, mitoxantrone is both a potent DNA intercalator and Top2cc poison. Its reduced potential to undergo redox reactions compared to doxorubicin may explain its reduced cardiotoxicity.78 It is used in first line therapy for pediatric and adult acute leukemia79 and second line therapy for breast, prostate cancers and hematological malignancies.79 Mitoxantrone is also approved for worsening forms of multiple sclerosis since 2000. Mitoxantrone has to be used with caution because of its risks of cardiotoxicity and secondary leukemia in relationship with Top2β poisoning.94
Four Top2-targeted drugs are presently in clinical development: F14512, vesaroxin, C-1311 and XK469.11 F14512 is a demethylepipodophyllotoxin derivative with a spermine side chain (Fig. 3D) targeting cells overexpressing the polyamine transport system (PTS).11 F14512 also binds Top2cc more persistently than etoposide probably because of the DNA binding of its spermine moiety. Voreloxin is an intercalative quinolone derivative in Phase II–III clinical development in combination with cytarabine for relapsing and refractory acute myeloblastic leukemia.95 DNA intercalation is important for its activity. C-1311 (Symadex; Fig. 3F) is an iminodazoacridinone derivative with tight DNA interactions both by DNA intercalation and possibly alkylation (reviewed in 11). The fourth Top2-targeted drug in clinical trials is the quinoxaline R(+)-XK469 (Fig. 3F), which has been reported to target Top2β specifically.96 However, its mechanism of action is complex with reported inhibition of protein kinases such as MEK, ERK and cdk1 (see 11).
ICRF-187 (Dexrazoxane) (Fig. 3G) differs from the other Top2-targeted drugs because it acts as a catalytic inhibitor rather than by trapping Top2cc.1,5,12,97 It is not used as a cytotoxic anticancer agent but as a modulator of anthracyclines’ cardiotoxicity78 and to treat extravasations resulting from intravenous anthracycline injections. Merbarone (Fig. 3G) is another catalytic inhibitor of Top2 useful for cellular and mechanistic studies.12 Finally, recent studies suggested that sodium salicylate, aspirine’s active component can act as catalytic Top2 inhibitor.98
7. Top2-targeted antibiotics
Bacterial type IIA topoisomerases (Fig. 1 and Table 1) [for details see 5,9,99,100] have been the target of antibiotics since the discovery of the antibacterial activity of novobiocin, coumermycin and nalidixic acid in the 1960s. Quinolones target the GyrA subunit of gyrase and the ParC subunit of Topo IV5 by interfacial inhibition39–41 (see section 36). Since coumermycin was never developed for clinical use and novobiocin has been withdrawn from the market, there are presently no clinical antibiotic targeting the GyrB subunit of gyrase (Table 1).5.
Prokaryotic Top2s are excellent targets because: 1) they are essential in all bacteria; 2) cleavage complexes are bactericidal (not just bacteriostatic); 3) their targeting does not affect host human enzymes (their selectivity is at least three order of magnitude higher for prokaryotic over eukaryotic enzymes); 4) the high degree of homology between gyrase and Topo IV5 enables the targeting of both enzymes and therefore the killing of a broad spectrum of bacteria with a single drug.
Quinolones (Fig. 3H) are totally synthetic and are among the most successful antimicrobials both clinically and economically.9 The first quinolone was discovered 50 years ago as an impurity in a batch of chloroquine,101 14 years before the identification gyrase as its molecular target.102 Several generations of fluoroquinolones have evolved since the 1960s to extend their activity from Gram-negative urinary infections to Gram-positive bacteria and to a broad range of infections including anaerobic infections and multidrug-resistant tuberculosis (Fig. 3H).9
8. Challenges and speculations
This last section is a selected list of questions, challenges and possible answers regarding topoisomerase biology and drug targeting. Topics are treated independently from each other and can be read one at a time. They are also outlined in Table 2.
8.1. Lack of drugs targeting type IA topoisomerases (Table 2 # 1)
All bacteria contain type IA (Topo I and Topo III) along with type IIA topoisomerases (see Table 1). It is generally accepted that bacterial Topo I primary removes hypernegative supercoiling, while Topo III decatenates newly replicated intertwined daughter DNA molecules.15 A similar division of labor may apply to the two human topoisomerase III. As Top3α as a prevalent “post-replicative DNA hemicatenane resolvase” in association with BLM helicase (see section 2), one might speculate that Top3β is the prevalent “hypernegative DNA supercoiling relaxase”. The rationale for targeting bacterial type IA topoisomerases stems from the fact that Topo I trapping by genetic alterations,103 similar to the trapping of bacterial type IIA by interfacial inhibitor antibiotics, produces rapid bacterial cell death.104 However, there is presently no clinical drug available to target bacterial type IA topoisomerases and efforts have been limited, primarily spearheaded by Yuk-Chin Tse-Dinh.105 The query for such a novel class of antibiotics can benefit from the recent crystal structures of a covalent Topo I-DNA intermediate103 and Topo I and III complexes with single-stranded DNA.106,107
It would also be useful to have small molecule inhibitors for eukaryotic type IA enzymes to explore the biology of Top3α and Top3β in human cells (see Table 1) and potentially develop Top3 inhibitors as anticancer agents (Table 2, sections 1–2).
8.2. Selective targeting type IIA topoisomerases: Top2α versus Top2β (Table 2 #2)
The currently used Top2-targeted anticancer drugs are dual inhibitors of Top2α and β.1,59,90,91 Yet, Top2β rather than Top2α is implicated in the adverse effects of Top2-targeted drugs, namely the therapy-related acute myeloid leukemia (t-AML) resulting from balanced chromosome translocations involving the mixed-lineage leukemia locus (MLL) at chromosome 11q23 in one third of t-AML patients following etoposide and mitoxantrone treatments.93,94,108 A recent study also showed that Top2β poisoning is implicated in the cardiotoxicity of anthracyclines.83
In addition to these toxic side effects related to Top2β, the fact that Top2α (TOP2A) is highly expressed and sometimes amplified in tumors such as breast and colon cancers along with ERB2,109,110 and that Top2β is acting as a housekeeping gene with broad roles at promoter sites,108,111 legitimates the discovery and design of drugs selective for Top2α versus Top2β. This conclusion questions the validity of further developing the Top2β-specific inhibitor R(+)XK469 (see Fig. 3F and section 6).96 The challenge of finding Top2α-specific inhibitors should be relatively easily achievable as both Top2 enzymes are readily available for biochemical and screening assays. Second, rationale drug development could take into consideration the prior knowledge that anthracyclines (Fig. 3C) tend to be Top2α-specific whereas demethylepipodophyllotoxin derivatives (Fig. 3D) are very effective against Top2β. Third, recent drug-enzyme-DNA co-crystal structures could be used to rationalize the chemical design of Top2α-specific drugs.24,42
8.3. Targeted drug delivery and therapeutic index enhancement (Table 2 #2)
Topoisomerase inhibitors are effective but suffer from limited tumor selectivity. Their side effects and dose-limiting toxicities are due to the poisoning of normal cells, which, like cancer cells, require topoisomerases for survival and growth, and are often primed for apoptosis.64 This is especially the case of bone marrow progenitor and rapidly dividing intestinal cells. In fact, only doubling the maximal-tolerated drug dose might be sufficient to markedly improve the therapeutic index of topoisomerase inhibitors. For instance, mice, whose bone marrow tolerate camptothecin better than humans can be cured with camptothecins because they tolerate higher drug exposures. 65 A similarly enhanced therapeutic index might also account for the activity of camptothecins in pediatric tumors because children tend to tolerate relatively higher drug exposures than adults.
