Abstract
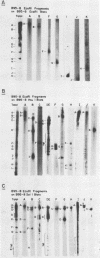
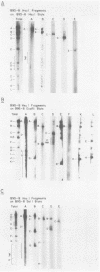
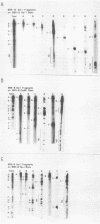
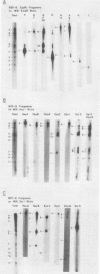
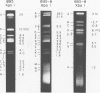
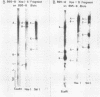
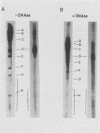
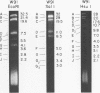
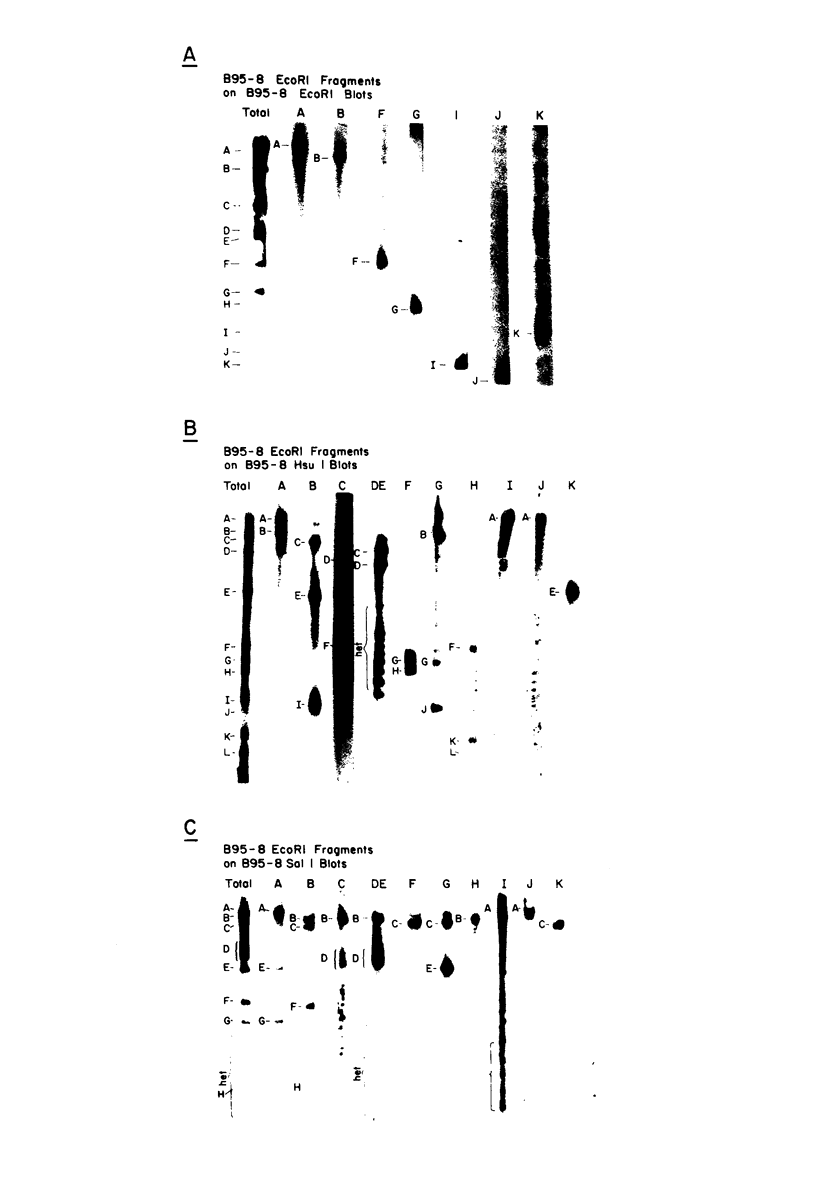
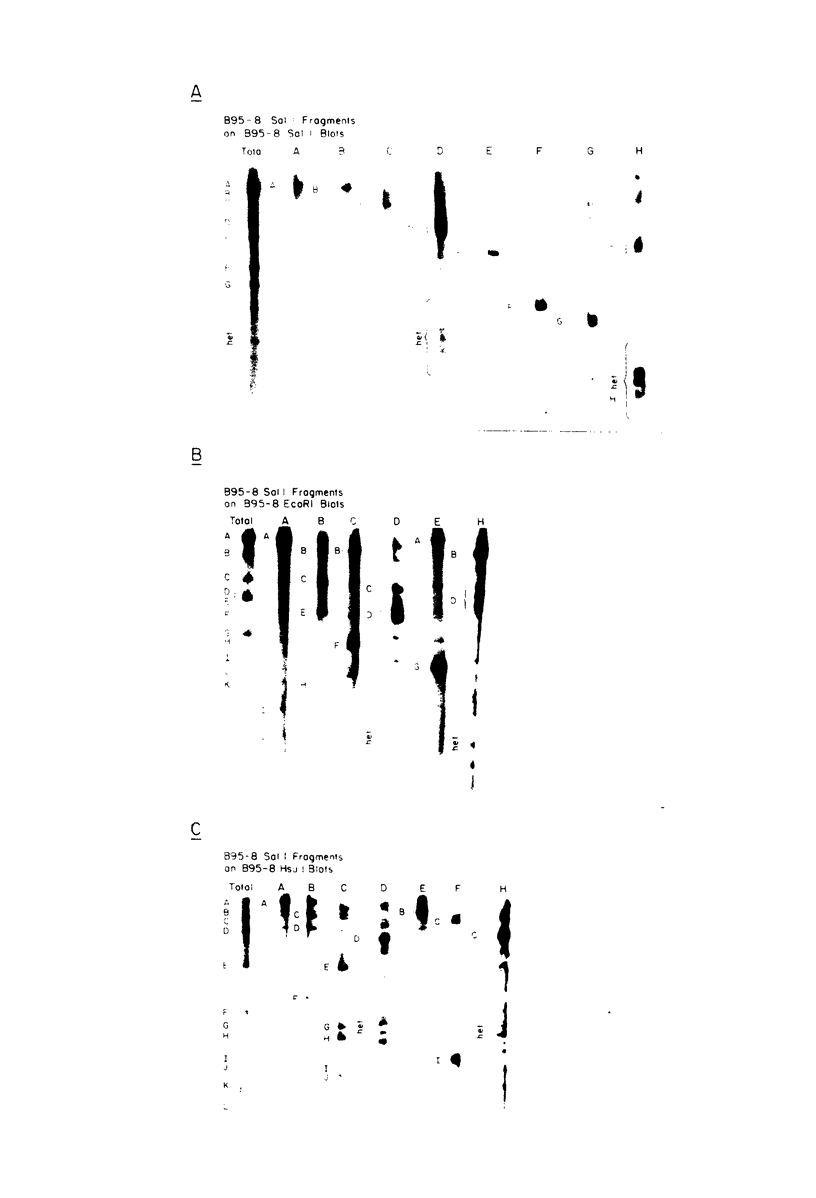
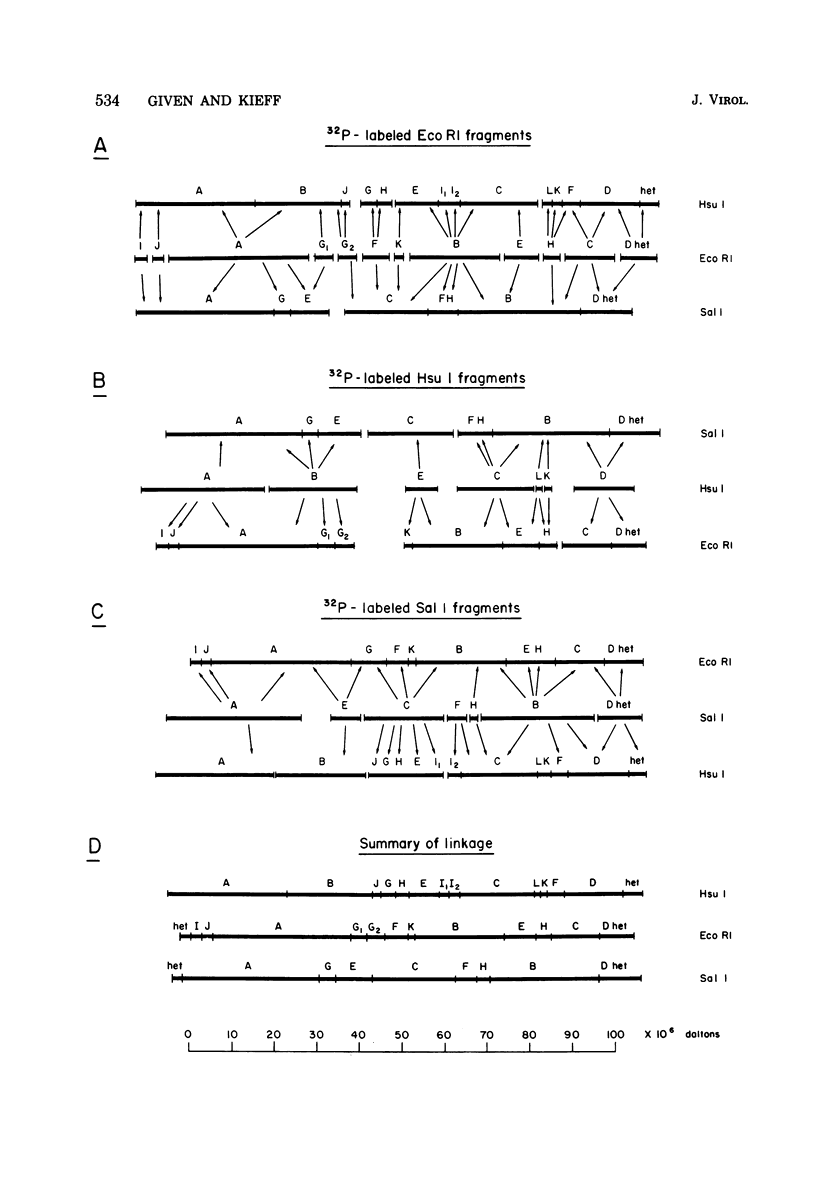
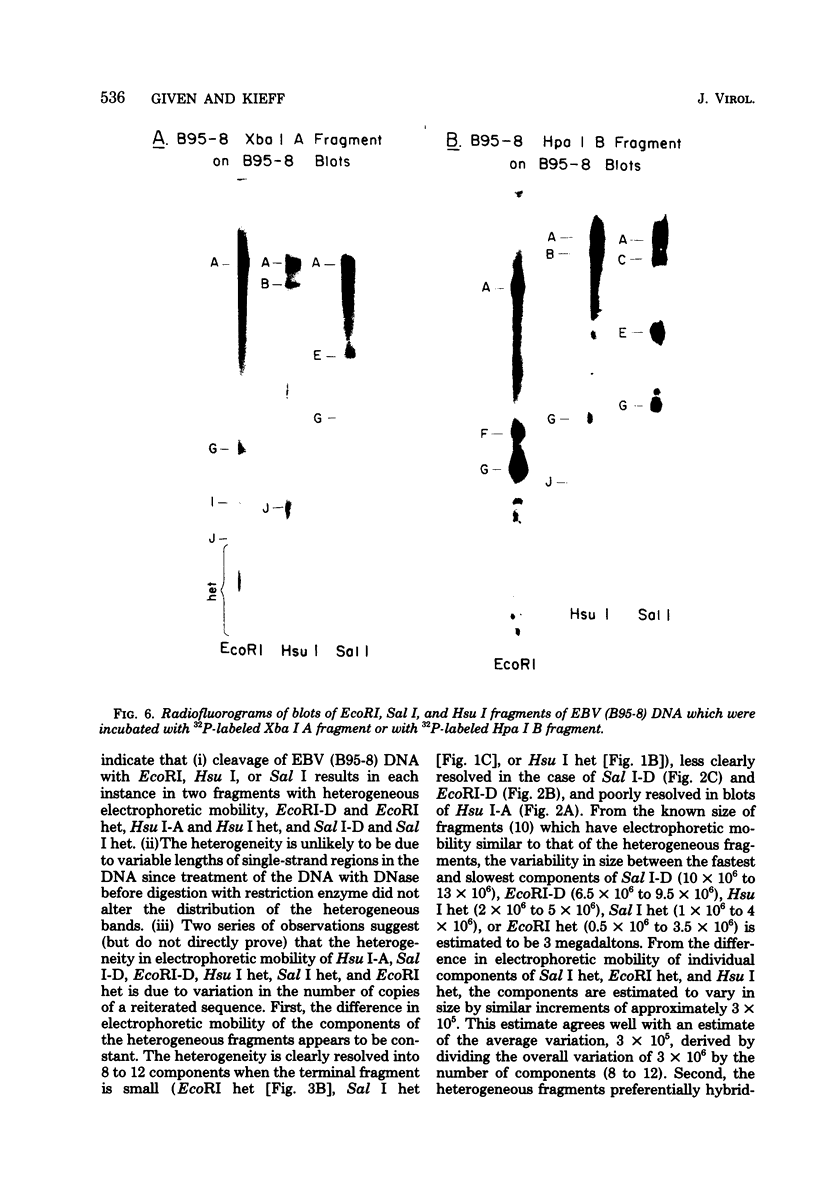
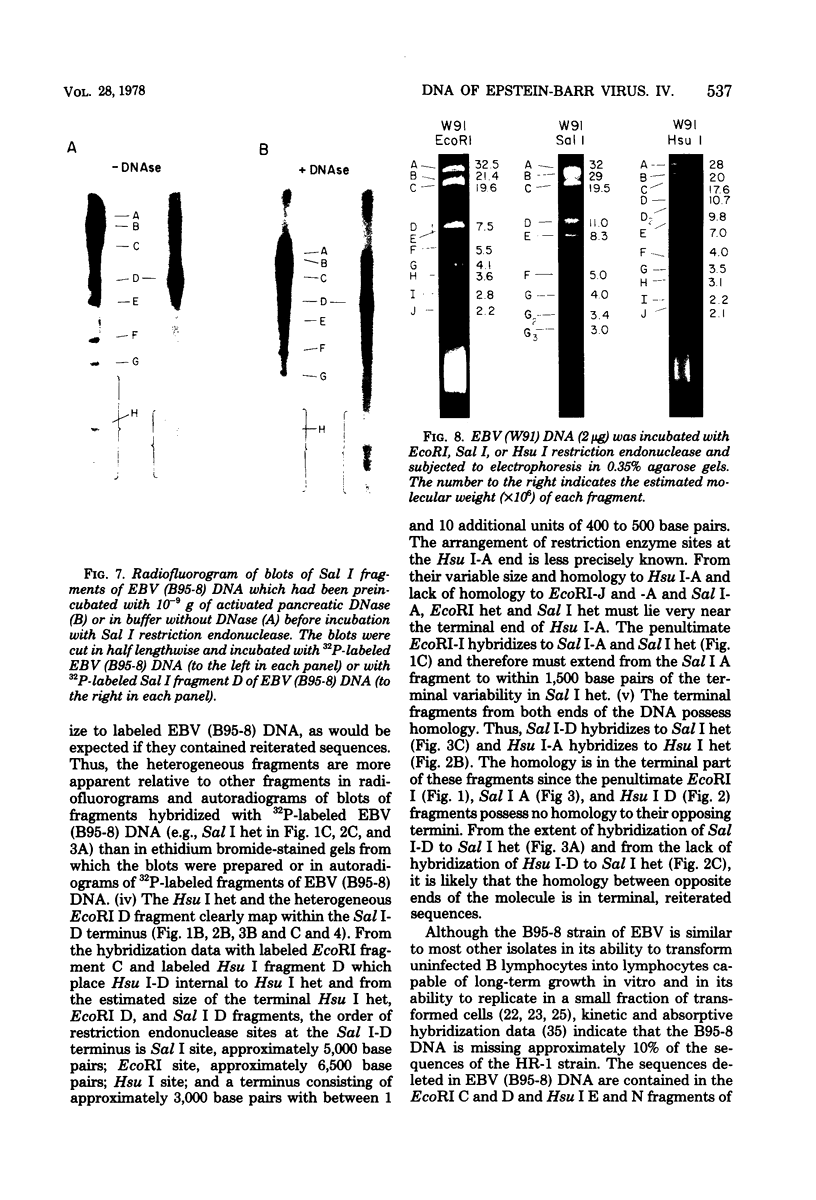
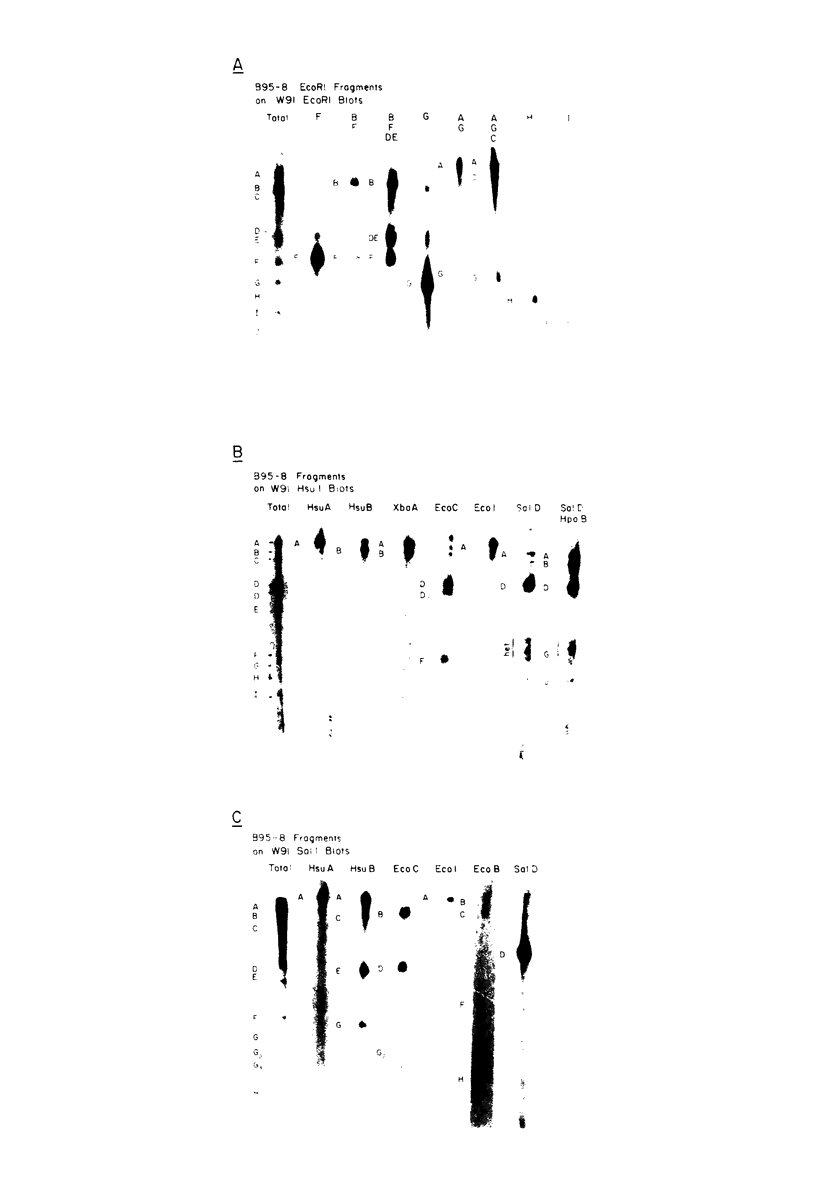
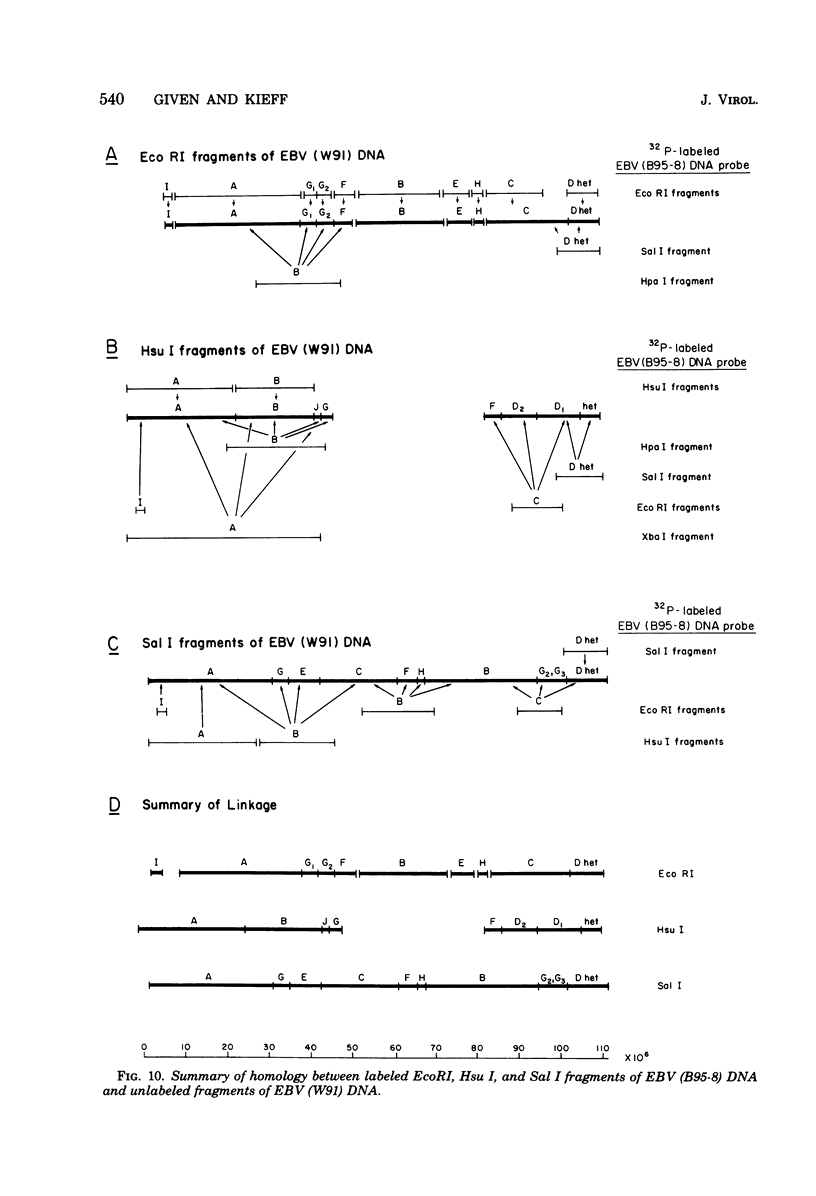
The arrangement of EcoRI, Hsu I, and Sal I restriction enzyme sites in the DNA of the B95-8 and W91 isolates of Epstein-Barr virus (EBV) has been determined from the size of the single-enzyme-cleaved fragments and from blot hybridizations that identify which fragments cut from the DNA with one enzyme contain nucleotide sequences in common with fragments cut from the DNA with a second enzyme. The DNA of the B95-8 isolate was the prototype for this study. The data indicate that (i) approximately 95 X 10(6) to 100 X 10(6) daltons of EBV (B95-8) DNA is in a consistent and unique sequence arrangement. (ii) Both termini are variable in length. One end of the molecule after Hsu I endonuclease cleavage consists of approximately 3,000 base pairs, with as many as 10 additional 500-base pair segments. The opposite end of the molecule after Sal I endonuclease cleavage consists of approximately 1,500 base pairs, with as many as 10 additional 500-base pair segments. (iii) The opposite ends of the molecule contain homologous sequences. The high degree of homology between the opposite ends of the molecule and the similarity in size of the "additional" 500-base pair segments suggests that