Several approaches are being implemented to target topoisomerase inhibitors to tumors while sparing normal tissues. Etirinotean pegol (NKTR-102) couples the active metabolite of irinotecan (SN-38) to polyethylene glycol, limiting the release of SN-38 to normal tissues with tight vasculature, whereas the drug is released into tumors by their intrinsically leaky blood vessels. A comparable approach is to encapsulate topoisomerase inhibitors into nanoparticles, which are preferentially sequestered in tumors with uneven blood flow and taken up by tumor cells. The ultimate approach would be to take advantage of tumor-specific components selectively expressed at the surface of tumor cells. In which case, the nanoparticles could be designed to dock with such cell surface receptors to deliver their drug payload to tumor cells. An original approach is being pursued for the Top2-targeted demethylepipodophyllotxin derivative, F14512 (see section 6). Attaching a polyamine (spermine) via a glycine linked (Fig. 3D) drives the update of the drug to cells overexpressing the polyamine transport system (PTS). This “Trojan horse” approach11,112 is giving promising results in AML clinical trials where the patients with leukemic cells with enhanced PTS are being identified with a (99m)Tc-HYNIC-spermine scintigraphic probe. 113 Finally, to our knowledge, no attempt is being made to use topoisomerase inhibitors as cytotoxins in antibody-drug conjugates (ADC).114. This is probably justified by the fact that topoisomerase-targeted drugs are active at micromolar rather than picomolar concentrations and therefore would require a heavy payload for the ADC approach to work.
8.4. Elucidating and targeting the repair pathways for topoisomerase-induced DNA damage: tyrosyl-DNA-phosphodiesterases (Table 2 #3–5)
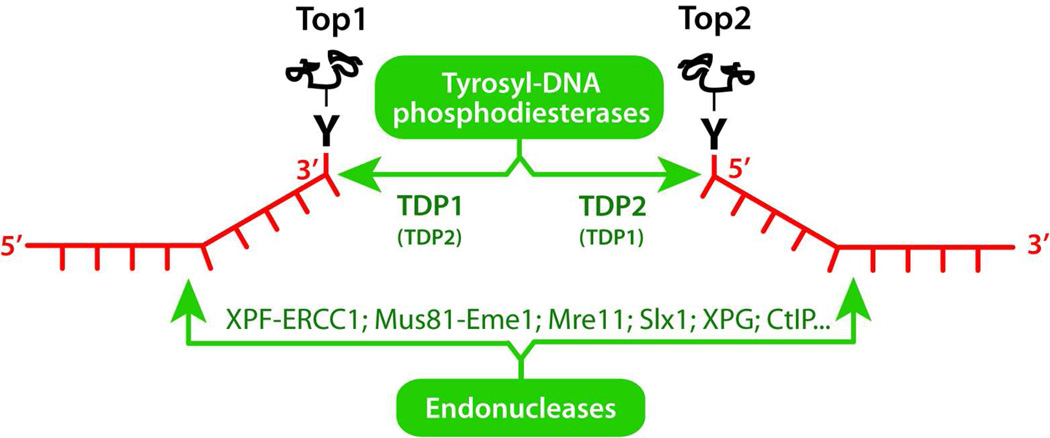
Because of the covalent attachment of topoisomerases to one of the cleaved DNA ends (see Figs. 1 & 2), cells need to remove Top1 from the 3’-ends and Top2 from the 5’-ends. Two main mechanisms are used (Fig. 4): precise cleavage of the tyrosyl-DNA bond by phosphodiesterases or endonuclease cleavage and elimination of the DNA strand attached to the topoisomerase.
Figure 4.
Schematic representation of the two main repair pathways removing topoisomerase-DNA complexes.
Tyrosyl-DNA-phosphodiesterase I (TDP1)115,116 was discovered by Nash and coworkers.117,118 Although TDP1 is conserved from yeast to humans, it is dispensable for the repair of Top1-mediated DNA damage because parallel pathways mostly represented by endonucleases (Fig. 4) are used to excise the Top1-DNA adducts. This explains why cells are selectively sensitive to TDP1 inactivation when they are also deficient in endonuclease pathways such as XPF/ERCC1 (Rad1/Rad10 in yeast) 119,120 or Mus81/Eme1 (Mus81/Mms4 in yeast) 121 or when they are deficient in cell cycle checkpoints.122 TDP1 functions in coordination with other repair complexes. Before TDP1 can process the tyrosyl-DNA bond, Top1 needs to be denatured or/and felled by the proteasome.123–126 TDP1 acts in a complex with XRCC1, PNK, ligase III and PARP.127 PNK is required to remove the 3’-phosphate left by TDP1 before DNA ligase(s) and polymerase(s) can process the 3’ terminus. Recent studies suggest that the coordinated functions of TDP1 and PARP can account for the potentiating effect of PARP inhibitors in combination with Top1 inhibitors.120 The discovery of TDP1 inhibitors is ongoing.116 They would be particularly suited in combination with Top1 inhibitors for patients whose tumors are deficient in the endonuclease pathways (ERCC1-XPF, Mre11, Mus81-Eme1, CtIP).120 The existence of robust biochemical assays and crystal structures of TDP1 bound to its tyrosyl-DNA substrate128–130 should facilitate the optimization and discovery of TDP1 inhibitors.
Tyrosyl-DNA-phosphodiesterase 2 (TDP2) was discovered more recently by Caldecott and coworkers after finding that the polypeptide encoded by TTRAP (TRAF and TNF receptor-associated protein), and previously associated with cellular stress responses and inhibition of NFkB activation, was the prevalent cellular 5’-tyrosyl phosphodiesterase responsible for resistance to Top2cc-targeted drugs.131 TDP2 has also been linked to viral replication.132–134 Because TDP2 generates 5’-phosphate termini, it is conceivable that TDP2 functions in coordination with the NHEJ repair pathways including Ku and DNA-PK. The screening of TDP2 inhibitors has just begun, and, as in the case of TDP1 should be facilitated by the recent elucidation of TDP2 crystal structures.135,136
It is important to stress that TDP1 and TDP2 are mechanistically and structurally very different despite they both function as monomers, prefer single-stranded DNA substrates,137,138 and can serve as a backup for each other.131,137,139,140 TDP1 functions without divalent metal in a two-step reaction involving a transient covalent intermediate between the DNA 3’-end and its catalytic histidine (H263 in humans). The release of TDP1 from the 3’-DNA end requires a second histidine (H493 in humans) whose mutation is the cause of the neurodegenerative disease SCAN1.141,142 (for scheme see 116) TDP1 structure shows a pseudo-dimeric fold with a catalytic site formed by the juxtaposition of 2 HKD motifs.116,129,143 On the other hand, TDP2 belongs to the Ape1, DNase I superfamily and uses Mg2+/Mn2+ coordination to hydrolyze 5’-phospho-tyrosyl bonds in one step (without covalent intermediate).138
In spite of the detailed knowledge of TDP1 and TDP2 molecular biology, less is known regarding the integration of TDP’s in the cellular repair pathways; i.e. which repair components are upstream and downstream of and parallel to their activities. More is presently known for TDP1, which is part of the XRCC1 repair complex with ligase III, PNK and PARP (see above), than for TDP2. Yet, both enzymes appear to function downstream from the proteasome. The relationship between the TDP’s and the non-homologous end-joining (NHEJ) and homologous recombination (HR) pathways remains to be clarified. The impact of proteasome inhibitors on the repair of topoisomerase cleavage complexes is also a potentially interesting avenue.120,126,144–147 Finally, further investigations are needed to elucidate the cellular cofactors and regulators of TDP2.