there are identical repeating units at both ends of the DNA. The arrangement of restriction endonuclease fragments of the DNA of the W91 isolate of EBV is similar to that of the B95-8 isolate and differs from the latter in the presence of approximately 7 X 10(6) daltons of "extra" DNA at a single site. Thus, the size of almost all EcoRI, Hsu I, and Sal I fragments of EBV (W91) DNA is identical to that of fragments of EBV (B95-8) DNA. A single EcoRI fragment, C, of EBV (W91) DNA is approximately 7 X 10(6) daltons larger than the corresponding EcoRI fragment of EBV (B95-8) DNA. Digestion of EBV (W91) DNA with Hsu I or Sal I restriction endonucleases produces two fragments (Hsu I D1 and D2 or Sal I G2 and G3) which differ in total size by approximately 7 X 10(6) daltons from the fragments of EBV (B95-8) DNA. Furthermore, the EcoRI, Hsu I, and Sal I fragments of EBV (W91) and (B95-8) DNAs, which are of similar molecular weight, have homologous nucleotide sequences. Moreover, the W91 fragments contain only sequences from a single region of the B95-8 genome. Two lines of evidence indicate that the "extra" sequences present in W91 EcoRI fragment C are viral DNA and not cellular. (i) The molecular weight of the "enlarged" EcoRI C fragment of EBV (W91) DNA is identical to that of the EcoRI C fragment of another isolate of EBV (Jijoye), (ii) The HR-1 clone of Jijoye has previously been shown to contain DNA which is not present in the B95-8 strain but is present in the EcoRI C and Hsu I D2 and D1 fragments of EBV (W91) DNA (N. Raab-Traub, R. Pritchett, and E. Kieff, J. Virol. 27:388-398, 1978).