8.5. Role of poly(ADP-ribose) polymerases (PARP) in the repair of topoisomerase-induced DNA damage and rational for combination therapy (Table 2 #5)
PARP inhibitors are highly synergistic with Top1 inhibitors but not with Top2 inhibitors, which fits with PARP activation by Top1 but not by Top2 inhibitors.120,148,149 PARP activation by Top1cc is both transcription- and replication-dependent120 and tightly coupled with TDP1 activity (our unpublished data). One possible model is that conversion of Top1cc into DNA damage by transcription and replication collisions recruits the proteasome, which prepares the DNA ends for processing by the TDP1-PARP complex, in which PARP acts as a cofactor of TDP1 to facilitate its stability and recruitment to the DNA damage sites along with XRCC1, PNK and ligase III. Combination of both PARP and TDP1 inhibitors together with Top1 inhibitors is unlikely to be synergistic because of the overlapping (epistatic) roles of TDP1 and PARP in the repair of Top1cc. On the other hand, either TDP1 or PARP inhibitors are likely to be beneficial in tumors with endonuclease (ERCC1) defects (see Fig. 4 and section 8.4 above).
8.6. Clinical determinants of response to anticancer Top1 and Top2 inhibitors and precision medicine (Table 2 #3–5)
Topoisomerase inhibitors are effective chemotherapies that should only be prescribed to patients who should benefit from the drugs. Otherwise, ineffective regimens delay access to the correct treatment, select for drug resistance and produce costly side effects. Because of the redundant repair pathways involved in the survival of cancer cells targeted by topoisomerase inhibitors, it has been difficult to pinpoint single determinants of response to anticancer topoisomerase inhibitors. Topoisomerases are required,34,44–46,52–54 yet there is no simple linear correlation between topoisomerase levels and drug response.60,61 Two main approaches are should define cancer-related defects predicting drug response (or lack of). First, step-by-step molecular biology analyses of the DNA repair (TDP’s, endonucleases, double-strand break repair; see above) and stress response (cell cycle checkpoints, survival and death) pathways should build the cellular response network and identify the most critical parameters that determine the cellular response to topoisomerase inhibitors. The second approach is to use the genomic analyses of tumors as in the TCGA programa and cell lines 68,150,151. For instance, siRNA screening recently identified the protein kinase TAK1152 and gene expression correlations identified the potential helicase and cell cycle regulator SLFN11 as critical determinants of response to topoisomerase inhibitors.150,153
In the future, precision medicineb with topoisomerase inhibitors will require the establishment of a genomic (or molecular biology) signature of the tumor that matches the activity of the drugs (Table 2 #4). In parallel, it is critical to set up pharmacodynamic biomarkers to monitor the response of the tumor within a few days after initiating the treatment. Such biomarkers could be related to topoisomerase and DNA damage response.60,61,72,154 Pursuit of therapy would then be based on quantitative tumor response.
Acknowledgements
We wish to thank all the members of the Laboratory of Molecular Pharmacology, past and present for their contribution and constructive suggestions. A special thank to C. Marchand for figure 2. Our studies are supported by the Center for Cancer Research (Z01 BC 0006161), National Cancer Institute.
Footnotes
References
- 1.Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer. 2009;9:338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nitiss JL. DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer. 2009;9:327–337. doi: 10.1038/nrc2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pommier Y. DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition. Chem Rev. 2009;109:2894–2902. doi: 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pommier Y, Cushman M. The indenoisoquinoline noncamptothecin topoisomerase I inhibitors: update and perspectives. Mol Cancer Ther. 2009;8:1008–1014. doi: 10.1158/1535-7163.MCT-08-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol. 2010;17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pommier Y, Marchand C. Interfacial inhibitors: targeting macromolecular complexes. Nat Rev Drug Discov. 2012;11:25–36. doi: 10.1038/nrd3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deweese JE, Osheroff N. The DNA cleavage reaction of topoisomerase II: wolf in sheep's clothing. Nucleic Acids Res. 2009;37:738–748. doi: 10.1093/nar/gkn937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q Rev Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 9.Mitscher LA. Bacterial topoisomerase inhibitors: quinolone and pyridone antibacterial agents. Chemical reviews. 2005;105:559–592. doi: 10.1021/cr030101q. [DOI] [PubMed] [Google Scholar]
- 10.Dalhoff A. Global fluoroquinolone resistance epidemiology and implictions for clinical use. Interdiscip Perspect Infect Dis. 2012;2012 doi: 10.1155/2012/976273. 976273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bailly C. Contemporary challenges in the design of topoisomerase II inhibitors for cancer chemotherapy. Chemical reviews. 2012;112:3611–3640. doi: 10.1021/cr200325f. [DOI] [PubMed] [Google Scholar]
- 12.Fortune JM, Osheroff N. Topoisomerase II as a target for anticancer drugs: when enzymes stop being nice. Prog Nucleic Acid Res Mol Biol. 2000;64:221–253. doi: 10.1016/s0079-6603(00)64006-0. [DOI] [PubMed] [Google Scholar]
- 13.Forterre P, Gribaldo S, Gadelle D, Serre MC. Origin and evolution of DNA topoisomerases. Biochimie. 2007;89:427–446. doi: 10.1016/j.biochi.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 14.Forterre P. In: DNA Topoisomerases and Cancer. Pommier Y, editor. Springer; 2012. [Google Scholar]
- 15.Viard T, de la Tour CB. Type IA topoisomerases: a simple puzzle? Biochimie. 2007;89:456–467. doi: 10.1016/j.biochi.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 16.Gellert M. DNA topoisomerases. Annu. Rev. Biochem. 1981;50:879–910. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- 17.Wang JC. DNA topoisomerases. Annu. Rev. Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 18.Champoux JJ. DNA topoisomerases: Structure, Function, and Mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 19.Wang JC. Untangling the double helix. Cold Spring Harbor, New York: Cold Spring Harbor Press; 2009. [Google Scholar]
- 20.Redinbo MR, Stewart L, Champoux JJ, Hol WG. Structural Flexibility in Human Topoisomerase I Revealed in Multiple Non-isomorphous Crystal Structures. J Mol Biol. 1999;292:685–696. doi: 10.1006/jmbi.1999.3065. [DOI] [PubMed] [Google Scholar]
- 21.Koster DA, Croquette V, Dekker C, Shuman S, Dekker NH. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature. 2005;434:671–674. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- 22.Koster DA, Palle K, Bot ES, Bjornsti MA, Dekker NH. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature. 2007;448:213–217. doi: 10.1038/nature05938. [DOI] [PubMed] [Google Scholar]
- 23.Seol Y, Zhang H, Pommier Y, Neuman KC. A kinetic clutch governs religation by type IB topoisomerases and determines camptothecin sensitivity. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1206480109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt BH, Osheroff N, Berger JM. Structure of a topoisomerase II-DNA-nucleotide complex reveals a new control mechanism for ATPase activity. Nature structural & molecular biology. 2012;19:1147–1154. doi: 10.1038/nsmb.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schoeffler AJ, Berger JM. Recent advances in understanding structure-function relationships in the type II topoisomerase mechanism. Biochem. Soc. Trans. 2005;33:1465–1470. doi: 10.1042/BST0331465. [DOI] [PubMed] [Google Scholar]
- 26.Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 27.Pommier Y, Cherfils J. Interfacial protein inhibition: a nature's paradigm for drug discovery. Trends Pharmacol. Sci. 2005;28:136–145. doi: 10.1016/j.tips.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 28.Pommier Y, Marchand C. Interfacial inhibitors of protein-nucleic acid interactions. Curr. Med. Chem. Anti-Canc. Agents. 2005;5:421–429. doi: 10.2174/1568011054222337. [DOI] [PubMed] [Google Scholar]
- 29.Capranico G, Kohn KW, Pommier Y. Local sequence requirements for DNA cleavage by mammalian topoisomerase II in the presence of doxorubicin. Nucleic Acids Res. 1990;18:6611–6619. doi: 10.1093/nar/18.22.6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pommier Y, Capranico G, Orr A, Kohn KW. Local base sequence preferences for DNA cleavage by mammalian topoisomerase II in the presence of amsacrine or teniposide. Nucleic Acids Res. 1991;19:5973–5980. doi: 10.1093/nar/19.21.5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Capranico G, Binaschi M. DNA sequence selectivity of topoisomerases and topoisomerase poisons. Biochim. Biophys. Acta. 1998;1400:185–194. doi: 10.1016/s0167-4781(98)00135-3. [DOI] [PubMed] [Google Scholar]
- 32.Jaxel C, Capranico G, Kerrigan D, Kohn KW, Pommier Y. Effect of local DNA sequence on topoisomerase I cleavage in the presence or absence of camptothecin. J. Biol. Chem. 1991;266:20418–20423. [PubMed] [Google Scholar]
- 33.Wall ME, Wani MC. Camptothecin and taxol: discovery to clinic -- thirteenth Bruce F. Cain Memorial Award lecture. Cancer Res. 1995;55:753–760. [PubMed] [Google Scholar]
- 34.Pommier Y, Pourquier P, Urasaki Y, Wu J, Laco G. Topoisomerase I inhibitors: selectivity and cellular resistance. Drug Resist. Updat. 1999;2:307–318. doi: 10.1054/drup.1999.0102. [DOI] [PubMed] [Google Scholar]
- 35.Staker BL, Hjerrild K, Feese MD, Behnke CA, Burgin AB, Jr, Stewart L. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc. Natl. Acad. Sci. U S A. 2002;99:15387–15392. doi: 10.1073/pnas.242259599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staker BL, Feese MD, Cushman M, Pommier Y, Zembower D, Stewart L, Burgin AB. Structures of three classes of anticancer agents bound to the human topoisomerase I-DNA covalent complex. J. Med. Chem. 2005;48:2336–2345. doi: 10.1021/jm049146p. [DOI] [PubMed] [Google Scholar]
- 37.Ioanoviciu A, Antony S, Pommier Y, Staker BL, Stewart L, Cushman M. Synthesis and Mechanism of Action Studies of a Series of Norindenoisoquinoline Topoisomerase I Poisons Reveal an Inhibitor with a Flipped Orientation in the Ternary DNA-Enzyme-Inhibitor Complex As Determined by X-ray Crystallographic Analysis. J. Med. Chem. 2005;48:4803–4814. doi: 10.1021/jm050076b. [DOI] [PubMed] [Google Scholar]
- 38.Marchand C, Antony S, Kohn KW, Cushman M, Ioanoviciu A, Staker BL, Burgin AB, Stewart L, Pommier Y. A novel norindenoisoquinoline structure reveals a common interfacial inhibitor paradigm for ternary trapping of topoisomerase I-DNA covalent complexes. Mol Cancer Ther. 2006;5:287–295. doi: 10.1158/1535-7163.MCT-05-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laponogov I, Sohi MK, Veselkov DA, Pan XS, Sawhney R, Thompson AW, McAuley KE, Fisher LM, Sanderson MR. Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat Struct Mol Biol. 2009;16:667–669. doi: 10.1038/nsmb.1604. [DOI] [PubMed] [Google Scholar]
- 40.Laponogov I, Pan XS, Veselkov DA, McAuley KE, Fisher LM, Sanderson MR. Structural Basis of Gate-DNA Breakage and Resealing by Type II Topoisomerases. PLoS ONE. 2010;5:8. doi: 10.1371/journal.pone.0011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bax BD, Chan PF, Eggleston DS, Fosberry A, Gentry DR, Gorrec F, Giordano I, Hann MM, Hennessy A, Hibbs M, Huang J, Jones E, Jones J, Brown KK, Lewis CJ, May EW, Saunders MR, Singh O, Spitzfaden CE, Shen C, Shillings A, Theobald AJ, Wohlkonig A, Pearson ND, Gwynn MN. Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature. 2010;466:935–940. doi: 10.1038/nature09197. [DOI] [PubMed] [Google Scholar]
- 42.Wu CC, Li TK, Farh L, Lin LY, Lin TS, Yu YJ, Yen TJ, Chiang CW, Chan NL. Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide. Science. 2011;333:459–462. doi: 10.1126/science.1204117. [DOI] [PubMed] [Google Scholar]
- 43.Hsiang YH, Hertzberg R, Hecht S, Liu LF. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J. Biol. Chem. 1985;260:14873–14878. [PubMed] [Google Scholar]
- 44.Eng WK, Faucette L, Johnson RK, Sternglanz R. Evidence that DNA topoisomerase I is necessary for the cytotoxic effects of camptothecin. Mol. Pharmacol. 1988;34:755–760. [PubMed] [Google Scholar]
- 45.Nitiss J, Wang JC. DNA topoisomerase-targeting antitumor drugs can be studied in yeast. Proc. Natl. Acad. Sci. U S A. 1988;85:7501–7505. doi: 10.1073/pnas.85.20.7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miao ZH, Player A, Shankavaram U, Wang YH, Zimonjic DB, Lorenzi PL, Liao ZY, Liu H, Shimura T, Zhang HL, Meng LH, Zhang YW, Kawasaki ES, Popescu NC, Aladjem MI, Goldstein DJ, Weinstein JN, Pommier Y. Nonclassic Functions of Human Topoisomerase I: Genome-Wide and Pharmacologic Analyses. Cancer Res. 2007;67:8752–8761. doi: 10.1158/0008-5472.CAN-06-4554. [DOI] [PubMed] [Google Scholar]
- 47.Covey JM, Jaxel C, Kohn KW, Pommier Y. Protein-linked DNA strand breaks induced in Mammalian cells by camptothecin, an inhibitor of topoisomerase I. Cancer Res. 1989;49:5016–5022. [PubMed] [Google Scholar]
- 48.Subramanian D, Kraut E, Staubus A, Young DC, Muller MT. Analysis of topoisomerase I/DNA complexes in patients administered topotecan. Cancer Res. 1995;55:2097–2103. [PubMed] [Google Scholar]
- 49.Padget K, Carr R, Pearson AD, Tilby MJ, Austin CA. Camptothecin-stabilised topoisomerase I-DNA complexes in leukaemia cells visualised and quantified in situ by the TARDIS assay (trapped in agarose DNA immunostaining) Biochem Pharmacol. 2000;59:629–638. doi: 10.1016/s0006-2952(99)00372-x. [DOI] [PubMed] [Google Scholar]