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams A., Bjursell G., Kaschka-Dierich C., Lindahl T. Circular Epstein-Barr virus genomes of reduced size in a human lymphoid cell line of infectious mononucleosis origin. J Virol. 1977 May;22(2):373–380. doi: 10.1128/jvi.22.2.373-380.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornkamm G. W., Delius H., Fleckenstein B., Werner F. J., Mulder C. Structure of Herpesvirus saimiri genomes: arrangement of heavy and light sequences in the M genome. J Virol. 1976 Jul;19(1):154–161. doi: 10.1128/jvi.19.1.154-161.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman T. G., Roizman B. Anatomy of bovine mammillitis DNA. I Restriction endonuclease maps of four populations of molecules that differ in the relative orientation of their long and short components. J Virol. 1978 Jan;25(1):395–407. doi: 10.1128/jvi.25.1.395-407.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolyniuk M., Pritchett R., Kieff E. Proteins of Epstein-Barr virus. I. Analysis of the polypeptides of purified enveloped Epstein-Barr virus. J Virol. 1976 Mar;17(3):935–949. doi: 10.1128/jvi.17.3.935-949.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckenstein B., Bornkamm G. W., Mulder C., Werner F. J., Daniel M. D., Falk L. A., Delius H. Herpesvirus ateles DNA and its homology with Herpesvirus saimiri nucleic acid. J Virol. 1978 Jan;25(1):361–373. doi: 10.1128/jvi.25.1.361-373.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber P., Nkrumah F. K., Pritchett R., Kieff E. Comparative studies of Epstein-Barr virus strains from Ghana and the United States. Int J Cancer. 1976 Jan 15;17(1):71–81. doi: 10.1002/ijc.2910170111. [DOI] [PubMed] [Google Scholar]
- Gerper P., Whang-Peng J., Monroe J. H. Transformation and chromosome changes induced by Epstein-Barr virus in normal human leukocyte cultures. Proc Natl Acad Sci U S A. 1969 Jul;63(3):740–747. doi: 10.1073/pnas.63.3.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward G. S., Jacob R. J., Wadsworth S. C., Roizman B. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward G. S., Smith M. G. The chromosome of bacteriophage T5. I. Analysis of the single-stranded DNA fragments by agarose gel electrophoresis. J Mol Biol. 1972 Feb 14;63(3):383–395. doi: 10.1016/0022-2836(72)90435-4. [DOI] [PubMed] [Google Scholar]
- Hayward S. D., Kieff E. DNA of Epstein-Barr virus. II. Comparison of the molecular weights of restriction endonuclease fragments of the DNA of Epstein-Barr virus strains and identification of end fragments of the B95-8 strain. J Virol. 1977 Aug;23(2):421–429. doi: 10.1128/jvi.23.2.421-429.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henle W., Diehl V., Kohn G., Zur Hausen H., Henle G. Herpes-type virus and chromosome marker in normal leukocytes after growth with irradiated Burkitt cells. Science. 1967 Sep 1;157(3792):1064–1065. doi: 10.1126/science.157.3792.1064. [DOI] [PubMed] [Google Scholar]
- Hinuma Y., Konn M., Yamaguchi J., Wudarski D. J., Blakeslee J. R., Jr, Grace J. T., Jr Immunofluorescence and herpes-type virus particles in the P3HR-1 Burkitt lymphoma cell line. J Virol. 1967 Oct;1(5):1045–1051. doi: 10.1128/jvi.1.5.1045-1051.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehn U., Lindahl T., Klein C. Fate of virus DNA in the abortive infection of human lymphoid cell lines by Epstein-Barr virus. J Gen Virol. 1972 Sep;16(3):409–412. doi: 10.1099/0022-1317-16-3-409. [DOI] [PubMed] [Google Scholar]
- Jondal M., Klein G. Surface markers on human B and T lymphocytes. II. Presence of Epstein-Barr virus receptors on B lymphocytes. J Exp Med. 1973 Dec 1;138(6):1365–1378. doi: 10.1084/jem.138.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai Y., Nonoyama M., Pagano J. S. Reassociation kinetics for Epstein-Barr virus DNA: nonhomology to mammalian DNA and homology of viral DNA in various diseases. J Virol. 