- 50.Dexheimer TS, Pommier Y. DNA cleavage assay for the identification of topoisomerase I inhibitors. Nat. Protoc. 2008;3:1736–1750. doi: 10.1038/nprot.2008.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Antony S, Agama KK, Miao ZH, Takagi K, Wright MH, Robles AI, Varticovski L, Nagarajan M, Morrell A, Cushman M, Pommier Y. Novel indenoisoquinolines NSC 725776 and NSC 724998 produce persistent topoisomerase I cleavage complexes and overcome multidrug resistance. Cancer Res. 2007;67:10397–10405. doi: 10.1158/0008-5472.CAN-07-0938. [DOI] [PubMed] [Google Scholar]
- 52.Rasheed ZA, Rubin EH. Mechanisms of resistance to topoisomerase I-targeting drugs. Oncogene. 2003;22:7296–7304. doi: 10.1038/sj.onc.1206935. [DOI] [PubMed] [Google Scholar]
- 53.Beretta GL, Perego P, Zunino F. Mechanisms of cellular resistance to camptothecins. Current medicinal chemistry. 2006;13:3291–3305. doi: 10.2174/092986706778773121. [DOI] [PubMed] [Google Scholar]
- 54.Burgess DJ, Doles J, Zender L, Xue W, Ma B, McCombie WR, Hannon GJ, Lowe SW, Hemann MT. Topoisomerase levels determine chemotherapy response in vitro and in vivo. Proc Natl Acad Sci U S A. 2008;105:9053–9058. doi: 10.1073/pnas.0803513105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O'Malley FP, Chia S, Tu D, Shepherd LE, Levine MN, Bramwell VH, Andrulis IL, Pritchard KI. Topoisomerase II Alpha and Responsiveness of Breast Cancer to Adjuvant Chemotherapy. J. Natl. Cancer Inst. 2009;101:644–650. doi: 10.1093/jnci/djp067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tewey KM, Rowe TC, Yang L, Halligan BD, Liu LF. Adriamycin-induced DNA damage mediated by mammalian DNA topoisomerase II. Science. 1984;226:466–468. doi: 10.1126/science.6093249. [DOI] [PubMed] [Google Scholar]
- 57.Capranico G, Riva A, Tinelli S, Dasdia T, Zunino F. Markedly reduced levels of anthracycline-induced DNA strand breaks in resistant P388 leukemia cells and isolated nuclei. Cancer Research. 1987;47:3752–3756. [PubMed] [Google Scholar]
- 58.Kohn KW. Beyond DNA cross-linking: history and prospects of DNA-targeted cancer treatment--fifteenth Bruce F. Cain Memorial Award Lecture. Cancer Res. 1996;56:5533–5546. [PubMed] [Google Scholar]
- 59.Willmore E, Frank AJ, Padget K, Tilby MJ, Austin CA. Etoposide targets topoisomerase IIalpha and IIbeta in leukemic cells: isoform-specific cleavable complexes visualized and quantified in situ by a novel immunofluorescence technique. Mol Pharmacol. 1998;54:78–85. doi: 10.1124/mol.54.1.78. [DOI] [PubMed] [Google Scholar]
- 60.Pfister TD, Reinhold WC, Agama K, Gupta S, Khin SA, Kinders RJ, Parchment RE, Tomaszewski JE, Doroshow JH, Pommier Y. Topoisomerase I levels in the NCI-60 cancer cell line panel determined by validated ELISA and microarray analysis and correlation with indenoisoquinoline sensitivity. Molecular Cancer Therapeutics. 2009;8:1878–1884. doi: 10.1158/1535-7163.MCT-09-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pfister TD, Hollingshead M, Kinders RJ, Zhang Y, Evrard YA, Ji J, Khin SA, Borgel S, Stotler H, Carter J, Divelbiss R, Kummar S, Pommier Y, Parchment R, Tomaszewski JE, Doroshow JH. Development and Validation of a Pharmacodynamic Immunoassay for Topoisomerase I in Mouse Xenografts for Preclinical Modeling and Clinical Trials. PLoS ONE. 2012 doi: 10.1371/journal.pone.0050494. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murai J, Huang S-y N, Das BB, Renaud A, Zhang Y, Doroshow JH, Ji J, Takeda S, Pommier Y. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Research. 2012;72:5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Makeyev Y, Muggia FM, Rajan A, Giaccone G, Furuta T, Rougier P. In: DNA Topoisomerases and Cancer. Pommier Y, editor. New York: Springer; 2012. [Google Scholar]
- 64.Vo TT, Ryan J, Carrasco R, Neuberg D, Rossi DJ, Stone RM, Deangelo DJ, Frattini MG, Letai A. Relative Mitochondrial Priming of Myeloblasts and Normal HSCs Determines Chemotherapeutic Success in AML. Cell. 2012;151:344–355. doi: 10.1016/j.cell.2012.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Giovanella BC, Stehlin JS, Wall ME, Wani MC, Nicholas AW, Liu LF, Silber R, Potmesil M. DNA topoisomerase I-targeted chemotherapy of human colon cancer in xenografts. Science. 1989;246:1046–1048. doi: 10.1126/science.2555920. [DOI] [PubMed] [Google Scholar]
- 66.Brangi M, Litman T, Ciotti M, Nishiyama K, Kohlhagen G, Takimoto C, Robey R, Pommier Y, Fojo T, Bates SE. Camptothecin resistance: role of the ATP-binding cassette (ABC), mitoxantrone-resistance half-transporter (MXR), and potential for glucuronidation in MXR-expressing cells. Cancer Res. 1999;59:5938–5946. [PubMed] [Google Scholar]
- 67.Teicher BA. Next generation topoisomerase I inhibitors: Rationale and biomarker strategies. Biochem Pharmacol. 2008;75:1262–1271. doi: 10.1016/j.bcp.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 68.Reinhold WC, Sunshine M, Liu H, Varma S, Kohn KW, Morris J, Doroshow J, Pommier Y. CellMiner: A Web-Based Suite of Genomic and Pharmacologic Tools to Explore Transcript and Drug Patterns in the NCI-60 Cell Line Set. Cancer Research. 2012;72:3499–3511. doi: 10.1158/0008-5472.CAN-12-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kohlhagen G, Paull K, Cushman M, Nagafufuji P, Pommier Y. Protein-linked DNA strand breaks induced by NSC 314622, a non-camptothecin topoisomerase I poison. Mol. Pharmacol. 1998;54:50–58. doi: 10.1124/mol.54.1.50. [DOI] [PubMed] [Google Scholar]
- 70.Pommier Y, Cushman M. 2009:1008–1014. doi: 10.1158/1535-7163.MCT-08-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tanizawa A, Fujimori A, Fujimori Y, Pommier Y. Comparison of topoisomerase I inhibition, DNA damage, and cytotoxicity of camptothecin derivatives presently in clinical trials. J. Natl. Cancer Inst. 1994;86:836–842. doi: 10.1093/jnci/86.11.836. [DOI] [PubMed] [Google Scholar]
- 72.Kinders R, Hollingshead MG, Lawrence S, Ji J, Tabb B, Bonner WM, Pommier Y, Rubinstein L, Evrard YA, Parchment RE, Tomaszewski JE, Doroshow JH. Development of a Validated Immunofluorescence Assay for {gamma}H2AX as a Pharmacodynamic Marker of Topoisomerase I Inhibitor Activity. Clin Cancer Res. 2010 doi: 10.1158/1078-0432.CCR-09-3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sooryakumar D, Dexheimer TS, Teicher BA, Pommier Y. Molecular and cellular pharmacology of the novel noncamptothecin topoisomerase I inhibitor genz-644282. Mol Cancer Ther. 2011;10:1490–1499. doi: 10.1158/1535-7163.MCT-10-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Burden DA, Osheroff N. Mechanism of action of eukaryotic topoisomerase II and drugs targeted to the enzyme. Biochim. Biophys. Acta. 1998;1400:139–154. doi: 10.1016/s0167-4781(98)00132-8. [DOI] [PubMed] [Google Scholar]
- 75.Zwelling LA, Michaels S, Erickson LC, Ungerleider RS, Nichols M, Kohn KW. Protein-associated deoxyribonucleic acid strand breaks in L1210 cells treated with the deoxyribonucleic acid intercalating agents 4'-(9-acridinylamino) methanesulfon-m-anisidide and adriamycin. Biochemistry. 1981;20:6553–6563. doi: 10.1021/bi00526a006. [DOI] [PubMed] [Google Scholar]