1973 Nov;12(5):1006–1012. doi: 10.1128/jvi.12.5.1006-1012.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly R. B., Cozzarelli N. R., Deutscher M. P., Lehman I. R., Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. XXXII. Replication of duplex deoxyribonucleic acid by polymerase at a single strand break. J Biol Chem. 1970 Jan 10;245(1):39–45. [PubMed] [Google Scholar]
- Kieff E. D., Bachenheimer S. L., Roizman B. Size, composition, and structure of the deoxyribonucleic acid of herpes simplex virus subtypes 1 and 2. J Virol. 1971 Aug;8(2):125–132. doi: 10.1128/jvi.8.2.125-132.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieff E., Levine J. Homology between Burkitt herpes viral DNA and DNA in continuous lymphoblastoid cells from patients with infectious mononucleosis. Proc Natl Acad Sci U S A. 1974 Feb;71(2):355–358. doi: 10.1073/pnas.71.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick B. A., Huang E. S. Human cytomegalovirus genome: partial denaturation map and organization of genome sequences. J Virol. 1977 Oct;24(1):261–276. doi: 10.1128/jvi.24.1.261-276.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig H., Biswal N., Benyesh-Melnick M. Characterization of DNA isolated from metaphase chromosomes of cells containing Epstein-Barr virus. Biochim Biophys Acta. 1971 Mar 11;232(2):261–270. doi: 10.1016/0005-2787(71)90579-x. [DOI] [PubMed] [Google Scholar]
- Menezes J., Leibold W., Klein G. Biological differences between Epstein-Barr virus (EBV) strains with regard to lymphocyte transforming ability, superinfection and antigen induction. Exp Cell Res. 1975 May;92(2):478–484. doi: 10.1016/0014-4827(75)90404-8. [DOI] [PubMed] [Google Scholar]
- Miller G., Coope D., Niederman J., Pagano J. Biological properties and viral surface antigens of Burkitt lymphoma- and mononucleosis- derived strains of Epstein-Barr virus released from transformed marmoset cells. J Virol. 1976 Jun;18(3):1071–1080. doi: 10.1128/jvi.18.3.1071-1080.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Lipman M. Comparison of the yield of infectious virus from clones of human and simian lymphoblastoid lines transformed by Epstein-Barr virus. J Exp Med. 1973 Dec 1;138(6):1398–1412. doi: 10.1084/jem.138.6.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Robinson J., Heston L., Lipman M. Differences between laboratory strains of Epstein-Barr virus based on immortalization, abortive infection, and interference. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4006–4010. doi: 10.1073/pnas.71.10.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Shope T., Lisco H., Stitt D., Lipman M. Epstein-Barr virus: transformation, cytopathic changes, and viral antigens in squirrel monkey and marmoset leukocytes. Proc Natl Acad Sci U S A. 1972 Feb;69(2):383–387. doi: 10.1073/pnas.69.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse L. S., Buchman T. G., Roizman B., Schaffer P. A. Anatomy of herpes simplex virus DNA. IX. Apparent exclusion of some parental DNA arrangements in the generation of intertypic (HSV-1 X HSV-2) recombinants. J Virol. 1977 Oct;24(1):231–248. doi: 10.1128/jvi.24.1.231-248.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T. R., Hudson J. B. Some properties of the genome of murine cytomegalovirus (MCV). Virology. 1973 Jul;54(1):135–149. doi: 10.1016/0042-6822(73)90123-2. [DOI] [PubMed] [Google Scholar]
- Nilsson K. High-frequency establishment of human immunoglobulin-producing lymphoblastoid lines from normal and malignant lymphoid tissue and peripheral blood. Int J Cancer. 1971 Nov 15;8(3):432–442. doi: 10.1002/ijc.2910080311. [DOI] [PubMed] [Google Scholar]