- 76.Long BH, Musial ST, Brattain MG. Comparison of cytotoxicity and DNA breakage activity of congeners of podophyllotoxin including VP16-213 and VM26: a quantitative structure-activity relationship. Biochemistry. 1984;23:1183–1188. doi: 10.1021/bi00301a024. [DOI] [PubMed] [Google Scholar]
- 77.Zechiedrich EL, Christiansen K, Andersen AH, Westergaard O, Osheroff N. Double-stranded DNA cleavage/religation of eukaryotic topoisomerase II: evidence for a nicked DNA intermediate. Biochemistry. 1989;28:6229–6236. doi: 10.1021/bi00441a014. [DOI] [PubMed] [Google Scholar]
- 78.Doroshow JH. In: Cancer chemotherapy and biotherapy. second ed. Chabner BA, Longo DL, editors. Philadelphia: Lippincott-Raven; 1996. [Google Scholar]
- 79.Mir O, Dahut W, Goldwasser F, Heery C. In: DNA Topoisomerases and Cancer. Pommier Y, editor. New York: Springer, Humana Press; 2012. [Google Scholar]
- 80.Tewey KM, Chen GL, Nelson EM, Liu LF. Intercalative antitumor drugs interfere with the breakage-reunion reaction of mammalian DNA topoisomerase II. J Biol Chem. 1984;259:9182–9187. [PubMed] [Google Scholar]
- 81.Ross WE, Glaubiger DL, Kohn KW. Protein-associated DNA breaks in cells treated with adriamycin or ellipticine. Biochim. Biophys. Acta. 1978;519:23–30. doi: 10.1016/0005-2787(78)90059-x. [DOI] [PubMed] [Google Scholar]
- 82.Pommier Y, Schwartz RE, Kohn KW, Zwelling LA. Formation and rejoining of deoxyribonucleic acid double-strand breaks induced in isolated cell nuclei by antineoplastic intercalating agents. Biochemistry. 1984;23:3194–3201. doi: 10.1021/bi00309a013. [DOI] [PubMed] [Google Scholar]
- 83.Zhang S, Liu X, Bawa-Khalfe T, Lu LS, Lyu YL, Liu LF, Yeh ET. Identification of the molecular basis of doxorubicin-induced cardiotoxicity. Nat Med. 2012;18:1639–1642. doi: 10.1038/nm.2919. [DOI] [PubMed] [Google Scholar]
- 84.Low RL, Orton S, Friedman DB. A truncated form of DNA topoisomerase IIbeta associates with the mtDNA genome in mammalian mitochondria. Eur J Biochem. 2003;270:4173–4186. doi: 10.1046/j.1432-1033.2003.03814.x. [DOI] [PubMed] [Google Scholar]
- 85.Chen GL, Yang L, Rowe TC, Halligan BD, Tewey KM, Liu LF. Nonintercalative antitumor drugs interfere with the breakage-reunion reaction of mammalian DNA topoisomerase II. The Journal of biological chemistry. 1984;259:13560–13566. [PubMed] [Google Scholar]
- 86.Long BH, Musial ST, Brattain MG. Single- and double-strand DNA breakage and repair in human lung adenocarcinoma cells exposed to etoposide and teniposide. Cancer Res. 1985;45:3106–3112. [PubMed] [Google Scholar]
- 87.Kerrigan D, Pommier Y, Kohn KW. Protein-linked DNA strand breaks produced by etoposide and teniposide in Mouse L1210 and Human VA-13 and HT-29 cell lines: relationship to cytotoxicity. NCI Monographs. 1987;4:117–121. [PubMed] [Google Scholar]
- 88.Pommier Y, Capranico G, Orr A, Kohn KW. Distribution of topoisomerase II cleavage sites in simian virus 40 DNA and the effects of drugs. J Mol Biol. 1991;222:909–924. doi: 10.1016/0022-2836(91)90585-t. [DOI] [PubMed] [Google Scholar]
- 89.Pommier Y, Orr A, Kohn KW, Riou JF. Differential effects of amsacrine and epipodophyllotoxins on topoisomerase II cleavage in the human c-myc proto-oncogene. Cancer Research. 1992;52:3125–3130. [PubMed] [Google Scholar]
- 90.Cornarotti M, Tinelli S, Willmore E, Zunino F, Fisher LM, Austin CA, Capranico G. Drug sensitivity and sequence specificity of human recombinant DNA topoisomerases IIalpha (p170) and IIbeta (p180) Mol Pharmacol. 1996;50:1463–1471. [PubMed] [Google Scholar]
- 91.Willmore E, Errington F, Tilby MJ, Austin CA. Formation and longevity of idarubicin-induced DNA topoisomerase II cleavable complexes in K562 human leukaemia cells. Biochemical Pharmacology. 2002;63:1807–1815. doi: 10.1016/s0006-2952(02)00920-6. [DOI] [PubMed] [Google Scholar]
- 92.Felix CA, Kolaris CP, Osheroff N. Topoisomerase II and the etiology of chromosomal translocations. DNA Repair (Amst) 2006;5:1093–1108. doi: 10.1016/j.dnarep.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 93.Azarova AM, Lyu YL, Lin CP, Tsai YC, Lau JY, Wang JC, Liu LF. From the Cover: Roles of DNA topoisomerase II isozymes in chemotherapy and secondary malignancies. Proc Natl Acad Sci U S A. 2007;104:11014–11019. doi: 10.1073/pnas.0704002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cowell IG, Sondka Z, Smith K, Lee KC, Manville CM, Sidorczuk-Lesthuruge M, Rance HA, Padget K, Jackson GH, Adachi N, Austin CA. Model for MLL translocations in therapy-related leukemia involving topoisomerase IIbeta-mediated DNA strand breaks and gene proximity. Proc. Natl. Acad. Sci. U.S.A. 2012;109:8989–8994. doi: 10.1073/pnas.1204406109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hawtin RE, Stockett DE, Byl JA, McDowell RS, Nguyen T, Arkin MR, Conroy A, Yang W, Osheroff N, Fox JA. Voreloxin is an anticancer quinolone derivative that intercalates DNA and poisons topoisomerase II. PLoS ONE. 2010;5:e10186. doi: 10.1371/journal.pone.0010186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gao H, Huang KC, Yamasaki EF, Chan KK, Chohan L, Snapka RM. XK469, a selective topoisomerase IIbeta poison. Proc Natl Acad Sci U S A. 1999;96:12168–12173. doi: 10.1073/pnas.96.21.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Andoh T. Bis(2,6-dioxopiperazines), catalytic inhibitors of DNA topoisomerase II, as molecular probes, cardioprotectors and antitumor drugs. Biochimie. 1998;80:235–246. doi: 10.1016/s0300-9084(98)80006-0. [DOI] [PubMed] [Google Scholar]
- 98.Bau JT, Kurz EU. Sodium salicylate is a novel catalytic inhibitor of human DNA topoisomerase II alpha. Biochemical Pharmacology. 2011;81:345–354. doi: 10.1016/j.bcp.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 99.Maxwell A. DNA gyrase as a drug target. Biochem Soc Trans. 1999;27:48–53. doi: 10.1042/bst0270048. [DOI] [PubMed] [Google Scholar]
- 100.Drlica K, Hiasa H, Kerns R, Malik M, Mustaev A, Zhao X. Quinolones: action and resistance updated. Curr Top Med Chem. 2009;9:981–998. doi: 10.2174/156802609789630947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lesher GY, Froelich EJ, Gruett MD, Bailey JH, Brundage RP. 1,8-Naphthyridine Derivatives. A New Class of Chemotherapeutic Agents. J Med Pharm Chem. 1962;91:1063–1065. doi: 10.1021/jm01240a021. [DOI] [PubMed] [Google Scholar]