- Nilsson K., Klein G., Henle W., Henle G. The establishment of lymphoblastoid lines from adult and fetal human lymphoid tissue and its dependence on EBV. Int J Cancer. 1971 Nov 15;8(3):443–450. doi: 10.1002/ijc.2910080312. [DOI] [PubMed] [Google Scholar]
- Pagano J. S., Huang C. H., Levine P. Absence of Epstein-Barr viral DNA in Amercian Burkitt's lymphoma. N Engl J Med. 1973 Dec 27;289(26):1395–1399. doi: 10.1056/NEJM197312272892604. [DOI] [PubMed] [Google Scholar]
- Pattengale P. K., Smith R. W., Gerber P. Selective transformation of B lymphocytes by E.B. virus. Lancet. 1973 Jul 14;2(7820):93–94. doi: 10.1016/s0140-6736(73)93286-8. [DOI] [PubMed] [Google Scholar]
- Pope J. H., Horne M. K., Scott W. Transformation of foetal human keukocytes in vitro by filtrates of a human leukaemic cell line containing herpes-like virus. Int J Cancer. 1968 Nov 15;3(6):857–866. doi: 10.1002/ijc.2910030619. [DOI] [PubMed] [Google Scholar]
- Pritchett R. F., Hayward S. D., Kieff E. D. DNA of Epstein-Barr virus. I. Comparative studies of the DNA of Epstein-Barr virus from HR-1 and B95-8 cells: size, structure, and relatedness. J Virol. 1975 Mar;15(3):556–559. doi: 10.1128/jvi.15.3.556-559.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab-Traub N., Pritchett R., Kieff E. DNA of Epstein-Barr virus. III. Identification of restriction enzyme fragments that contain DNA sequences which differ among strains of Epstein-Barr virus. J Virol. 1978 Aug;27(2):388–398. doi: 10.1128/jvi.27.2.388-398.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reedman B. M., Klein G. Cellular localization of an Epstein-Barr virus (EBV)-associated complement-fixing antigen in producer and non-producer lymphoblastoid cell lines. Int J Cancer. 