- 102.Gellert M, Mizuuchi K, O'Dea MH, Nash HA. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976;73:3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang Z, Cheng B, Tse-Dinh YC. Crystal structure of a covalent intermediate in DNA cleavage and rejoining by Escherichia coli DNA topoisomerase I. Proc Natl Acad Sci U S A. 2011;108:6939–6944. doi: 10.1073/pnas.1100300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cheng B, Shukla S, Vasunilashorn S, Mukhopadhyay S, Tse-Dinh YC. Bacterial cell killing mediated by topoisomerase I DNA cleavage activity. J Biol Chem. 2005;280:38489–38495. doi: 10.1074/jbc.M509722200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tse-Dinh YC. Bacterial topoisomerase I as a target for discovery of antibacterial compounds. Nucleic Acids Res. 2009;37:731–737. doi: 10.1093/nar/gkn936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Perry K, Mondragon A. Structure of a complex between E. coli DNA topoisomerase I and single-stranded DNA. Structure. 2003;11:1349–1358. doi: 10.1016/j.str.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 107.Changela A, DiGate RJ, Mondragon A. Structural studies of E. coli topoisomerase III-DNA complexes reveal a novel type IA topoisomerase-DNA conformational intermediate. J. Mol. Biol. 2007;368:105–118. doi: 10.1016/j.jmb.2007.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Haffner MC, Aryee MJ, Toubaji A, Esopi DM, Albadine R, Gurel B, Isaacs WB, Bova GS, Liu W, Xu J, Meeker AK, Netto G, De Marzo AM, Nelson WG, Yegnasubramanian S. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat Genet. 2010;42:668–675. doi: 10.1038/ng.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jarvinen TA, Liu ET. Simultaneous amplification of HER-2 (ERBB2) and topoisomerase IIalpha (TOP2A) genes--molecular basis for combination chemotherapy in cancer. Current cancer drug targets. 2006;6:579–602. doi: 10.2174/156800906778742497. [DOI] [PubMed] [Google Scholar]
- 110.Al-Kuraya K, Novotny H, Bavi P, Siraj AK, Uddin S, Ezzat A, Sanea NA, Al-Dayel F, Al-Mana H, Sheikh SS, Mirlacher M, Tapia C, Simon R, Sauter G, Terracciano L, Tornillo L. HER2, TOP2A, CCND1, EGFR and C-MYC oncogene amplification in colorectal cancer. J Clin Pathol. 2007;60:768–772. doi: 10.1136/jcp.2006.038281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 112.Kruczynski A, Vandenberghe I, Pillon A, Pesnel S, Goetsch L, Barret JM, Guminski Y, Le Pape A, Imbert T, Bailly C, Guilbaud N. Preclinical activity of F14512, designed to target tumors expressing an active polyamine transport system. Investigational new drugs. 2011;29:9–21. doi: 10.1007/s10637-009-9328-3. [DOI] [PubMed] [Google Scholar]
- 113.Pesnel S, Guminski Y, Pillon A, Lerondel S, Imbert T, Guilbaud N, Kruczynski A, Bailly C, Le Pape A. 99mTc-HYNIC-spermine for imaging polyamine transport system-positive tumours: preclinical evaluation. Eur J Nucl Med Mol Imaging. 2011;38:1832–1841. doi: 10.1007/s00259-011-1857-2. [DOI] [PubMed] [Google Scholar]
- 114.Hughes B. Antibody-drug conjugates for cancer: poised to deliver? Nature reviews. Drug discovery. 2010;9:665–667. doi: 10.1038/nrd3270. [DOI] [PubMed] [Google Scholar]
- 115.Dexheimer TS, Antony S, Marchand C, Pommier Y. Tyrosyl-DNA phosphodiesterase as a target for anticancer therapy. Anticancer Agents Med. Chem. 2008;8:381–389. doi: 10.2174/187152008784220357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Huang SY, Pommier Y, Marchand C. Tyrosyl-DNA Phosphodiesterase 1 (Tdp1) inhibitors. Expert Opin Ther Pat. 2011;21:1285–1292. doi: 10.1517/13543776.2011.604314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yang S-W, Burgin AB, Huizenga BN, Robertson CA, Yao KC, Nash HA. A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc. Natl. Acad. Sci. U.S.A. 1996;93:11534–11539. doi: 10.1073/pnas.93.21.11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pouliot JJ, Yao KC, Robertson CA, Nash HA. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topo I covalent complexes. Science. 1999;286:552–555. doi: 10.1126/science.286.5439.552. [DOI] [PubMed] [Google Scholar]
- 119.Vance JR, Wilson TE. Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc. Natl. Acad. Sci. U S A. 2002;99:13669–13674. doi: 10.1073/pnas.202242599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang YW, Regairaz M, Seiler JA, Agama KK, Doroshow JH, Pommier Y. Poly(ADP-ribose) polymerase and XPF-ERCC1 participate in distinct pathways for the repair of topoisomerase I-induced DNA damage in mammalian cells. Nucleic Acids Res. 2011;39:3607–3620. doi: 10.1093/nar/gkq1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Regairaz M, Zhang Y-W, Fu H, Agama KK, Tata N, Agrawal S, Aladjem MI, Pommier Y. Mus81-mediated DNA cleavage resolves replication forks stalled by topoisomerase I,ÄìDNA complexes. The Journal of Cell Biology. 2011;195:739–749. doi: 10.1083/jcb.201104003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pouliot JJ, Robertson CA, Nash HA. Pathways for repair of topoisomerase I covalent complexes in Saccharomyces cerevisiae. Genes Cells. 2001;6:677–687. doi: 10.1046/j.1365-2443.2001.00452.x. [DOI] [PubMed] [Google Scholar]
- 123.Beidler DR, Cheng YC. Camptothecin induction of a time- and concentration-dependent decrease of topoisomerase I and its implication in camptothecin activity. Mol Pharmacol. 1995;47:907–914. [PubMed] [Google Scholar]
- 124.Debethune L, Kohlhagen G, Grandas A, Pommier Y. Processing of nucleopeptides mimicking the topoisomerase I-DNA covalent complex by tyrosyl-DNA phosphodiesterase. Nucleic Acids Res. 2002;30:1198–1204. doi: 10.1093/nar/30.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang HF, Tomida A, Koshimizu R, Ogiso Y, Lei S, Tsuruo T. Cullin 3 promotes proteasomal degradation of the topoisomerase I-DNA covalent complex. Cancer Res. 2004;64:1114–1121. doi: 10.1158/0008-5472.can-03-2858. [DOI] [PubMed] [Google Scholar]
- 126.Lin CP, Ban Y, Lyu YL, Desai SD, Liu LF. A Ubiquitin-Proteasome Pathway for the Repair of Topoisomerase I-DNA Covalent Complexes. J. Biol. Chem. 2008;283:21074–21083. doi: 10.1074/jbc.M803493200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pommier Y, Barcelo JM, Rao VA, Sordet O, Jobson AG, Thibaut L, Miao ZH, Seiler JA, Zhang H, Marchand C, Agama K, Nitiss JL, Redon C. Repair of topoisomerase I-mediated DNA damage. Prog. Nucleic Acid Res. Mol. Biol. 2006;81:179–229. doi: 10.1016/S0079-6603(06)81005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Davies DR, Interthal H, Champoux JJ, Hol WG. Insights into substrate binding and catalytic mechanism of human tyrosyl-DNA phosphodiesterase (Tdp1) from vanadate and tungstate-inhibited structures. J Mol Biol. 2002;324:917–932. doi: 10.1016/s0022-2836(02)01154-3. [DOI] [PubMed] [Google Scholar]
- 129.Davies DR, Interthal H, Champoux JJ, Hol WG. The crystal structure of human tyrosyl-DNA phosphodiesterase, Tdp1. Structure (Camb) 2002;10:237–248. doi: 10.1016/s0969-2126(02)00707-4. [DOI] [PubMed] [Google Scholar]
- 130.Davies DR, Interthal H, Champoux JJ, Hol WG. Crystal structure of a transition state mimic for tdp1 assembled from vanadate, DNA, and a topoisomerase I-derived Peptide. Chem Biol. 2003;10:139–147. doi: 10.1016/s1074-5521(03)00021-8. [DOI] [PubMed] [Google Scholar]
- 131.Cortes Ledesma F, El Khamisy SF, Zuma MC, Osborn K, Caldecott KW. A human 5'-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009;461:674–678. doi: 10.1038/nature08444. [DOI] [PubMed] [Google Scholar]
- 132.Virgen-Slane R, Rozovics JM, Fitzgerald KD, Ngo T, Chou W, van der Heden van Noort GJ, Filippov DV, Gershon PD, Semler BL. An RNA virus hijacks an incognito function of a DNA repair enzyme. Proc. Natl. Acad. Sci. U. S. A. 2012 doi: 10.1073/pnas.1208096109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jones SA, Boregowda R, Spratt TE, Hu J. In vitro epsilon RNA-dependent protein priming activity of human hepatitis B virus polymerase. J Virol. 2012;86:5134–5150. doi: 10.1128/JVI.07137-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang JQ, Wang JJ, Li WJ, Huang L, Tian L, Xue JL, Chen JZ, Jia W. Cellular protein TTRAP interacts with HIV-1 integrase to facilitate viral integration. Biochemical and biophysical research communications. 2009;387:256–260. doi: 10.1016/j.bbrc.2009.06.153. [DOI] [PubMed] [Google Scholar]
- 135.Shi K, Kurahashi K, Gao R, Tsutakawa SE, Tainer JA, Pommier Y, Aihara H. Structural basis for recognition of 5'-phosphotyrosine adducts by Tdp2. Nature structural & molecular biology. 2012 doi: 10.1038/nsmb.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Schellenberg MJ, Appel CD, Adhikari S, Robertson PD, Ramsden DA, Williams RS. Mechanism of repair of 5'-topoisomerase II-DNA adducts by mammalian tyrosyl-DNA phosphodiesterase 2. Nature structural & molecular biology. 2012 doi: 10.1038/nsmb.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Murai J, Huang SY, Das BB, Dexheimer TS, Takeda S, Pommier Y. Tyrosyl-DNA phosphodiesterase 1 (TDP1) repairs DNA damage induced by topoisomerases I and II and base alkylation in vertebrate cells. The Journal of biological chemistry. 2012;287:12848–12857. doi: 10.1074/jbc.M111.333963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gao R, Huang SY, Marchand C, Pommier Y. Biochemical Characterization of Human Tyrosyl-DNA Phosphodiesterase 2 (TDP2/TTRAP): A Mg2+/Mn2+-DEPENDENT PHOSPHODIESTERASE SPECIFIC FOR THE REPAIR OF TOPOISOMERASE CLEAVAGE COMPLEXES. The Journal of biological chemistry. 2012;287:30842–30852. doi: 10.1074/jbc.M112.393983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nitiss KC, Malik M, He X, White SW, Nitiss JL. Tyrosyl-DNA phosphodiesterase (Tdp1) participates in the repair of Top2-mediated DNA damage. Proc Natl Acad Sci U S A. 2006;103:8953–8958. doi: 10.1073/pnas.0603455103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zeng Z, Sharma A, Ju L, Murai J, Umans L, Vermeire L, Pommier Y, Takeda S, Huylebroeck D, Caldecott KW, El-Khamisy SF. TDP2 promotes repair of topoisomerase I-mediated DNA damage in the absence of TDP1. Nucleic acids research. 2012 doi: 10.1093/nar/gks622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Takashima H, Boerkoel CF, John J, Saifi GM, Salih MA, Armstrong D, Mao Y, Quiocho FA, Roa BB, Nakagawa M, Stockton DW, Lupski JR. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat. Genet. 2002;32:267–272. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- 142.Interthal H, Chen HJ, Kehl-Fie TE, Zotzmann J, Leppard JB, Champoux JJ. SCAN1 mutant Tdp1 accumulates the enzyme--DNA intermediate and causes camptothecin hypersensitivity. Embo J. 2005;24:2224–2233. doi: 10.1038/sj.emboj.7600694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Interthal H, Pouliot JJ, Champoux JJ. The tyrosyl-DNA phosphodiesterase Tdp1 is a member of the phospholipase D superfamily. Proc. Natl. Acad. Sci. U S A. 2001;98:12009–12014. doi: 10.1073/pnas.211429198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Cusack JC, Jr, Liu R, Houston M, Abendroth K, Elliott PJ, Adams J, Baldwin AS., Jr Enhanced chemosensitivity to CPT-11 with proteasome inhibitor PS-341: implications for systemic nuclear factor-kappaB inhibition. Cancer Res. 2001;61:3535–3540. [PubMed] [Google Scholar]
- 145.Zhang A, Lyu YL, Lin CP, Zhou N, Azarova AM, Wood LM, Liu LF. A protease pathway for the repair of topoisomerase II-DNA covalent complexes. J Biol Chem. 2006;281:35997–36003. doi: 10.1074/jbc.M604149200. [DOI] [PubMed] [Google Scholar]
- 146.Lin CP, Ban Y, Lyu YL, Liu LF. Proteasome-dependent processing of topoisomerase I-DNA adducts into DNA double-strand breaks at arrested replication forks. J Biol Chem. 2009;284:28084–28092. doi: 10.1074/jbc.M109.030601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Takeshita T, Wu W, Koike A, Fukuda M, Ohta T. Perturbation of DNA repair pathways by proteasome inhibitors corresponds to enhanced chemosensitivity of cells to DNA damage-inducing agents. Cancer chemotherapy and pharmacology. 2009;64:1039–1046. doi: 10.1007/s00280-009-0961-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zwelling LA, Kerrigan D, Pommier Y, Michaels S, Steren A, Kohn KW. Formation and resealing of intercalator-induced DNA strand breaks in permeabilized L1210 cells without the stimulated synthesis of poly(ADP-ribose) J Biol Chem. 1982;257:8957–8963. [PubMed] [Google Scholar]
- 149.Bowman KJ, Newell DR, Calvert AH, Curtin NJ. Differential effects of the poly (ADP-ribose) polymerase (PARP) inhibitor NU1025 on topoisomerase I and II inhibitor cytotoxicity in L1210 cells in vitro. Br J Cancer. 2001;84:106–112. doi: 10.1054/bjoc.2000.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jane-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–307. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J, Liu Q, Iorio F, Surdez D, Chen L, Milano RJ, Bignell GR, Tam AT, Davies H, Stevenson JA, Barthorpe S, Lutz SR, Kogera F, Lawrence K, McLaren-Douglas A, Mitropoulos X, Mironenko T, Thi H, Richardson L, Zhou W, Jewitt F, Zhang T, O'Brien P, Boisvert JL, Price S, Hur W, Yang W, Deng X, Butler A, Choi HG, Chang JW, Baselga J, Stamenkovic I, Engelman JA, Sharma SV, Delattre O, Saez-Rodriguez J, Gray NS, Settleman J, Futreal PA, Haber DA, Stratton MR, Ramaswamy S, McDermott U, Benes CH. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Martin SE, Wu ZH, Gehlhaus K, Jones TL, Zhang YW, Guha R, Miyamoto S, Pommier Y, Caplen NJ. RNAi Screening Identifies TAK1 as a Potential Target for the Enhanced Efficacy of Topoisomerase Inhibitors. Curr Cancer Drug Targets. 2011;11:976–986. doi: 10.2174/156800911797264734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zoppoli G, Regairaz M, Leo E, Reinhold WC, Varma S, Ballestrero A, Doroshow JH, Pommier Y. Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc Natl Acad Sci U S A. 2012;109:15030–15035. doi: 10.1073/pnas.1205943109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kinders RJ, Hollingshead M, Khin S, Rubinstein L, Tomaszewski JE, Doroshow JH, Parchment RE. Preclinical modeling of a phase 0 clinical trial: qualification of a pharmacodynamic assay of poly (ADP-ribose) polymerase in tumor biopsies of mouse xenografts. Clin Cancer Res. 2008;14:6877–6885. doi: 10.1158/1078-0432.CCR-08-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Stewart L, Redinbo MR, Qiu X, Hol WG, Champoux JJ. A model for the mechanism of human topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]