1973 May;11(3):499–520. doi: 10.1002/ijc.2910110302. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Schulte-Holthausen H., zur Hausen H. Partial purification of the Epstein-Barr virus and some properties of its DNA. Virology. 1970 Mar;40(3):776–779. doi: 10.1016/0042-6822(70)90229-1. [DOI] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stevely W. S. Inverted repetition in the chromosome of pseudorabies virus. J Virol. 1977 Apr;22(1):232–234. doi: 10.1128/jvi.22.1.232-234.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden B., Summers W. C., Klein G. Nucleic acid renaturation and restriction endonuclease cleavage analyses show that the DNAs of a transforming and a nontransforming strain of Epstein-Barr virus share approximately 90% of their nucleotide sequences. J Virol. 1976 May;18(2):765–775. doi: 10.1128/jvi.18.2.765-775.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]
- Wadsworth S., Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J Virol. 1975 Jun;15(6):1487–1497. doi: 10.1128/jvi.15.6.1487-1497.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E. K., Roizman B., Savage T., Spear P. G., Mizell M., Durr F. E., Sypowicz D. Characterization of the DNA of herpesviruses associated with Lucké adenocarcinoma of the frog and Burkitt lymphoma of man. Virology. 1970 Sep;42(1):257–261. doi: 10.1016/0042-6822(70)90265-5. [DOI] [PubMed] [Google Scholar]
- Weinberg A., Becker Y. Studies on EB virus of Burkitt's lymphoblasts. Virology. 1969 Oct;39(2):312–321. doi: 10.1016/0042-6822(69)90051-8. [DOI] [PubMed] [Google Scholar]
- Wigler M., Silverstein S., Lee L. S., Pellicer A., Cheng Y. c., Axel R. Transfer of purified herpes virus thymidine kinase gene to cultured mouse cells. Cell. 1977 May;11(1):223–232. doi: 10.1016/0092-8674(77)90333-6. [DOI] [PubMed] [Google Scholar]